Abstract
Mineral nutrient deficiencies constitute major limitations for plant growth on agricultural soils around the world. To identify genes that possibly play roles in plant mineral nutrition, we recently generated a high-density array consisting of 1,280 genes from tomato (Lycopersicon esculentum) roots for expression profiling in nitrogen (N) nutrition. In the current study, we used the same array to search for genes induced by phosphorus (P), potassium (K+), and iron (Fe) deficiencies. RNA gel-blot analysis was conducted to study the time-dependent kinetics for expression of these genes in response to withholding P, K, or Fe. Genes previously not associated with P, K, and Fe nutrition were identified, such as transcription factor, mitogen-activated protein (MAP) kinase, MAP kinase kinase, and 14-3-3 proteins. Many of these genes were induced within 1 h after withholding the specific nutrient from roots of intact plants; thus, RNA gel-blot analysis was repeated for specific genes (transcription factor and MAP kinase) in roots of decapitated plants to investigate the tissue-specific location of the signal triggering gene induction. Both genes were induced just as rapidly in decapitated plants, suggesting that the rapid response to the absence of P, K, or Fe in the root-bathing medium is triggered either by a root-localized signal or because of root sensing of the mineral environment surrounding the roots. We also show that expression of Pi, K, and Fe transporter genes were up-regulated by all three treatments, suggesting coordination and coregulation of the uptake of these three essential mineral nutrients.
Mineral nutrient deficiencies constitute major limitations for crop plant growth on agricultural soils around the world. Among the essential mineral nutrients, P and K are the macronutrients (along with N) that require the greatest agricultural investment with regard to fertilizer inputs, and Fe is the micronutrient that is most limiting to agricultural production worldwide (Kochian, 2000). Hence, there has been considerable research over the past decade investigating molecular and physiological mechanisms of P, K, and Fe acquisition and use. Much of the progress from this research has involved the identification of structural genes of primary importance for mineral nutrition, including mineral ion transporters and enzymes involved in nutrient assimilation. As researchers have gained a more detailed understanding of these mineral nutrition-related genes and the proteins they encode, there has been a growing awareness that mineral nutrient acquisition and homeostasis is a highly regulated and complex set of processes. It is becoming clear that changes in plant mineral nutrient status result in signals that ultimately are transduced into alterations in expression of mineral nutrition-related genes and proteins, resulting in changes in plant mineral nutrition that are beneficial to the plant.
For example, P (primarily as phosphate [Pi]) is an important structural component of nucleic acids, phospholipids, and ATP. It acts as both a substrate and regulatory factor in photosynthesis and oxidative metabolism and participates in signal transduction by way of protein phosphorylation/dephosphorylation. Its low solubility and high sorption capacity in soils make it relatively unavailable to plant roots. As a consequence, Pi supply is one of the most important constraints to crop production worldwide (Raghothama, 1999). It has long been known that P deficiency triggers a large number of physiological changes in plants presumed to enhance Pi acquisition from the soil and to deal with suboptimal levels of P within the plant. These include changes in root architecture and root to shoot ratio, stimulation of root Pi absorption, increases in the release of phosphatases and RNases from roots, and changes in respiration that reduce the dependency in plant metabolism on P in high energy phosphate bonds (e.g. ATP). Molecular research into P nutrition has more recently indicated that P deficiency triggers changes in gene and protein expression in a way that indicates that a number of genes and proteins involved in P nutrition are regulated by plant P status (Liu et al., 1998; Muchhal and Raghothama, 1999; Raghothama, 1999; Mukatira et al., 2001).
The challenge now facing researchers in the field of molecular plant nutrition is to begin to identify and characterize the components of signaling cascades that plants use to sense changes in both their internal mineral status and the rhizosphere mineral environment and subsequently to transduce these signals to facilitate mineral nutrient homeostasis. In this paper, we have employed a cDNA array as a preliminary screening tool to rapidly identify genes involved in P, K, and Fe nutrition that had not previously been identified as such, with particular interest in candidate genes involved in mineral nutrient homeostasis. We recently generated a high-density array consisting of 1,280 mineral nutrition-related genes from tomato (Lycopersicon esculentum) roots that was used for gene expression profiling and resulted in the identification of a number of novel nitrate-induced genes that play roles in N nutrition (Wang et al., 2001). In the current study, we used the same cDNA array as a preliminary screening tool to identify mainly regulatory or signaling genes induced by specific mineral nutrient deficiencies (P, K, and Fe). More detailed investigation using RNA gel-blot analysis was subsequently used to study the time courses for expression of these genes in response to withholding P, K, or Fe. A set of putative regulatory genes associated with P, K, and Fe nutrition were identified, including genes that could play a role in mineral nutrition signal transduction such as transcription factor, mitogen-activated protein (MAP) kinase, and 14-3-3 proteins, as well as ion transporters up-regulated by all three deficiencies. Selected genes were also studied for their expression in decapitated plants to begin to determine whether the signal(s) triggering gene induction are localized to the shoot or root, or are attributable to root sending of the rhizosphere mineral status.
RESULTS
The high-density array containing the 1,280 mineral nutrition-related genes was screened with cDNA probes made from mRNA isolated from roots of tomato plants exposed to −Pi, −K, or −Fe hydroponic medium for 0, 1, 3, 6, 12, 24, or 48 h to identify candidate genes that were then studied in more detail via RNA gel-blot analysis for their possible role(s) in mineral nutrient homeostasis. To date, we have sequenced and annotated 657 of the 1,280 cDNAs on this array. The annotated list of these sequenced genes is contained in supplemental Table I by Wang et al. (2001; this supplemental data can be viewed at www.plantphysiol.org). When unsequenced cDNAs are found to exhibit strong changes in gene expression in response to changes in plant mineral status, these clones are then sequenced as part of the subsequent analysis. The screening of the array was used as a tool to rapidly identify candidate genes up-regulated by withholding P, K, or Fe from the nutrient medium. Candidate genes were chosen as ones that were rapidly (within hours) induced by any (or all) of the three nutrient treatments compared with full-nutrient control plants. These candidate genes were then used for more detailed analysis using RNA gel-blot analysis. In our initial differential screening of the mineral nutrition cDNA array, a total of 195 genes exhibited significant changes in expression either in response to alterations in plant status of a specific mineral nutrient (P, K, or Fe) or in response to more than one of these mineral status alterations. The great majority of the overall pool of 197 cDNA clones exhibited increased expression, which should be expected because the original subtractive libraries were designed to be enriched in genes up-regulated by changes in plant mineral status (many of these genes are also metabolism related). Within this set of clones were a number of genes that would be expected to be up-regulated by withholding a specific mineral nutrient, such as the Pi transporters LePT1 and LePT2 in response to −Pi conditions, the K+ transporters LeKC1 and LeHAK5 in response to −K+ conditions, and the Fe transporter LeIRT1 in response to −Fe conditions. Recovery of genes previously reported to be induced by these same specific changes in plant mineral status was viewed as validation of the strategy used in this research.
