Abstract
Mutualistic fungal endophytes infect many grass species and often confer benefits to the hosts such as reduced herbivory by insects and animals. The physiological interactions between the endophytes and their hosts have not been well characterized. Fungal-secreted proteins are likely to be important components of the interaction. In the interaction between Poa ampla and the endophyte Neotyphodium sp., a fungal β-1,6-glucanase is secreted into the apoplast, and activity of the enzyme is detectable in endophyte-infected plants. Sequence analysis indicates the β-1,6-glucanase is homologous to enzymes secreted by the mycoparasitic fungi Trichoderma harzianum and Trichoderma virens. DNA gel-blot analysis indicated the β-1,6-glucanase was encoded by a single gene. As a secreted protein, the β-1,6-glucanase may have a nutritional role for the fungus. In culture, β-1,6-glucanase activity was induced in the presence of β-1,6-glucans. From RNA gel blots, similar β-1,6-glucanases were expressed in tall fescue (Festuca arundinacea Schreb.) and Chewings fescue (Festuca rubra L. subsp. fallax [Thuill] Nyman) infected with the endophyte species Neotyphodium coenophialum and Epichloë festucae, respectively.
Fungal endophytes of the genus Neotyphodium (formerly Acremonium; Glenn et al., 1996) infect many grass species, some of which are important turf and forage grasses. The fungi colonize the intercellular spaces of the aerial plant parts but do not invade the plant cells. The endophyte-grass associations are generally considered to be mutualistic symbioses (Clay, 1988). In many associations, the production of alkaloids by the fungus results in reduced herbivory by insects and animals, thus benefiting the host (Breen, 1994; Bush et al., 1997). The fungi benefit from the access to nutrients provided by the plants.
Within the past 20 years, considerable knowledge has been gained on the synthesis and effects of alkaloids, the genetics and taxonomic relationships of endophytes, and the ecological effects of endophyte infection (Clay, 1990; Siegel and Schardl, 1991; Schardl, 1996; Bush et al., 1997). The physiological aspects of the endophyte-grass interactions have not, however, been well characterized in any system. We are investigating the physiology of the fungus-grass interaction with the long-range objective of eventually being able to manipulate agriculturally important interactions. We are using the Poa ampla cv Service (big bluegrass)/Neotyphodium sp. interaction as a model system for the grass/fungus interaction (Lindstrom et al., 1993). P. ampla is apomictic, so we have a ready supply of plants of identical genotype. We also have uninfected plants of the identical genotype, which were identified in older seed lots in which the endophyte had lost viability.
Almost nothing is known of the proteins relevant to the interaction between the plant hosts and the fungal endophytes. We are interested in fungal-secreted proteins because they are likely to be important components of the mutualistic interaction because they are located at the interface of the two species. We have previously reported on a fungal subtilisin-like proteinase and an invertase detected in endophyte-infected plants (Lindstrom et al., 1993; Lindstrom and Belanger, 1994; Lam et al., 1995; Reddy et al., 1996). Both of these enzymes are fungal-secreted proteins. Invertase is certainly involved in nutrient acquisition from the apoplast. The physiological role of the proteinase is not yet known, but homologous proteinases are considered to play an important role in the pathogenicity of entomopathogenic, nematophagous, and mycoparasitic fungi by degrading the protein linkages in the hosts' outer integument (Geremia et al., 1993; Bonants et al., 1995; St. Leger, 1995). From protein sequencing, we have identified additional fungal-secreted proteins expressed in the infected plants. Here, we report the characterization of a fungal β-1,6-glucanase that is found in the apoplast of endophyte-infected (E+) P. ampla.
RESULTS
Detection of β-1,6-Glucanase Sequences from an Apoplastic Protein
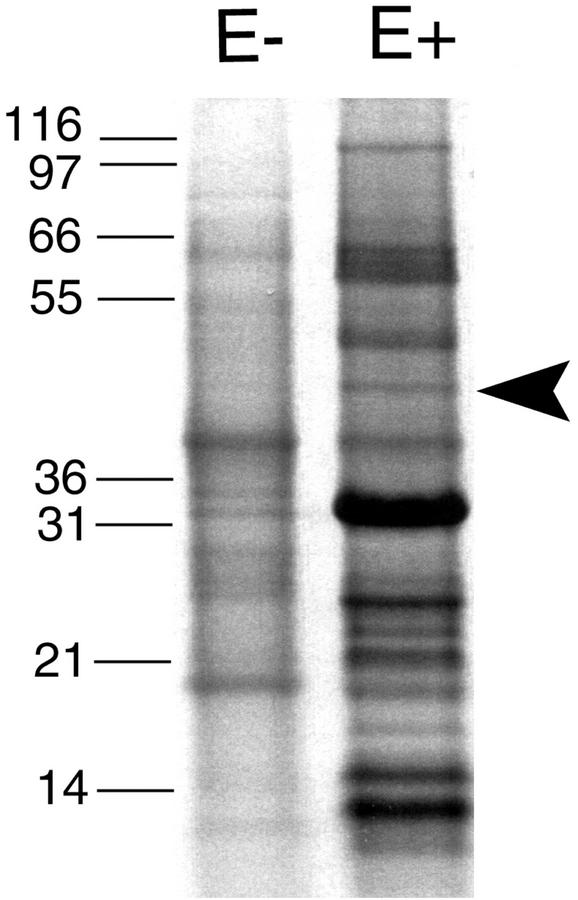
Apoplastic proteins were isolated from endophyte-free (E−) and endophyte-infected (E+) plants and were separated by SDS-PAGE. A number of protein bands were prominent in the sample from the E+ plants but not in the sample from the E− plants (Fig. 1). The prominent band at approximately 34 kD in the E+ sample is the fungal subtilisin-like proteinase, which we previously characterized (Lindstrom and Belanger, 1994; Reddy et al., 1996).
Figure 1.
SDS-PAGE analysis of apoplastic proteins isolated from E+ and E− leaf sheaths. The molecular masses of the protein standards are indicated in kilodaltons. Arrow indicates the 47-kD band containing the β-1,6-glucanase.
Peptides from the 47-kD band in the E+ sample were subjected to both tandem mass spectrometry (MS/MS) and Edman degradation. Four peptide sequences (WDSGDPR, TELNDPR, NVYQDVCANYR, and DAGNQKFETHWR) were obtained that were similar to predicted trypsin peptides from an endo-β-1,6-glucanase from the fungus Trichoderma harzianum (Lora et al., 1995). Degenerate oligonucleotides based on the peptide sequence NVYQDVC and on a sequence conserved among the T. harzianum β-1,6-glucanase and three exo-β-1,3-glucanases (Lora et al., 1995) were used in PCR of a cDNA library prepared from E+ P. ampla leaf sheath tissue. A 900-bp amplified band was cloned whose sequence was homologous to the T. harzianum endo-β-1,6-glucanase. Because the sequence was similar to a reported fungal sequence, it was considered to have originated from the fungal endophyte rather than from the host plant and to likely encode a β-1,6-glucanase. These assumptions were confirmed by subsequent analyses described below. The PCR clone was used to screen the cDNA library, and a full-length clone was obtained.
The nucleotide and deduced amino acid sequences of the full-length clone are shown in Figure 2. A 429-amino acid protein with an Mr of 48,043 is predicted from the cDNA sequence. The two peptide sequences (DAGNQKFETHWR and NVYQDVCANYR) obtained from Edman degradation of peptides generated from the 47-kD apoplastic protein band are precisely predicted from the cDNA sequence. MS/MS sequencing predicted two peptide sequences (WDSGDPR and TELNDPR) which are encoded by the cDNA but with single-amino acid differences in each sequence (WDSGNPR and TQLNDPR). These discrepancies are likely attributable to the nature of the MS/MS technique, which compares the mass of the peptide with a database of predicted trypsin peptides from GenBank entries. In both cases, there was only a single mass unit difference between the peptide sequences predicted by the MS/MS sequencing and those predicted by the cDNA sequence.
Figure 2.
