Abstract
Expression of nuclear genes that encode the A and B subunits of chloroplast glyceraldehyde-3-phosphate dehydrogenase (GAPA and GAPB) of Arabidopsis is known to be regulated by light. We used a negative selection approach to isolate mutants that were defective in light-regulated expression of the GAPA gene. Two dominant mutants belonging to the same complementation group, uga1-1 and uga1-2, were then characterized. These two mutants showed a dramatic reduction in GAPA mRNA level in both mature plants and seedlings. Surprisingly, mutations in uga1-1 and uga1-2 had no effect on the expression of GAPB and several other light-regulated genes. In addition, we found that the chloroplast glyceraldehyde-3-phosphate dehydrogenase enzyme activity of the mutants was only slightly lower than that of the wild type. Western-blot analysis showed that the GAPA protein level was nearly indistinguishable between the wild-type and the uga mutants. These results suggested that posttranscriptional control was involved in the up-regulation of the GAPA protein in the mutants. The uga1-1 mutation was mapped to the bottom arm of chromosome V of the Arabidopsis genome.
Transcription is one of the primary steps at which light regulates gene expression in plants (Terzaghi and Cashmore, 1995). Two classes of photoreceptors, phytochrome and blue light/UV-A receptor (cryptochrome), are involved in the regulation of photosynthetic genes (Batschauer, 1998; Briggs and Huala, 1999; Deng and Quail, 1999; Fankhauser and Chory, 1999). It has been suggested that eukaryotic phytochromes are Ser/Thr kinases with a two-component His kinase ancestry (Yeh et al., 1997; Yeh and Lagarias, 1998; Fankhauser and Chory, 1999; Fankhauser et al., 1999). Five phytochrome genes have been identified in Arabidopsis (Clack et al., 1994; Quail et al., 1995; Quail, 1997). Current evidence indicates that the different phytochromes may have distinct functions (Quail et al., 1995; Quail, 1997). Genetic and molecular studies have led to the identification of four blue-light photoreceptors in Arabidopsis (Briggs et al., 2001). CRY1 (HY4) and CRY2/PHH1 have partial overlapping functions in promoting anthocyanin formation and inhibiting hypocotyl elongation (Ahmad and Cashmore, 1993; Ahmad et al., 1995; Lin, 2000), whereas PHOT1/NPH1 and PHOT2 regulate phototropism, stomatal opening, and chloroplast movement (Liscum and Briggs, 1995; Briggs and Huala, 1999; Kinoshita et al., 2001; Sakai et al., 2001). In addition, the mutations in CRY1 and CRY2 genes affect blue-light-mediated regulation of photosynthetic gene expression (Ahmad et al., 1995; Conley and Shih, 1995; Mazzella et al., 2001).
Several mutants affecting the light signal transduction pathway appear to be defective in genes that encode transcription factors. PIF3 was found not only to interact directly with PhyB but also with the promoters of many light-regulated genes (Ni et al., 1999; Martinez-Garcia et al., 2000). The hfr1/rsf1/rep1 mutants, on the other hand, appeared to be specific for PhyA pathway (Fairchild et al., 2000; Fankhauser and Chory, 2000; Soh et al., 2000). The HFR1 gene product is a bHLH protein and, therefore, a putative DNA-binding protein (Fairchild et al., 2000; Soh et al., 2000). It was also found to interact with PIF3 (Ni et al., 1999; Fairchild et al., 2000; Martinez-Garcia et al., 2000). Other phytochrome-specific intermediates have also been cloned via the isolation of mutants. The HY5 gene product was shown to be a basic Leu Zipper transcription factor that interacts with light-responsive promoters (Chattopadhyay et al., 1998b).
A number of cis-acting elements, including GT elements, G boxes, I boxes, CGF element, and CCA element, have been characterized from several photosynthetic genes, including RBCS and LHCB, the nuclear genes encoding the small subunit of Rubisco and light harvest complex proteins, respectively (Donald and Cashmore, 1990; Gilmartin et al., 1990; Anderson et al., 1994; Kenigsbuch and Tobin, 1995; Terzaghi and Cashmore, 1995; Wang et al., 1997b). Based on in vitro-binding assays, genes that encode GBF, GT1, and CCA1 factors have been identified in Arabidopsis. A survey of the Arabidopsis genomic sequences indicated that each of these genes belongs to a small gene family, with a highly conserved sequence in the putative DNA-binding domains. To show which member(s) in the gene family is involved in light regulation, it is essential to establish a direct link between the in vitro-binding activities and the in vivo function of transcription activation. This line of evidence is mostly lacking, with the exception of CCA1, in which it was shown that transgenic Arabidopsis plants expressing antisense RNA for CCA1 showed reduced phytochrome induction of the endogenous LHCB1-3 gene (Wang et al., 1997b).
We have been studying light regulation of two nuclear genes (GAPA and GAPB) that encode chloroplast glyceraldehyde-3-phosphate dehydrogenase (GAPDH) from Arabidopsis. In higher plants, there are two chloroplast GAPDH isozymes, with subunit structures of A4 and A2B2, which are key enzymes in the photosynthetic carbon fixation cycle (Cerff, 1982). In previous studies, we showed that the expression of these two genes is coordinately regulated by light at the transcriptional level in tobacco (Nicotiana tabacum) and Arabidopsis (Shih and Goodman, 1988; Dewdney et al., 1993). Several cis-acting elements and their cognate binding factors of both GAPA and GAPB genes were identified (Conley et al., 1994; Kwon et al., 1994; Park et al., 1996; Chan et al., 2001). In etiolated seedlings, a short light pulse can induce transient increases of GAPA and GAPB mRNA levels. However, this induction cannot be reversed by subsequent far-red light treatment (Dewdney et al., 1993). These regulatory patterns are distinct from those of the pea (Pisum sativum) RBCS genes (Kaufman et al., 1984) and Arabidopsis LHCB genes (Karlin-Neumann et al., 1988), in which the effect of a short red light pulse can be reversed by a subsequent far-red light treatment. Continuous exposure of dark-treated mature plants or etiolated seedlings to red, blue, or white light is required for sustained high-level expression of GAPA and GAPB genes in Arabidopsis, with blue and white light much more efficient than red light (Dewdney et al., 1993; Conley and Shih, 1995). Our results indicated that this effect is mediated by a combination of phytochromes and the blue light photoreceptor encoded by the CRY1 (HY4) gene (Conley and Shih, 1995). Results from saturation linker scan mutagenesis of the GAPB promoter constructs in transgenic Arabidopsis suggest that a single cis-acting element may respond to more than one photoreceptor (Chan et al., 2001).
