Abstract
Previously, we have studied the expression and regulation of four α- and 14 β-expansin genes in deepwater rice (Oryza sativa). We now report on the structure, expression, and regulation of 22 additional α-expansin (Os-EXP) genes, four expansin-like (Os-EXPL) genes, and one expansin-related (Os-EXPR) gene, which have recently been identified in the expressed sequence tag and genomic databases of rice. α-Expansins are characterized by a series of conserved Cys residues in the N-terminal half of the protein, a histidine-phenylalanine-aspartate (HFD) motif in the central region, and a series of tryptophan residues near the carboxyl terminus. Of the 22 additional α-expansin genes, five are expressed in internodes and leaves, three in coleoptiles, and nine in roots, with high transcript levels in the growing regions of these organs. Transcripts of five α-expansin genes were found in roots only. Expression of five α-expansin genes was induced in the internode by treatment with gibberellin (GA) and by wounding. The wound response resulted from excising stem sections or from piercing pinholes into the stem of intact plants. EXPL proteins lack the HFD motif and have two additional Cys residues in their C- and N-terminal regions. The positions of conserved tryptophan residues at the C-terminal region are different from those of α- and β-expansins. Expression of the Os-EXPL3 gene is correlated with elongation and slightly induced by applied GA. However, the expression of the Os-EXPL1 and Os-EXPL2 genes showed limited correlation with cell elongation and was not induced by GA. We found no expression of the Os-EXPR1 gene in the organs examined.
Deepwater rice (Oryza sativa) is a subsistence crop in regions of Southeast Asia that are flooded during the monsoon season. To avoid drowning, deepwater rice has evolved the capacity to elongate very rapidly when it becomes submerged. The growth response of submerged deepwater rice plants is elicited by GA and is based on enhanced cell division activity in the intercalary meristem of the youngest internode and on increased elongation of the newly formed cells. Besides its importance as a crop plant, deepwater rice is also a well-suited model organism to study plant growth (for review, see Kende et al., 1998).
Expansins are proteins that mediate long-term extension of isolated cell walls. Originally, they were grouped into two related families, the α- and β-expansins (for review, see Cosgrove, 2000). Four α-expansins have been studied in deepwater rice (Cho and Kende, 1997a, 1997b). The expression of the corresponding genes is organ specific and correlated with growth and acid-inducible cell wall extensibility. Expression of two α-expansin genes, Os-EXP2 and Os-EXP4, is induced by submergence and treatment with GA (Cho and Kende, 1997b). As the expressed sequence tag (EST) databases expanded and the rice genome was sequenced, it became clear that α- and β-expansin genes belong to a large superfamily of genes (Lee et al., 2001; Li et al., 2002). Genes encoding proteins with significant amino acid identities to α- and β-expansins but lacking some of their conserved motifs were also recognized in the databases, and phylogenetic analysis indicates that they belong to the expansin superfamily of genes (D.J. Cosgrove, Pennsylvania State University, University Park, http://www.bio.psu.edu/expansins; Lee et al., 2001; Li et al., 2002). The large number of expansins poses intriguing questions regarding the function of these proteins and the significance of their redundancy. Determining the pattern and control of expansin gene expression is the first step in elucidating the function of individual expansins.
Fourteen β-expansin genes have been identified in rice (Lee and Kende, 2001). Five of these are expressed in the internode, and their expression is induced by GA and wounding. Here, we report on the identification of 22 additional α-expansin (EXP) genes, four expansin-like (EXPL) genes, and one expansin-related (EXPR) gene of rice in the genomic and EST databases. We studied their expression patterns in various organs and tested the effect of GA and wounding on the level of their transcripts.
RESULTS
Sequence Analysis of α-Expansins of Rice
A search of the rice EST and genomic databases yielded 26 putative α-expansin genes (Lee et al., 2001). Four of them (Os-EXP1–Os-EXP4) had already been studied extensively with respect to their sequence, expression, and regulation (Cho and Kende, 1997a, 1997b, 1998). Analysis with the PSORT program (Nakai and Kanehisa, 1992) predicts that α-expansins have a signal peptide for entry into the secretory pathway and secretion to the cell wall. The molecular masses of the mature α-expansin proteins range from 23.1 to 28.4 kD. They possess the characteristic expansin motifs, namely conserved Cys residues in the N-terminal region of the protein, a putative catalytic domain with the His-Phe-Asp (HFD) motif in the central portion of the protein, and conserved Trp residues in the putative cellulose-binding domain in the C-terminal region (Fig. 1). Rice α-expansins are somewhat less divergent from each other (55% average amino acid identity between mature proteins; Table I) than are rice β-expansins (51% average amino acid identity between mature proteins; Table I; Lee and Kende, 2001).
Figure 1.
Alignment of the amino acid sequences of α-expansin, β-expansin, EXPL, and EXPR proteins. Positions of the highly conserved sequences, the conserved Cys (C) and Trp (W) residues, and the HFD motif are indicated by shading or by letters above the top sequence. In this alignment, the HFD motif of β-expansins is shifted by seven amino acids toward the amino terminus when compared with α-expansins. Signal peptide sequences are given as predicted by the PSORT program (Nakai and Kanehisa, 1992) and are represented in lowercase. The alignment was performed using the ClustalW program and was printed with the BOXSHADE program.
Table I.
Average percent identity of amino acid sequences between members of each expansin family of rice
| Family | Os-EXP | Os-EXPB | Os-EXPL | Os-EXPR |
|---|---|---|---|---|
| Os-EXP | 55 | – | – | – |
| Os-EXPB | 21 | 51 | – | – |
| Os-EXPL | 15 | 20 | 50 | – |
| Os-EXPR | 17 | 23 | 28 | NA |
Values were calculated using the amino acid sequences without the signal peptides. Nos. of the proteins are 26, 14, 4, and 1 for Os-EXP, Os-EXPB (Lee and Kende, 2001), Os-EXPL, and Os-EXPR, respectively. NA, Not available because there is only one EXPR gene.