Nucleotide sequence and deduced amino acid sequence of the β-1,6-glucanase cDNA clone. The protein-coding regions are in uppercase letters and the 5′- and 3′-flanking regions are in lowercase letters. The sequences corresponding to the experimentally determined amino acid sequences are underlined with a solid line. The sequence corresponding to the conserved region used for one of the degenerate oligonucleotides is underlined with a dashed line. The predicted signal sequence cleavage site and the proenzyme cleavage site are indicated by arrows.
Because the protein band was isolated from apoplastic proteins, it was expected to be a secreted protein with an N-terminal signal sequence. Analysis of the predicted N-terminal amino acid sequence using a neural network method identified a signal sequence cleavage site between amino acids 17 (A) and 18 (W; Nielsen et al., 1997; http://www.cbs.dtu.dk/services/SignalP/index.html). There are no potential N-glycosylation sites of NXT/S in the amino acid sequence of the mature protein.
Similarity of the Neotyphodium sp. β-1,6-Glucanase to Other Sequences
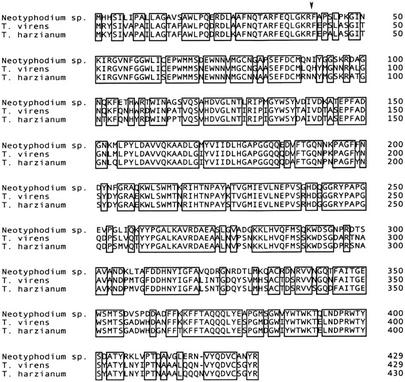
Only two other sequences similar to the Neotyphodium sp. β-1,6-glucanase amino acid sequence have been reported. Both are from the biocontrol fungi T. harzianum and Trichoderma virens (Lora et al., 1995; Kim et al., 2002). Both Trichoderma spp. are soil-borne filamentous fungi that are potent mycoparasites of a many-plant pathogenic fungi (Papavizas, 1985; Chet, 1987). A comparison of the deduced amino acid sequences of the Neotyphodium sp. β-1,6-glucanase and those from the Trichoderma spp. is shown in Figure 3. The endophyte amino acid sequence is 76% identical to that of T. virens and 73% identical to that of T. harzianum. At the DNA level, the Neotyphodium sp. sequence is 72% and 74% identical to the sequences of T. virens and T. harzianum, respectively.
Figure 3.
Comparison of the deduced amino acid sequences of the Neotyphodium sp. β-1,6-glucanase and the T. virens and T. harzianum homologs. Boxes enclose identical amino acids. The position of the N terminus of the mature proteins, experimentally determined for the Neotyphodium sp. and T. harzianum sequences, is indicated by an arrow. GenBank accession numbers for the corresponding DNA sequences are: Neotyphodium sp., AF535131; T. virens, AF395757; and T. harzianum, X79197.
On the basis of sequence comparisons, the relationship of the Trichoderma spp. β-1,6-glucanases to family 5 glycosyl hydrolases was recognized previously (Lora et al., 1995; Kim et al., 2002). Most members of family 5 are exo-β-1,3-glucanases or endo-β-1,4-glucanases (Henrissat and Davies, 1997; http://www.expasy.ch/cgi-bin/lists?glycosid.txt). The active site residues of the Candida albicans exo-β-1,3-glucanase have been determined from crystallography (Cutfield et al., 1999). Although there is only 22% overall sequence identity, six of the eight amino acid residues in the active site of the exo-β-1,3-glucanase are conserved in the Neotyphodium and Trichoderma β-1,6-glucanases, including both catalytic residues E-192 and E-292 (Fig. 4).
Figure 4.
Comparison of the amino acid sequences of the Neotyphodium sp. β-1,6-glucanase with the C. albicans exo-β-1,3-glucanase. The pre-pro regions of the proteins are not included, and the numbering begins with the first amino acid of the mature enzymes. Spaces were inserted to maximize alignment. The experimentally determined active site residues in the exo-1,3-glucanase are indicated by asterisks. The GenBank accession number for the C. albicans enzyme is X56556.
Expression of the Endophyte β-1,6-Glucanase in Brewer's Yeast (Saccharomyces cerevisiae)
The endophyte β-1,6-glucanase was expressed in Brewer's yeast with the aim of purifying it from the medium and confirming the enzymatic activity. No β-1,6-glucanase activity could be detected in the medium, but activity could be detected in cell extracts. Crude extracts of Brewer's yeast cells transformed with the endophyte β-1,6-glucanase expression vector had twice the β-1,6-glucanase activity when compared with extracts from cells transformed with the vector alone (Table I). These results confirmed that the Neotyphodium sp. cDNA clone encoded a β-1,6-glucanase. Brewer's yeast does not have a homologous β-1,6-glucanase sequence in its genome (Goffeau et al., 1996), and no enzyme preferentially cleaving β-1,6-glucans has been reported. Brewer's yeast cytoplasmic β-1,3-glucanases that preferentially cleave laminarin, predominantly a β-1,3-glucan, but that also cleave pustulan, a β-1,6-glucan, have been reported (Hien and Fleet, 1983). The background activity seen in extracts of Brewer's yeast cells transformed with the vector only likely originates from these endogenous enzymes.
Table I.
β-1,6-Glucanase activity of yeast cells transformed with the pYES2-glucanase expression vector or pYES2 vector-only control
| Experiment | β-1,6-Glucanase Activity
|
|
|---|---|---|
| Glucanase transformant | Vector control | |
| units mg−1 | ||
| 1 | 0.346 ± 0.01 a | 0.153 ± 0.03 b |
| 2 | 0.286 ± 0.02 a | 0.150 ± 0.03 b |
Activity was assayed in two separate experiments. Data shown are the means and ses of three replicates. One unit is the amount of enzyme that catalyzes the release of 1 μmol Glc min−1. Within each experiment, different letters signify statistical significance at P < 0.05.
We had expected that the signal sequence of the Neotyphodium sp. would be recognized in Brewer's yeast and target the protein for secretion. Expression of the T. harzianum enzyme in Brewer's yeast was reported to result in secretion, as assessed by a clearing zone surrounding the yeast colony when grown on pustulan (Lora et al., 1995). The T. harzianum enzyme has also been expressed in the yeast Pichia pastoris and was secreted into the medium (Bom et al., 1998).
Because the Neotyphodium sp. and the T. harzianum β-1,6-glucanase signal peptide sequences are similar and the predicted cleavage sites are identical, it was surprising that the Neotyphodium sp. enzyme was not secreted into the medium. To confirm that the sequence surrounding the presumed signal peptide cleavage site in the yeast expression vector had not undergone a mutation, we isolated the plasmid from the yeast transformant and sequenced the N-terminal region of the coding sequence. No mutation in the sequence had occurred. The reason the enzyme was retained within the cell rather than secreted is not known.
β-1,6-Glucanase Induction in the Presence of β-1,6-Glucans
The β-1,6-glucanase was secreted into the medium of the Neotyphodium sp. endophyte when grown in culture. The level of β-1,6-glucanase activity secreted into the medium was determined in four different culture formulations to determine optimal conditions for production of the enzyme. Regulation of β-1,6-glucanase expression has been reported for a number of fungal species (Lora et al., 1995; Pitson et al., 1997).
Autoclaved yeast cells and potato dextrose broth were used as undefined rich media, which differ regarding the presence of β-1,6-glucans. Yeast cell walls contain β-1,6-glucans (Manners et al., 1973), which represent approximately 12% to 15% of the cell wall carbohydrate polymers (Magnelli et al., 2002). None would be expected in potato dextrose broth because plant cells do not contain β-1,6-glucans (Varner and Lin, 1989). β-1,6-Glucanase activity in a semidefined medium, tryptone-0.1% (w/v) Suc (Lam et al., 1995), was compared with that in the same medium also containing 1% (w/v) pustulan. A low Suc concentration was used to simulate the low nutrient conditions found in the plant apoplast.