In addition to the identification of cis-acting elements, we are interested in obtaining mutations that affect light regulation of GAPA and GAPB genes. Although a variety of photomorphogenic mutants are available in Arabidopsis, most of these mutants are defective in early steps in light-signaling pathways or are not defective in GAP gene expression (Conley and Shih, 1995; M.-C. Shih, unpublished data). Therefore, we used a negative selection scheme to isolate regulatory mutants that are defective in light activation of the GAPA gene. Here, we report the characterization of two of these mutants. Our results indicated that these two mutations affect very downstream steps in light signal transduction pathways leading to the activation of the GAPA gene.
RESULTS
Selection of Mutants Affecting GAPA Gene Expression
In the presence of allyl alcohol, wild-type plants with functional ADH enzyme will die because of the conversion of allyl alcohol to toxic aldehyde by ADH. In contrast, plants without functional ADH can survive allyl alcohol treatment. Negative selection schemes using ADH as a selectable marker were used to isolate aar mutants, which are defective in hypoxic induction of ADH (Conley et al., 1999), and cue mutants, which are defective in controlling the expression of LHCB3 (Li et al., 1995; Lopez-Juez et al., 1998). We designed a similar selection scheme to isolate regulatory mutants that are defective in light activation of the GAPA gene.
In the current scheme, we first transformed an Arabidopsis ADH null mutant, adh1-2, with a construct that puts ADH and β-glucuronidase (GUS) coding sequences under the control of separate GAPA promoters (see “Materials and Methods” for details). Several independent transgenic lines that have GUS and ADH activity were obtained. In all of these lines, the expression of ADH and GUS transgenes was regulated by light similar to that of the GAPA gene. One of these lines, AG-5G, was chosen for mutagenesis. In 5-d-old etiolated AG-5G seedlings, the accumulation of ADH activity reached a steady-state level after 12 to 24 h of white light treatment, similar to that of the endogenous GAPA gene (Dewdney et al., 1993; Conley and Shih, 1995). Titration experiments indicated that 7.5 mm allyl alcohol is needed to cause 100% lethality of the 24-h light-treated seedlings.
To obtain mutants that underexpress ADH, a total of 50,000 M2 seeds of AG-5G were germinated on filter papers in the dark for 5 d and then subjected to 24 h of white light treatment. The filters were then transferred onto medium containing 7.5 mm allyl alcohol. After 2 h, filters were moved onto a fresh agar medium. The surviving plants, which must have lacked ADH activity, were assayed for GUS activity in leaves. Among the 99 plants that survived allyl alcohol selection, 77 were GUS positive and 22 were GUS negative. Only seven of the latter mutants survived long enough to produce seeds, whereas the other 15 died or failed to set seeds after transfer to the soil. The lethality could be because of mutations in essential genes or the occurrence of multiple mutations in these plants. The surviving adh−gus− plants, designated as uga (underexpressor of GAPA), are presumably defective in regulatory genes that control the expression of GAPA. We characterized two of these mutants, ugab3 and ugab9, as described below.
Table I showed that the F1 progeny from crosses between line AG-5G and each of the two uga mutants had low GUS activity (GUS−), indicating that all of them exhibited the mutant phenotype. These results suggested that both ugab3 and ugab9 mutations are dominant. However, the F2 progeny from both crosses deviated significantly from the expected 1:3 ratio. GUS+ and GUS− plants in the F2 progeny from the cross between AG-5G and ugab3 showed a 1:6 ratio, whereas the cross between AG-5G and ugab9 produced a 1:8 ratio. One possible explanation for this observation could be that the presence of the transgene resulted in the expression of ADH at abnormally high level in leaves. This may have caused a high rate of lethality to the individual plants that show the wild-type phenotype, hence yielding a lower than expected wild-type progeny. Table I also shows that the F2 progeny of the ugab3 × ugab9 cross gave a GUS+:GUS− ratio of 0:132, i.e. all the F2 progeny were mutant. This indicated that the mutations in b3 and b9 belonged to the same complementation group. The ugab3 and ugab9 mutants were hence renamed uga1-1 and uga1-2, respectively.
Table I.
Genetic analysis of uga mutants
| Cross | GUS+:GUS− in F1 | GUS+:GUS− in F2 |
|---|---|---|
| AG-5G × ugab3 | 0 :13 | 6 :37 |
| AG-5G × ugab9 | 0 :28 | 5 :40 |
| ugab3 × ugab9 | 0 :15 | 0 :132 |
GUS enzymatic assays were used to assess GUS+ and GUS− phenotype. F1 and F2 progeny with GUS activity comparable with the homozygous AG-5G line as shown in Fig. 1B were assigned as GUS+, whereas those with GUS activity similar to homozygous mutants were assigned as GUS−.