To select a reasonable number of genes for further study, we focused on genes that: (a) exhibited the strongest increase in expression; (b) exhibited the most rapid increase in expression to begin to identify candidate genes for roles in signaling aspects of mineral nutrient homeostasis; and (c) exhibited increases in expression in response to changes in plant status for all three mineral nutrients, to identify candidate genes that might play a general role in mineral nutrition or represent signaling components common to regulation of all three mineral nutrients. On the basis of these criteria, we narrowed our focus to a set of 17 genes identified by array analysis and subsequently confirmed via RNA gel blots (Table I). In this group are a number of putative regulatory genes that responded to all three deficiency treatments. These include a Leu-zipper transcription factor (nutrient-induced transcription factor [Nitf]), MAP kinase, MAP kinase kinase (MEK1), and 14-3-3 proteins. They exhibited the strongest and most rapid increase in expression in response to changes in plant status for all three mineral nutrients. We also found that a specific transporter for one mineral nutrient often was induced by the absence of the other nutrients. This “cross talk” phenomenon was also reported in our previous study on nitrate-induced genes in tomato roots in that we found that withholding P, K, or Fe induced root nitrate transporters (Wang et al., 2001).
Table I.
Genes induced by Pi, K, or Fe deficiencies in tomato roots and confirmed by northern analysis
| Clone ID | Gene (Induction) | Homology/Function | Accession No. | GenBank Hit | Reference |
|---|---|---|---|---|---|
| D22-1 | MAP kinase (up 1–3 h) | Tobacco WIPK/signaling | AW979631 | D61377 | Seo et al. (1995) |
| H10-1 | Nitf (up 1–6 h) | Tobacco TGA1a/regulatory | AAK71287 | X16450 | Katagiri et al. (1989) |
| K20-1 | MEK1 (up 1 h) | Tomato gene/signaling | AW220008 | CAA04261 | Hackett et al. (1998) |
| O22-1 | 14-3-3 (up 3–12 h) | Tomato gene/signaling | AW218217 | AF079450X95905 | Roberts and Bowles (1999); Pnueli et al. (2001) |
| 27L1 | LePT1 (up 3–12 h) | Tomato gene/Pi transporter | AF022873 | AF022873 | Liu et al. (1998) |
| E13-1 | LeHAK5 (up 1 h) | Arabidopsis HAK5 /K+ transporter | AW979921 | AF129478 | |
| G22-1 | LeKC1 (up 3–12 h) | Carrot Kdc1/K+ channel | AW218955 | AJ249962 | Downey et al. (2000) |
| I20-1 | LeIRT2 (3–24 h) | Tomato gene/iron transporter | AW219515 | AF246266 | Eckhardt et al. (2001) |
| 1A16 | Nicotianamine synthase (up 3–6 h) | Tomato gene /nicotianamine biosynthesis | BG791292 | CAB42052 | Ling et al. (1999) |
| N5-2 | Glu decarboxylase (up 1–6 h) | Tobacco GAD/GABA biosynthesis | BG713805 | AAB40608 | |
| I17-2 | Enolase (up 1–12 h) | Tomato gene/glycolyis | BG791245 | X58108 | Van der Straeten et al. (1991) |
| A4-4 | ATHP3 (up 1–6 h) | Arabidopsis ATHP3 /phosphorelay mediator | BG791212 | BAA37112 | Miyata et al. (1998) |
| D11-1 | ERF3 (up 1–3 h) | Tobacco ERF3 /regulatory | AW979604 | AB024575 | |
| G20-2 | SKD1 (up 3–24 h) | Human SKD1 /membrane transport | AW621245 | AAF17203 | Scheuring et al. (1999) |
| G19-1 | bet3 (up 12–48 h) | Mouse bet3/vesicular transport | AW218945 | AF041433 | Jiang et al. (1998) |
| N3-1 | dwarf1 (down after 1 h) | Arabidopsis dwarf1/cell elongation | BG791226 | U12400 | Takahashi et al. (1995) |
| B2-1 | MAP kinase 6 (up 3–6 h) | Tobacco MAPK 6/signaling | AW980007 | X83879 | Wilson et al. (1995) |
Except for dwarf1, all other genes were up-regulated by nutrient treatments. See text for details.
Regulatory and Signaling Genes
Several genes encoding proteins that could play a role in signal transduction pathways linking signals resulting from changes in plant nutrient status to altered expression and activity of mineral ion transporters and assimilation enzymes were found to be up-regulated very rapidly (between 1 and 3 h) after withholding Pi, K, or Fe. These genes include MAP kinases, a MAP kinase kinase (MEK1), a putative transcription factor (Nitf), and a 14-3-3 protein that were investigated in more detail via RNA gel-blot analysis.
MAP Kinases
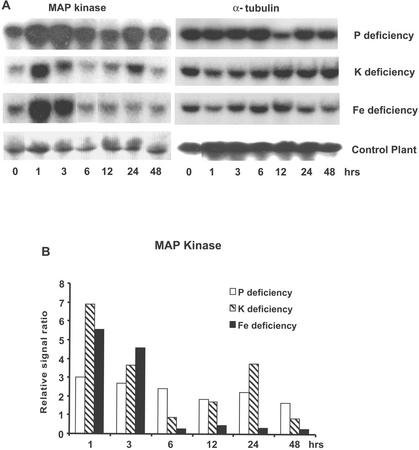
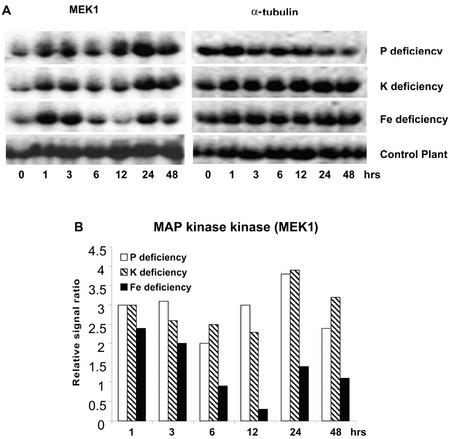
In response to Pi, K, and Fe deficiency treatments, a gene exhibiting significant sequence homology to a tobacco (Nicotiana tabacum) wound-inducible protein kinase (WIPK) was induced very rapidly and strongly (within 1 h after exposure to −P, −K, or −Fe nutrient solution; Fig. 1). This WIPK homolog belongs to the family of MAP kinases. This gene exhibited a stronger (5- to 7-fold) increase in expression in response to K and Fe deficiency that was transient in nature, lasting approximately 3 h, whereas its response to Pi deficiency was more sustained (Fig. 1). A MAP kinase kinase, MEK1 was also rapidly and transiently induced within 1 h after exposure to the deficiency conditions (Fig. 2). This gene has previously been found to be induced by leaf senescence and wounding in tomato (Hackett et al., 1998).
Figure 1.