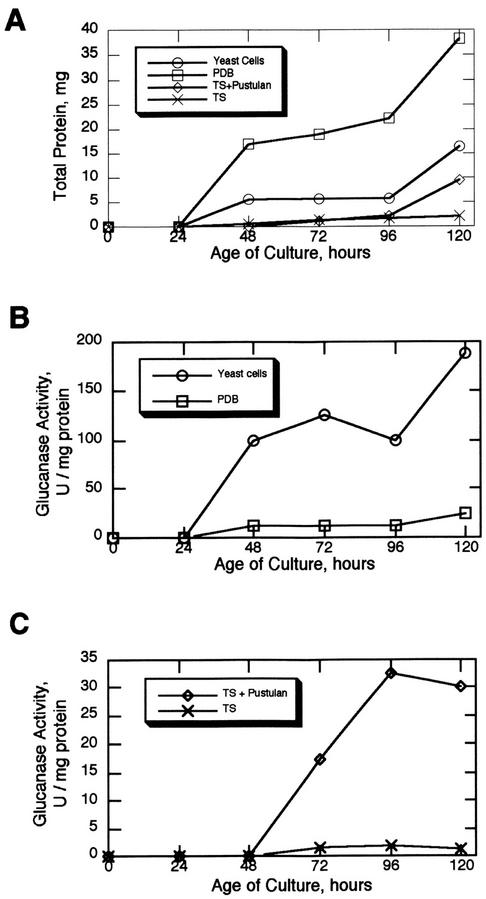
The amount of total secreted proteins and β-1,6-glucanase specific activity in the four media were determined over a period of 5 d (Fig. 5). The total protein data presented in Figure 5 is the protein level in the media after subtracting the protein contributed from the media itself, and so represents the protein secreted by the endophyte. β-1,6-Glucanase activity could be detected in the media from all four formulations but was dramatically higher in the two containing β-1,6-glucans. At the 120-h point, the β-1,6-glucanase specific activity was 7.5-fold higher in the yeast cell medium relative to the potato dextrose broth and 24-fold higher in the tryptone-Suc + pustulan relative to the tryptone-Suc alone. In the comparison of the two undefined rich media, the amount of secreted proteins produced was actually lower in the yeast cell media compared with the potato dextrose broth, but the β-1,6-glucanase specific activity was higher. With the two tryptone-Suc media, the amount of secreted proteins produced was essentially the same up to the 96-h time point, but the β-1,6-glucanase specific activity was higher in the presence of pustulan beginning at 72 h in culture. Taken together, these results indicate that expression of the Neotyphodium sp. β-1,6-glucanase is dependent on culture conditions and may be induced in the presence of β-1,6-glucans.
Figure 5.
Total secreted protein and β-1,6-glucanase specific activity in the media of Neotyphodium sp. grown in different culture formulations. PDB, Potato dextrose broth; TS, tryptone-Suc.
Partial Purification of the Endophyte β-1,6-Glucanase
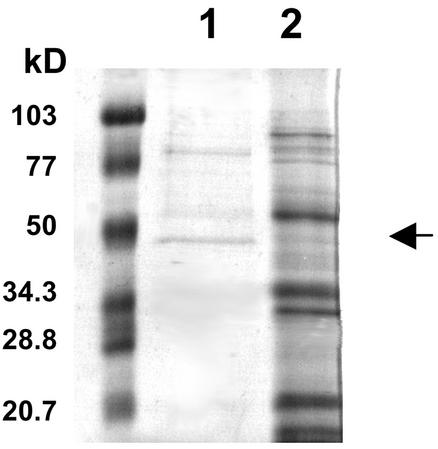
Because the highest β-1,6-glucanase specific activity was obtained when the endophyte was grown in the yeast cell medium, that medium was chosen as the source of enzyme for purification. The β-1,6-glucanase was partially purified by affinity binding to its substrate, pustulan. With this method, a 28-fold purification could be achieved in one step (Table II). The partially purified enzyme had two major protein bands at 88 and 47 kD when subjected to SDS-PAGE (Fig. 6). The 47-kD band corresponded to the size of the apoplastic protein band from which the β-1,6-glucanase peptide sequences were obtained. The N-terminal sequence of the 47-kD protein band was determined to be FAPSLPKG, which is predicted by the cDNA from amino acids 41 to 48. This implies that there is another proteolytic processing step after cleavage of the signal peptide that removes 23 additional amino acids. The calculated Mr of the mature protein based on the sequence data is 43,603, slightly smaller than that predicted from SDS-PAGE. The N-terminal sequence of the mature enzyme is at the same relative position and is similar to that determined for the T. harzianum enzyme (Lora et al., 1995). Both N-terminal sequences are immediately preceded by the amino acid sequence KR, a KEX2 protease recognition site (Julius et al., 1984).
Table II.
Partial purification of β-1,6-glucanase from the culture filtrate of the Neotyphodium sp. grown in yeast cell medium
| Fraction | Total Protein | Total Activity | Specific Activity | Purification |
|---|---|---|---|---|
| mg | units | units mg−1 | ||
| Culture filtrate | 400 | 69 | 0.017 | 1 |
| Pustulan adsorption | 0.06 | 0.28 | 0.48 | 28.2 |
Figure 6.
SDS-PAGE analysis of the partially purified β-1,6-glucanase. Lane 1, Partially purified β-1,6-glucanase obtained after pustulan adsorption, 4.5 μg; lane 2, crude culture filtrate from the Neotyphodium sp. grown in yeast cell medium, 15 μg. The arrow indicates the position of the 47-kD band. The molecular masses of the protein standards on the left are indicated in kilodaltons.
Substrate Specificity of the Endophyte β-1,6-Glucanase
The activity of the partially purified β-1,6-glu-canase was determined against glucan substrates with different linkages (Table III). The enzyme was most active against pustulan, a β-1,6-glucan, and had weaker activity against laminarin, which has predominantly β-1,3-linkages with some β-1,6-linkages. The enzyme had no activity against carboxymethylcellulose or dextran that have β-1,4 and α-1,6 linkages, respectively.
Table III.
Substrate specificity of the partially purified β-1,6-glucanase from Neotyphodium sp. after affinity adsorption to pustulan
| Substrate, Linkage | Relative Activity |
|---|---|
| % | |
| Pustulan, β-1,6 | 100 |
| Laminarin, β-1,3; β-1,6 | 12 |
| Carboxymethylcellulose, β-1,4 | 0 |
| Dextran, α-1,6 | 0 |
β-1,6-Glucanase Activity in E+ Plants
β-1,6-glucanase enzymatic activity (0.8 ± 0.1 μmol Glc released mg−1 protein) was detected in E+ leaf sheath crude extracts. No activity was detected in E− leaf sheaths, so all of the activity detected in the E+ plants can be attributed to the endophyte. No β-1,6-glucanase enzymes have been reported for any plant species. Because the endophytic hyphae are a small component of the leaf sheath mass, the endophytic proteins are expected to be a small component of a leaf sheath extract. Even so, the fungal β-1,6-glucanase was present in detectable levels.
β-1,6-Glucanase Message in Infected Plant Tissue and in Cultured Fungus
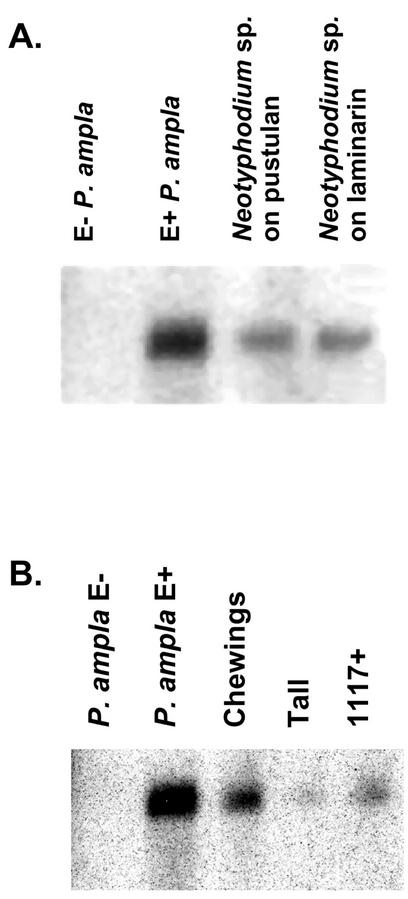
The β-1,6-glucanase message could be detected in total RNA extracted from infected P. ampla leaf sheaths and from the fungus cultured with pustulan or laminarin (Fig. 7A). No hybridization was detectable in RNA from endophyte-free plants, indicating that the host, P. ampla, does not express a highly similar β-1,6-glucanase.