Effects of uga Mutations on the Expression of GAPA
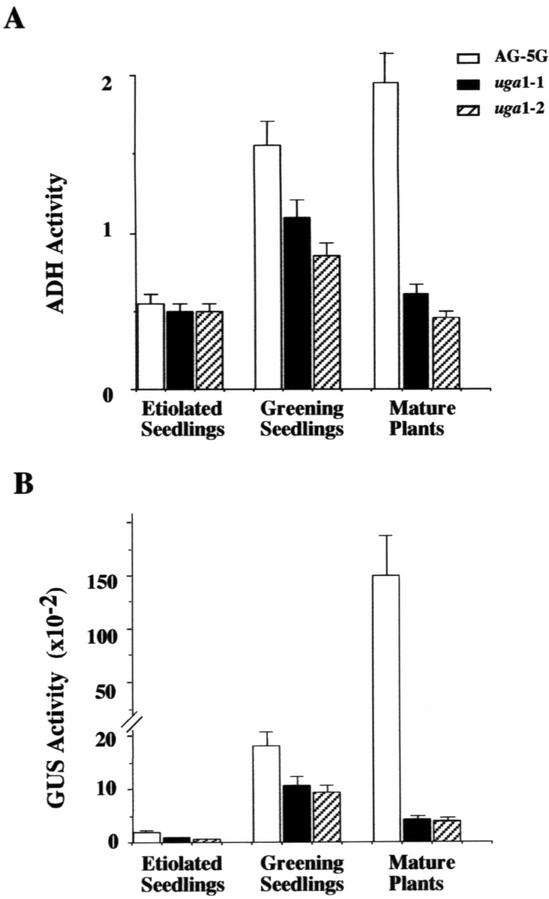
To quantify the effect of uga mutations on the expression of GAPA::GUS and GAPA::ADH transgenes, we compared the levels of ADH and GUS activities in wild-type AG-5G and uga mutants in 5-d-old etiolated seedlings subjected to 24 h of white light treatment. Both mutants showed a moderate reduction in ADH and GUS activity compared with the AG-5G line (Fig. 1A). When 4-week-old light-grown plants were assayed for reporter gene activities, the difference between the wild type and the mutants was far more dramatic. As shown in Figure 1B, the two mutants exhibited 30% and 23% of the wild-type level of steady-state ADH activity, whereas their GUS activity was reduced to 3% and 5% of that in the wild type. These results indicated that both mutants are impaired in the expression of both the ADH and GUS reporter genes, especially so in mature plants.
Figure 1.
Effects of uga mutations on GAPA::ADH and GAPA::GUS transgenes. GUS and ADH activities of AG-5G and uga mutants in 5-d-old etiolated seedlings, greening seedlings, and 4-week-old plants were determined as described in “Materials and Methods.” A, Unit of ADH enzyme is defined as an increase in A340 of 1 min mg protein−1. B, GUS activity is expressed as pmol 4-methylumbelliferone min−1 mg−1 protein. The data presented are the average of three independent treatments. Plants grown at different times were used for replicated treatments. For each treatment, a total of about 500 plants was pooled and used for protein extracts preparation. Error bars = sds.
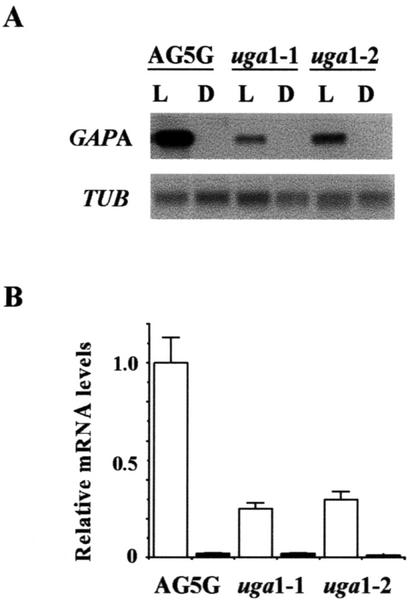
To determine the effects of uga mutations on the expression of the endogenous GAPA gene, mRNA levels from 5-d-old light-grown Arabidopsis seedlings were compared with those from 5-d-old etiolated seedlings. The results from one set of representative northern blots were illustrated in Figure 2A. These experiments were repeated three times and the resulting blots were quantified using the GAPA mRNA levels from light-grown AG5G as 100% (Fig. 2B). In greening seedlings, GAPA mRNA levels in uga1-1 and uga1-2 were 2- to 3-fold lower than the level in AG-5G (Fig. 2).
Figure 2.
Effects of uga mutations on the expression of GAPA in seedlings. A, Total RNAs from seedlings grown in continuous light (L) or complete darkness (D) for 5 d were isolated and analyzed by northern-blot analysis. Representative data are from gels loaded with 5 μg RNA lane−1 and probed with radiolabeled GAPA or TUB. B, Each northern-blot analysis was repeated three times using RNA samples from plants grown at different times. Relative densitometric values were obtained by first taking the ratio of GAPA signal intensity over that of the corresponding TUB signal for each lane, and then dividing that by the ratio to obtain obtained for light-grown AG-5G. Therefore, relative densitometric value for light-grown AG5G is taken as 1. Error bars = sds.
