Abstract
Light stress and salt stress are major environmental factors that limit the efficiency of photosynthesis. However, we have found that the effects of light and salt stress on photosystem II (PSII) in the cyanobacterium Synechocystis sp. PCC 6803 are completely different. Strong light induced photodamage to PSII, whereas salt stress inhibited the repair of the photodamaged PSII and did not accelerate damage to PSII directly. The combination of light and salt stress appeared to inactivate PSII very rapidly as a consequence of their synergistic effects. Radioactive labeling of cells revealed that salt stress inhibited the synthesis of proteins de novo and, in particular, the synthesis of the D1 protein. Northern- and western-blotting analyses demonstrated that salt stress inhibited the transcription and the translation of psbA genes, which encode D1 protein. DNA microarray analysis indicated that the light-induced expression of various genes was suppressed by salt stress. Thus, our results suggest that salt stress inhibits the repair of PSII via suppression of the activities of the transcriptional and translational machinery.
Light stress and salt stress are important environmental factors that limit plant growth and productivity (Berry and Björkman, 1980; Boyer, 1982; Powles, 1984). Strong light impairs the activity of the photosynthetic apparatus, in particular that of photosystem II (PSII), via a process known as photodamage or photoinhibition (for review, see Kok, 1956; Jones and Kok, 1966a, 1966b; Barber and Andersson, 1992; Aro et al., 1993). Kyle et al. (1984) suggested that the primary damaging effect of light might be the impairment of the quinone-binding protein, which is now known as the D1 protein (hereafter D1), in the PSII complex (Ohad et al., 1984; Aro et al., 1993). Impairment of D1 results in disruption of the light-dependent separation of charge between P680 and pheophytin a, and this phenomenon is associated with interruption of the transport of electrons that is mediated by PSII. However, photodamaged PSII can be repaired, and the repair process involves the rapid turnover of D1, with degradation of damaged D1 (Lindahl et al., 2000; Haussühl et al., 2001) and subsequent light-dependent synthesis de novo of the precursor to D1 (hereafter pre-D1; Aro et al., 1993). The damaged D1 is replaced by newly synthesized pre-D1 (Marder et al., 1984; Mattoo et al., 1984, 1988; Ohad et al., 1984; Schuster et al., 1988) from which a carboxy-terminal sequence is then removed by specific lumenal proteases (Reisfeld et al., 1982; Taylor et al., 1988; Inagaki et al., 1989; Taguchi et al., 1995).
In the field, under natural conditions, salt stress very often occurs in combination with light stress, and several reports have appeared on the effects of salt stress on PSII under light stress. Salt stress apparently enhances the inhibition by strong light of PSII in Chlamydomonas reinhardtii (Neale and Melis, 1989), in leaves of barley (Hordeum vulgare; Sharma and Hall, 1991), sorghum (Sorghum bicolor; Sharma and Hall, 1991), and rye (Secale cereale; Hertwig et al., 1992), and in Spirulina platensis (Lu and Zhang, 1999). However, the mechanisms by which salt stress enhances the photodamage to PSII remain to be clarified.
In the cyanobacterium Synechocystis sp. PCC 6803 (hereafter Synechocystis), PSII is resistant to salt stress alone. Thus, the activity of PSII is unaffected in cells that have been incubated for 15 h in the presence of 0.5 m NaCl in darkness (Allakhverdiev et al., 1999). However, the effects of salt stress on PSII under strong light remain to be clarified in this organism.
In the present study, we investigated the interaction between the effects of light stress and salt stress on PSII in Synechocystis. We found that the combination of light and salt stress has a strong synergistic and damaging effect on PSII and, moreover, that salt stress inhibited the recovery of PSII from light-induced inactivation. Labeling of proteins in vivo and western- and northern-blotting analyses suggested that salt stress inhibited the expression of the psbA genes for pre-D1 at of transcriptional and the translational level.
RESULTS
Synergistic Effects of Light Stress and Salt Stress on PSII
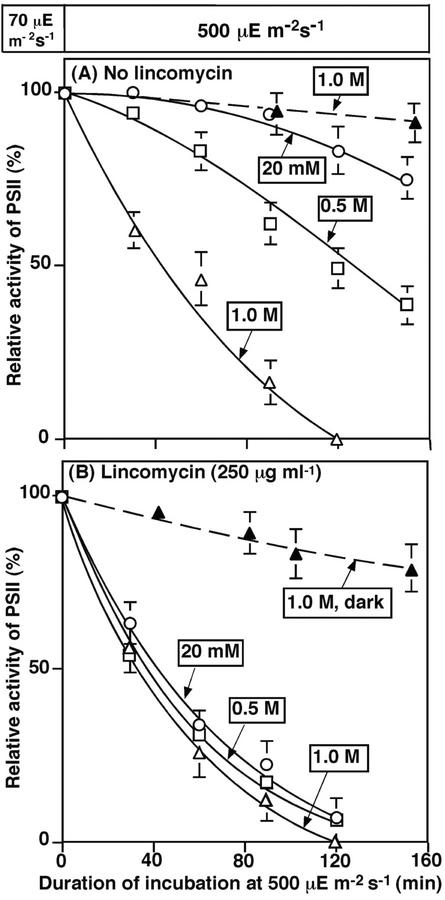
We examined the effects of NaCl at various concentrations on changes in the PSII activity of Synechocystis during exposure of cells to light stress (Fig. 1). Exposure to light at 500 μE m−2 s−1 under low-salt conditions (20 mm NaCl) resulted in minimal inactivation of PSII: After incubation for 120 min, only about 10% of the original activity disappeared. In the presence of 0.5 m NaCl, in contrast, inactivation occurred more rapidly, and 50% of the original activity had disappeared after incubation for 120 min. In the presence of 1.0 m NaCl, the activity of PSII declined even more rapidly, and no activity was detectable after 120 min (Fig. 1A). In darkness, exposure of cells to 1.0 m NaCl did not result in any inactivation over the entire duration of the experiment. These results demonstrated that, whereas exposure of cells to light stress or salt stress resulted in minimal inactivation of PSII, the combination of the two kinds of stress induced marked inactivation of PSII, with apparent synergism between the effects of strong light and high salt.
Figure 1.
Effects of NaCl and lincomycin on PSII activity during incubation of Synechocystis cells in light. Cells were incubated in light at 500 μE m−2 s−1 in the presence of NaCl at various concentrations. At designated times, a portion of the cell suspension was withdrawn and, after the addition of 1.0 mm 1,4-benzoquinone to the suspension, PSII activity was examined by monitoring the light-dependent evolution of oxygen. The activity that corresponded to 100% was 614 ± 56 μmol O2 mg−1 chlorophyll (Chl) h−1. A, Cells were incubated in the absence of lincomycin. B, Cells were incubated in the presence of 250 μg mL−1 lincomycin. ○, 20 mm NaCl; □, 0.5 m NaCl; ▵, 1.0 m NaCl; ▴, 1.0 m NaCl in darkness. Each point and bar represent the average ± se of results from four independent experiments.
To examine the contribution of protein synthesis de novo to the stress-induced inactivation of PSII, we incubated cells in darkness for 10 min in the presence of 250 μg mL−1 lincomycin, an inhibitor of protein synthesis, prior to exposure of cells to light at 500 μE m−2 s−1 in the presence of 20 mm, 0.5 m, or 1.0 m NaCl. Figure 1B shows that the inhibition of protein synthesis by lincomycin markedly accelerated the inactivation of PSII. The inactivation observed in the presence of lincomycin was unaffected by NaCl. However, the extent of inactivation in the presence of lincomycin was only minimal when cells were incubated in the presence of 1.0 m NaCl in darkness. These observations suggest that protein synthesis de novo might be involved in the synergistic effects of light stress and salt stress during the inactivation of PSII.
We performed the same set of experiments as those for which the results are shown in Figure 1 with light at 250 and 2,000 μE m−2 s−1. The rate of inactivation depended on the intensity of light, but essentially the same results were obtained with respect to the synergistic effects of light stress and the salt stress (data not shown).