RNA gel-blot profile for tomato root MAP kinase induced by deprivation of Pi, K, or Fe. The MAP kinase is homologous to tobacco WIPK and is rapidly induced by withholding Pi, K, and Fe from the plant. A, RNA gel blot. In this and subsequent experiments, RNA abundance for roots of treated plants was compared with gel blots for roots of control (nutrient-sufficient) plants analyzed at the same time points to ensure that the changes in transcript abundance were not attributable to a diurnal response. In none of the experiments did the control plants show significant changes in gene expression over the 48-h time period used for these experiments. B, Quantitative expression data for the RNA gel blot depicted in A. In Figure 1B and the subsequent figures (except Fig. 4), the autoradiograph of the RNA gel blot was scanned, and the image was computer digitized. The gel-blot intensities were quantified using the NIH ImageJ program. Changes in gene expression were quantified as a relative signal ratio, which was determined by dividing the quantified gel-blot intensity at each time point by the RNA-blot intensity for the 0-h nutrient-sufficient plants.
Figure 2.
Up-regulation of the tomato MAP kinase kinase, MEK1, induced by Pi, K, or Fe deprivation. A, RNA gel blot. B, Quantitative expression data for the RNA gel blot depicted in A.
Transcription Factor
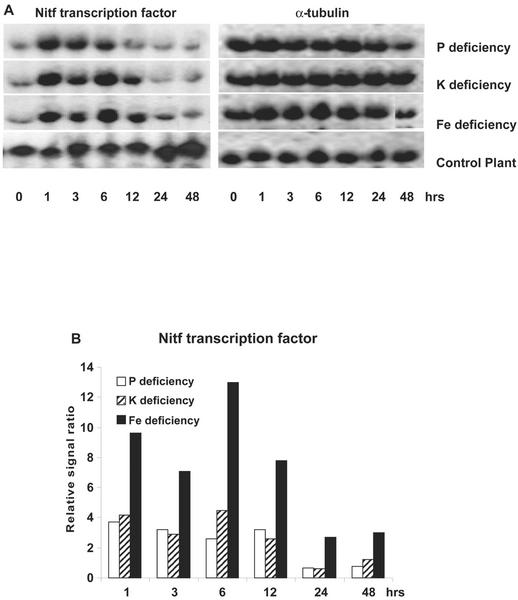
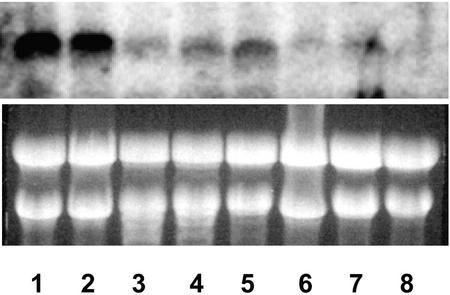
A putative transcription factor, Nitf, was found to be very strongly induced by all three nutrient deficiencies (Fig. 3). Nitf exhibited a 4- to 10-fold increase in expression within 1 h after withholding Pi, K, or Fe. In a previous study, we also found this gene to be strongly induced by nitrate resupply (Wang et al., 2001). Its expression was not altered by exposure of tomato plants to salt or cold stress (data not shown), suggesting that its enhanced expression may be specific to changes in plant mineral nutrition. Furthermore, an analysis of its tissue-specific expression showed that Nitf is preferentially expressed in roots, with a decreased level in flowers and younger leaves and a much lower expression in stems, older leaves, and unripe fruits (Fig. 4). Comparison of the amino acid sequence for Nitf with homologs from other plant species indicated it was 80% identical to the tobacco bZIP transcription factor, TGA1a (Katagiri et al., 1989). It has been reported that TGA1a can selectively activate transcription of target genes in response to auxins and auxin analogs, and TGA1a has been implicated as playing a role in the expression of plant genes involved in chemical defense (Pascuzzi et al., 1998). TGA1a has also been found to be highly expressed in tobacco root (Katagiri et al., 1989), especially in the root tip meristems (Klinedinst et al., 2000).
Figure 3.
The transcription factor Nitf was induced by Pi, K, and Fe deprivation in tomato roots. A, RNA gel blot. B, Quantitative expression data for the RNA gel blot depicted in A.
Figure 4.
Tissue-specific expression of the transcription factor, Nitf, in different tissues and organs of tomato seedlings grown under nutrient-sufficient hydroponical conditions. Lanes 1 and 2, Roots. Because the root system was relatively large, different parts of the roots were harvested and bulked. Lane 3, Older leaves harvested from below middle trusse of the plants. These are completely functioning leaves, healthy, not showing any aging symptoms. Lane 4, Young leaves harvested from the top of the plants. They were barely fully expanded. Lane 5, Flowers. Lane 6, Old stems. Main stems harvested from bottom part of the plants, directly above the root. Lane 7, Young stems. Main stems harvested from the top part of the plants. Lane 8, Unripe fruits. Green tomato fruits of various sizes. All tissues were harvested from the same plants at the same physiological age.
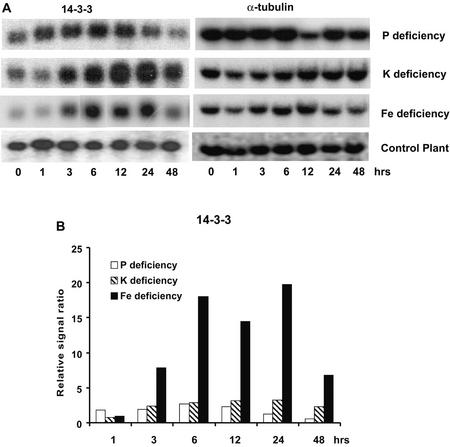
14-3-3 Protein
In eukaryotes, 14-3-3 proteins play important roles in the import of nuclear-encoded chloroplast proteins, assembly of transcription factor complexes, and regulation of enzyme activity in response to intracellular signal transduction cascades (Roberts, 2000). In plants, 14-3-3 proteins have been shown to play a role in regulating important metabolic enzymes such as nitrate reductase and Suc phosphate synthase, in addition to activation of the plasma membrane H+-ATPase (for review, see Chung et al., 1999). In this study, a putative 14-3-3 protein was found to respond to Pi, K, and Fe deprivation (Fig. 5). This tomato 14-3-3 protein responded most strongly to K and Fe deprivation, whereas its response to −Pi treatment is more moderate.
Figure 5.
RNA gel blot for a gene encoding a 14-3-3 protein induced by Pi, K, and Fe deprivation. A, RNA gel blot. B, Quantitative expression data for the RNA gel blot depicted in A.