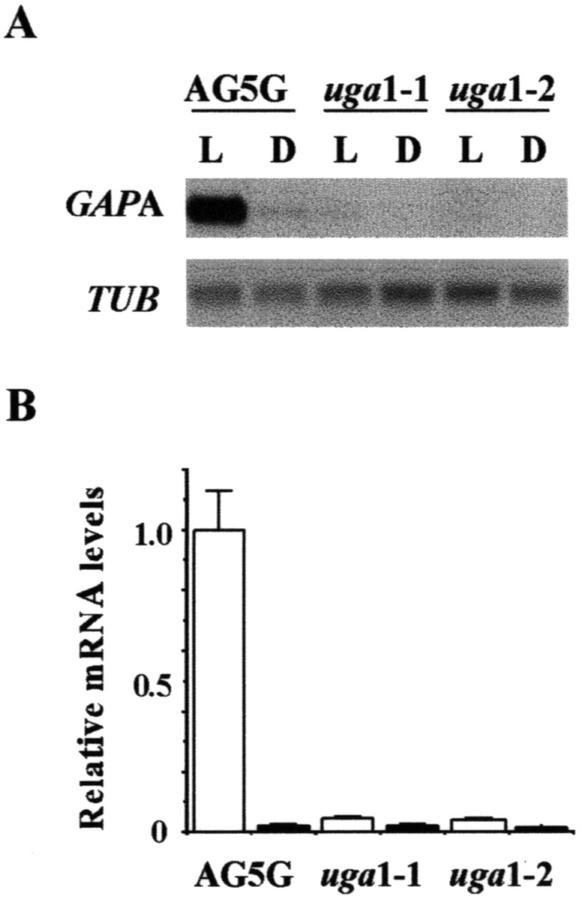
To determine the effects of uga mutations on the expression of GAPA in mature plants, mRNAs from light-grown 4-week-old plants were compared with those of plants that were light grown for 4 weeks and then dark adapted for 5 d (Fig. 3A). The quantification data showed that levels of GAPA mRNA in uga1-1 and uga1-2 were more than 20-fold lower than that of AG-5G in 3-week-old plants (Fig. 3B). Consistent with our prior results (Dewdney et al., 1993; Conley and Shih, 1995), the data also showed that there was barely detectable GAPA mRNA in both etiolated seedlings (Fig. 2) and dark-adapted mature plants (Fig. 3). The combined results demonstrated that uga1-1 and uga1-2 mutations affect the expression of both the endogenous GAPA gene and the GAPA::GUS and GAPA::ADH transgenes. Therefore, it is likely that these mutations are defective in a regulatory gene that controls the expression of GAPA. However, the observation that the uga mutations had more severe effects on the mRNA levels of GAPA in mature plants than in seedlings suggested that the transcription complexes required for GAPA activation are not identical in these two stages.
Figure 3.
Effects of uga mutations on the expression of GAPA in mature plants. A, Total RNAs from light-grown 4-week-old plants (L) were compared with those of plants that were light grown for 4 weeks and then dark adapted for 5 d (D) were isolated and analyzed by northern-blot analysis. Representative data are from gels loaded with 5 μg RNA lane−1 and probed with radiolabeled GAPA or TUB. B, Each northern-blot analysis was repeated three times and quantified as described in Figure 2. The average densitometric value for light-grown AG5G is taken as 1. Error bars = sds.
Effects of uga Mutations on the Expression of Other Light-Regulated Genes
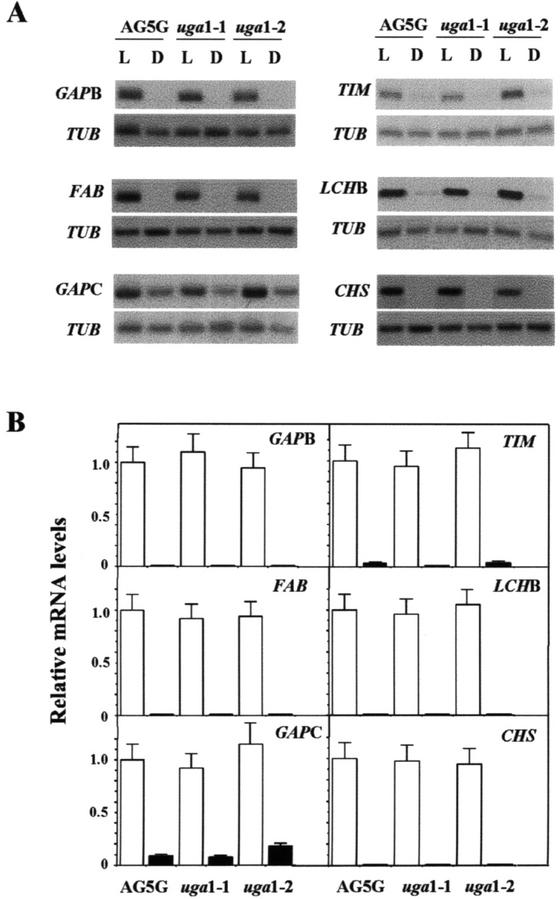
Because GAPA and GAPB gene products constitute subunits of the GAPDH holoenzyme (Cerff, 1982), it is reasonable to expect that these two genes are controlled by the same regulatory mechanism. Therefore, we compared the GAPB mRNA levels between wild-type and the two uga mutants in mature light-grown and dark-adapted plants by northern-blot analysis (Fig. 4A). Surprisingly, the levels of GAPB mRNA in uga1-1 and 1-2 were similar to that of wild type (Fig. 4, A and B). Next, we determined the effects of uga mutations on the expression of two other carbon fixation genes, TIM and FBA. We found that the kinetics of mRNA accumulation for these two genes during light induction were identical to those of GAPA and GAPB (M.-C. Shih, unpublished data). However, no significant difference in the transcription of these genes was observed between the uga mutants and AG-5G (Fig. 4).
Figure 4.
Effects of uga mutations on the expression of other light-regulated genes in mature plants. A, Northern-blot analyses of RNAs from 4-week-old light-grown (L) or dark-adapted (D) AG-5G, uga1-1, and uga1-2 were performed as described in Figure 3 with radiolabeled GAPB, TIM, FAB, LCHB, GAPC, CHS, and TUB probes. B, Each northern-blot analysis was repeated three times and quantified. The average densitometric value of each gene from light-grown AG5G is taken as 1. Error bars = sds.
Because combinatorial cis-acting elements are required to confer light responsiveness of light-regulated promoters in plants (Terzaghi and Cashmore, 1995; Puente et al., 1996; Chattopadhyay et al., 1998a), it is possible that uga mutations affect the expression of genes from different metabolic pathways. Therefore, we performed northern-blot analyses for three other genes, including GAPC, LHCB3, and CHS (Feinbaum and Ausubel, 1988; Yang et al., 1993; Li et al., 1995), which are known to be regulated by light. In addition to light, the transcription of GAPC, which encodes the C subunit of GAPDH, could also be regulated by Suc (Shih and Goodman, 1988; Yang et al., 1993). As shown in Figure 4, A and B, there were no observable differences in the mRNA levels of these genes in either light-grown or dark-adapted mature plants between the two uga mutants and the AG-5G line. These data suggested that the uga mutations specifically affect the expression of GAPA.