Sequence Analysis of EXPL and EXPR Genes of Rice
We identified five rice genes that show significant amino acid identity to α- and β-expansins but lack some of the conserved features of expansins (Fig. 1; Table I). The derived amino acid sequences of these five rice genes are similar to those of four Arabidopsis genes. Phylogenetic analysis indicates that the Arabidopsis genes belong to two new subfamilies of the expansin superfamily. They were named EXPL (EXPANSIN-LIKE) and EXPR (EXPANSIN-RELATED; D.J. Cosgrove, http://www.bio.psu.edu/expansins; Lee et al., 2001). In analogy to the Arabidopsis genes, we designated four of the rice genes as expansin like (Os-EXPL1–Os-EXPL4) and one as expansin related (Os-EXPR1). The deduced Os-EXPL proteins show 50% average amino acid identity to each other (Table I). They contain conserved Cys residues in the N-terminal and conserved Trp residues in the C-terminal regions, and an ambiguous signal peptide. In addition, all EXPL proteins have from one to three NXT/S motifs, which may be N-linked glycosylation sites. They lack, however, the HFD motif in the putative catalytic domain of the protein; one otherwise conserved Trp is missing, and the positions of the other Trp residues are different from those in α- and β-expansins. EXPL proteins have additional Trp residue in the C-terminal and two additional Cys residues in the N- and C-terminal regions (Fig. 1).
The EXPR gene in the rice genome shows 40% amino acid identity to the only EXPR gene of Arabidopsis (protein accession no. CAB80974.1). The deduced Os-EXPR protein has the conserved Cys residues in the N-terminal and the conserved Trp residues in the C-terminal region, three NXT/S motifs, and a signal peptide for secretion to the cell wall (Fig. 1). It lacks the HFD motif in the center of the protein, and two otherwise conserved Trp are missing in the C-terminal region. The major difference between EXPL and EXPR proteins is in the C- and N-terminal regions. EXPR of both Arabidopsis and rice lack the amino acid extensions that are present at the C and N terminus of EXPL proteins and that contain two conserved Cys residues each (Fig. 1).
Organ-Specific Expression of α-Expansin and EXPL Genes in Deepwater Rice
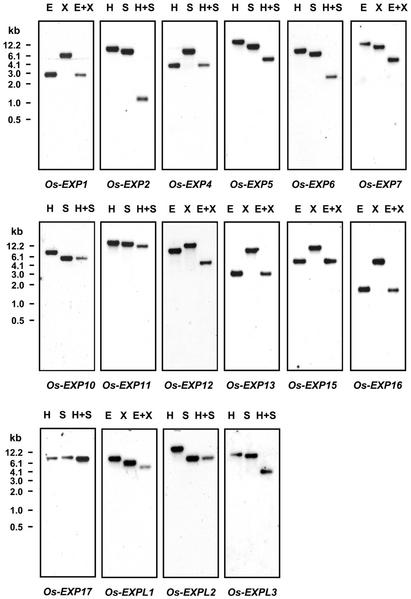
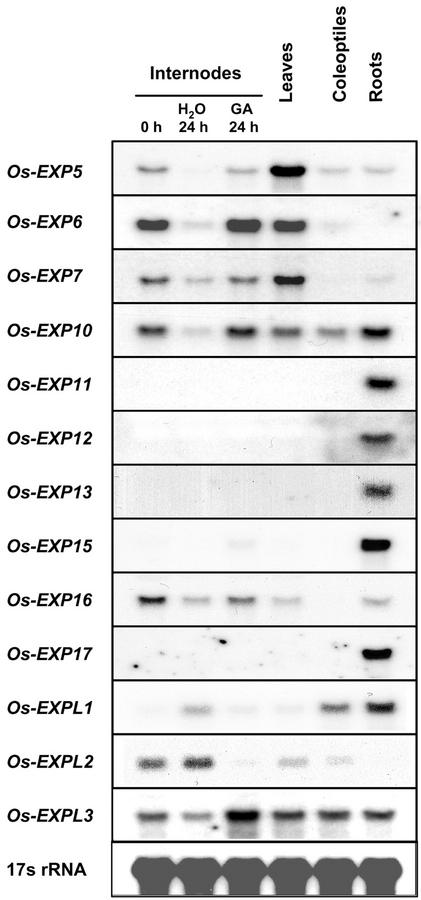
To determine which of the recently identified α-expansin, EXPL, and EXPR genes are expressed in deepwater rice, we prepared gene-specific probes consisting mainly of the 3′-untranslated regions (UTRs) of the respective genes (Table II). When no cDNA was available, the gene-specific regions were amplified by RT-PCR from mRNA or by PCR from genomic DNA. DNA gel-blot analyses performed under the same stringency conditions as used for the RNA gel blots showed that, with one exception, the probes were specific and that no detectable cross hybridization to other genes occurred (Fig. 2). A probe corresponding to the 3′-UTR region of Os-EXPL4 (accession no. AP003624) cross-reacted with multiple bands on a DNA gel blot (results not shown). Because of this lack of specificity, no expression studies for Os-EXPL4 were performed. However, the presence of two ESTs in the database of Indica rice indicates that the Os-EXPL4 gene is expressed (Yu et al., 2002; http://210.83.138.53/rice). The transcript levels of the recently identified 22 α-expansin, three EXPL, and one EXPR genes were examined in the basal 1-cm region of the uppermost internode, which contains the intercalary meristem and the elongation zone; in the basal 1-cm region of GA-treated and control internodes; in the basal 5-cm region of elongating leaves; in the apical 1-cm region of coleoptiles from 3-d-old seedlings; and in the apical 0.5-cm region of roots from 3-d-old seedlings (Fig. 3).
Table II.
Primer sets used for the PCR amplifications of gene-specific probes of α-expansins, EXPL, and EXPR genes of rice
| Gene Name | Primer Sequence (5′ to 3′) | Template | Product Size | Accession No.