Ion Transporters
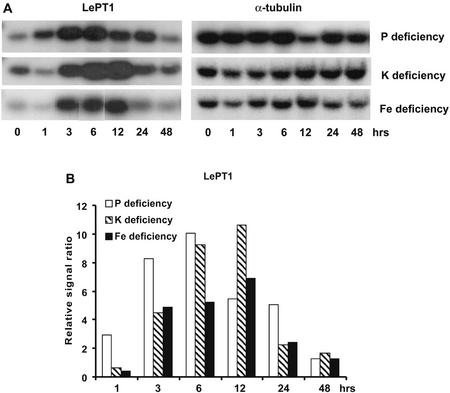
The expression of the tomato Pi transporter, LePT1, was up-regulated in response to all three deficiency treatments in a similar time-dependent manner in that it was up-regulated between 3 to 12 h after imposition of the specific mineral deficiency (Fig. 6). It was interesting to note that LePT1 expression was increased more strongly in response to K and to Fe deficiencies than it was in response to Pi deficiency. An earlier study of LePT1 expression did not find a change in its expression in response to K and Fe deficiencies when the deficiency was imposed for 5 d (Liu et al., 1998), suggesting that K and Fe deficiency induction of the gene was transient in nature. LePT2 was found to be strongly induced by Pi deficiency, but induction by K and Fe deficiencies was not significant (data not shown). Overall, transcript level of LePT2 was markedly lower than that of LePT1 based on intensity of hybridization signal.
Figure 6.
Expression of the tomato Pi transporter, LePT1, was strongly up-regulated in roots of plants subjected to Pi, K, and Fe deprivation. A, RNA gel blot. B, Quantitative expression data for the RNA gel blot depicted in A.
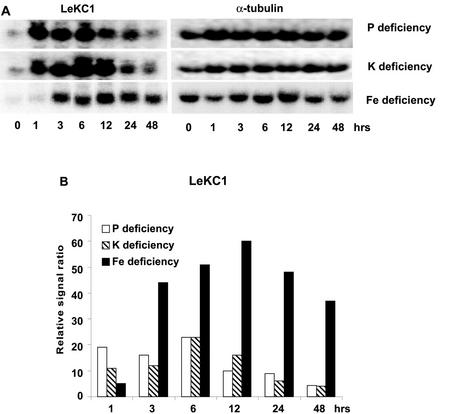
Expression of a K+ channel homolog (LeKC1) of the carrot (Daucus carota) K+ transporter, Kdc1 (Downey et al., 2000), was increased rapidly in response to withholding all three mineral nutrients (Fig. 7). Expression of LeKC1 was increased most rapidly in response to Pi and K deficiencies (1 h), whereas increased expression in response to −Fe conditions was seen after 3 h of treatment. Kdc1 was recently shown to be a voltage- and pH-dependent inwardly rectifying K+ channel that is expressed in carrot roots (Downey et al., 2000). Another K+ transporter, LeHAK5, was also rapidly induced by all three deficiencies (data not shown).
Figure 7.
Expression of a K+ channel homolog of carrot Kdc1, LeKC1 was up-regulated by Pi, K, and Fe deprivation. A, RNA gel blot. B, Quantitative expression data for the RNA gel blot depicted in A.
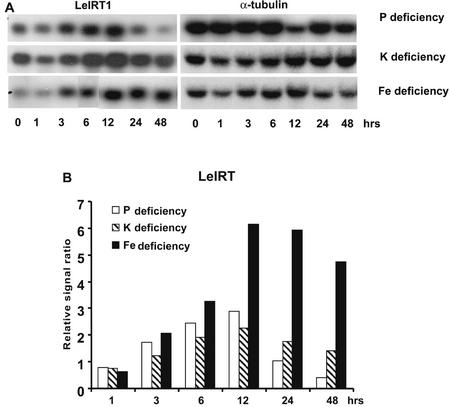
Expression of the tomato Fe transporter, LeIRT1, was also up-regulated by all three mineral deficiencies (Fig. 8). It has been previously shown that LeIRT1 expression was induced most strongly in response to Fe deficiency (Eckhardt et al., 2001). Compared with other transporter genes (LeKC1, LePT1, and LeHAK5), the overall transcript abundance of LeIRT1 was also considerably lower.
Figure 8.
Expression of the tomato Fe transporter, LeIRT1, was up-regulated by Pi, K, and Fe deprivation. A, RNA gel blot. B, Quantitative expression data for the RNA gel blot depicted in A.
Gene Induction Experiments Using Decapitated Plants
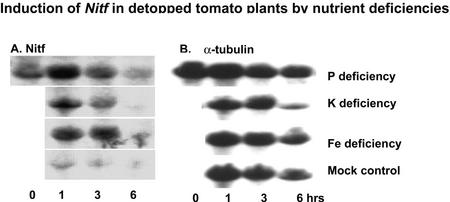
The induction of many of these genes within 1 h after withholding P, K, and Fe suggests that the signal triggering this response may be root localized and may not arise from the shoot as often hypothesized (Liu et al., 1998; Burleigh and Harrison, 1999). It would not be expected for these rather large, 1-month-old tomato plants grown under luxury nutrient conditions that withholding P, K, or Fe for 1 h would cause a significant decline in shoot mineral nutrient status. To investigate this further, RNA gel-blot analysis was repeated for Nitf, because it was the gene most strongly and rapidly induced by all three nutrient deficiencies. Because we were using root systems separated from the shoot and thus removed from their primary source of energy in photosynthates transported from the roots, the experiments were limited to 0-, 1-, 3-, and 6-h time points. As shown in Figure 9, Nitf was strongly induced over the first 3 h with a pattern similar to that seen for roots of intact plants (Fig. 3). At the 6-h time point, a decline in Nitf transcript abundance was seen for all treatments, suggesting a general decline in root metabolism. It is interesting to note that in the nutrient-sufficient decapitated root systems, Nitf expression declined rapidly after excision, compared with no change in expression in intact, nutrient-sufficient roots (compare Fig. 3 with 9).
Figure 9.
Expression of Nitf to Pi, K, and Fe deprivation in decapitated plants. Control plant is the same RNA blot run with RNA from roots of decapitated plants under nutrient-sufficient conditions. A, RNA gel blot. B, α-Tubulin loading control.
Genes Induced by Specific Mineral Deficiencies
Pi Deficiency
A number of additional genes were up-regulated specifically in response to withholding Pi from the nutrient solution (Table I). Several of these play roles in plant metabolism and or plant stress responses. A Glu decarboxylase gene homologous to petunia (Petunia hybrida) GAD (Baum et al., 1993) was induced 1 to 6 h after imposition of −Pi conditions. GAD is a calmodulin-binding protein and catalyzes the conversion of Glu to γ-aminobutyric acid, a four-carbon, non-protein amino acid. Levels of γ-aminobutyric acid in plant can be increased severalfold in response to many diverse stimuli, including heat shock, mechanical stimulation, hypoxia, and phytohormones (Shelp et al., 1999). A homolog of barley (Hordeum vulgare) nicotianamine synthase, a gene involved in biosynthesis of phytosiderophores in grasses (Herbik et al., 1999; Higuchi et al., 1999; Ling et al., 1999), was up-regulated in roots of Pi-deprived seedlings. The gene was expressed at a high level (13-fold increase) 3 to 24 h after withholding Pi. Nicotianamine synthase was previously shown to be regulated by Fe in monocots (Higuchi et al., 2001). Finally, a gene encoding enolase (2-phospho-d-glycerate hydratase) was also induced rapidly, between 1 and 12 h after withholding Pi. Enolase is an key enzyme in glycolysis and catalyzes the interconversion of 2-phosphoglycerate to PEP. In maize (Zea mays), one of the enolase genes (ENO1) was shown to be induced after 24 h of anaerobic treatment (Lal et al., 1998).