Biochemical Characterization of uga Mutants
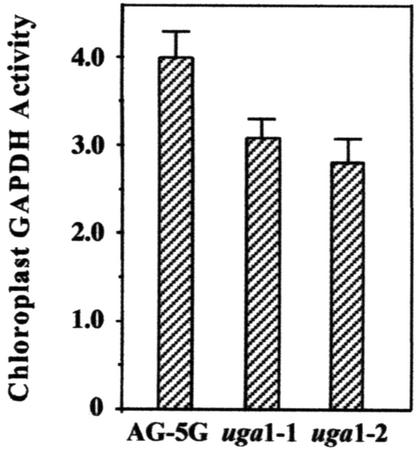
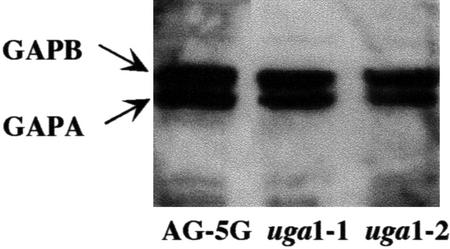
Because the GAPA mRNA levels decreased drastically in both uga1-1 and uga1-2 mutants, we decided to examine whether the chloroplast GAPDH activity in these mutants was similarly affected. As seen in Figure 5, the two mutants showed only slightly lower chloroplast GAPDH activities compared with AG-5G. This result suggested that posttranscriptional regulation of GAPA mRNA or posttranslational modification of the GAPDH enzyme could have occurred to compensate for the reduced GAPA mRNA level in the uga mutants. To distinguish between these possibilities, western-blot analysis was performed to quantify the protein levels of the A and B subunits (Fig. 6). The data showed that there were similar amounts of A and B polypeptides in leaf extracts from wild type, uga1-1, and uga1-2. These findings suggested that translational control of GAPA must have occurred in the uga mutants to compensate for their reduced levels of GAPA mRNA.
Figure 5.
Chloroplast GAPDH activity of mature plants. Chloroplast GAPDH activity of 5-week-old plants from AG-5G and uga mutants was assayed as described by Cerff (1982). Each reading was obtained from the pooling of the aerial portions of 10 individual plants per line. The data shown are the average of two independent measurements from plants grown at different times. Specific activity is calculated as the rate of decrease of A366 per milligram protein extract. Error bars = sds.
Figure 6.
Western analysis of GAPA and GAPB in AG-5G and uga mutants. Total cellular proteins were isolated from leaves of mature AG-5G, uga1-1, and uga1-2 plants. Ten-microgram proteins from each sample were subjected to western-blot analysis using rabbit antibody raised against the GAPDH A2B2 tetramer. The arrows indicate the positions of A and B subunits of the chloroplast GAPDH.
The uga1-1 Mutation Maps to the Bottom Arm of Chromosome V
We used the simple sequence length polymorphism (SSLP) mapping method (Bell and Ecker, 1994) to determine the chromosomal location of the uga1-1 mutation. The transgenic line AG-5G, the parental strain of uga mutants, is derived from Columbia ecotype (Col-O). We performed crosses between uga1-1 and Landsberg erecta (Ler) to generate F2 progeny as mapping populations. To score F2 progeny, we needed a suitable marker. Unfortunately, uga mutants lack any visible phenotype and the cross to Ler resulted in a loss of one or two copies of the transgene in some of the F2 progeny. As a result, GUS activity could not be used as a scoreable phenotype. However, knowing that the GAPA mRNA level differs by almost 20-fold between wild type and uga1-1 (Fig. 3), we used the GAPA mRNA levels to assess the genotype of the F2 progeny.
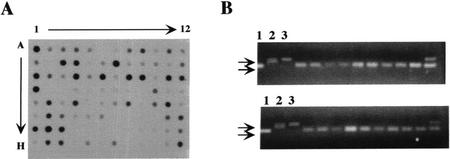
Dot-blot analyses were used to compare GAPA mRNA levels of a population of F2 progeny. We isolated total RNA from leaves of 94 F2 progeny of uga1-1 × Ler. Genomic DNA was isolated from each of these plants by the method of Edwards et al. (1991). RNA from F2 progeny was subjected to slot-blot analyses using a P32-labeled cDNA fragment of GAPA as the hybridizing probe. We found that 20 of 94 F2 progeny had GAPA mRNA levels comparable with those of wild type and that the remaining 74 samples had very low levels of GAPA mRNA (Fig. 7A). Because of the dominant nature of uga1-1 mutation, the 74 plants with low mRNA levels should be either UGA1+/uga1-1 or uga1-1/uga1-1 and the 20 plants with high GAPA mRNA should be homozygous UGA1+. As opposed to the skewed ratios obtained using GUS expression as the phenotypic marker, the RNA dot blot gave a 1:3.3 ratio, which was close to the expected 1:3 ratio. This confirmed that the uga1-1 mutation is a dominant, monogenic mutation.
Figure 7.
SSLP analyses of uga1-1 × Ler F2 progeny. A, Dot-blot analysis was performed as described in “Materials and Methods” to compare GAPA mRNA levels of 94 F2 plants. RNA from AG-5G line was loaded on two corners of the filter (A1 and H12) to be used as a quantification standard. B, Gel electrophoresis of PCR products for the ciw9 SSLP marker. Genomic DNA from UGA3+/UGA3+ F2 plants was used in PCR with ciw9 as the primer pair. PCR conditions were identical to those described by Bell and Ecker (1994). Lanes 1 through 3 were PCR products from reactions using genomic DNA from (1) Ler (2), Col-O, and (3) AG-5G, respectively, as templates.