|
|
|---|---|---|---|---|---|
| cDNA | Genomic DNA | ||||
| bp | |||||
| Os-EXP1 | CTCGACCTCACAGCAGTTCTCTTA | S5074a | 291 | AF261270 | AF394543 |
| TCATCGATTGGCAAGCACCTC | |||||
| Os-EXP2 | GGCTAATTCCGTTTTTCAGTT | R1518a | 369 | U30477 | AF394544 |
| TGCTCCCCCACAATCTCC | |||||
| Os-EXP3 | GTCGCCCCGTCCAACTGGTTC | R0448a | 268 | U30479 | CL006919.165b |
| AATTGGTGGGCAAAACATTCA | |||||
| Os-EXP4 | CCAGTTCTAGCCGCCACCGACATC | cDNA clone | 388 | U85246 | AF394545 |
| ATTCCGTTGCAAGGCCATCACTCC | |||||
| Os-EXP5 | TTCCGCTCAATTTTACTCG | E31402a | 505 | AF247162 | AF394546 |
| TTTTTTTTTTTTTTTTTTTTTTTA | |||||
| Os-EXP6 | TAGATTAGTTTAGCCAAGAGG | E60552a | 429 | AF247163 | CL014794.24b |
| TTTTTTTTTTTTTTTTTTTTTTTA | |||||
| Os-EXP7 | AAAACTGCAATATACCCTCTTA | E31204a | 335 | AF247164 | AF394547 |
| TTTTTTTTTTTTTTTTTTTTTTTC | |||||
| Os-EXP8 | CATTAATTGCAAGCCTATCTCA GAGAGGGAGTATGCCGAATGT | Reverse transcriptase (RT) product of mRNA | 320 | NAc | AC007789 |
| Os-EXP9 | CAAATTGGGCTTAGTTCAGGT | RT product of mRNA | 198 | NA | AC007789 |
| CTCTGAAAACCGTTGCAAATC | |||||
| Os-EXP10 | GAGAGGAAAGGGTACCAATAGCA | C61881a | 332 | AF247165 | AL662987 |
| TTTTTTTTTTTTTTTTTTTTTTTA | |||||
| Os-EXP11 | GCCAGTTCTAGCCGTTCC | RT product of mRNA | 276 | NA | AP000837 |
| CATTGCGCTCGTCACCACT | |||||
| Os-EXP12 | ACCAGCGGCGTGCAGTTCTAC | Genomic DNA | 296 | AY046929 | AF394548 |
| AGGCATGGCGGGCTATCG | |||||
| Os-EXP13 | TTCGCGAGCAACATACAG | Genomic DNA | 254 | NA | AF394549 |
| TACATCGCACATACATACAAAAG | |||||
| Os-EXP14 | CAAGCTGCAATTCAAGTAA | Genomic DNA | 376 | NA | AF394550 |
| GATAGTTCGGGAGGGTAA | |||||
| Os-EXP15 | ATAAGGCTGCAGATTGAAGAAG | Genomic DNA | 303 | NA | AF394551 |
| CACGGTAGAAATGACTGGTAGC | |||||
| Os-EXP16 | AAGGCAAGCAGTTCGTC | Genomic DNA | 334 | NA | AF394552 |
| AGTTTATGCACCTCTATTATGTC | |||||
| Os-EXP17 | CCAGGGATCCAACAACTTCTACTA | Genomic DNA | 498 | NA | AP000616 |
| CGGATCGGATATACATAAACCAAT | |||||
| Os-EXP18 | TTCGGGCAAACCTTCAGC | Genomic DNA | 233 | NA | AF394553 |
| TTGCAACTTAATTTACATCCATCA | |||||
| Os-EXP19 | CGGCCAAACATTCAGCACCTACCA | Genomic DNA | 297 | NA | AF394554 |
| CAAACGGGGCCGAAGAAACCACTA | |||||
| Os-EXP20 | CGGCCAAACCTTCAGCACCTACCA | Genomic DNA | 365 | NA | AF394555 |
| GCCTTTAATTGCCACTCTGCTTCC | |||||
| Os-EXP21 | CGCGCCTAGAACGATGATGTATGT | Genomic DNA | 404 | NA | AF394556 |
| GGTGGTTGGGGTAAAATGGAAAGA | |||||
| Os-EXP22 | TTGGGTTTCTACTGCCTGACTGAT | Genomic DNA | 493 | NA | AF394557 |
| CGCGTTGCAACTGTTTTTCTTAT | |||||
| Os-EXP23 | TGATACCGGAGCGTTTCTTTTG | Genomic DNA | 316 | NA | AF394558 |
| CCTGCGTGGTGTGGACTATGA | |||||
| Os-EXP24 | TTCTACGGCCTGCGACTGATT | Genomic DNA | 280 | NA | AF394559 |
| TGTTCCCCTTTTATTTATGATTTG | |||||
| Os-EXP25 | GACATTCACCAGCAACCAG | Genomic DNA | 337 | NA | AF394560 |
| GTACGTAACAGCAGTCCTCTCCT | |||||
| Os-EXP26 | CTACCAGGCCAAGAAGAA | Genomic DNA | 171 | AY100692 | AF394561 |
| TTAGTAAATACGCAGAGGAT | |||||
| Os-EXPL1 | ACGCACGAGTGGAAGTAGAAGC | Genomic DNA | 188 | AY100693 | AY039022 |
| GAAGAAGAAGAAACGGAGGAGGAA | |||||
| Os-EXPL2 | ACGCAGGAGTGGAAGTGACA | Genomic DNA | 214 | NA | AC051633 |
| CACCCCGAGTCGAGTAAACAACAC | |||||
| Os-EXPL3 | CTGGAAGTGAAAGGTGTGCTA | Genomic DNA | 344 | AY100694 | AP004315 |
| TAGCAATTAGTATGGGGGTAGC | |||||
| Os-EXPR1 | CTTGGCTTGTTTCACCTCTCCTAT | Genomic DNA | 317 | NA | AP003956 |
| CTTTATGCATTTCCGAGTTTA | |||||
Expressed sequence tag clone no.
Contig no. of genomic DNA from Torrey Mesa Research Institute database (Goff et al., 2002; http://portal.tmri.org/rice/).
NA, Not available.
Figure 2.
DNA gel-blot analyses showing the specificity of the gene-specific probes. Genomic DNA was digested with EcoRI (E), XbaI (X), HindIII (H), SacI (S), EcoRI and XbaI (E+X), or with HindIII and SacI (H+S). The digested DNA was separated by gel electrophoresis, blotted onto a Hybond N+ membrane, and hybridized to the gene-specific probes indicated under each blot under the same conditions as described for RNA gel-blot analysis. We are only showing the DNA gel-blot analysis of those genes whose expression was detected in deepwater rice.
Figure 3.
RNA gel-blot analysis of α-expansin and EXPL gene expression in internodes, leaves, coleoptiles, and roots of deepwater rice. Each lane contained 20 μg of total RNA isolated from the organs indicated above the lanes. Internodes, 0 to 1 cm above the second highest node of the youngest, growing internode of adult plants; to test the effect of GA, excised 20-cm-long stem sections were incubated with or without 50 μm GA3 for 24 h; leaves, 0 to 5 cm above the basal collar of growing leaves of adult plants; coleoptiles, 0 to 1 cm below the tip of growing coleoptiles of 3-d-old seedlings; roots, 0 to 0.5 cm above the tip of growing roots of 3-d-old seedlings. 17s rRNA was used as internal loading control.