There were several other genes that could play a role in nutrient regulation/signaling that were induced by Pi deficiency. A cDNA clone that is 63% identical and 78% similar to Arabidopsis ATHP3 was up-regulated 1 to 6 h after Pi deprivation (Table I). ATHP3 functions as a two-component phosphorelay mediator between a sensor His kinase and response regulators and was previously found to be expressed more strongly in roots than in other tissues (Miyata et al., 1998). Also, a homolog of an Arabidopsis ethylene-responsive element binding factor (ERF) was rapidly induced in response to Pi deficiency. ERF is a member of a novel family of transcription factors that are specific to plants. It is interesting to note that the Arabidopsis ERF 3 gene (ATERF3) was not induced by ethylene, but was moderately induced by drought, wounding, and salt (NaCl) stress and by exposure to the protein synthesis inhibitor, cycloheximide (Fujimoto et al., 2000).
K and Fe Deficiencies
There were several genes, the expression of which was increased specifically to K or Fe deprivation. In response to −K conditions, a homolog of the mouse SKD1 gene was up-regulated very rapidly (within 1 h) and strongly (8- to 18-fold; Table I). This tomato gene shares 75% identity and 85% similarity to SKD1 in mice, where SKD1 has been implicated in the transport of membrane vesicles from endosomes to the vacuole and is thought to regulate membrane traffic through endomembrane compartments. SKD1 is a “housekeeping” gene in mice, and in mice, it is expressed ubiquitously (Scheuring et al., 1999). Another cDNA clone specifically up-regulated by K deprivation was a homolog of the bet3 gene (56% identity and 77% similarity). Bet3 expression was found to be highest at the end of the −K treatment (48 h). BET3 is required for vesicular transport between the endoplasmic reticulum and Golgi complex in yeast (Jiang et al., 1998).
In response to Fe deficiency, a cDNA clone homologous to the tobacco MAP kinase 6 (91% identical) was induced 3 to 6 h after initiation of the −Fe conditions (Table I). Finally, a homolog of the dwarf1 gene, which is required for cell elongation in Arabidopsis (Takahashi et al., 1995), was found to be rapidly down-regulated within 1 h in response to Fe deficiency.
DISCUSSION
Previous studies strongly suggest that changes in plant status for different mineral nutrients are linked to expression of mineral nutrition-related genes in a manner that facilitates mineral nutrient homeostasis (for reviews, see Grossman and Takahashi, 2001; Kochian, 2000; Raghothama, 2000). Therefore, we are interested in identifying genes that may play a role in plant responses to changes in mineral status. Seventeen different mineral deficiency-induced genes were identified and confirmed by RNA gel-blot analysis, along with genes previously reported to be induced specifically by P, K, and Fe deficiency in plants. These previously reported genes include phosphate transporters (LePT1 and LePT2; Liu et al., 1998) induced in response to −P conditions, K transporters (LeHAK5 and LeKC1) induced by K deprivation, and an Fe transporter (LeIRT1, Eckhardt et al., 2001) induced by −Fe conditions. LeKC1 reported here is a different gene from LKT1 (accession no. CAA65254) reported previously by Hartje et al. (2000) in tomato and is 48% identical to Arabidopsis K+ inward-rectifying channel AtKC1 (AAC98810). LeHAK5 and LeKC1 have not been reported previously in tomato.
Genes that could play roles in signaling and/or regulation were induced very rapidly (1–3 h) by withholding P, K, or Fe. These include MAP kinase and MAP kinase kinase (MEK1) genes, a transcription factor, and a gene encoding a 14-3-3 protein. This is the first report, to our knowledge, in plants for induction of MAP kinases (a WIPK homolog and MEK1) by changes in plant mineral status. In both Brewer's yeast (Saccharomyces cerevisiae) and fission yeast (Schizosaccharomyces pombe), MAP kinase pathways have been found to be activated in response to limited nutrient availability, and have been implicated in survival responses to nutrient limitation (Widmann et al., 1999). In tobacco, expression of WIPK has been shown to be induced by other stresses such as wounding, viral infection, and fungal elicitors (Seo et al., 1995; Zhang and Klessig, 1998; Zhang et al., 2000), whereas induction of MEK1 has been associated with leaf senescence and wounding (Hackett et al., 1998). In plants, MAP kinase pathways have previously been found to function in signal transduction in response to mechanical perturbation, wounding, abiotic stresses (heat, cold, drought, and osmotic), and pathogen attack (Meskiene and Hirt, 2000).
A transcription factor (Nitf, a homolog of tobacco TGA1a) was induced rapidly (in less than 1 h) and most strongly by all three nutrient deficiencies (Fig. 3). Nitf exhibits strong amino acid sequence identity (80% identical) to the tobacco TGA1a transcription factor. TGA1a has been found to interact with PR-1, a group of pathogenesis-related proteins strongly induced in plants by pathogen attack, salicylic acid, and developmental cues, and by changes in nutrient status (Strompen et al., 1998). We previously found that the Nitf transcription factor that was shown here to be induced rapidly by P, K, or Fe deprivation, also was induced by nitrate resupply (Wang et al., 2001). This is coincidental with the observation that nitrate resupply (Wang et al., 2001) and P, K, and Fe deprivation all were found to induce expression of nitrate, Pi, K, and Fe transporters. Furthermore, we found that Nitf was not induced by salt or cold stress, suggesting that its expression may be mineral nutrient specific. The induced expression of Nitf appears to precede the expression of the mineral ion transporters, and leads us to speculate that it may play a role in the early mineral nutrient response pathway. This possibility is the subject of current research.
With regard to the identification of signaling genes involved in linking changes in plant mineral status to nutrient homeostasis, two very interesting observations arose. First, it was intriguing to see that many of these genes were induced by all three mineral deficiencies. This suggests that there is cross talk at the molecular level between the plant's systems for sensing changes in P, K, and Fe status that previously have not been recognized. For the MAP kinase and transcription factor that were induced by exposing roots to −P, −K, or −Fe conditions, it is possible that these genes all play a general role in mineral nutrient homeostasis and signaling. It could alternatively be that there are separate signal transduction pathways for P, K, and Fe nutrition that share elements that are common to all three pathways. As researchers gain a better understanding of signaling pathways triggered by abiotic stresses, it is becoming clear that there are a number of examples of cross talk between different stresses (Knight and Knight, 2001). Many abiotic stresses trigger increase in levels of free cytosolic Ca2+ as a primary signal and involve protein phosphatases and kinases (including MAP kinases). Thus, it might not be surprising to find that signals arising from changes in the plant status of different mineral nutrients may share common elements.