Next, we performed PCR analysis of the 20 UGA1+/UGA1+ plants using 14 primer pairs corresponding to 14 SSLP markers that span the Arabidopsis genome over its five chromosomes, with at least one marker on each arm (see “Materials and Methods” for the list). Our results showed that the markers on chromosomes I through IV had no association with the Ler/Ler ecotype. However, in the case of the marker ciw9 that lies on the bottom arm of chromosome V, 18 of 20 F2 progeny had a Ler/Ler ecotype at this locus, indicating that the UGA1 gene was linked to this marker (Fig. 7B). Confirming the linkage of the UGA1 gene to this marker, it was found that the ciw10 marker, which is also located on the bottom arm of chromosome V, was also linked to the UGA1 gene, but not as tightly. Here, 11 of 20 samples showed the Ler/Ler ecotype (data not shown). The uga1-1 mutation, therefore, is mapped to the bottom arm of chromosome V in the Arabidopsis genome in the vicinity of the ciw9 marker (at 88 cM).
DISCUSSION
We have identified two allelic mutations that affect the expression of the GAPA gene in Arabidopsis. Our results showed that the mRNA levels of both GAPA::GUS and GAPA::ADH transgenes and the endogenous GAPA gene in light-grown uga1-1 and uga1-2 mutants are greatly reduced. One possible explanation for this observation is that the effect of uga mutations on light induction of the GAPA gene is mediated at the transcriptional level. However, there were examples that light affects mRNA stability and this effect often involved the 5′- or 3′-untranslated region (UTR) sequences (Dickey et al., 1998; Anderson et al., 1999). The fact that the GAPA::GUS and GAPA::ADH transgenes in the AG5G line contain all or part of the 5′-UTR of GAPA (see “Materials and Methods”) raised the possibility that the uga mutations might affect the mRNA stability of GAPA. We are in favor of the first interpretation, because results from our nuclear run-on experiments indicated that light effect on the steady-state GAPA mRNA level occurred mainly at the transcription level in both tobacco and Arabidopsis (Shih and Goodman, 1988; see also supplemental data). In addition, we have identified two cis-acting elements that are required for light induction of GAPA by deletional analyses of promoter constructs in transgenic plants (Conley et al., 1994; Park et al., 1996). A combination of these two elements could confer light responsiveness on a basal promoter that was not regulated by light (Park et al., 1996).
Results from our genetic analysis showed that both uga1-1 and uga1-2 mutations are dominant (Table I). With some exceptions, e.g. shy2 (Kim et al., 1996; 1998), most photomorphogenic mutants that have been isolated, such as phyA, phyB, red1, fhy1, fhy3, and cue1, are all recessive (Parks and Quail, 1993; Whitelam et al., 1993; Li et al., 1995; Wagner et al., 1997). There are a few ways to explain how this dominant phenotype could occur. First, the UGA1 gene in its normal wild-type state could function as a positively acting intermediate in the signaling pathway leading to the light-activated transcription of GAPA. This would mean that the uga mutant gene product must act in a dominant negative manner. One possibility is that the resulting functional UGA1 gene product is a multimeric protein comprising several subunits of the UGA1 gene product. The binding of a mutated subunit could cause the entire protein structure to lose its function and, therefore, fail to effect the light-activated transcription of GAPA. Alternatively, we could propose that the UGA1 gene product in its wild-type state normally represses GAPA transcription. Along with the action of other positively acting transcription factors, the UGA1 gene product would maintain an acceptable level of GAPA mRNA under a given set of environmental conditions. Repression could be achieved either by interaction with other light-signaling molecules in the pathway or by direct interaction with the GAPA promoter. The mutation could have resulted in a much tighter interaction and, therefore, a more dramatic repression effect.
It should be pointed out that, although the RNA dot blot of the F2 progeny generated from the mapping cross (uga1-1 × Ler) showed the expected 1:3 segregation ratio (Fig. 7A), the data obtained from F2 progeny of the backcross (uga1-1 × AG-5G) showed a 1:6 ratio as determined by the GUS assay (Table I). One possible explanation for this abnormal segregation ratio in the latter cross is that the AG-5G line contains the GAPA::ADH transgene. The regulation of endogenous ADH levels in a plant is tightly controlled in terms of tissue specificity and in its response to hypoxia and other environmental stresses (Dolferus et al., 1994; Chung and Ferl, 1999; Conley et al., 1999; Ellis et al., 1999). In AG-5G, however, where the ADH gene is driven by a GAPA promoter, the ADH activity is expressed about 70-fold higher than that in the Col wild-type plant (C.S. Chan and M.-C. Shih, unpublished data). Furthermore, its expression occurs throughout the entire plant instead of being tissue specific, which could have resulted in physiological abnormality in AG-5G. In fact, AG-5G plants were observed as slow-growing compared with the true wild type, Col. Assuming that the overexpression and misexpression of ADH in AG-5G had increased lethality, the introduction of uga mutations, which decrease ADH expression, might have actually increased the survival rate of AG-5G.
Specificity of the uga Mutations
Among a number of light-regulated genes examined here, uga1-1 and uga1-2 affect only the expression of GAPA. This was seen in the dramatic reduction in the steady-state GAPA mRNA level in mature plants (Fig. 3), whereas little or no effect was seen in the expression of GAPB and several other light-regulated genes, namely LHCB3, FBA1, and TIM (Fig. 4). Furthermore, we found that several photomorphogenetic phenotypes, including hypocotyl length, chlorophyll content, and chloroplast, appeared to be normal in the uga mutants (data not shown). In contrast, in most other light-signaling mutants, more than one gene or class of genes is affected. For example, the cue1 mutant (now known to be a mutation in the PPT gene), which was isolated based on defective LHCB3-promoter driven reporter activity, was not only defective in endogenous LHCB3 transcription but also in the transcription of RBCS and RBCL (Li et al., 1995). The psi2 mutant, which was isolated based on elevated LHCB2-LUC (luciferase) activity, was found to be hypersensitive in LHCB1, LHCB2, CHS, and RBCS expression when compared with the wild-type equivalent (Genoud et al., 1998). However, we cannot eliminate the possibility that other genes that have not been examined in this study are unaffected by the uga mutations. The implications of this specificity are severalfold. First, the UGA1 gene is likely to lie downstream in the light-signaling pathway leading to the transcriptional activation of GAPA. The second implication is that although GAPA and GAPB may be coordinately regulated at the transcriptional level (Dewdney et al., 1993), there probably exist distinct portions of their pathways that are independent of each other.