Figure 7.
RNA gel-blot analysis of β-1,6-glucanase transcripts. A, Lane 1, E− P. ampla; lane 2, E+ P. ampla; lane 3, Neotyphodium sp. grown on 0.5% (w/v) pustulan; lane 4, Neotyphodium sp. grown on 0.5% (w/v) laminarin. Fifteen micrograms of total RNA was used from E− and E+ P. ampla leaf sheaths. Ten micrograms of total RNA was used from the Neotyphodium sp. fungal cultures. B, Lane 1, E− P. ampla; lane 2, E+ P. ampla; lane 3, E+ Chewings fescue; lane 4, E+ tall fescue; lane 5, Chewings fescue 1117 artificially infected with the Neotyphodium sp. Four micrograms of poly(A+) RNA was used for each sample.
β-1,6-Glucanase message was also detected in RNA extracted from other endophyte-infected grass species (Fig. 7B). Hybridization with RNA extracted from tall fescue infected with Neotyphodium coenophialum Morgan-Jones and Gams, and Chewings fescue (Festuca rubra L. subsp. fallax [Thuill] Nyman) infected with Epichloë festucae (Leuchtmann et al., 1994) indicates that a homologous glucanase is also expressed by other endophyte species in their host plants. Hybridization was also detected in RNA from a Chewings fescue artificially inoculated with the Neotyphodium sp. endophyte that infects P. ampla (Johnson-Cicalese et al., 2000). The hybridization intensities varied among the four RNA samples from the different infected plant species. It is impossible to know whether this is attributable to different levels of expression in the different fungal species or to different amounts of fungal tissue within the infected plants.
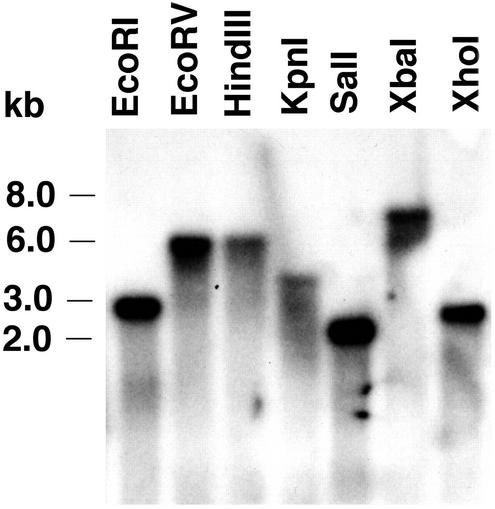
β-1,6-Glucanase Is Encoded by a Single Gene
Gel-blot analysis of the Neotyphodium sp. DNA hybridized with the β-1,6-glucanase 900-bp PCR fragment indicated that the glucanase is encoded by a single gene. When the DNA was digested with restriction enzymes for which there are no sites in the glucanase cDNA EcoRV, HindIII, KpnI, XbaI, and XhoI, only single hybridizing bands were detected (Fig. 8). When the DNA was digested with enzymes that have single restriction sites within the glucanase cDNA EcoRI and SalI, single hybridizing bands were also seen because the restriction sites were either outside of or close to the 5′ end of the 900-bp glucanase fragment used as a probe. These data indicate that there are no additional closely related glucanase genes in the Neotyphodium sp. genome. Two distinct β-1,6-glucanases have been reported from T. harzianum (de la Cruz et al., 1995; de la Cruz and Llobell, 1999).
Figure 8.
DNA gel-blot analysis of Neotyphodium sp. DNA. Twelve micrograms of total DNA per sample was digested with the indicated restriction enzymes, fractionated by electrophoresis on an agarose gel, transferred onto a nylon membrane, and hybridized with the radiolabeled β-1,6-glucanase 900-bp PCR clone.
DISCUSSION
This is the first report, to our knowledge, of expression of a fungal β-1,6-glucanase in an endophyte-infected grass. The glucanase was originally identified from peptide sequencing of an apoplastic protein isolated from infected leaf sheaths, indicating that the enzyme accumulates to detectable levels in the plant. The enzymatic activity and fungal origin of the β-1,6-glucanase were confirmed by partial purification from the medium of the endophyte grown in culture. The homologous β-1,6-glucanase from T. harzianum was determined be an endoglucanase (de la Cruz et al., 1995). Considering the level of identity between the two enzymes, the Neotyphodium sp. enzyme is likely also to act as an endoglucanase. In culture, the level of the enzyme was induced up to 24-fold in the presence of β-1,6-glucans.
The cell walls of fungi generally contain chitin, β-1,3-glucans, and β-1,6-glucans in varying proportions (Wessels, 1994). The steady-state model of hyphal wall growth describes the deposition of wall material and secretion of hydrolytic enzymes (Wessels, 1994, 1999). Hyphal growth occurs from the tip where β-1,3-glucans and chitin are deposited. At the plastic tip area of growing hyphae, hydrolytic enzymes are secreted by bulk flow and are crucial for the breakdown of environmental polymers that are nutrient sources for the fungi. Behind the growing tip, cross-linking of the wall components by β-1,6-glucans contributes to rigidification of the wall. In this model, supported by considerable experimental data, enzymatic wall loosening is not necessary for tip growth, and secretion of the hydrolytic enzymes required for nutrition is not impeded by a rigid wall (Sietsma et al., 1995). Wall loosening by the action of hydrolytic enzymes would be required only for processes such as hyphal branching and sporulation, which require modification to the mature, rigid fungal wall (Wessels, 1999).
Although the production of β-1,6-glucanases by fungi has been known for decades (Reese et al., 1962), their physiological function has not been conclusively established in any species. β-1,6-Glucanases have been purified from a number of fungal species including Acremonium persicinum, Neurospora crassa, Rhizopus chinensis, Penicillium spp., and Saccharomycopsis fibuligera (Yamamoto et al., 1974; Santos et al., 1979; Schep et al., 1984; Hiura et al., 1987; Mulenga and Berry, 1994; Pitson et al., 1996). The only DNA sequences reported to date, however, are from two mycoparasitic fungi, T. harzianum and T. virens (Lora et al., 1995; Kim et al., 2002).
As secreted hydrolytic enzymes, β-1,6-glucanases likely have a role in fungal nutrition by degrading glucan polymers in their environment. The soil fungus A. persicinum secretes an extracellular β-1,6-glucanase that may participate synergistically with other enzymes in the degradation of an extracellular storage glucan synthesized by this species (Stasinopoulos and Seviour, 1989; Pitson et al., 1991). Mycoparasitic fungi secrete enzymes capable of degrading the cell walls of their hosts. β-1,6-Glucanase secretion by T. harzianum is considered to be a component that acts synergistically with other hydrolytic enzymes in the mycoparasitism of that species (de la Cruz et al., 1995).
Secretion of the Neotyphodium sp. β-1,6-glucanase into the apoplast of the infected plants suggests it has a role in nutrition of the endophyte. Plant cell walls, however, do not contain β-1,6-glucans (Varner and Lin, 1989), so the substrate for the β-1,6-glucanase is likely of fungal origin. The β-1,6-glucanase may function in degradation and reassimilation of the endophytic cell wall. It is not known whether the endophytes synthesize extracellular storage glucans that could be substrates for the enzyme. The β-1,6-glucanase may function in the branching of the fungal hyphae. Within the infected plants, the hyphae generally run parallel to the leaf axis but are occasionally branched (Siegel et al., 1987). It is also possible the β-1,6-glucanase could function in the degradation of the walls of other fungi that may be encountered within the host plants. This last possibility is interesting because in some plant-fungus associations, endophyte infection results in improved disease resistance (Clarke et al., 2002). The endophyte would gain access to nutrients with the result of protecting the plants from potential pathogens. The remarkable similarity of the amino acid sequence of the Neotyphodium sp. enzyme to that of T. harzianum and T. virens suggests that the enzymes have similar substrate affinities.