With regard to this topic, it was interesting to see that withholding Pi, K, or Fe all induced expression of Pi, K, and Fe transporters. In a recent study, we also found that resupplying nitrate to N-starved tomato plants induced Pi transporter and K+ channel genes, in addition to nitrate transporters, and that imposition of −P or −K conditions increased the expression of nitrate transporters and Pi and K+ transporters (Wang et al., 2001). These findings provide further circumstantial evidence for the synergistic regulation of the nutrition of different mineral elements. Other examples of this type of response have recently been reported in the literature. Huang et al. (2000) found in barley that Zn deficiency induced the expression of Pi transporters in both P-sufficient and -deficient seedlings, which may help in explaining the long-held observation of Zn deficiency-induced P toxicity in plants. Also in barley, K deficiency was found to increase the expression of a high-affinity P transporter (Smith et al., 1999).
The second surprising observation concerning plant signaling and mineral nutrient homeostasis was the rapidity with which these genes were induced by withholding specific mineral nutrients from the root system. For several of these genes, such as Nitf and MAP kinase, a strong increase in expression was seen at the 1-h time point for nutrient deprivation. Previous published studies focusing on changes in the expression of specific mineral ion transporters by alterations in plant status for the same mineral nutrient have all employed longer periods of nutrient deprivation. For example, in previous studies of tomato Pi transporter gene expression, Pi is usually withheld from the plant for a period of days, with the shortest time period for P deprivation being 12 h (Liu et al., 1998). The organ- or tissue-specific localization of the signal triggering nutrient deficiency-induced responses is still poorly understood. P is probably the best studied nutrient for this topic, because P deficiency is known to trigger a wide range of physiological responses and changes in gene expression (Raghothama, 2000). There have been several studies that have presented evidence in support of the hypothesis that the signal triggering increased P transporter expression in response to P deficiency arises from the shoot (Liu et al., 1998; Burleigh and Harrison, 1999). What is envisaged is that under continuing growth under low P conditions, decrease in shoot P status triggers the transport of a signal from the shoot to root, triggering P transporter gene induction. The findings presented here and those presented in our previous report on nitrate-induced genes (Wang et al., 2001) suggest that plants respond very rapidly to the lack of a mineral nutrient in the solution bathing the root. This raises the possibility that roots may be able to “sense” changes in external availability of mineral nutrients via currently unexplained mechanisms. It is alternatively possible that changes in the mineral status of cells of the root epidermis may provide the signal triggering changes in gene expression, because this is the only plant tissue that possibly could experience a significant change in mineral status after 1 to 3 h of nutrient deprivation. Preliminary evidence in support of either of these possibilities comes from the experiment with decapitated plants. Here, we saw that Nitf, which was the gene most rapidly and strongly induced by deprivation of all three mineral nutrients in intact plants, showed a similar response in roots of the decapitated plants. These findings do suggest that induction of mineral nutrition-related genes by nutrient deprivation can occur independent of shoot-derived signals. If root-derived signals (either inside or external to the root) do play a key role in regulating plant mineral nutrition, we speculate that this would occur in concert with shoot-derived signals. Thus, it is tempting to suggest that signals from the shoot would communicate the internal mineral status and those generated from the root signal external nutrient availability. This scenario fits with a current model on systemic (which involves signals from the shoot) and localized (such as lateral root formation) responses for a plant's need for mineral nutrients as well as in response to their availability in the environment (Forde, 2002). Further experiments are necessary to test this hypothesis.
We also found in a separate study that a tomato hemoglobin gene was rapidly and strongly induced by Pi, K, and Fe deprivation and by nitrate resupply (Y.-H. Wang, L.V. Kochian, J.F. Doyle, and D.F. Garvin, unpublished data). This gene was most homologous to a swam oak hemoglobin II (GenBank accession no. P23244) as well as a soybean (Glycine max) non-symbiotic hemoglobin (accession no. AAA97887). It also exhibits a strong homology (71% identity and 82% similarity) to Arabidopsis AHB1, which is a class 1 non-symbiotic hemoglobin gene. Interestingly, Arabidopsis AHB1 was also found to be induced by nitrate resupply (Wang et al., 2000). The possible role of this hemoglobin gene in plant mineral nutrition is not well understood and is also the subject of current investigations in our lab.
CONCLUSION
In summary, we have identified a number of genes that respond rapidly to P, K, or Fe deprivation that previously had not been identified as associated with P, K, or Fe nutrition. Several of these genes, including those encoding a Leu-zipper transcription factor, MAP kinase, MAP kinase kinase, and a 14-3-3 protein, may play a role in signal transduction pathways linking changes in mineral status to alterations in gene expression facilitating mineral homeostasis. These genes were all induced rapidly (within 1–3 h) by changes in external P, K, or Fe status, even in decapitated nutrient-stressed plants. Furthermore, we found that expression of Pi, K, and Fe transporters was also rapidly induced by deprivation of all three mineral nutrients. These findings suggest that: (a) there is cross talk between signaling pathways for plant responses to different mineral nutrients; (b) coordination and regulation of the uptake and transport of different mineral nutrients quite possibly occurs; and (c) the rapid induction of these genes suggests that there may be systems in plant roots enabling this organ to sense changes in the mineral status of the soil environment in close proximity to the root.
MATERIALS AND METHODS
Plant Materials
Tomato (Lycopersicon esculentum) plants (TA496) were grown hydroponically as described in Wang et al. (2001). In brief, plants were grown in black plastic pots containing 2 L of modified one-fifth Johnson's solution, which consists of the following macronutrients: 1.2 mm KNO3, 0.8 mm Ca(NO3)2, 0.2 mm NH4H2PO4, and 0.2 mm MgSO4; and the following micronutrients: 50 μm KCl, 12.5 μm H3BO3, 1 μm MnSO4, 1 μm ZnSO4, 0.5 μm CuSO4, 0.1 μm H2MoO4, 0.1 μm NiSO4, and 10 μm Fe-EDDHA). Five plants were grown in each pot in a controlled environment growth chamber with a 16-h light period at 24°C and a 8-h dark period at 20°C. Modest aeration was provided to minimize perturbation to the roots. The nutrient solution was replaced with fresh solution after first 2 weeks and then was replaced weekly for the following 2 weeks and then every other day after that. After 5 weeks of growth, the control nutrient solutions were replaced by the same solutions lacking either Pi, K, or Fe. For the −Pi solution, the 0.2 mm NH4H2PO4 was replaced with 0.2 mm (NH4)2SO4. For the −K solution, the 1.2 mm KNO3 and 0.8 mm Ca(NO3)2 in the nutrient solution were replaced by 1.4 mm Ca(NO3)2 and the 50 μm KCl in the micronutrient solution was replaced with 50 μm CaCl2. For the −Fe solution, the 10 μm Fe-EDDHA was simply left out of the nutrient solution. The five plants in a single pot were harvested at 0, 1, 3, 6, 12, 24, and 48 h after the plants were exposed to nutrient solutions lacking Pi, K, or Fe. Control plants grown under nutrient-sufficient conditions were harvested at the same 0-, 1-, 3-, 6-, 12-, 24-, and 48-h time points, and roots were harvested for RNA extraction in an identical fashion to the nutrient-deprived plants. During the 48 h of the experiment, the nutrient solution was replaced with fresh, identical solution after 24 h. For the experiment with decapitated plants, the shoots were excised at the shoot base and the entire decapitated root system was handled as described above for the experiments with intact plants. Roots and leaves were separated, frozen in liquid N2, and stored in −80°C for RNA isolation.
mRNA Isolation, cDNA Arrays, and RNA Gel-Blot Analysis
mRNA was isolated from root tissue using the Poly(A) Pure Kit (Ambion, Austin, TX) following the manufacturer's protocol as in the previous study (Wang et al., 2001).