Importance of Translational Control of the GAPA Protein
Although the uga mutants showed drastic reduction in steady-state GAPA mRNA levels, the uga mutants appeared to survive very well, even though the A4 isozyme accounts for 80% of the total chloroplast GAPDH activity in the plant. The assay of chloroplast GAPDH enzyme activities in 5-week-old plants revealed that chloroplast GAPDH activity was only modestly reduced in the mutants compared with AG-5G (Fig. 5). Furthermore, western-blot analysis also revealed that the GAPA protein levels in the uga mutants were indistinguishable from that in AG-5G (Fig. 6). These results suggested that step(s) between the end of transcription and the completion of translation is the critical step in determining the final GAPDH levels in the uga mutants.
There are two ways in which such posttranscriptional regulation could have been achieved. First, one can envision a system whereby differential rate of transcription does not play any significant role in the regulation of the final GAPA protein levels. This could occur if the GAPA mRNA were always made in excess of what is required by the cell. Alternatively, because GAPA mRNA degradation is relatively fast (Dewdney et al., 1993; Conley and Shih, 1995), it is possible that the differences in transcription rates between AG-5G and the uga mutants may have little significance. Instead, translational control led to nearly equal levels of the GAPA protein between wild type and mutants. We could not argue in favor of either model based on our current results.
Many nuclear genes in plants are regulated at the posttranscriptional level (Gallie and Bailey-Serres, 1997). In Arabidopsis, ACS5, which encodes 1-aminocyclopropane-1-carboxylic acid synthase, is shown to be regulated posttranscriptionally (Woeste et al., 1999). The cytokinin-inducible soybean (Glycine max) CIM1 gene is regulated by the stability of the CIM1 mRNA rather than by the CIM1 transcriptional level per se (Downes and Crowell, 1998). In a study of the thylakoid peptide plastocyanin and the Rieske polypeptides, mRNA transcript levels may have increased 10-fold upon illumination, but association of transcripts with polysomes only increased 2- to 3-fold, suggesting that mRNA uptake into polysomes is an important step of posttranscriptional control (Palomares et al., 1993). In the case of the proton-ATPase gene, regulation by translation rate in response to developmental and environmental cues is signified by the presence of a long 5′-UTR that contained an upstream open reading frame (Michelet et al., 1994). There is a 47-bp UTR in the GAPA transcript, suggesting translation as a possible mechanism of control. As reviewed by Bailey-Serres (1999), translation of mRNA is emerging as an important mode of gene regulation where initiation is frequently the step at which regulation is achieved. Some features that influence translation rate include the interactions between the 5′ and 3′ ends of the message, and variation in the cap-binding protein of which there are three types in Arabidopsis, as triggered by developmental and other environmental cues (Gallie and Bailey-Serres, 1997).
MATERIALS AND METHODS
Generation of Transgenic Arabidopsis Plants
An Arabidopsis adh1-2 mutant in a Col background obtained from Dr. Dan Voytas (Department of Genetics, Iowa State University) was used as the starting strain. A binary construct carrying two consecutive reporters, ADH and GUS, each driven by the GAPA promoter, was constructed as follows. The GAPA::GUS/pBI101, which linked about a 1-kb promoter sequence and the complete 47-bp 5′-UTR of GAPA to the GUS coding sequence (Conley et al., 1994), was used as the starting plasmid. A DNA fragment that contains the −1,045 to +30 of GAPA was generated by PCR and linked to a DNA fragment containing the complete coding sequence of ADH. The GAPA::ADH DNA fragment was then cloned into the BamHI site of the GAPA::GUS/pBI101. The resulting pBI101 derivative was mobilized into Agrobacterium tumefaciens by triparental mating (Bevan, 1984) and then transformed into the adh1-2/adh1-2 starting strain using the floral dip method as described by Clough and Bent (1998). T1 progeny containing at least one copy of the transgene were selected by kanamycin resistance. Each transgenic line was carried on to the T2 generation, where a transgenic line with a single transgene insertion was selected based on a 3:1 segregation of the kanamycin resistance phenotype at the T2 generation. Within the transgenic T2 population, a homozygous line, designated as AG-5G, was selected based on a 4:0 segregation pattern at the F3 and then again at the F4 generation. The bulked seeds of this line constitute the parental strain, which has an adh1-2/adh1-2/Col background and carries two reporters, ADH and GUS, each driven by the GAPA promoter.
Mutagenesis and Generation of M2 Progeny
Twenty thousand seeds of the AG-5G line were subjected to ethane methane sulfonate mutagenesis according to the method described by Somerville and Ogren (1982), with a few modifications. In brief, seeds were soaked in 0.1 mm ethane methane sulfonate for 16 h with rocking at room temperature, washed with 100 mm sodium thiosulphate, and rinsed thoroughly with water. Seeds were then treated at 4°C for 3 d before being sown onto soil at a density of 0.5 cm−2 and maintained in a growth chamber at 22°C. M1 plants were carried on to the next generation by selfing, after which M2 seeds were harvested into four separate pools.
Allyl Alcohol Selection
M2 seeds were imbibed on Whatman No. 1 filter paper (Whatman, Clifton, NJ) soaked in 3.5 mL of Murashige and Skoog liquid medium containing 2% (w/v) Suc in glass petri dishes at a density of about 500 seeds per plate, carefully spread out using a sterile plastic pipette tip. Control plates containing AG-5G seeds as well as seeds of the adh1-2 mutant were similarly prepared to be later used for comparison of lethality in allyl alcohol. Seeds were cold incubated at 4°C in the dark for 3 d and then transferred to a dark growth chamber at 22°C for 5 d. The etiolated seedlings were then subjected to 24 h of white light treatment as described in the following section. We found that the expression of the GAPA gene reached a maximal level after 24 h of white light treatment (Dewdney et al., 1993; Conley and Shih, 1995). The filters were then transferred onto medium containing 7.5 mm allyl alcohol in Murashige and Skoog + 2% (w/v) Suc. This concentration was the minimum concentration of allyl alcohol that would cause >99% lethality to AG-5G seedlings. After 2 h, the filters were transferred onto fresh agar Murashige and Skoog medium containing 2% (w/v) Suc. Allyl alcohol resistant mutants were isolated on d 4 to 5. These allyl alcohol resistant plants were then subjected to GUS histochemical staining over a 24-h staining period.