In addition to the intercellular endophytic location, the Neotyphodium sp. infecting P. ampla and some other endophyte species also are found epiphytically (Moy et al., 2000). The epiphyllous mycelium was proposed to function as a defensive net preventing colonization of the leaf surface by other organisms. If hydrolytic enzymes such as β-1,6-glucanase and the subtilisin-like protease are also secreted on the leaf surface, they could be components of the proposed niche exclusion (Moy et al., 2000).
MATERIALS AND METHODS
Plant Material
Neotyphodium sp.-infected (E+) Poa ampla cv Service (PI 387931) plants were used in this study. This cultivar was released by the Alaska Department of Natural Resources as an improved roadside grass. Neotyphodium sp.-free (E−) plants were obtained from infected seed that lost viable endophytes as a result of long-term storage. Because of apomixis, all plants were genetically identical. P. ampla leaf sheaths were examined microscopically for the presence or absence of endophyte infection by aniline blue staining (Bacon and White, 1994).
Fungal Culture
Cultures of Neotyphodium sp. were obtained from infected P. ampla plants. Leaf sheaths from infected plants were surface sterilized for 15 min in 1.25% (w/v) sodium hypochlorite (20% [v/v] bleach), rinsed in sterile water, and then placed on potato dextrose agar (PDA) medium (Difco Laboratories, Detroit). After incubation at 24°C for 2 to 3 weeks in darkness, fungal mycelia emerged from plant tissue. Fungal cultures were subsequently subcultured and maintained on PDA plates. If the culture was to be used for nucleic acid extraction, PDA plates overlaid with cellophane sheets were used.
For growth on pustulan or laminarin, the endophyte was grown on plates of a minimal salts medium (M9; Maniatis et al., 1982) without Glc but supplemented with 0.25% (w/v) yeast extract and either 0.5% (w/v) pustulan (Calbiochem, La Jolla, CA) or 0.5% (w/v) laminarin (Sigma-Aldrich, St. Louis).
Protein Electrophoresis
SDS-PAGE was performed in gels containing 10% (w/v) polyacrylamide using a minigel apparatus (Bio-Rad, Hercules, CA). SDS sample buffer (2×) contained 125 mm Tris-HCl, pH 6.8, 4.6% (w/v) SDS, 20% (v/v) glycerol, and 0.002% (w/v) bromphenol blue (Laemmli, 1970). Gels were run at 200 V and stained with Coomassie Brilliant Blue.
Isolation and Peptide Sequencing of Apoplastic Proteins
Leaf sheaths were cut to 2-cm lengths, cleaned, and vacuum infiltrated for 30 min with 100 mm Tris-HCl, pH 8.0, 50 mm dithiothreitol, 10 mm ascorbic acid, and 5 mm phenylmethylsulfonyl fluoride. The leaf sheaths were washed with water three times, blotted dry, and collected in a 3-cc syringe. The syringe was placed in a 50-mL tube and centrifuged at 2,000g for 10 min at 4°C. The apoplastic fluid was collected and concentrated with a Microcon YM-30 (Millipore Corporation, Bedford, MA). Thirty micrograms of protein was mixed with an equal volume of 2× SDS sample buffer, heated at 100°C for 5 min, and subjected to SDS-PAGE. The bands of interest were excised, and gel slices were washed with 50% (v/v) acetonitrile in water. Sequence analysis of peptides generated by trypsin digestion was performed by the Harvard Microchemistry Facility (Harvard University, Cambridge, MA) by microcapillary reverse-phase HPLC nano-electrospray/MS/MS on a Finnigan LCQ quadrupole ion trap mass spectrometer.
Nucleic Acid Isolation
Neotyphodium sp. DNA was isolated from 2-week-old cultures grown on PDA overlaid with cellophane. The fungal mycelium was ground to a fine powder in liquid nitrogen. The powdered mycelium was extracted in 500 mm NaCl, 100 mm Tris-HCl, pH 8.0, 50 mm EDTA, 1% (w/v) SDS, 10 mm 1,10-phenanthroline, and 0.07% (w/v) β-mercaptoethanol (Dellaporta et al., 1983) in a ratio of 5 mL buffer g−1 tissue. The sample was extracted with an equal volume of phenol, incubated at room temperature for 5 min, and centrifuged at 5,000g for 10 min. The aqueous layer was then extracted once with an equal volume of phenol:chloroform:isoamyl alcohol (25:24:1, v/v) and once with chloroform:isoamyl alcohol (24:1, v/v). The DNA in the final aqueous layer was precipitated by adding an equal volume of isopropanol. The sample was incubated overnight at −20°C, and the DNA was collected by centrifugation at 5,000g for 10 min. The DNA pellet was dissolved in 10 mm Tris-HCl, pH 8.0, and 1 mm EDTA and treated with 50 μg mL−1 RNase for 30 min at 37°C. The DNA solution was extracted with phenol:chloroform:isoamyl alcohol as above and ethanol precipitated.
Total RNA was isolated from endophyte-infected and uninfected plants and Neotyphodium sp. cultures using Tri-Reagent (Sigma-Aldrich). Fungal and plant tissue were ground to a fine powder with liquid nitrogen and resuspended in Tri-Reagent (10 mL g−1). Debris was pelleted by centrifugation and the supernatant was extracted once with chloroform. The aqueous layer was precipitated with isopropanol, and the RNA pellet was washed once with ethanol and dissolved in water. Poly(A+) RNA was isolated from total RNA using a commercial kit (Oligotex mRNA Purification Midi Kit, Qiagen USA, Valencia, CA).
Gel-Blot Analyses
For DNA gel-blot analysis, 12 μg of DNA from the Neotyphodium sp. endophyte was digested with EcoRI, EcoRV, HindIII, KpnI, SalI, XbaI, or XhoI in a 50-μL total reaction volume at 37°C for 12 h. The DNA was subjected to electrophoresis through a 0.8% (w/v) agarose gel. DNA in the gel was depurinated by washing in 0.25 n HCl for 12 min. The gel was then washed in water, and the DNA was transferred to a nylon membrane (Zeta-Probe, Bio-Rad) overnight with 0.4 m NaOH (Reed and Mann, 1985). The membrane was washed in 2× SSC and fixed by drying completely.
For RNA gel-blot analyses, RNA was subjected to electrophoresis in formaldehyde agarose gels and transferred to nylon membranes (Magnagraph, Osmonics, Minnetonka, MN) as described by Selden (1987). RNA was fixed to the membrane with a UV Crosslinker (Fisher Scientific, Pittsburgh).
The 900-bp β-1,6-glucanase PCR clone was labeled with [α32P]dCTP using a commercial kit (Prime-It II Random Primer Labeling Kit, Stratagene, La Jolla, CA) for use as a probe for all hybridization reactions. For both DNA and RNA gel blots, filters were prehybridized at 42°C in 50% (v/v) formamide, 5× SSC, 5× Denhardt's solution (1× Denhardt's solution is 0.02% [w/v] Ficoll, 0.02% [w/v] PVP, and 0.02% [w/v] bovine serum albumin), 50 mm sodium phosphate, pH 6.8, 1% (w/v) SDS, 100 μg mL−1 calf thymus DNA, and 2.5% (w/v) dextran sulfate. The hybridization solution was 5 × 105 cpm mL−1 of 32P-labeled fragment, 50% (v/v) formamide, 5× SSC, 1× Denhardt's solution, 20 mm sodium phosphate, pH 6.8, 1% (w/v) SDS, 100 μg mL−1 calf thymus DNA, and 5% (w/v) dextran sulfate. Hybridized membranes were washed with 2× SSPE, 0.5% (w/v) SDS for 15 min at room temperature, 2× SSPE, 0.5% (w/v) SDS for 15 min at 65°C, and 0.2× SSPE, 0.2% (w/v) SDS for 15 min at 65°C. The washed membranes were exposed to x-ray film (XOMAT-AR, Kodak, Rochester, NY) with an intensifying screen.