Inhibition of the Repair of PSII by NaCl
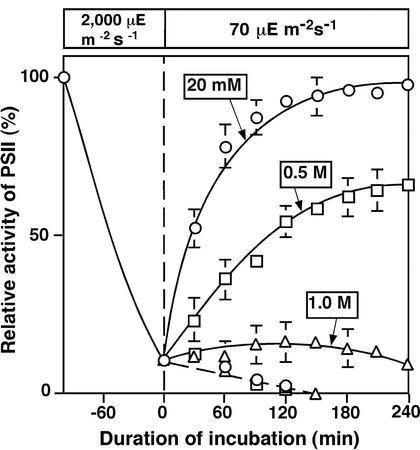
Figure 2 shows the effects of NaCl on the recovery of PSII activity after cells had been exposed to light at 2,000 μE m−2 s−1 for 100 min, a treatment that reduced the activity of PSII to approximately 10% of the original level. To monitor the recovery of PSII, we then incubated the cells in light at 70 μE m−2 s−1 for 4 h in the presence of various concentrations of NaCl. In low-salt medium (20 mm NaCl), the activity of PSII returned to 90% of the initial value within 2 h, and recovery was complete within 3 h. When cells were incubated with 0.5 m NaCl, recovery was slow and only 60% of the original activity was restored after 4 h. However, in the presence of 1.0 m NaCl or 250 μg mL−1 lincomycin, recovery was completely blocked. These results, together with those in Figure 1, demonstrate that NaCl at high concentrations inhibits the repair of PSII. This phenomenon might explain the apparent ability of NaCl to accelerate the light-induced damage to PSII, as seen in Figure 1A.
Figure 2.
Effects of NaCl and lincomycin on the recovery of the PSII activity of Synechocystis cells from light-induced inactivation. Cells were incubated for 100 min in low-salt medium (20 mm NaCl) in light at 2,000 μE m−2 s−1 to induce 90% inactivation of PSII. Cells were then incubated in light at 70 μE m−2 s−1 in the presence of NaCl at various concentrations and in the presence of 250 μg mL−1 lincomycin or in its absence. At designated times, a portion of the cell suspension was withdrawn, and PSII activity was examined as described in the legend to Figure 1. ○, 20 mm NaCl; □, 0.5 m NaCl; ▵, 1.0 m NaCl. Solid lines, in the absence of lincomycin; dashed line, in the presence of 250 μg mL−1 lincomycin. The activity that corresponded to 100% was 562 ± 49 μmol O2 mg−1 Chl h−1. Each point and bar represent the average ± se of results from five independent experiments.
Inhibition by NaCl of Recovery of the Light-Induced Quenching of Chl Fluorescence
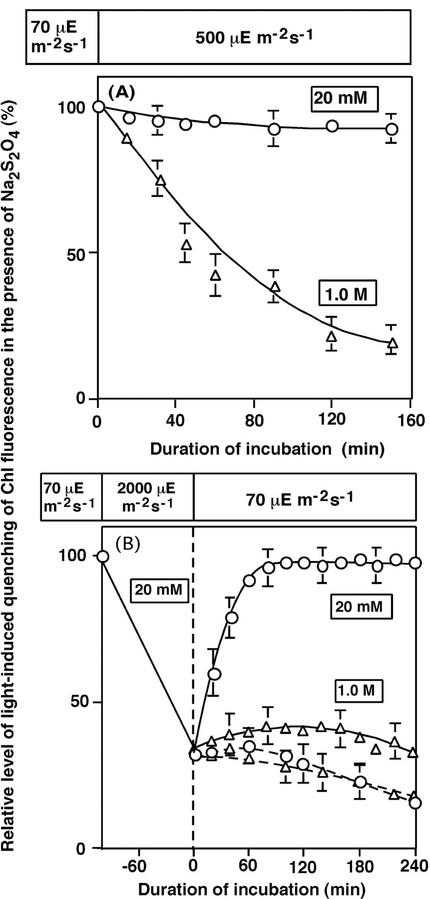
To identify the site of damage to PSII, we monitored the light-induced quenching of Chl fluorescence in the presence of sodium dithionite. Such quenching corresponds to the reduction of pheophytin a in the photochemical reaction center complex in intact cells (Klimov et al., 1986; Allakhverdiev et al., 1988; Ke, 2001). When Synechocystis cells were exposed for 150 min to light at 500 μE m−2 s−1 in the presence of 1.0 m NaCl, the extent of the light-induced quenching decreased to 20% of the original level; in low-salt medium (20 mm NaCl), there was no detectable decrease in light-induced quenching, as shown in Figure 3A.
Figure 3.
Effects of NaCl on changes in the light-induced quenching of Chl fluorescence during the light-induced inactivation of PSII and its recovery in Synechocystis cells. A, Cells were incubated in light at 500 μE m−2 s−1 at 34°C in the presence of 20 mm or 1.0 m NaCl. At designated times, a portion of the cell suspension was withdrawn and, after the addition of 1 mg mL−1 sodium dithionite, the light-induced quenching of Chl fluorescence was examined at 34°C. ○, 20 mm NaCl; ▵, 1.0 m NaCl. B, Cells were incubated for 100 min at 34°C in low-salt medium (20 mm NaCl) in light at 2,000 μE m−2 s−1, which decreased the light-induced quenching of Chl fluorescence to 65% of the original value. Cells were then incubated at 34°C in light at 70 μE m−2 s−1 in the presence of 20 mm or 1.0 m NaCl and in the presence of 250 μg mL−1 lincomycin or in its absence. At designated times, a portion of the cell suspension was withdrawn and the light-induced quenching of Chl fluorescence was examined at 34°C after the addition of 1 mg mL−1 sodium dithionite. ○, 20 mm NaCl; ▵, 1.0 m NaCl. Solid lines, in the absence of lincomycin; dashed lines in the presence of lincomycin. Each point and bar represent the average ± se of results from four independent experiments.
We also examined the effects of NaCl on the recovery of the light-induced quenching of Chl fluorescence after cells had been exposed to light at 2,000 μE m−2 s−1 for 100 min. In low-salt medium, the light-induced quenching returned to normal within 2 h. However, in the presence of 1.0 m NaCl, such recovery was completely suppressed (Fig. 3B). These results suggested that the site of damage to PSII under light and salt stress might be the photochemical reaction center.
Inhibition by NaCl of Protein Synthesis
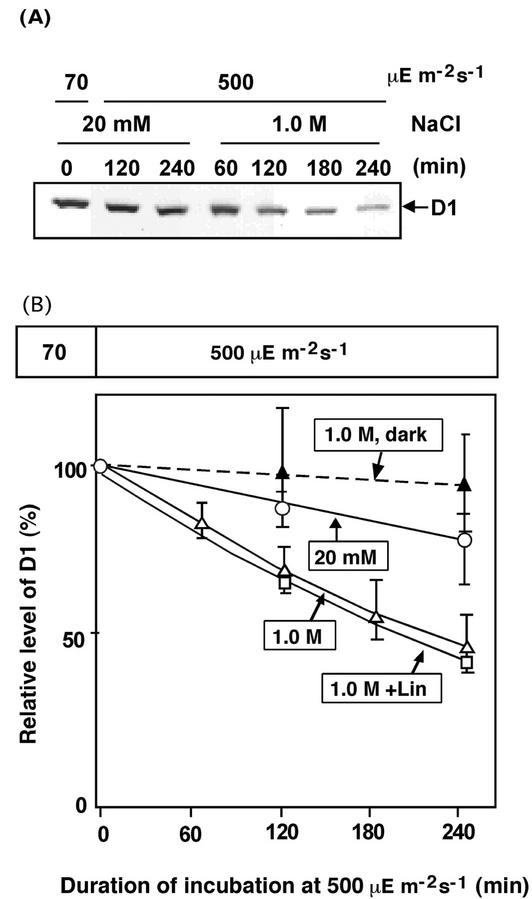
We used western-blotting analysis to examine the effects of NaCl on the level of D1 during incubation of Synechocystis cells in light at 500 μE m−2 s−1 (Fig. 4). The level of D1 decreased slowly in low-salt medium. High-salt conditions (1.0 m NaCl) accelerated the decrease in the level of D1, but lincomycin did not accelerate this decrease. However, the level of D1 was still close to 50% of the original level after incubation of cells in light in the presence of 1.0 m NaCl for 4 h, conditions that completely abolished the activity of PSII. This discrepancy might be explained by the fact that immunoblotting analysis revealed the impaired form of D1 in addition to the active form (Barber and Andersson, 1992; Aro et al., 1993).
Figure 4.
Changes in the level of D1 during the light-induced inactivation of PSII. A, Results of western-blotting analysis. B, Quantitation of the results shown in A. Cells were incubated in light at 500 μE m−2 s−1 in the presence of 20 mm NaCl (○), 1.0 m NaCl (▵), or 1.0 m NaCl plus 250 μg mL−1 lincomycin (□). Cells were also incubated in darkness in the presence of 1.0 m NaCl (▴). At designated times, a portion of the cell suspension was withdrawn and thylakoid membranes were isolated. Proteins were analyzed by PAGE as described in “Materials and Methods.” Each point and bar represent the average ± se of results from four independent experiments.