Of the 22 recently identified α-expansin genes, 10 (Os-EXP5, Os-EXP6, Os-EXP7, Os-EXP10, Os-EXP11, Os-EXP12, Os-EXP13, Os-EXP15, Os-EXP16, and Os-EXPB17) are expressed in deepwater rice. Os-EXP5 and Os-EXP10 are expressed in all organs tested; Os-EXP6 is expressed in all organs tested except in roots; Os-EXP7 and Os-EXP16 are expressed in all organs tested except in coleoptiles; and Os-EXP11, Os-EXP12, Os-EXP13, Os-EXP15, and Os-EXP17 are expressed in the roots only. Os-EXPL1 is highly expressed in the root and coleoptile, Os-EXPL2 in the internode, and Os-EXPL3 in the internode, leaf, coleoptile, and root. We could not find any transcripts of the Os-EXPR1 gene, and no Os-EXPR1 cDNA exists in the database.
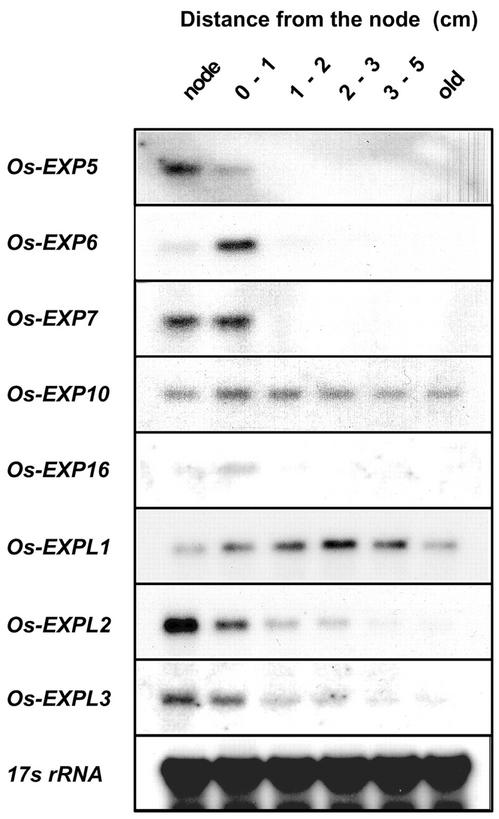
Expression of α-Expansin and EXPL Genes in Different Developmental Regions of Internodes
In rice, as in other grasses, stem elongation occurs at the base of the highest internode, just above the second highest node. Five of the recently identified α-expansin genes—Os-EXP5, Os-EXP6, Os-EXP7, Os-EXP10, and Os-EXP16—are expressed in the highest internode and in the subtending node of intact plants (Fig. 4). The transcript level of three α-expansins—Os-EXP6, Os-EXP7, and Os-EXP16—is highest in the basal region of the internode, 0 to 1 cm above the second highest node. This region contains the intercalary meristem and the elongation zone (Kende et al., 1998). Os-EXP5 is expressed at the highest level in the second highest node. Except for Os-EXP10, α-expansin mRNA was not detected beyond the 1-cm zone above the node. Os-EXP10 is evenly expressed in all regions of the internode. Os-EXPL1 showed highest expression in the nongrowing regions of the internode, whereas expression of Os-EXPL2 and Os-EXPL3 was most pronounced in the second highest node and decreased with distance from the node (Fig. 4).
Figure 4.
RNA gel-blot analysis of α-expansin and EXPL gene expression in the second highest node and in different regions of the uppermost internode. Each lane contained 20 μg of total RNA isolated from the node and the internodal regions indicated above the lanes. Node, Second highest node; 0 to 1 cm, internodal region containing the intercalary meristem and most of the elongation zone; 1 to 2 cm, internodal region containing the upper part of the elongation zone and the differentiation zone; 2 to 3 cm and 3 to 5 cm, internodal regions containing the differentiation zone; old, oldest part of the internode 5 cm above the node. 17s rRNA was used as internal loading control.
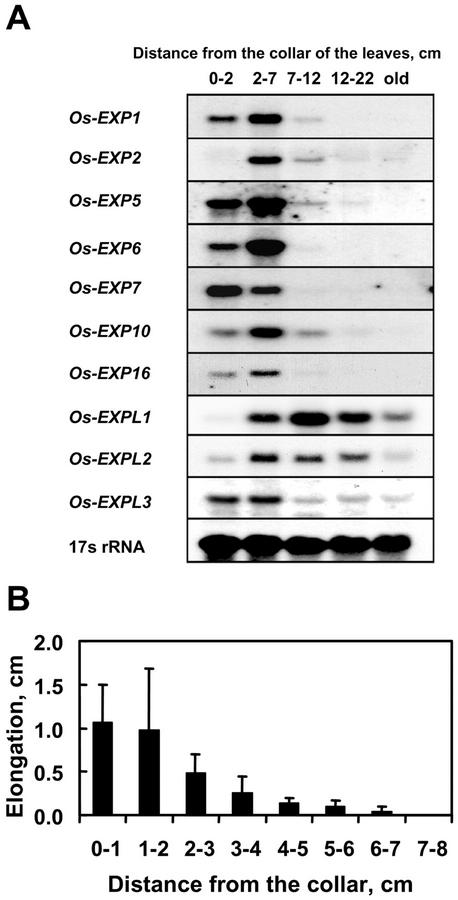
Expression of α-Expansin and EXPL Genes and Elongation in Different Developmental Regions of Leaves
In grasses, such as rice, only the youngest leaves grow, and elongation occurs mainly at the base of the leaf, just above the leaf collar. Seven α-expansin genes—Os-EXP1, Os-EXP2, Os-EXP5, Os-EXP6, Os-EXP7, Os-EXP10, and Os-EXP16—were expressed in the youngest leaf of intact plants (Fig. 5A). Except for Os-EXP7, all α-expansins showed highest expressions in the 2- to 7-cm region above the collar. The expression of Os-EXP7 was highest in the 0- to 2-cm region above the collar. The expression level of α-expansin was very low or undetectable in the nonelongating region of the leaf. The expression pattern of Os-EXPL3 was similar to that of α-expansins. In contrast, the transcript level of Os-EXPL1 was highest in the nonelongating regions and that of Os-EXPL2 remained high in the nonelongating regions.
Figure 5.