Library Construction and Screening
A cDNA library was constructed using poly(A+) RNA from endophyte-infected P. ampla leaf sheath tissue. cDNA synthesis and phage packaging was carried out using a commercial kit (λ ZAP-Express cDNA Library Construction Kit, Stratagene). The primary cDNA library contained 4 × 106 plaque-forming units. Two-hundred thousand plaque-forming units from the amplified library were screened using the 32P-labeled 900-bp β-1,6-glucanase PCR clone as a probe. Hybridization conditions were the same as for the gel blots. The cDNA inserts from positive plaques were excised from the λ-vector as recombinant pBK-CMV phagemids (Short et al., 1988). The phagemids were sequenced using an ABI 373A automated sequencer. Sequences were assembled into contigs using AutoAssembler software (Applied Biosystems, Foster City, CA).
PCR Amplification of a β-1,6-Glucanase cDNA Fragment
Degenerate oligonucleotide primers for PCR were designed based on one of the peptide sequences obtained from the 47-kD apoplastic protein band and on a region conserved in a number of glucanases (Lora et al., 1995). The sequences of the degenerate primers were: primer A, 5′-GIATHCCCA-THGGITAYTGG-3′, designed from the conserved sequence RIPIGYW; primer B, 5′-CAIACRTCYTGRTAIACRTT-3′, designed from the peptide sequence NVYQDVC. The symbols used for the mixed bases are: I, deoxyinosine; R, A+G; H, A+C+T; and Y, C+T.
The degenerate oligonucleotide primers were used in PCR amplification of the cDNA library prepared from Neotyphodium sp. infected P. ampla. PCR reactions contained 10 mm Tris-HCl, pH 8.3, 50 mm KCl, 2.5 mm MgCl2, 12.5 pmol of each dNTP, 0.5 μg of each primer, and 2 units of Taq polymerase. PCR was carried out in a GeneAmp 9600 thermocycler (PerkinElmer Life Sciences, Boston). Touchdown PCR (Don et al., 1991) cycling parameters were used. Initial denaturation was conducted at 94°C for 5 min. Cycle 1 consisted of denaturation at 94°C for 30 s, annealing at 57°C for 30 s, and extension at 72°C for 2 min. Every two subsequent cycles, the annealing temperature was decreased by 1°C until 47°C was reached. An additional 30 cycles at an annealing temperature of 47°C were performed, followed by a final extension at 72°C for 10 min. PCR products were resolved on a 1% (w/v) agarose gel, and a single 900-bp band was detected. The DNA band was excised and purified using a commercial kit (QIAquick Gel Extraction Kit, Qiagen USA). The purified band was ligated into the pGEM-T Easy vector (Promega, Madison, WI) and transformed into DH5α Escherichia coli competent cells (PGC Scientifics, Frederick, MD). Plasmids from E. coli transformants were purified and used as templates in AmpliTaq FS cycle sequencing reactions (PerkinElmer Life Sciences). The 900-bp insert was completely sequenced by primer walking. Reactions were analyzed on an ABI 373A Automated DNA Sequencer (PerkinElmer Life Sciences).
β-1,6-Glucanase Activity Assay
Glucanase activity was determined by measuring the amount of reducing sugars released from a glucan substrate (Dygert et al., 1965; Zheng and Wozniak, 1997). Substrates used were pustulan, a linear β-1,6-glucan; laminarin, a β-1,3-glucan with some β-1,6-linkages; carboxymethylcellulose (Sigma-Aldrich), a β-1,4-glucan; and dextran (Sigma-Aldrich), an α-1,6-glucan. Reduced substrates were prepared by incubating 1% (w/v) substrate with 0.1 volume of 0.4 m sodium borohydride at 4°C overnight. The substrate was precipitated with 3 volumes of 100% ethanol, pelleted, air-dried, and resuspended in 50 mm potassium acetate, pH 5.5% to 0.4% (w/v).
For determination of activity from the yeast extracts and the partially purified protein, 20 μL of substrate was incubated with 10 μL of enzyme for 30 min at 40°C. After the incubation, 100 μL of solution A (4 g of Na2CO3, 1.6 g of Gly, and 45 mg CuSO4 in 100 mL of water) and 100 μL of solution B (120 mg of neocuproine HCl in 100 mL of water) were added. The mixture was boiled for 10 min, and the absorbance was measured at 450 nm. Glc was used as a standard. One unit of enzyme activity was defined as the amount of enzyme that catalyzes the release of 1 μmol Glc min−1.
For determination of activity in plant crude extracts, 0.1 g of leaf sheaths was ground in liquid N2 and extracted in 3 mL of 10 mm Tris-HCl, pH 8.0. The assay volume was 400 μL and contained 85 μg of protein, 200 μL of 0.4% (w/v) pustulan, and 20 μL of 1 m potassium acetate, pH 5.5. The reactions were incubated at 40°C for 18 h. They were stopped with 400 μL each of solutions A and B and boiled for 10 min. Protein concentrations were determined using the Bio-Rad Protein Assay Reagent with bovine serum albumin as a standard.
Expression of the β-1,6-Glucanase in Brewer's Yeast (Saccharomyces cerevisiae)
The β-1,6-glucanase cDNA was cloned into the vector pYES2 (Invitrogen, Carlsbad, CA) for inducible expression in Brewer's yeast. The β-1,6-glucanase cDNA phagemid and the pYES2 plasmid were digested with the restriction enzymes SacI and XbaI at 37°C for 1 h. The restriction fragments were separated by electrophoresis through a 1% (w/v) agarose gel, and the vector and cDNA fragments were excised and purified using Gene Clean (Bio 101, Carlsbad, CA). The cDNA fragment was ligated to the pYES2 vector at room temperature for 3 h and transformed into DH10B E. coli cells (Invitrogen) by electroporation (Cell-Porator, Invitrogen). Plasmids from E. coli transformants were purified and analyzed for the presence of the β-1,6-glucanase cDNA insert by SacI/XbaI restriction digestion.
A positive clone was transformed into INVSc1 Brewer's yeast cells (MATa his3Δ1 leu2 trp1-289 ura3-52/MATα his3Δ1 leu2 trp1-289 ura3-52; Invitrogen). As a control, yeast cells were also transformed with the vector pYES2. INVSc1 cells were grown in a 50-mL culture of yeast peptone dextrose medium (1% [w/v] yeast extract, 2% [w/v] peptone, and 2% [w/v] dextrose) at 30°C and 250 rpm to OD600 = 0.7. Cells were pelleted and resuspended in 2 mL of 100 mm lithium acetate, pH 7.5, 5 mm Tris, pH 7.5, and 0.5 mm EDTA. Resuspended cells (100 μL) were then mixed with 1 μg of plasmid DNA, 100 μg of calf thymus DNA, and 700 μL of 100 mm lithium acetate, pH 7.5, 40% (w/v) PEG-3350, 10 mm Tris-HCl, pH 7.5, and 1 mm EDTA and incubated at 30°C for 30 min. Dimethyl sulfoxide (88 μl) was added, and the cell mixture was then heat shocked at 42°C for 7 min. The transformed yeast cells were then pelleted, resuspended in 10 mm Tris-HCl, pH 7.5, and 1 mm EDTA, and plated on SC minimal medium plates without uracil (SC −U; 0.67% [w/v] nitrogen base without amino acids, 2% [w/v] raffinose, 0.01% [w/v] adenine, Arg, Cys, Leu, Lys, Thr, and Trp, and 0.005% [w/v] Asp, His, Ile, Met, Phe, Pro, Ser, Tyr, and Val) for selection. After incubation at 30°C for 3 d, a positive clone was isolated and maintained on SC −U medium with 2% (w/v) raffinose.
To induce β-1,6-glucanase expression, the transformed yeast cells were suspended in SC −U culture media supplemented with 2% (w/v) Gal to 0.4 mg cells mL−1 and incubated at 30°C for 24 h. Cell extracts containing the recombinant protein were obtained by resuspending the induced cells in breaking buffer (50 mm sodium phosphate, pH 7.4, 1 mm EDTA, and 5% [v/v] glycerol) and then lysing the cells with glass beads using a Mini-Bead Beater-8 (BioSpec Products, Bartlesville, OK).