To monitor the synthesis of D1 de novo during the repair of PSII, we incubated cells for 100 min under strong light (2,000 μE m−2 s−1), which reduced the activity of PSII to 10% of the original level (see Fig. 2), and we then incubated the cells under weak light (70 μE m−2 s−1) for 4 h in the presence of NaCl at various concentrations. The level of D1 decreased by 50% during the exposure of cells to strong light (Fig. 4). During subsequent repair in weak light, the level of D1 returned to normal in low-salt medium (data not shown), reflecting the repair of PSII. In the presence of 1.0 m NaCl, there was no increase in the level of D1. Therefore, we postulated that NaCl inhibited the synthesis of D1 de novo.
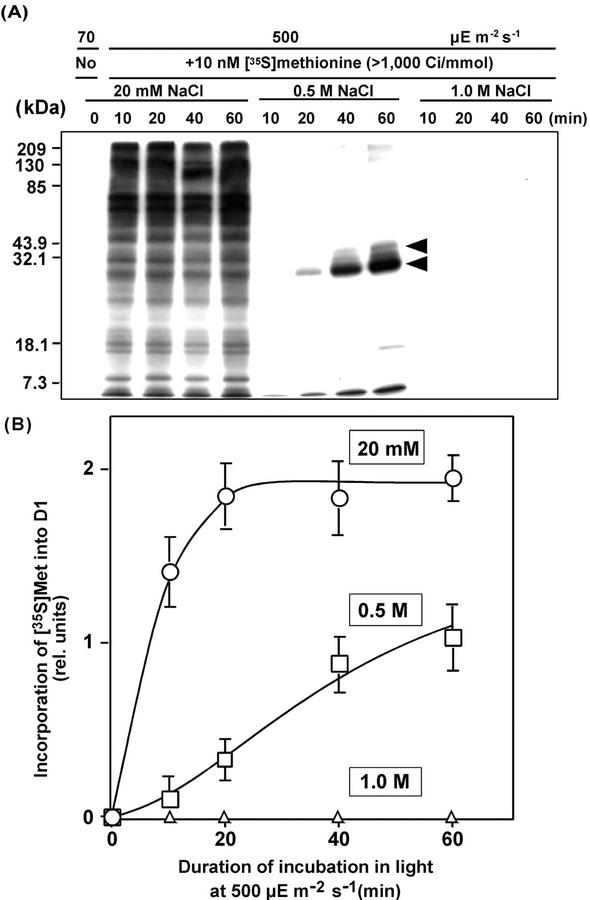
We examined the effects of NaCl on protein synthesis de novo by monitoring the incorporation of [35S]Met into proteins in thylakoid membranes. Figure 5A shows that the presence of 0.5 m NaCl markedly suppressed the synthesis of almost all proteins. However, these conditions also induced the expression of a specific protein of approximately 25 kD. No similar induction of this protein was observed in the presence of 20 mm NaCl. Identification and characterization of this protein will be the focus of future research. The presence of 1.0 m NaCl totally inhibited the synthesis of all proteins (Fig. 5A).
Figure 5.
Effects of NaCl on the synthesis of membrane-bound proteins during exposure of Synechocystis cells to light. Cells were incubated with 10 nm [35S]Met in light at 500 μE m−2 s−1 in the presence of 20 mm, 0.5 m, or 1.0 m NaCl. At designated times, a portion of the cell suspension was withdrawn for preparation of thylakoid membranes. Proteins from thylakoid membranes were analyzed by PAGE as described in “Materials and Methods.” Proteins from thylakoid membranes corresponding to 0.8 μg of Chl were applied to each lane. A, Patterns of radiolabeled proteins after PAGE. The top and bottom arrows indicate the positions of D1 (32 kD) and of the NaCl-induced protein of 25 kD, respectively. The results shown are representative of the results of four independent experiments, each of which gave similar results. B, The time course of incorporation of [35S]Met into D1. Each point and bar represent the average ± se of results from four independent experiments. Other details are the same as those described in the legend to Figure 1.
We further examined quantitatively the effect of NaCl on the synthesis of D1 de novo (Fig. 5B). Under normal conditions, i.e. in the presence of 20 mm NaCl, incorporation of radioactive Met was rapid and reached a maximum level at 20 min. However, the incorporation in 0.5 m NaCl was distinct but at a low rate. This might correspond to the relatively slow decline of PSII activity in 0.5 m NaCl at 500 μE m−2s−1 (see Fig. 1A). At 1.0 m of NaCl, no incorporation of radioactive Met was observed, which might correspond to the rapid inactivation of PSII (see Fig. 1A).
Inhibition by NaCl of the Synthesis of Pre-D1
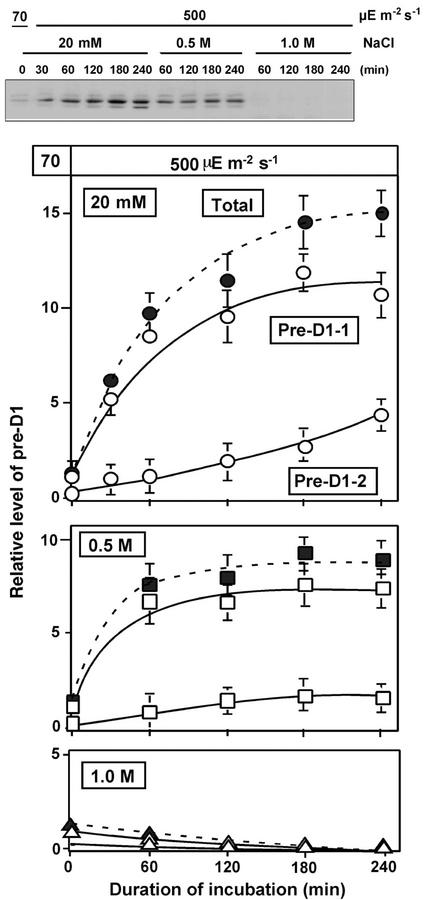
We examined the effects of NaCl on the level of pre-D1 in further detail by western blotting. Figure 6 (top panel) shows that specific antibodies raised against a peptide of 16 amino acid residues that corresponded to the carboxy-terminal extension of pre-D1 (products of the psbAII and psbAIII genes) detected two proteins with molecular masses of 34 to 35 and 32 to 33 kD, respectively. We postulated that the top and bottom bands on the gel corresponded to pre-D1 and an intermediate in the processing of pre-D1, respectively, as proposed by Inagaki et al. (2001), and we designated these proteins pre-D1-1 and pre-D1-2, respectively. By contrast, antibodies against D1 detected a protein of 31 kD.
Figure 6.
Effects of NaCl on levels of pre-D1 during incubation of Synechocystis cells in light. Cells were incubated in light at 500 μE m−2 s−1 in the presence of 20 mm, 0.5 m, or 1.0 m NaCl. At designated times, a portion of the cell suspension was withdrawn for preparation of thylakoid membranes, which were subjected to western-blotting analysis as described in the text. The results are shown quantitatively in the bottom panels. ○, 20 mm; □, 0.5 m; ▵, 1.0 m NaCl. Open symbols, pre-D1-1 and pre-D1-2 (the top and bottom bands, respectively, on the gel); closed symbols, total pre-D1 (pre-D1-1 plus pre-D1-2). Each point and bar represent the average ± se of results from four independent experiments. Other details are the same as those described in the legend to Figure 1.
Figure 6 (bottom panels) shows changes in the levels of pre-D1-1 and pre-D1-2 during exposure of cells to light at 500 μE m−2 s−1. In low-salt medium, levels of pre-D1-1 and pre-D1-2 increased with time. In the presence of 0.5 m NaCl, the increases in levels of both proteins were suppressed by more than 50%. In the presence of 1.0 m NaCl, there was no increase at all in the level of either form of pre-D1.