RNA gel-blot analysis of α-expansin and EXPL gene expression in the leaves and growth of leaves. A, RNA gel-blot analysis of α-expansin and EXPL gene expression in growing rice leaves. Each lane contained 20 μg of total RNA isolated from the regions indicated above the lanes. Old, Oldest part of the leaves 22 cm above the collar. 17s rRNA was used as internal loading control. B, Elongation of each region of the leaves during a 48-h period.
We also measured the elongation of various regions of rice leaves (Fig. 5B). Most elongation occurred in the region 0 to 1 cm above the collar and gradually decreased with distance from the collar. No growth was recorded 7 cm above the collar.
Expression of α-Expansin and EXPL Genes in Response to GA Treatment
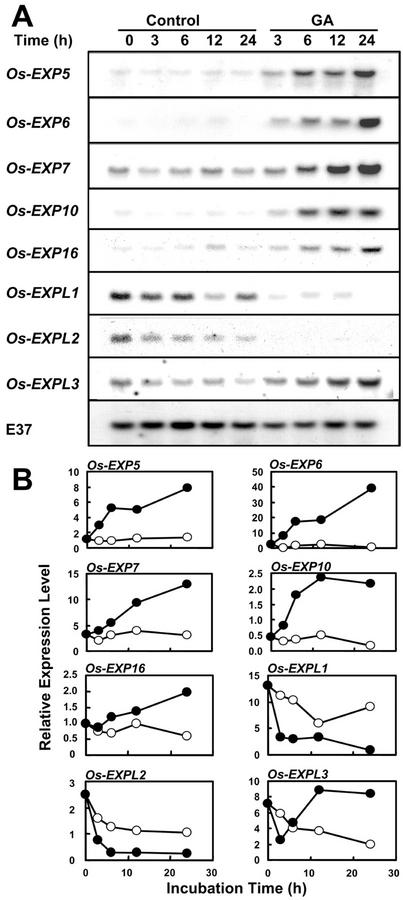
We tested the effect of GA on the expression of five α-expansin genes (Os-EXP5, Os-EXP6, Os-EXP7, Os-EXP10, and Os-EXP16) and three EXPL genes (Os-EXPL1, Os-EXPL2, and Os-EXPL3) in the internode (Fig. 6, A and B). The time course and magnitude of induction by GA varied according to genes. Transcripts of the above α-expansin genes, except those of Os-EXP10, accumulated gradually during the 24 h of incubation. The level of Os-EXP10 mRNA increased rapidly during the first 12 h of incubation and leveled off thereafter. The expression level of Os-EXPL3 increased slightly as a result of treatment with GA, but, interestingly, the expression levels of Os-EXPL1 and Os-EXPL2 decreased after treatment with GA. These results confirm those of an independently performed experiment shown in Figure 3. The effect of GA on the expression of individual α-expansin and EXPL genes was confirmed in two to four independent experiments.
Figure 6.
Expression of α-expansin and EXPL genes in GA-treated stem sections. Stem sections were first incubated for 8 h in distilled water to dissipate the wound effect and then transferred to 50 μm GA3 or distilled water (control) for the times indicated above the lanes. A, RNA gel-blot analysis. Each lane contained 20 μg of total RNA isolated from the 0- to 2-cm region above the second highest node, which includes the intercalary meristem, the elongation zone, and the lower part of the differentiation zone. E37 was used as internal loading control. B, Quantification of transcript levels of each gene. Expression levels are shown in PhosphorImager values × 10−3 after 24 h of exposure. ○, Control; ●, GA treatment.
The Time Course of α-Expansin Gene Expression in Internodes during Incubation of Stem Sections in Water
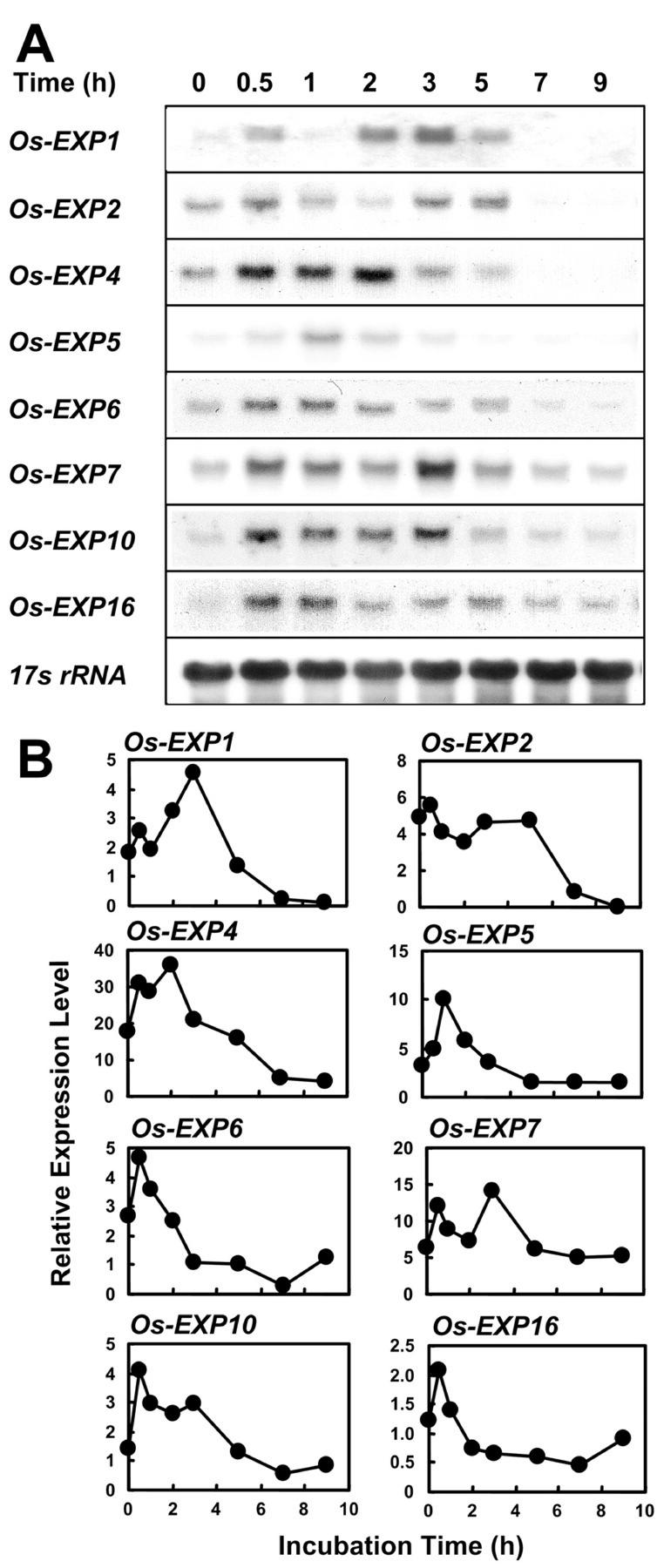
The expression of α-expansin genes increased in internodes after excision of stem sections. We determined the time course of this increase during incubation in water and found that the expression pattern differed among the eight genes that are expressed in the internode after isolation of the sections (Fig. 7, A and B). The stem sections used to investigate the effect of GA on α-expansin and EXPL transcript accumulation were first incubated in water for 8 h to dissipate the effect of excision before GA was applied. The effect of excising stem sections on the expression of individual α-expansin genes was confirmed in two to three independent experiments.