Isolation of the β-1,6-Glucanase Expression Plasmid from Brewer's Yeast
Two loops full of the Brewer's yeast transformant cells growing on yeast peptone dextrose medium were suspended in 200 μL of lysis buffer (10 mm Tris-HCl, pH 8.0, 1 mm Na2EDTA, 100 mm NaCl, and 0.1% [w/v] SDS). Glass beads were added, and the cells were lysed in a Bead Beater for 75 s. An equal volume of phenol:chloroform:isoamyl alcohol (25:24:1, v/v) was added and mixed in the Bead Beater for 60 s. The mixture was centrifuged, and the aqueous layer was extracted with an equal volume of chloroform. The plasmid in the aqueous layer was purified using Gene Clean (Bio 101) and used to transform DH10B E. coli cells by electroporation. The plasmid was purified from an E. coli transformant and sequenced.
Secreted β-1,6-Glucanase Activity from the Cultured Endophyte
The endophyte was grown in four culture formulations, and the secreted β-1,6-glucanase activity in the media was determined. The four media were autoclaved yeast cells, potato dextrose broth, tryptone-Suc medium (Lam et al., 1995), and tryptone-Suc medium containing 1% (w/v) pustulan. The yeast cell medium was prepared by resuspending 50 g of washed Bakers yeast (ICN, Costa Mesa, CA) in 1 L of water and autoclaving the media for 20 min. The tryptone-Suc medium consisted of Murashige and Skoog basal salt micronutrients (Sigma-Aldrich) supplemented with 332.2 mg L−1 CaCl2, 180.7 mg L−1 MgSO4, 3.0 g L−1 KH2PO4, 2.0 g L−1 K2HPO4, 2.0 g L−1 tryptone, 200 μg L−1 thiamine, and 0.1% (w/v) Suc.
Partial Purification of the β-1,6-Glucanase
For purification of the β-1,6-glucanase, the endophyte was grown in the yeast cell medium for 5 d. The fungal mycelia and yeast cells were separated from the culture filtrate by centrifuging the culture at 5,000g for 10 min and then filtering the culture supernatant through Miracloth (Calbiochem). Eight-hundred milliliters of the culture filtrate was then stirred with 600 mg of ethanol-precipitated pustulan at 4°C overnight to allow adsorption of the β-1,6-glucanase. Ethanol-precipitated pustulan was prepared by dissolving pustulan in hot water (1 mg mL−1), cooling, adding 2 volumes of 95% (v/v) ethanol, centrifuging at 25,000g for 15 min, and then drying in a vacuum centrifuge. After adsorption, the pustulan was pelleted and washed twice with 1 m NaCl, 70 mm MES, pH 6.0. Bound protein was then eluted by resuspending the pustulan pellet in 2 m NaCl and 70 mm MES, pH 6.0. The eluted protein was concentrated, and the buffer changed to 10 mm MES, pH 6.0, using Centricon-30 concentrators (Amicon, Beverly, MA).
The partially purified protein was subjected to SDS-PAGE (Laemmli, 1970) and electroblotted to a polyvinylidene difluoride membrane (ProBlott, Applied Biosystems) for protein sequencing. Transferred proteins were visualized by staining the membrane with 0.2% (w/v) Ponceau S (Sigma-Aldrich) in 1% (v/v) acetic acid. The membrane was destained with water, and the 47-kD band was excised. N-terminal sequencing was performed by the Texas Microchemistry Facility (University of Texas Medical Branch, Galveston).
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Footnotes
This work was supported by the National Science Foundation (grant no. IBN 96–04537 to F.C.B.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010108.
LITERATURE CITED
- Bacon CW, White JF., Jr . Stains, media, and procedures for analyzing endophytes. In: Bacon C, White Jr JF, editors. Biotechnology of Endophytic Fungi. Boca Raton, FL: CRC Press; 1994. pp. 47–56. [Google Scholar]
- Bom IJ, Dielbandhoesing SK, Harvey KN, Oomes SJCM, Klis FM, Brul S. A new tool for studying the molecular architecture of the fungal cell wall: one-step purification of recombinant Trichoderma β-(1–6)-glucanase expressed in Pichia pastoris. Biochim Biophys Acta. 1998;1425:419–424. doi: 10.1016/s0304-4165(98)00096-8. [DOI] [PubMed] [Google Scholar]
- Bonants PJM, Fitters PFL, Thijs H, den Belder E, Waalwijk C, Henfling JWDM. A basic serine protease from Paecilomyces lilacinus with biological activity against Meloidogyne hapla eggs. Microbiology. 1995;141:775–784. doi: 10.1099/13500872-141-4-775. [DOI] [PubMed] [Google Scholar]
- Breen JP. Acremonium endophyte interactions with enhanced plant resistance to insects. Annu Rev Entomol. 1994;39:401–423. [Google Scholar]
- Bush LP, Wilkinson HH, Schardl CL. Bioprotective alkaloids of grass-fungal endophyte symbioses. Plant Physiol. 1997;114:1–7. doi: 10.1104/pp.114.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chet I. Trichoderma: application, mode of action, and potential as a biocontrol agent of soil borne plant pathogenic fungi. In: Chet I, editor. Innovative Approaches to Plant Disease Control. New York: John Wiley & Sons; 1987. pp. 137–160. [Google Scholar]
- Clarke BB, White JF Jr, Funk CR, Sun S, Huff DR, Hurley RH (2002) Enhanced resistance to dollar spot in endophyte-infected fine fescues. Plant Dis (in press) [DOI] [PubMed]
- Clay K. Fungal endophytes of grasses: a defensive mutualism between plants and fungi. Ecology. 1988;69:10–16. [Google Scholar]
- Clay K. Fungal endophytes of grasses. Annu Rev Ecol Syst. 1990;21:275–297. [Google Scholar]
- Cutfield SM, Davies GJ, Murshudov G, Anderson BF, Moody PCE, Sullivan PA, Cutfield JF. The structure of the exo-beta-(1,3)-glucanase from Candida albicans in native and bound forms: relationship between a pocket and groove in family 5 glycosyl hydrolases. J Mol Biol. 1999;294:771–783. doi: 10.1006/jmbi.1999.3287. [DOI] [PubMed] [Google Scholar]
- de la Cruz J, Llobell A. Purification and properties of a basic endo-beta-1,6-glucanase (BGN16.1) from the antagonistic fungus Trichoderma harzianum. Eur J Biochem. 1999;265:145–151. doi: 10.1046/j.1432-1327.1999.00698.x. [DOI] [PubMed] [Google Scholar]
- de la Cruz J, Pintor-Toro JA, Benitez T, Llobell A. Purification and characterization of an endo-beta-1,6-glucanase from Trichoderma harzianum that is related to its mycoparasitism. J Bacteriol. 1995;177:1864–1871. doi: 10.1128/jb.177.7.1864-1871.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: version II. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS. “Touchdown” PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dygert SL, Li H, Florida D, Thomas JA. Determination of reducing sugar with improved precision. Anal Biochem. 1965;13:367–374. doi: 10.1016/0003-2697(65)90327-1. [DOI] [PubMed] [Google Scholar]
- Geremia RA, Goldman GH, Jacobs D, Ardiles W, Vila SB, Van Montagu M, Herrera-Estrella A. Molecular characterization of the proteinase-encoding gene, prb1, related to mycoparasitism by Trichoderma harzianum. Mol Microbiol. 1993;8:603–613. doi: 10.1111/j.1365-2958.1993.tb01604.x. [DOI] [PubMed] [Google Scholar]
- Glenn AE, Bacon CW, Price R, Hanlin RT. Molecular phylogeny of Acremonium and its taxonomic implications. Mycologia. 1996;88:369–383. [Google Scholar]
- Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M et al. Life with 6000 genes. Science. 1996;274:546–552. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- Henrissat B, Davies G. Structural and sequence-based classification of glycoside hydrolases. Curr Opin Struct Biol. 1997;7:637–644. doi: 10.1016/s0959-440x(97)80072-3. [DOI] [PubMed] [Google Scholar]