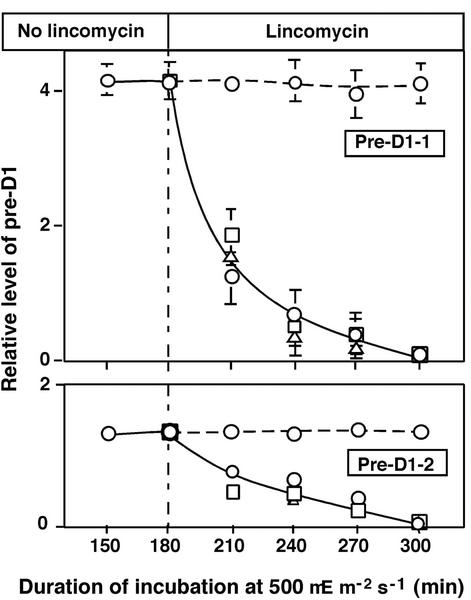
The levels of pre-D1-1 and pre-D1-2 reflect a balance between their synthesis (translation of psbA transcripts), processing, and degradation. To focus specifically on effects of NaCl on rates of processing and degradation, we exposed cells to light at 500 μE m−2 s−1 for 180 min to raise levels of pre-D1-1 and pre-D1-2 to a maximum (see Fig. 6), and then we added lincomycin to block any synthesis of pre-D1 de novo. Under these conditions, we were able to examine the effects of NaCl on the stability of pre-D1-1 and pre-D1-2. Figure 7 clearly demonstrates that NaCl had no effect on the stability of pre-D1-1 and pre-D1-2. These results, together with the results in Figure 6, suggest that the decreases in the levels of pre-D1-1 and pre-D1-2, as seen in Figure 5, might have been caused by inhibition of the synthesis de novo of pre-D1 and not by acceleration of the processing and/or degradation of the precursor proteins.
Figure 7.
Effects of NaCl on the stability of pre-D1 during incubation of Synechocystis cells in the presence of lincomycin. Cells were incubated for 180 min in light at 500 μE m−2 s−1 in 20 mm NaCl. Lincomycin at 250 μg mL−1 was then added together with 0.5 m or 1.0 m NaCl, and incubation was continued in the light at 500 μE m−2 s−1. At designated times, a portion of the cell suspension was withdrawn for preparation of thylakoid membranes, which were subjected to western-blotting analysis as described in the text. Quantitative results of western blotting are shown. ○, 20 mm NaCl in the absence (dashed line) or presence (uninterrupted line) of 250 μg mL−1 lincomycin; □, 0.5 m NaCl in the presence of 250 μg mL−1 lincomycin; ▵, 1.0 m NaCl in the presence of 250 μg mL−1 lincomycin. Each point and bar represent the average ± se of results from three independent experiments.
Inhibition by NaCl of the Transcription of psbA Genes
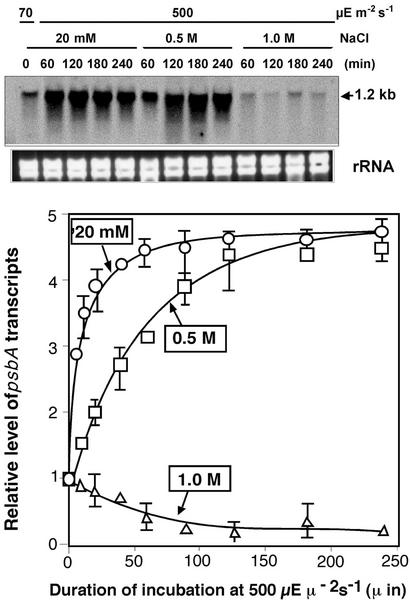
To identify the step(s) in the synthesis of D1 de novo that is inhibited by NaCl, we examined the effects of NaCl on the levels of transcripts of psbA genes, which encode pre-D1, during incubation of Synechocystis in light at 500 μE m−2 s−1 (Fig. 8). The level of psbA (psbAII and psbAIII) transcripts increased rapidly in low-salt medium. The presence of 0.5 m NaCl markedly delayed the increase in the level of these transcripts. However, the level of the transcripts at the stationary phase, namely, after a 180-min incubation in light at 500 μE m−2 s−1, was unaffected by 0.5 m NaCl. The presence of 1.0 m NaCl completely abolished any increase in the level of the transcripts.
Figure 8.
Effects of NaCl on levels of psbA transcripts during incubation of Synechocystis cells in light. Cells were incubated in light at 500 μE m−2 s−1 in the presence of 20 mm, 0.5 m, or 1.0 m NaCl. At designated times, a portion of the cell suspension was withdrawn for extraction of RNA, which was subjected to northern-blotting analysis as described in the text. The levels of transcripts were normalized by reference to levels of rRNA and the results are shown quantitatively in the bottom panel. ○, 20 mm; □, 0.5 m; ▵, 1.0 m NaCl. Each point and bar represent the average ± se of results from three independent experiments.
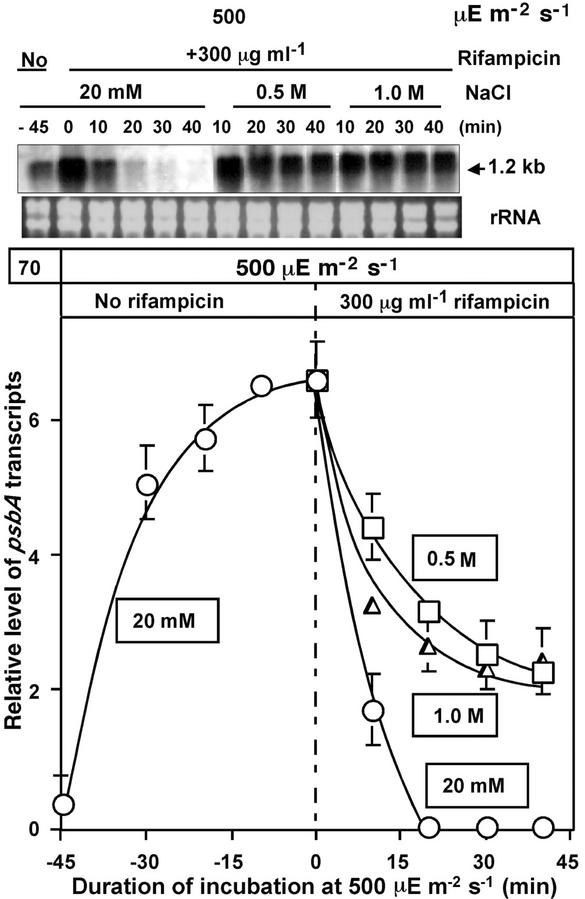
The level of psbA transcripts is a result of a balance between the rate of transcription of the psbA genes and the rate of degradation of the psbA transcripts. Therefore, a decrease in levels of psbA transcripts could be explained by the suppression of transcription or by destabilization of the transcripts. To identify the process that contributed to the inhibitory effect of NaCl, we designed an experiment in which the stability of psbA transcripts was separated from the rate of transcription by rifampicin, an inhibitor of transcription. In the experiment presented in Figure 9, rifampicin was added after the level of psbA transcripts reached a maximum level to observe the degradation of psbA transcripts. Under normal conditions, i.e. at 20 mm of NaCl, the transcripts decayed with a half-life time of about 5 min. In the presence of 0.5 m or 1.0 m NaCl, the decay of the psbA transcripts was significantly slower than in 20 mm NaCl. These results demonstrate clearly that NaCl did not destabilize the 1.2-kb psbA transcripts, but rather, stabilized them. These findings, together with the results in Figure 8, strongly suggest that NaCl inhibited transcription of the psbA genes.
Figure 9.
Effects of NaCl on the stability of psbA transcripts during incubation of Synechocystis cells in the presence of rifampicin. Cells were incubated for 45 min in light at 500 μE m−2 s−1 in the presence of 20 mm NaCl. Then, 300 μg mL−1 rifampicin was added together with 0.5 m or 1.0 m NaCl, and incubation was continued in light at 500 μE m−2 s−1. At designated times, a portion of the cell suspension was withdrawn for extraction of RNA, which was subjected to northern-blotting analysis as described in the text. The results are shown quantitatively in the bottom panel. The other experimental conditions were the same as those described in the legend to Figure 8. ○, 20 mm; □, 0.5 m; ▵, 1.0 m NaCl. Each point and bar represent the average ± se of results from four independent experiments.
Effects of NaCl on Overall Gene Expression in Synechocystis
We used a DNA microarray to examine the effects of NaCl on the expression of genes other than the psbA genes (Table I). The set of genes whose expression was induced by strong light alone was essentially the same as that reported by Hihara et al. (2001). However, the expression of psb genes for other components of PSII was not significantly induced by strong light. Table I shows the striking effects on gene expression of NaCl at 0.5 m. The inducibility by light of approximately 60% of light-inducible genes was strongly diminished by salt stress, and that of approximately 20% was moderately suppressed. The inducibility by light of a further 20% of light-inducible genes was enhanced by 0.5 m NaCl.
Table I.