Figure 7.
Expression of α-expansin genes in stem sections. Stem sections were excised and incubated in water for the times indicated above the lanes. A, RNA gel-blot analysis. Each lane contained 20 μg of total RNA isolated from the 0- to 2-cm region above the second highest node, which includes the intercalary meristem, the elongation zone, and the lower part of the differentiation zone. 17s rRNA was used as loading control. B, Quantification of the mRNA level of each gene. Expression levels are shown in PhosphorImager values × 10−3 after 24 h of exposure.
The Time Course of α-Expansin Expression in Internodes of Whole Plants in Response to Wounding
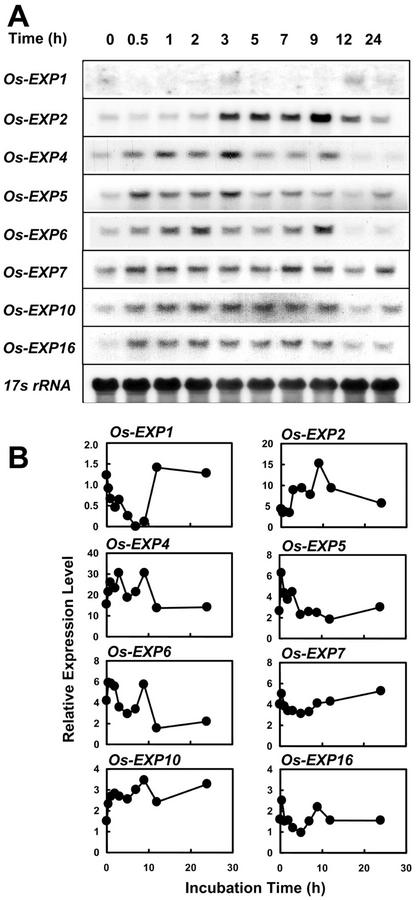
To examine whether the enhanced expression of α-expansin genes in excised stem sections was the result of wounding, we determined the level of Os-EXP transcripts in internodes of whole plants that had been wounded by piercing six pinholes into the stem 2 cm below the second highest node where the stem section would have been excised. Transcripts of all eight Os-EXP genes accumulated as a result of wounding, but again with varying time courses (Fig. 8, A and B). The expression of the Os-EXP genes declined after 9 h but at a slower rate than in stem sections. The effect of wounding on the expression of individual α-expansin genes was confirmed in two independent experiments.
Figure 8.
Expression of α-expansin genes in the internodes of wounded plants. The internodes were wounded by piercing pinholes around a circle into the stems 2 cm below the second highest node. The tissue samples were collected at the times indicated above the lanes. A, RNA gel-blot analysis. Each lane contained 20 μg of total RNA isolated from the 0- to 2-cm region above the second highest node, which includes the intercalary meristem, the elongation zone and the lower part of the differentiation zone. 17s rRNA was used as loading control. B, Quantification of the mRNA levels of each gene. Expression levels are shown in PhosphorImager values × 10−3 after 24 h of exposure.
DISCUSSION
In earlier work, we studied the expression pattern and regulation of four α- and 14 β-expansin genes in deepwater rice (Cho and Kende, 1997a, 1997b; Lee and Kende, 2001). Recently, we found 22 new α-expansins in the rice databases (Lee et al., 2001), as well as five genes, which, in analogy to the nomenclature in Arabidopsis (D.J. Cosgrove, http://www.bio.psu.edu/expansins; Lee et al., 2001), we named expansin like (Os-EXPL) and expansin related (Os-EXPR). These genes are grouped on two separate branches of the phylogenetic tree representing the expansin superfamily (D.J. Cosgrove, http://www.bio.psu.edu/expansins; Lee et al., 2001; Li et al., 2002). Li et al. (2002) proposed a new nomenclature for the expansins and assigned Arabidopsis EXPL and EXPR to the β-expansins. Although purified α- and β-expansins have been shown to possess wall-loosening activity (McQueen-Mason et al., 1992; Cosgrove et al., 1997), no expansin function has been demonstrated for EXPL and EXPR proteins. Because of their distinct amino acid sequences, their separate position on the phylogenetic tree, and because we do not know whether they have wall-loosening activity, we agree with the nomenclature of D.J. Cosgrove (http://www.bio.psu.edu/expansins), as also used by Lee et al. (2001), and refer to these proteins as expansin like and expansin related.
Eight expansin genes are expressed in the root only (Table III). It is interesting to note that there are no internode-, leaf-, or coleoptile-specific expansin transcripts. In our previous study, no expansin transcripts were detected in rice leaves (Cho and Kende, 1997b). In the experiments reported here, we found that seven α-expansin genes are expressed in leaves, including two (Os-EXP1 and Os-EXP2) for which no transcripts had been found in leaves before. The discrepancy between our earlier and present results is because of the differences in the leaf regions examined. In both instances, young, expanding leaves were analyzed. However, Cho and Kende (1997b) determined expansin mRNA levels in the tip zone of the leaf; in the present study, expansin mRNA levels were found to be highest at the base of the leaf.
Table III.