See Wang et al. (2001) for details concerning construction of the high-density arrays containing 1,280 mineral nutrition-related cDNAs, as well as array and northern hybridizations. To quantify the mRNA signal ratio from the RNA gel-blot analysis, exposed films were scanned and plotted using ImageJ software (National Institutes of Health) to obtain quantitative readings. Relative signal ratios were calculated as the ratio of the intensity of the treatment signal for a specific time compared with the signal intensity at time 0 (which is for a nutrient-sufficient plant). All RNA gel-blot results were from two different sets of treated tomato plants and repeated at least once within each set of plants. Thus, each RNA gel-blot experiment was replicated at least four times with two separate sets of plants, and representative gel blots are shown in the figures.
ACKNOWLEDGMENTS
We thank Dr. Donald Grieson of University of Nottingham for providing the tomato MEK1 clone and Nick Van Eck and Dr. Steve Tanksley of Cornell University for providing tomato seeds as well as stem, leaf, flower, and unripe fruit tissues for northern analysis. We would also like to thank Jon Shaff and Holly Manslank of the U.S. Plant, Soil, and Nutrition Laboratory for their expertise in setting up and maintaining the tomato hydroponic culture system.
Footnotes
This work was supported by the Agricultural Research Service Agricultural Genome Program (to L.V.K. and D.F.G.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.008854.
LITERATURE CITED
- Baum G, Chen Y, Arazi T, Takatsuji H, Fromm H. A plant glutamate decarboxylase containing a calmodulin binding domain: cloning, sequence, and functional analysis. J Biol Chem. 1993;268:19610–19617. [PubMed] [Google Scholar]
- Burleigh SH, Harrison MJ. The down-regulation of Mt4-like genes by phosphate fertilization occurs systemically and involves phosphate translocation to the shoots. Plant Physiol. 1999;119:241–248. doi: 10.1104/pp.119.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H-J, Sehnke PC, Ferl RJ. The 14-3-3 proteins: cellular regulators of plant metabolism. Trends Plant Sci. 1999;4:367–371. doi: 10.1016/s1360-1385(99)01462-4. [DOI] [PubMed] [Google Scholar]
- Downey P, Szabo I, Ivashikina N, Negro A, Guzzo F, Ache P, Hedrich R, Terzi T, Schiavo FL. Kdc1, a novel carrot root K+channel: cloning, characterization and expression in mammalian cells. J Biol Chem. 2000;275:39420–39426. doi: 10.1074/jbc.M002962200. [DOI] [PubMed] [Google Scholar]
- Eckhardt U, Marques AM, Buckhout TJ. Two iron-regulated cation transporters from tomato complement metal uptake-deficient yeast mutants. Plant Mol Biol. 2001;45:437–448. doi: 10.1023/a:1010620012803. [DOI] [PubMed] [Google Scholar]
- Forde BG. The role of long-distance signaling in plant responses to nitrate and other nutrients. J Exp Bot. 2002;53:39–43. [PubMed] [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M. Arabidopsisethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell. 2000;12:393–404. doi: 10.1105/tpc.12.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman A, Takahashi H. Macronutrient utilization by photosynthetic eukaryotes and the fabric of interactions. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:163–210. doi: 10.1146/annurev.arplant.52.1.163. [DOI] [PubMed] [Google Scholar]
- Hackett RM, Oh SA, Morris PC, Grierson D. A tomato MAP kinase kinase gene (accession no. AJ000728) differentially regulated during fruit development, leaf senescence, and wounding. Plant Physiol. 1998;117:1526. [Google Scholar]
- Hartje S, Zimmermann S, Klonus D, Mueller-Roeber B. Functional characterisation of LKT1, a K+ uptake channel from tomato root hairs, and comparison with the closely related potato inwardly rectifying K+ channel SKT1 after expression in Xenopusoocytes. Planta. 2000;210:723–731. doi: 10.1007/s004250050673. [DOI] [PubMed] [Google Scholar]
- Herbik A, Koch G, Mock HP, Dushkov D, Czihal A, Thielmann J, Stephan UW, Baumlein H. Isolation, characterization and cDNA cloning of nicotianamine synthase from barley: a key enzyme for iron homeostasis in plants. Eur J Biochem. 1999;265:231–239. doi: 10.1046/j.1432-1327.1999.00717.x. [DOI] [PubMed] [Google Scholar]
- Higuchi K, Suzuki K, Nakanishi H, Yamaguchi H, Nishizawa NK, Mori S. Cloning of nicotianamine synthase genes, novel genes involved in the biosynthesis of phytosiderophores. Plant Physiol. 1999;119:471–480. doi: 10.1104/pp.119.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi K, Watanabe S, Takahashi M, Kawasaki S, Nakanishi H, Nishizawa NK, Mori S. Nicotianamine synthase gene expression differs in barley and rice under Fe-deficient conditions. Plant J. 2001;25:159–167. doi: 10.1046/j.1365-313x.2001.00951.x. [DOI] [PubMed] [Google Scholar]
- Huang C, Barker SJ, Langridge P, Smith FW, Graham RD. Zinc deficiency up-regulates expression of high-affinity phosphate transporter genes in both phosphate-sufficient and -deficient barley roots. Plant Physiol. 2000;124:415–422. doi: 10.1104/pp.124.1.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Scarpa A, Zhang L, Stone S, Feliciano E, Ferro-Novick S. A high copy suppressor screen reveals genetic interactions between BET3and a new gene: evidence for a novel complex in ER-to-Golgi transport. Genetics. 1998;149:833–841. doi: 10.1093/genetics/149.2.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri F, Lam E, Chua NH. Two tobacco DNA-binding proteins with homology to the nuclear factor CREB. Nature. 1989;340:727–730. doi: 10.1038/340727a0. [DOI] [PubMed] [Google Scholar]
- Klinedinst S, Pascuzzi P, Redman J, Desai M, Arias J. A xenobiotic-stress-activated transcription factor and its cognate target genes are preferentially expressed in root tip meristems. Plant Mol Biol. 2000;42:679–688. doi: 10.1023/a:1006332708388. [DOI] [PubMed] [Google Scholar]
- Kochian LV. Molecular physiology of mineral nutrient acquisition, transport, and utilization. In: Buchanan BB, Gruissem W, Jones RL, editors. Biochemistry and Molecular Biology of Plants. Rockville, MD: American Society of Plant Physiologists; 2000. pp. 1204–1249. [Google Scholar]