Effects of salt stress on light-induced gene expression, as determined with a DNA microarray
| ORF | Gene | Product | Light Inducibility (Ratio)
|
||
|---|---|---|---|---|---|
| 20 mm NaCl | 0.5 m NaCl | 1.0 m NaCl | |||
| Genes whose light inducibility was diminished by NaCl at 0.5 m | |||||
| sll0218 | Protein of unknown function | 10.6 ± 3.0 | 1.2 ± 0.1 | 1.2 ± 0.1 | |
| slr1641 | clpB | ClpB protein | 10.2 ± 4.8 | 1.3 ± 0.2 | 1.0 ± 0.1 |
| sll0846 | Protein of unknown function | 9.9 ± 2.3 | 1.3 ± 0.2 | 0.7 ± 0.0 | |
| sll0430 | htpG | Heat-shock protein Hsp90 | 9.7 ± 1.1 | 0.7 ± 0.1 | 0.7 ± 0.0 |
| sll1732 | ndhF3 | NADH dehydrogenase I, chain L | 8.1 ± 5.8 | 1.1 ± 0.1 | 1.0 ± 0.1 |
| sll0219 | Potential FMN protein | 7.0 ± 1.5 | 1.0 ± 0.2 | 1.2 ± 0.0 | |
| slr1291 | ndhD2 | NADH dehydrogenase I, chain M | 6.8 ± 1.2 | 0.6 ± 0.1 | 0.6 ± 0.0 |
| sll1733 | ndhD3 | NADH dehydrogenase I, chain M | 6.0 ± 1.2 | 1.0 ± 0.1 | 0.8 ± 0.1 |
| slr2076 | groEL1 | 60-kDa chaperonin 1 | 5.7 ± 1.1 | 1.2 ± 0.2 | 0.6 ± 0.1 |
| sll1734 | cupA | 5.4 ± 1.0 | 0.8 ± 0.1 | 0.9 ± 0.0 | |
| slr2075 | groES | 10-kDa chaperonin | 5.1 ± 1.4 | 1.1 ± 0.2 | 0.5 ± 0.0 |
| sll0416 | groEL2 | 60-kDa chaperonin 2 | 4.5 ± 1.7 | 1.4 ± 0.2 | 0.6 ± 0.0 |
| slr1963 | H2O-soluble carotenoid protein | 4.5 ± 0.6 | 1.0 ± 0.1 | 0.6 ± 0.0 | |
| sll1028 | ccmK2 | CO2-concentrating mechanism protein | 4.2 ± 2.4 | 0.9 ± 0.1 | 0.8 ± 0.1 |
| slr0006 | Protein of unknown function | 4.2 ± 0.8 | 0.7 ± 0.0 | 0.7 ± 0.1 | |
| sll1041 | ABC transporter ATP-binding protein | 4.2 ± 0.4 | 0.7 ± 0.1 | 0.8 ± 0.0 | |
| sll1911 | Protein of unknown function | 4.1 ± 0.2 | 0.7 ± 0.0 | 0.8 ± 0.1 | |
| sll1030 | ccmL | CO2-concentrating mechanism protein | 3.8 ± 1.7 | 0.8 ± 0.1 | 0.7 ± 0.1 |
| ssl2971 | Protein of unknown function | 3.5 ± 1.0 | 1.2 ± 0.1 | 1.3 ± 0.0 | |
| slr1280 | ndhK | NADH dehydrogenase I, chain B | 3.3 ± 0.7 | 0.6 ± 0.0 | 0.7 ± 0.0 |
| slr0007 | Protein of unknown function | 3.2 ± 0.8 | 0.9 ± 0.2 | 0.8 ± 0.1 | |
| sll0217 | Potential FMN-protein | 3.2 ± 0.6 | 0.9 ± 0.1 | 0.7 ± 0.0 | |
| slr1281 | ndhJ | NADH dehydrogenase I, chain C | 3.2 ± 0.5 | 0.7 ± 0.1 | 0.8 ± 0.1 |
| sll0005 | Protein of unknown function | 3.1 ± 0.5 | 1.1 ± 0.2 | 1.1 ± 0.1 | |
| Genes whose light inducibility was moderately repressed by NaCl at 0.5 m | |||||
| ssr2016 | Protein of unknown function | 23.4 ± 6.5 | 9.7 ± 1.6 | 0.8 ± 0.0 | |
| sll1514 | hspA | 16.6-kDa small HSP | 11.0 ± 2.1 | 5.9 ± 0.6 | 0.5 ± 0.1 |
| ssl1633 | hliC | CAB/ELIP/HLIP superfamily | 5.7 ± 0.6 | 3.9 ± 0.4 | 0.5 ± 0.1 |
| slr1675 | hypA1 | Hydrogenase-expression/formation protein | 5.7 ± 1.3 | 3.7 ± 0.5 | 1.3 ± 0.1 |
| sll0170 | dnaK2 | DnaK protein 2 | 5.6 ± 1.5 | 2.2 ± 0.2 | 0.8 ± 0.0 |
| sll1483 | Periplasmic protein | 4.5 ± 0.9 | 2.1 ± 0.3 | 0.6 ± 0.0 | |
| slr1674 | Protein of unknown function | 4.0 ± 1.4 | 2.5 ± 0.5 | 0.7 ± 0.0 | |
| ssl2162 | Protein of unknown function | 3.5 ± 0.5 | 1.8 ± 0.5 | 1.1 ± 0.0 | |
| sll0306 | sigB | Sigma factor B | 3.5 ± 1.3 | 2.3 ± 0.1 | 0.8 ± 0.0 |
| Genes whose light inducibility was enhanced by NaCl at 0.5 m | |||||
| ssr2595 | hliB | High light-inducible protein B | 9.7 ± 1.8 | 24.9 ± 2.0 | 0.6 ± 0.1 |
| ssl2542 | hliA | High light-inducible protein A | 7.1 ± 0.6 | 9.3 ± 1.1 | 0.9 ± 0.1 |
| sll0528 | Protein of unknown function | 6.6 ± 0.8 | 14.6 ± 3.0 | 0.8 ± 0.0 | |
| slr1544 | Protein of unknown function | 6.1 ± 0.8 | 20.4 ± 3.3 | 0.7 ± 0.1 | |
| slr0959 | Protein of unknown function | 3.9 ± 1.0 | 5.5 ± 1.1 | 1.5 ± 0.2 | |
| slr1516 | sodB | Superoxide dismutase | 3.6 ± 0.2 | 5.5 ± 0.6 | 0.6 ± 0.0 |
| slr1915 | Protein of unknown function | 3.3 ± 1.9 | 4.4 ± 1.1 | 0.9 ± 0.0 | |
| ssl3044 | Hydrogenase component | 3.2 ± 0.2 | 4.8 ± 0.5 | 1.0 ± 0.2 | |
Cells that had been grown at 70 μE m−2 s−1 under normal conditions (control cells) were incubated in light at 500 μE m−2 s−1 for 10 min in 20 mm, 0.5 m, or 1.0 m NaCl. Then, mRNA was extracted from cells, cDNA was synthesized, and DNA microarray analysis was performed as described in “Materials and Methods.” The data presented here are ratios of levels of transcripts of individual genes from cells incubated in light to levels of those from control cells. This list includes the genes that yielded ratios of more than 3.0. The values are averages of four experiments with two samples from independent cultures.
At 1.0 m of NaCl, none of the light-inducible genes was induced by light. These observations indicated that the inducibility by light of transcription was depressed not only in the case of psbA genes, but also in the case of almost all of the light-inducible genes.
DISCUSSION
NaCl Inhibits the Repair of PSII
Previous studies of the photosynthetic machinery in vivo have demonstrated that salt stress enhances the light-induced inactivation of PSII (Neale and Melis, 1989; Lu and Zhang, 1999). In the present study, we confirmed the synergistic negative effects of light stress and salt stress on the PSII complex in Synechocystis. The extent of the light-induced inactivation of PSII reflects a balance between the rate at which damage is induced and the rate of repair of PSII (Greer et al., 1986). In our experimental system, the light-induced damage to PSII and the repair of PSII were clearly separate phenomena. Damage was inflicted by strong light (500 μE m−2 s−1) in the presence of lincomycin (Fig. 1), whereas repair was achieved in weak light (70 μE m−2 s−1) after PSII had been damaged by exposure of cells to very strong light (2,000 μE m−2 s−1; Figs. 2 and 3). Salt stress (1.0 m NaCl) strongly inhibited repair, but had no effect on the light-induced damage to PSII.
In natural habitats, photosynthetic organisms are often exposed to light stress and, in many instances, salt stress is combined with light stress. Thus, the combined effects of salt and light stress are of considerable importance in nature and agriculture.