Expression of expansin superfamily genes in various organs of rice
| Family | Organs
|
|||
|---|---|---|---|---|
| Internodes | Leaves | Coleoptiles | Roots | |
| α-Expansins | Os-EXP1a | Os-EXP1 | Os-EXP1a | Os-EXP1a |
| Os-EXP2a | Os-EXP2 | Os-EXP2a | Os-EXP2a | |
| Os-EXP4a | Os-EXP5 | Os-EXP4a | Os-EXP3ab | |
| Os-EXP5 | Os-EXP6 | Os-EXP5 | Os-EXP4a | |
| Os-EXP6 | Os-EXP7 | Os-EXP6 | Os-EXP5 | |
| Os-EXP7 | Os-EXP10 | Os-EXP10 | Os-EXP7 | |
| Os-EXP10 | Os-EXP16 | Os-EXP10 | ||
| Os-EXP16 | Os-EXP11b | |||
| Os-EXP12b | ||||
| Os-EXP13b | ||||
| Os-EXP15b | ||||
| Os-EXP16 | ||||
| Os-EXP17b | ||||
| β-Expansins | Os-EXPB3c | Os-EXPB4d | Os-EXPB3d | Os-EXPB2bd |
| Os-EXPB4c | Os-EXPB12d | Os-EXPB4d | Os-EXPB3d | |
| Os-EXPB6c | Os-EXPB6d | Os-EXPB4d | ||
| Os-EXPB11c | Os-EXPB11d | Os-EXPB6d | ||
| Os-EXPB12c | Os-EXPB8bd | |||
| Os-EXPB11d | ||||
| EXPL | Os-EXPL1 | Os-EXPL1 | Os-EXPL1 | Os-EXPL1 |
| Os-EXPL2 | Os-EXPL2 | Os-EXPL2 | Os-EXPL3 | |
| Os-EXPL3 | Os-EXPL3 | Os-EXPL3 | ||
Of the four organs tested, expressed only in roots.
Y. Lee and H. Kende (unpublished data).
In most instances, the highest levels of expansin transcripts were found in the most rapidly growing regions of tissues and organs of rice (Cho and Kende, 1997a, 1997b, 1998; Huang et al., 2000; Lee and Kende, 2001). A few exceptions to this correlation have been noted, however. For example, Os-EXP2 mRNA is also present in nongrowing regions of roots and internodes (Cho and Kende, 1997b). With the exception of Os-EXP7, whose transcript level in the leaf is highest 0 to 2 cm above the leaf collar, all other α-expansin genes showed highest expression in the region 2 to 7 cm above the collar (Fig. 5A). This region is still elongating but at a much slower rate than the region 0 to 2 cm above the collar (Fig. 5B). In leaves of the grass Festuca pratensis, expression of one α- and two β-expansin genes was also highest in the zone whose growth rate had diminished (Reidy et al., 2001). It appears likely that occurrence of expansin transcripts in slowly growing or nongrowing tissues reflects expansin functions related to cellular differentiation.
The expression of α-expansin genes in the growing internode of deepwater rice is enhanced by GA (Fig. 6, A and B), by excision of stem sections (Fig. 7, A and B), and by wounding of whole plants (Fig. 8, A and B). The accumulation of α- and β-expansin mRNA in GA-treated tissue is consistent with the notion that expansins are involved in mediating GA-induced rapid internodal elongation in deepwater rice (Cho and Kende, 1997b; Lee and Kende, 2001). Wound-induced expression of β-expansin genes was reported previously (Lee and Kende, 2001). The relative induction of α-expansin gene expression by wounding was not as pronounced as that of β-expansins. The significance of expansin mRNA accumulation in wounded deepwater rice internodes is not known.
The DNA-derived amino acid sequences of EXPL and EXPR genes show significant amino acid identity to α- and β-expansins, although some of the characteristic motifs of the expansins are missing (Table I; Fig. 1). We could not find any expression of the EXPR gene in the rice organs tested, and the expression patterns of the Os-EXPL1 and Os-EXPL2 genes are quite different from those of the α- and β-expansins. Expression of both expansin-like genes in rice internodes was down-regulated by GA and was high in the nonelongating region of leaves. These results indicate that the expansin-like proteins do not act in GA-regulated stem elongation of rice and that their function may be different from that of α- and β-expansins.
Our combined results indicate that at least eight α- and five β-expansin genes are expressed in deepwater rice internodes (Table III). With the exception of Os-EXP1 (Cho and Kende, 1997b), GA enhances their expression, and most, if not all, may be involved in mediating internodal elongation. It is not yet known whether each of the α- and β-expansins has a specific function in growth or whether their functions are largely overlapping. Individual expansins could have different substrate specificities, there may be differences in their biochemical modes of action, and they could act in different cell types of the internode. The significance of expansin gene activation by wounding and the role of the EXPL and EXPR proteins are entirely unknown.
MATERIALS AND METHODS
Growth and Treatment of Plants
Seeds of deepwater rice (Oryza sativa L. cv Pin Gaew 56) were obtained from the International Rice Research Institute (Los Baños, Philippines). Plants were grown as described by Stünzi and Kende (1989). The plant material was collected as described previously (Lee and Kende, 2001). Treatment of stem sections with GA3 and wounding of internodal tissue were performed as described by Lee and Kende (2001). To measure the elongation of leaves, pinholes were pierced through the leaf sheath into the underlying young leaf with a 26-gauge needle starting at the collar and moving up the leaf sheath at 1-cm intervals. The distance between the pinholes was measured 48 h later.
Isolation of Nucleic Acids
Genomic DNA was isolated according to Dellaporta et al. (1983), and total RNA according to Verwoerd et al. (1989). The PolyATtract kit (Promega, Madison, WI) was used to enrich poly(A+) RNA; the enriched product is referred to as poly(A+) RNA.
Preparation of Probes
For RT-PCR, total RNA was isolated from mature plants, and poly(A+) RNA was purified. One hundred nanograms of poly(A+) RNA was subjected to RT-PCR using Superscript II (Life Technologies, Rockville, MD) and 1 μL of oligo(dT)18 (500 μg mL−1) as a reverse primer. After incubation at 42°C for 50 min and inactivation of the reverse transcriptase for 15 min at 70°C, the reactions were subjected to 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min, in the presence of the gene-specific primer pairs (Table II). PCR was performed with Taq DNA polymerase (Promega) according to the manufacturer's instructions in a PTC200 thermal cycler (MJ Research, Watertown, MA). For PCR amplification from plasmid or plant genomic DNA, gene-specific primer pairs (Table II) were used to amplify the fragments containing the putative 3′-UTRs under the conditions given above. The PCR products were purified by gel electrophoresis and cloned into the pGEM-T Easy vector (Promega) for sequencing.