- Knight H, Knight M. Abiotic stress signaling pathways: specificity and cross talk. Trends Plant Sci. 2001;6:262–267. doi: 10.1016/s1360-1385(01)01946-x. [DOI] [PubMed] [Google Scholar]
- Lal SK, Chwenfang Lee C, Sachs MM. Differential regulation of enolase during anaerobiosis in maize. Plant Physiol. 1998;118:1285–1293. doi: 10.1104/pp.118.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling HQ, Koch G, Baumlein H, Ganal MW. Map-based cloning of chloronerva, a gene involved in iron uptake of higher plants encoding nicotianamine synthase. Proc Natl Acad Sci USA. 1999;96:7098–7103. doi: 10.1073/pnas.96.12.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Muchhal MS, Uthappa M, Kononowicz AK, Raghothama KG. Tomato phosphate transporter genes are differentially regulated in plant tissues by phosphorus. Plant Physiol. 1998;116:91–99. doi: 10.1104/pp.116.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meskiene I, Hirt H. MAP kinase pathways: molecular plug-and-play chips for the cell. Plant Mol Biol. 2000;42:791–806. doi: 10.1023/a:1006405929082. [DOI] [PubMed] [Google Scholar]
- Miyata S, Urao T, Yamaguchi-Shinozaki K, Shinozaki K. Characterization of genes for two-component phosphorelay mediators with a single HPt domain in Arabidopsis thaliana. FEBS Lett. 1998;437:11–14. doi: 10.1016/s0014-5793(98)01188-0. [DOI] [PubMed] [Google Scholar]
- Muchhal US, Raghothama KG. Transcriptional regulation of plant phosphate transporters. Proc Natl Acad Sci USA. 1999;96:5868–5872. doi: 10.1073/pnas.96.10.5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukatira U, Liu C, Varadarajan DK, Raghothama KG. Negative regulation of phosphate starvation induced genes. Plant Physiol. 2001;127:1854–1862. [PMC free article] [PubMed] [Google Scholar]
- Pascuzzi P, Hamilton D, Bodily K, Arias J. Auxin-induced stress potentiates trans-activation by a conserved plant basic/leucine-zipper factor. J Biol Chem. 1998;273:26631–26637. doi: 10.1074/jbc.273.41.26631. [DOI] [PubMed] [Google Scholar]
- Pnueli L, Gutfinger T, Hareven D, Ben-Naim O, Ron N, Adir N, Lifschitz E. Tomato SP-interacting proteins define a conserved signaling system that regulates shoot architecture and flowering. Plant Cell. 2001;13:2687–2702. doi: 10.1105/tpc.010293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama KG. Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:665–693. doi: 10.1146/annurev.arplant.50.1.665. [DOI] [PubMed] [Google Scholar]
- Raghothama KG. Phosphate transport and signaling. Curr Opin Plant Biol. 2000;3:182–187. [PubMed] [Google Scholar]
- Roberts MR. Regulatory 14-3-3 protein-protein interactions in plant cells. Curr Opin Plant Biol. 2000;3:400–405. doi: 10.1016/s1369-5266(00)00103-5. [DOI] [PubMed] [Google Scholar]
- Roberts MR, Bowles DJ. Fusicoccin, 14-3-3 proteins, and defense responses in tomato plants. Plant Physiol. 1999;119:1243–1250. doi: 10.1104/pp.119.4.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuring S, Bodor O, Rohricht RA, Muller S, Beyer A, Kohrer K. Cloning, characterisation, and functional expression of the Mus musculusSKD1 gene in yeast demonstrates that the mouse SKD1 and the yeast VPS4 genes are orthologues and involved in intracellular protein trafficking. Gene. 1999;234:149–159. doi: 10.1016/s0378-1119(99)00163-8. [DOI] [PubMed] [Google Scholar]
- Seo S, Okamoto M, Seto H, Ishizuka K, Sano H, Ohashi Y. Tobacco MAP kinase: a possible mediator in wound signal transduction pathways. Science. 1995;270:1988–1992. doi: 10.1126/science.270.5244.1988. [DOI] [PubMed] [Google Scholar]
- Shelp BJ, Bown AW, McLean MD. Metabolism and functions of gamma-aminobutyric acid. Trends Plant Sci. 1999;4:446–452. doi: 10.1016/s1360-1385(99)01486-7. [DOI] [PubMed] [Google Scholar]
- Smith FW, Cybinski DH, Rae AL. Regulation of expression of genes encoding phosphate transporters in barley roots. In: Gissel-Nielsen G, Jensen A, editors. Plant Nutrition-Molecular Biology and Genetics: Proceedings of the Sixth International Symposium on Genetics and Molecular Biology of Plant Nutrition. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1999. pp. 145–150. [Google Scholar]
- Strompen G, Gruner R, Pfitzner UM. An as-1-like motif controls the level of expression of the gene for the pathogenesis-related protein 1a from tobacco. Plant Mol Biol. 1998;37:871–883. doi: 10.1023/a:1006003916284. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Gasch A, Nishizawa N, Chua NH. The DIMINUTO gene of Arabidopsisis involved in regulating cell elongation. Genes Dev. 1995;9:97–107. doi: 10.1101/gad.9.1.97. [DOI] [PubMed] [Google Scholar]
- Van der Straeten D, Rodrigues-Pousada RA, Goodman HM, Van Montagu M. Plant enolase: gene structure, expression, and evolution. Plant Cell. 1991;3:719–735. doi: 10.1105/tpc.3.7.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Guegler K, LaBrie ST, Crawford NM. Genomic analysis of a nutrient response in Arabidopsisreveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. Plant Cell. 2000;12:1491–1509. doi: 10.1105/tpc.12.8.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-H, Garvin DF, Kochian LV. Nitrate-induced genes in tomato roots Array analysis reveals novel genes that may play a role in nitrogen nutrition. Plant Physiol. 2001;127:345–359. doi: 10.1104/pp.127.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmann C, Gibson S, Jarpe MB, Johnson GL. Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol Rev. 1999;79:143–180. doi: 10.1152/physrev.1999.79.1.143. [DOI] [PubMed] [Google Scholar]
- Wilson C, Anglmayer R, Vicente O, Heberle-Bors E. Molecular cloning, functional expression in Escherichia coli, and characterization of multiple mitogen-activated-protein kinases from tobacco. Eur J Biochem. 1995;233:249–257. doi: 10.1111/j.1432-1033.1995.249_1.x. [DOI] [PubMed] [Google Scholar]
- Zhang S, Klessig DF. Resistance gene N-mediated de novo synthesis and activation of a tobacco mitogen-activated protein kinase by tobacco mosaic virus infection. Proc Natl Acad Sci USA. 1998;95:7433–7438. doi: 10.1073/pnas.95.13.7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Liu Y, Klessig DF. Multiple levels of tobacco WIPK activation during the induction of cell death by fungal elicitins. Plant J. 2000;23:339–347. doi: 10.1046/j.1365-313x.2000.00780.x. [DOI] [PubMed] [Google Scholar]