NaCl Inhibits the Synthesis of Proteins de Novo
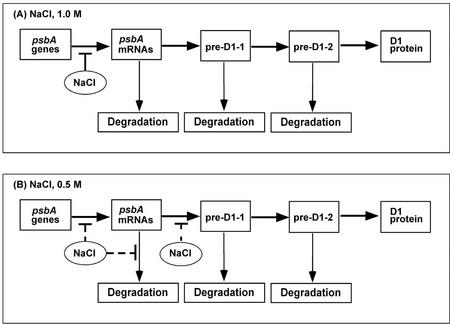
We attempted to determine whether the synthesis of D1 from the psbA genes was regulated at the level of transcription of the psbA genes, at the level of translation and stability of psbA transcripts, and/or at the level of processing and stability of pre-D1. Northern-blotting analysis (Figs. 8 and 9) indicated that 1.0 m NaCl abolished the accumulation of psbA transcripts by inhibiting transcription. Labeling of proteins in vivo provided direct evidence for the inhibition by NaCl of the synthesis of D1 de novo (Fig. 5). Thus, it seems likely that inhibition by 1.0 m NaCl of the synthesis of D1 de novo occurs primarily at the transcriptional level (Fig. 10). The specific step(s) in transcription that is inhibited by NaCl remains to be identified.
Figure 10.
A schematic representation of the proposed steps required for expression of psbA genes and the synthesis of D1, with sites of apparent inhibition by high levels of NaCl (T bars; weaker inhibition is indicated by broken T bars). A, 1.0 m NaCl. B, 0.5 m NaCl.
We observed two bands after western blotting with antibodies against the carboxy-terminal extension of pre-D1, namely, the amino acid sequence SGEGAPVALTAPAVNG. Shestakov et al. (1994) demonstrated that pre-D1 is converted to D1 by CtpA, a specific carboxy-terminal-processing protease. Inagaki et al. (2001) demonstrated that this processing involves two separate steps and, moreover, that the top and bottom bands observed after gel electrophoresis correspond to pre-D1 (pre-D1-1) and an intermediate in the processing of pre-D1-1, namely, pre-D1-2.
Western-blotting analysis of pre-D1 (Fig. 6) indicated that levels of pre-D1-1 and pre-D1-2 in cells that had been incubated in the presence of 0.5 m NaCl were about 50% of those in low-salt medium (20 mm NaCl), which might correspond to the 50% level of recovery of PSII activity shown in Figure 2. The level of pre-D1 is the result of a balance between the synthesis, processing, and degradation of the protein, and the results in Figure 7 indicate that NaCl had no effect on the processing and degradation of either form of pre-D1. Northern-blotting analysis, for which results are shown in Figures 8 and 9, demonstrated that in the presence of 0.5 m NaCl, the accumulation of psbA transcripts was delayed, but the maximum level of psbA transcripts was unaffected. These observations suggest that translation of psbA transcripts to yield pre-D1 was partially inhibited by 0.5 m NaCl.
Taken together, our results indicate that NaCl inhibited the transcription and translation of psbA genes (Fig. 10). However, inhibition of transcription was the salient factor that was primarily responsible for inhibition of the repair of the PSII complex at 1.0 m NaCl, whereas inhibition of translation was most responsible for the partial inhibition of repair at 0.5 m NaCl.
The Overall Transcription and Translation of Genes Is Affected by NaCl
The results of DNA microarray analysis (Table I) demonstrated that 1.0 m NaCl completely inhibited the light-induced accumulation of the transcripts of all the light-inducible genes, confirming the results of labeling with [35S]Met. These observations suggest that inhibition of transcription by 1.0 m NaCl was the primary cause of inhibition of the light-induced synthesis of light-inducible proteins (Fig. 10). At 0.5 m NaCl, transcription of most of the light-inducible genes ceased to be inducible by light. However, the light inducibility of some light-inducible genes was enhanced to some extent. These results might correspond to the synthesis of a protein of 25 kD (Fig. 5), whose light inducibility was enhanced in 0.5 m NaCl. However, it is unclear which gene encoded the 25-kD protein.
The results of labeling with [35S]Met (Fig. 5) demonstrated that 1.0 m NaCl inhibited the light-induced synthesis de novo not only of D1, but also of all other proteins. At 0.5 m, NaCl also inhibited the light-induced synthesis of all the light-inducible proteins, with a few exceptions, for example, the 25-kD protein. At 0.5 m NaCl, the light inducibility of the synthesis of D1 de novo was reduced and synthesis of the 25-kD protein appeared to be enhanced. Thus, the salt stress due to NaCl significantly depressed the light inducibility of the synthesis de novo of almost all of the light-inducible genes.
MATERIALS AND METHODS
Cells and Culture Conditions
The original sample of Synechocystis sp. PCC 6803 was kindly donated by Dr. John G. K. Williams (DuPont de Nemours & Co., Wilmington, DE). Cells were grown photoautotrophically in glass tubes (2.5 cm, i.d., × 20 cm; 100 mL) at 34°C under constant illumination from incandescent lamps at 70 μE m−2 s−1 (in which E indicates an Einstein, namely, 1 mole of photons) in BG-11 medium (Stanier et al., 1971) supplemented with 20 mm HEPES-NaOH, pH 7.5. This medium contained 20 mm Na+ ions and is referred to as low-salt medium. By contrast, medium that contained added NaCl is referred to as high-salt medium. Cultures were aerated with sterile air that contained 1% (v/v) CO2 (Ono and Murata, 1981).
Exposure of Cells to Light Stress and Salt Stress
Cells from 3-d-old cultures were harvested by centrifugation at 6,000g for 6 min at room temperature and were resuspended in fresh BG-11 medium at a Chl concentration of 5 ± 0.05 μg mL−1. Suspensions of cells were then incubated at 34°C for 2 h in 100-mL glass tubes in growth chambers under conditions identical to the original culture conditions. Salt stress was applied by addition of NaCl at 0.5 or 1.0 m, and light stress involved exposure to light at 500 or 2,000 μE m−2 s−1. In some experiments, protein synthesis was blocked by inclusion in the medium of 250 μg mL−1 lincomycin (Sigma Chemical, St. Louis), which was added to the culture medium 10 min before the start of incubation.
Measurement of Photosynthetic Activity
We measured the activity of PSII in intact cells by monitoring oxygen-evolving activity at 34°C with a Clark-type oxygen electrode (Hansatech Instruments, King's Lynn, UK) in the presence of 1.0 mm 1,4-benzoquinone, which accepts electrons from PSII and inhibits respiration (Ono and Murata, 1981; Tasaka et al., 1996), as described previously (Allakhverdiev et al., 1999; 2000a, 2000b). The sample, in a 3-mL cuvette, was illuminated by light that had been passed through a red optical filter (R-60; Toshiba, Tokyo) and an infrared-absorbing filter (HA-50; Hoya Glass, Tokyo). The intensity of light at the surface of the cuvette was 2,000 μE m−2 s−1.
Kinetics of Changes in the Fluorescence of Chl a
Light-induced quenching of Chl fluorescence due to the reduction of pheophytin (Klimov et al., 1986; Allakhverdiev et al., 1988; Ke, 2001) in intact cells was monitored with a fluorometer (PAM-101; Heinz Walz, Effeltrich, Germany) in the pulse-amplitude modulation mode. The light-induced quenching of Chl fluorescence was measured at 34°C in the presence of 1 mg mL−1 sodium dithionite after continuous exposure of the sample to actinic light (λ > 520 nm) from an incandescent lamp (KL-1500 Electronic; Schott Glasswerke, Wiesbaden, Germany) at 2,700 μE m−2 s−1. The concentration of Chl was determined as described by Arnon et al. (1974).
Labeling of Proteins in Vivo
A suspension of cells at a concentration corresponding to 5 ± 0.05 μg Chl mL−1 was supplemented with 10 nm [35S]Met (>1,000 Ci mmol−1; Amersham Pharmacia Biotech, Buckinghamshire, UK), as described previously (Nishiyama et al., 2001). Then the suspension was incubated at 34°C for designated periods of time in light at 500 μE m−2 s−1 in the presence of 20 mm, 0.5 m, or 1.0 m NaCl. The labeling was terminated by the addition of nonradioactive Met to a final concentration of 1.0 mm and immediate cooling of samples in iced water. Cells were collected by centrifugation at 5,000g for 6 min at 4°C, and thylakoid membranes were isolated from these cells as described previously (Allakhverdiev et al., 2000a). Thylakoid membranes were solubilized by incubation for 5 min at 65°C in 60 mm Tris[hydroxymethyl]-aminomethane (pH adjusted to 6.8 with HCl) that contained 2% (w/v) SDS, 5% (v/v) 2-mercaptoethanol, and 10% (v/v) glycerol, and then proteins were separated by PAGE (12.5% [w/v] polyacrylamide) in the presence of 0.08% (w/v) SDS and 6 m urea, as described previously (Laemmli, 1970; Taguchi et al., 1993, 1995). Solubilized thylakoid membranes corresponding to 0.8 μg of Chl a were loaded in each lane. Labeled proteins on the gel were visualized by exposure of the dried and fixed gel to x-ray film. Radioactivity of radiolabeled D1 was quantitated with a digital camera system (LAS-1000; Fuji Photo Film, Tokyo).