DNA fragments containing the inserts of gene-specific regions of α-expansin, EXPL, and EXPR genes, of E37, and of 17S rDNA were excised from the cloning vectors with restriction enzymes and isolated from agarose gels with a DNA purification system (Wizard PCR Preps, Promega). E37 is a truncated cDNA encoding parts of a chloroplast inner membrane protein; the E37 transcript is constitutively expressed in deepwater rice internodes (Van der Knaap and Kende, 1995). 17S rRNA (Zarembinski and Theologis, 1993) and E37 served as loading controls.
RNA Gel-Blot Analysis
Twenty micrograms of total RNA was separated electrophoretically in a 1.2% (w/v) formaldehyde-agarose gel (Ausubel et al., 1987) and transferred to Hybond N+ membrane (Amersham Pharmacia, Piscataway, NJ). Blots were prehybridized and hybridized as described previously (Lee and Kende, 2001). The radioactivity on blots was visualized by autoradiography using Hyperfilm MP (Amersham Pharmacia). The exposure time was adjusted to the intensity of the signals. The radioactivity associated with the transcripts was quantified by PhosphorImager analysis (Molecular Dynamics, Sunnyvale, CA) after 24 h of exposure.
DNA Gel-Blot Analysis
Four micrograms of genomic DNA was digested with EcoRI, HindIII, SacI, XbaI, EcoRI, and XbaI, or HindIII and SacI, separated in an agarose gel (0.8% [w/v]), and transferred to a Hybond N+ membrane. Preparation of the probes and hybridizations were performed as described previously (Lee and Kende, 2001). The radioactivity on blots was visualized by autoradiography using Hyperfilm MP (Amersham Pharmacia). The exposure time was adjusted to the intensity of the signals.
DNA and Amino Acid Sequence Analysis
The nucleotide and deduced amino acid sequences were analyzed with the DNASTAR program (DNASTAR, Madison, WI). The protein localization site was predicted using the PSORT program (Nakai and Kanehisa, 1992). Multiple sequence alignments were performed using the ClustalW (version 1.8) Multiple Sequence Alignment program (http://searchlauncher.bcm.tmc.edu) and printed using BOXSHADE 3.20 (http://www.ch.embnet.org).
ACKNOWLEDGMENTS
We thank the National Institute of Agrobiological Resources (Tsukuba, Japan) and Dr. Christine B. Michalowski (University of Arizona, Tucson) for providing rice α-expansin and EXPL ESTs; the Monsanto Company (St. Louis) and the Torrey Mesa Research Institute (San Diego) for giving us access to their respective rice genome databases; and Jacob Soule and Sarah Norris (both at Michigan State University, East Lansing) for technical help.
Footnotes
This work was supported by the National Science Foundation (grant no. IBN 0076524) and by the U.S. Department of Energy (grant no. DE–FG–02–91ER20021).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.008888.
LITERATURE CITED
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1987. [Google Scholar]
- Cho HT, Kende H. Expansins and internodal growth of deepwater rice. Plant Physiol. 1997a;113:1145–1151. doi: 10.1104/pp.113.4.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HT, Kende H. Expression of expansin genes is correlated with growth in deepwater rice. Plant Cell. 1997b;9:1661–1671. doi: 10.1105/tpc.9.9.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HT, Kende H. Tissue localization of expansins in deepwater rice. Plant J. 1998;15:805–812. doi: 10.1046/j.1365-313x.1998.00258.x. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Loosening of plant cell walls by expansins. Nature. 2000;407:321–326. doi: 10.1038/35030000. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ, Bedinger P, Durachko DM. Group I allergens of grass pollen as cell wall-loosening agents. Proc Natl Acad Sci USA. 1997;94:6559–6564. doi: 10.1073/pnas.94.12.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: version II. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- Goff SA, Ricke D, Lan T-H, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H et al. A draft sequence of the rice genome (Oryza sativa L. ssp. Japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- Huang J, Takano T, Akita S. Expression of α-expansin genes in young seedlings of rice (Oryza sativa L.) Planta. 2000;211:467–473. doi: 10.1007/s004250000311. [DOI] [PubMed] [Google Scholar]
- Kende H, van der Knaap E, Cho HT. Deepwater rice: a model plant to study stem elongation. Plant Physiol. 1998;118:1105–1110. doi: 10.1104/pp.118.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Choi D, Kende H. Expansins: ever-expanding numbers and functions. Curr Opin Plant Biol. 2001;4:527–532. doi: 10.1016/s1369-5266(00)00211-9. [DOI] [PubMed] [Google Scholar]
- Lee Y, Kende H. Expression of β-expansins is correlated with internodal elongation in deepwater rice. Plant Physiol. 2001;127:645–654. [PMC free article] [PubMed] [Google Scholar]
- Li Y, Darley CP, Ongaro V, Fleming A, Schipper O, Baldauf SL, McQueen-Mason SJ. Plant expansins are a complex multigene family with an ancient evolutionary origin. Plant Physiol. 2002;128:1–11. doi: 10.1104/pp.010658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason S, Durachko DM, Cosgrove DJ. Two endogenous proteins that induce cell wall extension in plants. Plant Cell. 1992;4:1425–1433. doi: 10.1105/tpc.4.11.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M. A knowledge base for prediction protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidy B, McQueen-Mason S, Nösberger J, Fleming J. Differential expression of α- and β-expansin genes in the elongating leaf of Festuca pratensis. Plant Mol Biol. 2001;46:491–504. doi: 10.1023/a:1010621417854. [DOI] [PubMed] [Google Scholar]
- Stünzi JT, Kende H. Gas composition in the internal air spaces of deepwater rice in relation to growth induced by submergence. Plant Cell Physiol. 1989;30:49–56. [Google Scholar]
- Van der Knaap E, Kende H. Identification of a gibberellin-induced gene in deepwater rice using differential display of mRNA. Plant Mol Biol. 1995;28:589–592. doi: 10.1007/BF00020405. [DOI] [PubMed] [Google Scholar]
- Verwoerd TC, Dekker BMM, Hoekema A. A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 1989;17:2362–2362. doi: 10.1093/nar/17.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK-S, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X et al. A draft sequence of the rice genome (Oryza sativa L. ssp. Indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- Zarembinski TI, Theologis A. Anaerobiosis and plant growth hormones induce two genes encoding 1-aminocyclopropane-1-carboxylate synthase in rice (Oryza sativaL.) Mol Biol Cell. 1993;4:363–373. doi: 10.1091/mbc.4.4.363. [DOI] [PMC free article] [PubMed] [Google Scholar]