- Hien NH, Fleet GH. Separation and characterization of six (1,3)-β-glucanases from Saccharomyces cerevisiae. J Bacteriol. 1983;156:1204–1213. doi: 10.1128/jb.156.3.1204-1213.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiura N, Nakajima T, Matsuda K. Purification and some properties of an endo-β-1,6-glucanase from Neurospora crassa. Agric Biol Chem. 1987;51:3315–3321. [Google Scholar]
- Johnson-Cicalese J, Secks ME, Lam CK, Meyer WA, Murphy JA, Belanger FC. Cross species inoculation of Chewings and strong creeping red fescues with fungal endophytes. Crop Sci. 2000;40:1485–1489. [Google Scholar]
- Julius D, Brake A, Blair L, Kunisawa R, Thorner J. Isolation of the putative structural gene for the lysine-arginine-cleaving endopeptidase required for processing of yeast prepro-alpha-factor. Cell. 1984;37:1075–1089. doi: 10.1016/0092-8674(84)90442-2. [DOI] [PubMed] [Google Scholar]
- Kim D-J, Baek J-M, Uribe P, Kenerley CM, Cook DR. Cloning and characterization of multiple glycosyl hydrolase genes from Trichoderma virens. Curr Genet. 2002;40:374–384. doi: 10.1007/s00294-001-0267-6. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lam CK, Belanger FC, White JF, Jr, Daie J. Invertase activity in Epichloe/Acremonium fungal endophytes and its possible role in choke disease. Mycol Res. 1995;99:867–873. [Google Scholar]
- Leuchtmann A, Schardl CL, Siegel MR. Sexual compatibility and taxonomy of a new species of Epichloe symbiotic with fine fescue grasses. Mycologia. 1994;86:802–812. [Google Scholar]
- Lindstrom JT, Belanger FC. Purification and characterization of an endophytic fungal proteinase that is abundantly expressed in the infected host grass. Plant Physiol. 1994;106:7–16. doi: 10.1104/pp.106.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom JT, Sun S, Belanger FC. A novel fungal protease expressed in endophytic infection of Poa species. Plant Physiol. 1993;102:645–650. doi: 10.1104/pp.102.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lora JM, de la Cruz J, Llobell A, Benitez T, Pintor-Toro JA. Molecular characterization and heterologous expression of an endo-beta-1,6-glucanase gene from the mycoparasitic fungus Trichoderma harzianum. Mol Gen Genet. 1995;247:639–645. doi: 10.1007/BF00290356. [DOI] [PubMed] [Google Scholar]
- Magnelli P, Cipollo JF, Abeijon C. A refined method for the determination of Saccharomyces cerevisiae cell wall composition and β-1,6-glucan fine structure. Anal Biochem. 2002;301:136–150. doi: 10.1006/abio.2001.5473. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1982. pp. 68–69. [Google Scholar]
- Manners DJ, Masson AJ, Patterson JC, Bjorndal H, Lindberg B. The structure of a β-(1–6)-d-glucan from yeast cell walls. Biochem J. 1973;135:31–36. doi: 10.1042/bj1350031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy M, Belanger FC, Duncan R, Freehoff A, Leary C, Meyer W, Sullivan R, White JF., Jr Identification of epiphyllous mycelial nets on leaves of grasses infected by clavicipitaceous endophytes. Symbiosis. 2000;28:291–302. [Google Scholar]
- Mulenga DK, Berry DR. Isolation and characterization of a unique endo-beta-1,6-glucanase from the yeast Saccharomycopis fibuligera NCYC 451. Microbios. 1994;80:143–154. [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Papavizas GC. Trichoderma and Gliocladium: biology, ecology, and potential for biocontrol. Annu Rev Phytopathol. 1985;23:23–54. [Google Scholar]
- Pitson SM, Seviour RJ, Bott J, Stasinopoulos SJ. Production and regulation of β-glucanases in Acremonium and Cephalosporium isolates. Mycol Res. 1991;95:352–356. [Google Scholar]
- Pitson SM, Seviour RJ, McDougall BM. Production of β-glucan degrading enzymes by Acremonium and Cephalosporium species. Mycol Res. 1997;101:153–158. [Google Scholar]
- Pitson SM, Seviour RJ, McDougall BM, Stone BA, Sadek M. Purification and characterization of an extracellular (1–6)-beta-glucanase from the filamentous fungus Acremonium persicinum. Biochem J. 1996;316:841–846. doi: 10.1042/bj3160841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy PV, Lam CK, Belanger FC. Mutualistic fungal endophytes express a proteinase that is homologous to proteases suspected to be important in fungal pathogenicity. Plant Physiol. 1996;111:1209–1218. doi: 10.1104/pp.111.4.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed KC, Mann DA. Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res. 1985;13:7207–7221. doi: 10.1093/nar/13.20.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese ET, Parrish FW, Mandels M. β-d-1,6-Glucanases in fungi. Can J Microbiol. 1962;8:327–334. doi: 10.1139/m62-045. [DOI] [PubMed] [Google Scholar]
- Santos T, Nombela C, Villanueva JR, Larriba G. Characterization and synthesis regulation of Penicillium italicum 1,6-beta-glucanase. Arch Microbiol. 1979;121:265–270. doi: 10.1007/BF00425066. [DOI] [PubMed] [Google Scholar]
- Schardl CL. Epichloe species: fungal symbionts of grasses. Annu Rev Phytopathol. 1996;34:109–130. doi: 10.1146/annurev.phyto.34.1.109. [DOI] [PubMed] [Google Scholar]
- Schep GP, Shepard MG, Sullivan PA. Purification and properties of a beta-1,6-glucanase from Penicillium brefeldianum. Biochem J. 1984;223:707–714. doi: 10.1042/bj2230707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selden RF. Analysis of RNA by northern hybridization. In: Ausubel FE, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. New York: John Wiley; 1987. pp. 4.9.1–4.9.8. [Google Scholar]
- Short U, Fernandez JM, Sorge JA, Huse WD. λZAP: a bacteriophage λ expression vector with in vivo excision properties. Nucleic Acids Res. 1988;16:7583–7599. doi: 10.1093/nar/16.15.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel MR, Latch GCM, Johnson MC. Fungal endophytes of grasses. Annu Rev Phytopathol. 1987;25:293–315. [Google Scholar]
- Siegel MR, Schardl CL. Fungal endophytes of grasses: detrimental and beneficial associations. In: Andrews JH, Hirano SS, editors. Microbial Ecology of Leaves. New York: Springer-Verlag; 1991. pp. 198–221. [Google Scholar]
- Sietsma JH, Wosten HAB, Wessels JGH. Cell wall growth and protein secretion in fungi. Can J Bot. 1995;73:S388–S395. [Google Scholar]
- Stasinopoulos SJ, Seviour RJ. Exopolysaccharide formation by isolates of Cephalosporium and Acremonium. Mycol Res. 1989;92:55–60. [Google Scholar]
- St. Leger RJ. The role of cuticle-degrading proteases in fungal pathogenesis of insects. Can J Bot. 1995;73:S1119–S1125. [Google Scholar]
- Varner JE, Lin LS. Plant cell wall architecture. Cell. 1989;56:231–239. doi: 10.1016/0092-8674(89)90896-9. [DOI] [PubMed] [Google Scholar]
- Wessels JGH. Developmental regulation of fungal cell wall formation. Annu Rev Phytopathol. 1994;32:413–437. [Google Scholar]
- Wessels JGH. Fungi in their own right. Fungal Genet Biol. 1999;27:134–145. doi: 10.1006/fgbi.1999.1125. [DOI] [PubMed] [Google Scholar]
- Yamamoto S, Kobayashi R, Nagasaki S. Purification and properties of an endo-β-1,6-glucanase from Rhizopus chinensis R-69. Agric Biol Chem. 1974;38:1493–1500. [Google Scholar]
- Zheng Y, Wozniak CA. Adaptation of a β-1,3-glucanase assay to microplate format. BioTechniques. 1997;22:922–926. doi: 10.2144/97225st06. [DOI] [PubMed] [Google Scholar]