Western-Blotting Analysis
Thylakoid membranes were isolated and solubilized as described previously (Allakhverdiev et al., 2000a) and as summarized above. After electrophoresis, the separated proteins were blotted onto a nitrocellulose membrane (Schleicher & Schuell, Keene, NH) in a semidry transfer apparatus (Atto Co., Tokyo). D1 and pre-D1 were then detected immunologically with an enhanced chemiluminescence western-blotting kit according to the protocol supplied with the kit (Amersham International, Buckinghamshire, UK). The D1 protein was detected with rabbit antibodies raised against amino acid residues 55 through 78 in the AB loop of D1 from spinach (Taguchi et al., 1995). These antibodies recognize the products (D1) of psbAI, psbAII, and psbAIII genes because the amino acid sequence of the AB loop is exactly the same among the three kinds of product. The pre-D1 protein was detected with rabbit antibodies raised against an oligopeptide of 16 amino acid residues (SGEGAPVALTAPAVNG) that corresponded to the carboxyl terminus of pre-D1 (the products of the psbAII and psbAIII genes) from Synechocystis. As second antibodies, we used horseradish peroxidase-linked antibodies raised in donkey against rabbit immunoglobulin G (Amersham International). The antibodies raised in rabbit against D1 were kindly provided by Prof. Kimiyuki Satoh (Department of Biology, Okayama University, Japan), and the antibodies against pre-D1 were generated in our laboratory. The digital camera system was used to monitor signals from blotted membranes and to quantify D1 and pre-D1.
Northern-Blotting Analysis
Total RNA was extracted from cells, and northern-blotting analysis was performed as described previously (Los et al., 1997). Rifampicin was used as an inhibitor of transcription to determine the stability of psbA transcripts. Equal amounts of RNA (4 μg) from each sample were loaded on the gel and rRNA was visualized by staining with ethidium bromide. A 1.0-kb fragment of DNA that included the coding region of the psbAII gene was amplified by the PCR with primers 5′-AACGACTCTCCAACAGCGCGAAA-3′and 5′-CGTTCGTGCATTACTTCAAAACCG-3′ and genomic DNA from Synechocystis as the template. The amplified fragment of DNA was ligated into the TA cloning vector pT7Blue-T (Novagen, Darmstadt, Germany). The plasmid was digested at the HincII and NcoI sites within the insert. The resultant 700-bp fragment of DNA was conjugated with alkaline phosphatase using an Alkphos Direct kit (Amersham Pharmacia Biotech, Piscataway, NJ) and the conjugate was used as the probe. This probe recognized the transcripts of psbAII and psbAIII genes. After hybridization, blots were soaked in CDP-star solution (Amersham Pharmacia Biotech), and signals from hybridized RNAs were detected with the digital camera system.
Preparation of cDNAs for DNA Microarray Analysis
Cells in culture were killed by the addition of an equal volume of an ice-cold mixture of phenol and ethanol (1:20, w/v) in an ice bath. Total RNA was then extracted as described previously (Los et al., 1997) and was treated with RNase-free DNase I (Nippon Gene, Tokyo) to remove contaminating genomic DNA. cDNAs, labeled with fluorescent dyes (Cy3 and cy5; Amersham Pharmacia Biotech), were prepared from 10 μg of total RNA with an RNA Fluorescence Labeling Core kit (M-MLV, version 2.0; Takara Co., Kyoto) according to the manufacturer's instructions.
DNA Microarray Analysis
Genome-wide analysis of transcription was performed with a DNA microarray, as described previously (Suzuki et al., 2001; Kanesaki et al., 2002). In brief, we used a Synechocystis DNA microarray (CyanoCHIP, v1.5; Takara Co.), which included 3,078 of a total of 3,169 genes for hybridization by incubation for 16 h at 65°C with Cy3- and Cy5-labeled cDNAs in 30 μL of 6× SSC (1× SSC contains 150 mm NaCl and 15 mm sodium citrate), 0.2% (w/v) SDS, 5× Denhardt's solution, and 100 ng μL−1 denatured salmon sperm DNA. After hybridization, the microarray was washed with 2× SSC at 60°C for 10 min, with 0.2× SSC that contained 0.1% (w/v) SDS at 60°C for 10 min, and finally with 0.2× SSC at room temperature. After the final rinse, all moisture was removed by evaporation under an air spray prior to analysis with an array scanner (GMS 418; Affymetrix, Woburn, MA). Signals were quantified with ImaGene, version 4.1 software (BioDiscovery, Los Angeles) with normalization by reference to the total intensity of signals from all genes with the exception of genes for rRNAs. This procedure allowed calculation of changes in the level of the transcript of each gene relative to the total amount of mRNA.
ACKNOWLEDGMENTS
We thank Prof. Kimiyuki Satoh, Okayama University, for his generous gift of antibodies against D1 and Prof. Itzhak Ohad, Hebrew University, for helpful discussions and comments on the manuscript.
Footnotes
This work was supported, in part, by the Ministry of Education, Science and Culture, Japan (Grant-in-Aid for Scientific Research no. 13854002), by the Cooperative Research Program of the National Institute for Basic Biology on the Stress Tolerance of Plants, and by the Japan Society for the Promotion of Science (Invitation Fellowship for Research in Japan to S.I.A.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.011114.
LITERATURE CITED
- Allakhverdiev SI, Klimov VV, Ladygin VG. Photoreduction of pheophytin in photosystem II reaction centers of intact cells of green algae and cyanobacteria under anaerobic conditions. Biofizika (Moscow) 1988;33:442–447. [PubMed] [Google Scholar]
- Allakhverdiev SI, Nishiyama Y, Suzuki I, Tasaka Y, Murata N. Genetic engineering of the unsaturation of fatty acids in membrane lipids alters the tolerance of Synechocystis to salt stress. Proc Natl Acad Sci USA. 1999;96:5862–5867. doi: 10.1073/pnas.96.10.5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allakhverdiev SI, Sakamoto A, Nishiyama Y, Inaba M, Murata N. Ionic and osmotic effects of NaCl-induced inactivation of photosystems I and II in Synechococcus sp. Plant Physiol. 2000b;123:1047–1056. doi: 10.1104/pp.123.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allakhverdiev SI, Sakamoto A, Nishiyama Y, Murata N. Inactivation of photosystems I and II in response to osmotic stress in Synechococcus: contribution of water channels. Plant Physiol. 2000a;122:1201–1208. doi: 10.1104/pp.122.4.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon DI, McSwain BD, Tsujimoto HY, Wada K. Photochemical activity and components of membrane preparations from blue-green algae: coexistence of two photosystems in relation to chlorophyll a and removal of phycocyanin. Biochim Biophys Acta. 1974;357:231–245. doi: 10.1016/0005-2728(74)90063-2. [DOI] [PubMed] [Google Scholar]
- Aro E-M, Virgin I, Andersson B. Photoinhibition of photosystem II: inactivation, protein damage and turnover. Biochim Biophys Acta. 1993;1143:113–134. doi: 10.1016/0005-2728(93)90134-2. [DOI] [PubMed] [Google Scholar]
- Barber J, Andersson B. Too much of a good thing: light can be bad for photosynthesis. Trends Biochem Sci. 1992;17:61–66. doi: 10.1016/0968-0004(92)90503-2. [DOI] [PubMed] [Google Scholar]
- Berry J, Björkman O. Photosynthetic response and adaptation to temperature in higher plants. Annu Rev Plant Physiol. 1980;31:491–543. [Google Scholar]
- Boyer JS. Plant productivity and environment. Science. 1982;218:443–448. doi: 10.1126/science.218.4571.443. [DOI] [PubMed] [Google Scholar]
- Greer DH, Berry JA, Björkman O. Photoinhibition of photosynthesis in intact bean leaves: role of light and temperature, and requirement for chloroplast-protein synthesis during recovery. Planta. 1986;168:253–260. doi: 10.1007/BF00402971. [DOI] [PubMed] [Google Scholar]
- Haussühl K, Andersson B, Adamska I. A chloroplast DegP2 protease performs the primary cleavage of the photodamaged D1 protein in plant photosystem II. EMBO J. 2001;20:713–722. doi: 10.1093/emboj/20.4.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertwig B, Streb P, Feierabend J. Light dependence of catalase synthesis and degradation in leaves and the influence of interfering stress conditions. Plant Physiol. 1992;100:1547–1553. doi: 10.1104/pp.100.3.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hihara Y, Kamei A, Kanehisa M, Kaplan A, Ikeuchi M. DNA microarray analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell. 2001;13:793–806. doi: 10.1105/tpc.13.4.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki N, Fujita S, Satoh K. Solubilization and partial purification of a thylakoid enzyme of spinach involved in the processing of D1 protein. FEBS Lett. 1989;246:218–222. [Google Scholar]
- Inagaki N, Yamamoto Y, Satoh K. A sequential two-step proteolytic process in the carboxyl-terminal truncation of precursor D1 protein in Synechocystis sp. PCC 6803. FEBS Lett. 2001;509:197–201. doi: 10.1016/s0014-5793(01)03180-5. [DOI] [PubMed] [Google Scholar]
- Jones LW, Kok B. Photoinhibition of chloroplast reactions: kinetics and action spectra. Plant Physiol. 1966a;41:1037–1043. doi: 10.1104/pp.41.6.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones LW, Kok B. Photoinhibition of chloroplast reactions: multiple effects. Plant Physiol. 1966b;41:1044–1049. doi: 10.1104/pp.41.6.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanesaki Y, Suzuki I, Allakhverdiev SI, Mikami K, Murata N. Salt stress and hyperosmotic stress regulate the expression of different sets of genes in Synechocystis sp. PCC 6803. Biochem Biophys Res Commun. 2002;290:339–348. doi: 10.1006/bbrc.2001.6201. [DOI] [PubMed] [Google Scholar]
- Ke B. The transient intermediate electron acceptor of photosystem II, pheophytin. In: Ke B, editor. Photosynthesis: Photobiochemistry and Photobiophysics. Vol. 10. Dordrecht, The Netherlands: Kluwer Academic Publishers; 2001. pp. 305–322. [Google Scholar]
- Klimov VV, Allakhverdiev SI, Ladygin VG. Photoreduction of pheophytin in photosystem II of the whole cells of green algae and cyanobacteria. Photosynth Res. 1986;10:355–361. doi: 10.1007/BF00118301. [DOI] [PubMed] [Google Scholar]
- Kok B. On the inhibition of photosynthesis by intense light. Biochim Biophys Acta. 1956;21:234–244. doi: 10.1016/0006-3002(56)90003-8. [DOI] [PubMed] [Google Scholar]
- Kyle DJ, Ohad I, Arntzen CJ. Membrane protein damage and repair: selective loss of a quinone-protein function in chloroplast membranes. Proc Natl Acad Sci USA. 1984;81:4070–4074. doi: 10.1073/pnas.81.13.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lindahl M, Spetea C, Hundal T, Oppenheim AB, Adam Z, Andersson B. The thylakoid FtsH protease plays a role in the light-induced turnover of the photosystem II D1 protein. Plant Cell. 2000;12:419–431. doi: 10.1105/tpc.12.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Los DA, Ray MK, Murata N. Differences in the control of the temperature-dependent expression of four genes for desaturases in Synechocystis sp. PCC 6803. Mol Microbiol. 1997;25:1167–1175. doi: 10.1046/j.1365-2958.1997.5641912.x. [DOI] [PubMed] [Google Scholar]
- Lu C-M, Zhang J-H. Effects of salt stress on PSII function and photoinhibition in the cyanobacterium Spirulina platensis. J Plant Physiol. 1999;155:740–745. [Google Scholar]
- Marder JB, Goloubinoff P, Edelman M. Molecular architecture of the rapidly metabolized 32-kilodalton protein of photosystem II: indications for COOH-terminal processing of a chloroplast membrane polypeptide. J Biol Chem. 1984;259:3900–3908. [PubMed] [Google Scholar]
- Mattoo AK, Hoffman-Falk H, Marder JB, Edelman M. Regulation of protein metabolism: coupling of photosynthetic electron transport to in vivo degradation of the rapidly metabolized 32-kilodalton protein of the chloroplast membranes. Proc Natl Acad Sci USA. 1984;81:1380–1384. doi: 10.1073/pnas.81.5.1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattoo AK, Marder JB, Edelman M. Dynamics of the photosystem II reaction center. Cell. 1988;56:241–246. doi: 10.1016/0092-8674(89)90897-0. [DOI] [PubMed] [Google Scholar]
- Neale PJ, Melis A. Salinity-stress enhances photoinhibition of photosystem II in Chlamydomonas reinhardtii. J Plant Physiol. 1989;134:619–622. doi: 10.1104/pp.92.4.1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama Y, Yamamoto H, Allakhverdiev SI, Inaba M, Yokota A, Murata N. Oxidative stress inhibits the repair of photodamage to the photosynthetic machinery. EMBO J. 2001;20:5587–5594. doi: 10.1093/emboj/20.20.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad I, Kyle DJ, Arntzen CJ. Membrane protein damage and repair: removal and replacement of inactivated 32-kilodalton polypeptide in chloroplast membranes. J Cell Biol. 1984;99:481–485. doi: 10.1083/jcb.99.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T, Murata N. Chilling susceptibility of the blue-green alga Anacystis nidulans: effect of growth temperature. Plant Physiol. 1981;67:176–181. doi: 10.1104/pp.67.1.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powles SB. Photoinhibition of photosynthesis induced by visible light. Annu Rev Plant Physiol. 1984;35:15–44. [Google Scholar]
- Reisfeld A, Mattoo AK, Edelman M. Processing of a chloroplast-translated membrane protein in vivo: analysis of the rapidly synthesized 32000-dalton shield protein and its precursor in Spirodela oligorrhiza. Eur J Biochem. 1982;124:125–129. doi: 10.1111/j.1432-1033.1982.tb05914.x. [DOI] [PubMed] [Google Scholar]
- Schuster G, Timberg R, Ohad I. Turnover of thylakoid photosystem II proteins during photoinhibition of Chlamydomonas reinhardtii. Eur J Biochem. 1988;177:403–410. doi: 10.1111/j.1432-1033.1988.tb14389.x. [DOI] [PubMed] [Google Scholar]
- Sharma PK, Hall DO. Interaction of salt stress and photoinhibition on photosynthesis in barley and sorghum. J Plant Physiol. 1991;138:614–619. [Google Scholar]
- Shestakov SV, Anbudurai PR, Stanbekova GE, Gadzhiev A, Lind LK, Pakrasi HB. Molecular cloning and characterization of the ctpA gene encoding a carboxyl-terminal processing protease: analysis of a spontaneous photosystem II-deficient mutant strain of the cyanobacterium Synechocystis sp. PCC 6803. J Biol Chem. 1994;269:19354–19359. [PubMed] [Google Scholar]
- Stanier RY, Kunisawa R, Mandel M, Cohen-Bazire G. Purification and properties of unicellular blue-green algae (order Chroococcales) Bacteriol Rev. 1971;35:171–205. doi: 10.1128/br.35.2.171-205.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki I, Kanesaki Y, Mikami K, Kanehisa M, Murata N. Cold-regulated genes under control of the cold sensor Hik33 in Synechocystis. Mol Microbiol. 2001;40:235–244. doi: 10.1046/j.1365-2958.2001.02379.x. [DOI] [PubMed] [Google Scholar]
- Taguchi F, Yamamoto Y, Inagaki N, Satoh K. Recognition signal for the C-terminal processing protease of D1 precursor protein in the photosystem II reaction center: an analysis using synthetic oligopeptides. FEBS Lett. 1993;326:227–231. doi: 10.1016/0014-5793(93)81796-3. [DOI] [PubMed] [Google Scholar]
- Taguchi F, Yamamoto Y, Satoh K. Recognition of the structure around the site of cleavage by the carboxyl-terminal processing protease for D1 precursor protein of the photosystem II reaction center. J Biol Chem. 1995;270:10711–10716. doi: 10.1074/jbc.270.18.10711. [DOI] [PubMed] [Google Scholar]
- Tasaka Y, Gombos Z, Nishiyama Y, Mohanty P, Ohba T, Ohki K, Murata N. Targeted mutagenesis of acyl-lipid desaturases in Synechocystis: evidence for the important roles of polyunsaturated membrane lipids in growth, respiration and photosynthesis. EMBO J. 1996;15:6416–6425. [PMC free article] [PubMed] [Google Scholar]
- Taylor MA, Packer JCL, Bowyer JR. Processing of the D1 polypeptide of the photosystem II reaction centre and photoactivation of a low fluorescence mutant (LF-1) of Scenedesmus obliquus. FEBS Lett. 1988;237:229–233. [Google Scholar]