Light and Growth Conditions
Plants on soil were kept at 22°C under 16-/8-h light/dark cycle. White light was provided by three cool-white 35-W fluorescent lamps at 50 μmol m−2s−1. Blue light was used at 5.5 μmol m−2s−1 supplied by four fluorescent lamps with a blue plexiglas 3-mm filter (Rohm-Haas no. 2423, Ditric Optics, Hudson, MA) as previously described (Conley and Shih, 1995). In the case of etiolated seedlings, about 200 (for enzyme assays) or 400 (for RNA extraction) seeds were imbibed on 3.5 mL of Murashige and Skoog + 2% (w/v) Suc liquid medium soaked on two pieces of Whatman No. 1 filter paper in a glass petri dish, vernalized for 3 d, and then transferred to a light-proof incubator at 22°C for etiolation.
RNA Isolation and Northern-Blot Analyses
Total RNA was isolated by the Triazol LS method (Life Technologies/Gibco-BRL, Cleveland) using about 50 to 200 mg of plant tissue for each extraction. Northern analysis was performed as previously described (Conley and Shih, 1995). Five micrograms of RNA was loaded per lane, unless otherwise stated. Gels were blotted overnight onto nylon Hybond N+ membranes (Amersham Pharmacia Biotech, Piscataway, NJ) with 10× SSC as the transfer buffer. The GAPA, GAPB, GAPC, RBCS, LCHB, and TUB cDNA probes were as described by Conley and Shih (1995). The TIM fragment was excised by SalI and NotI resulting in a 1.4-kb fragment; FBA1 was 1.1 kb in length after excision by HindIII. Normalization for loading was accomplished by stripping the original probe off the filter by dipping in deionized water at 80°C and checking for counts using a Geiger counter. The filter was then reprobed with TUB, the transcription of which is unaffected by light (Conley and Shih, 1995). The bands on the autoradiograph of each northern were quantified using Scion Image version 1.62 software (National Institutes of Health, Bethesda, MD). Relative mRNA levels were then determined by taking the ratio of the band intensity specific for the gene probe of interest minus the background intensity to that for TUB.
Enzymatic Assays
GUS enzyme assay and histochemical staining were performed as described by Jefferson et al. (1987). ADH enzyme assays were performed as described by Xie and Wu (1989). This assay uses ethanol as the substrate and measures the production of NADH. Measurement of NADH formation was performed in a DU 64 spectrophotometer (Beckman Instruments, Fullerton, CA). A unit of ADH is defined as the production of 1 nmol of NADH min−1 mg−1 protein. Chloroplast GAPDH assays were performed as described by Cerff (1982). To determine chloroplast-specific GAPDH activity, NADPH was used as the starting cofactor instead of NADH. Specific activity is calculated as the rate of decrease of A366 per milligram protein extract.
Western-Blot Analysis
The aerial portions of 10 to 15 plants from each mutant line were harvested and homogenized in liquid nitrogen using a mortar and pestle after which cold homogenization buffer (15 mm HEPES [pH 7.6], 40 mm KCl, 5 mm MgCl2, 1 mm dithiothreitol, and 0.1 mm phenylmethylsulfonyl fluoride) was added at 10 mL g−1 of fresh tissue. A 10-fold volume of 4 m ammonium sulfate was added drop-wise with stirring. The mixture was then centrifuged at 20,000 rpm in a SW41 swing bucket rotor (30,000g) at 4°C for 30 min. The supernatant was filtered through a Miracloth (Calbiochem, La Jolla, CA) after which freshly ground ammonium sulfate was added slowly at 0.33 g mL−1 to precipitate proteins. Proteins were then spun down at 19,000 rpm in an SW41 at 4°C for 30 min and resuspended in 1 mL of buffer (20 mm HEPES [pH 7.6], 40 mm KCl, 1 mm dithiothreitol, 0.1 mm EDTA, 0.5 mm phenylmethylsulfonyl fluoride, and 10% [w/v] glycerol). Aliquot (400 μL) of the extract was desalted using ultra-free low-bind (10-kD cutoff) filter apparatus (no. UFC3LGC00, Millipore, Bedford, MA). Ten micrograms of protein per sample was used for western analysis as previously described (Wang et al., 1997a) using the semiwet transfer system. The membrane was incubated with a 1:3,000 (w/v) dilution of the rabbit antibody generated against the GAPDH A2B2 isozyme in blocking buffer at room temperature with swirling for 1 h. Under these conditions, this antibody reacts specifically with A and B subunits (Wang et al., 1997a). The bands were visualized with ECL western-blotting detection solution (Amersham-Pharmacia Biotech) and quantified with National Institutes of Health Scion Image software version 1.62.
Mapping Cross and F2 Progeny
The uga1-1 mutant was crossed to the Ler wild type. The F1 seeds resulting from this cross were grown and selfed to produce F2 seeds. The homozygous recessive F2 progeny resulting from the mapping cross were selected based on the results from the RNA dot-blot analysis.
For dot blot analysis, a 9- by 12-cm nylon Hybond N+ membrane (Amersham) was prewet in 10× SSC, blotted dry on Whatman No. 1 filter paper, and assembled on the dot blot apparatus (Bio-Rad Laboratories, Hercules, CA) according to the manufacturer's manual. The 96 wells were then rehydrated by the addition of 500 μL of 10× SSC into each well and applying vacuum until dry. Three micrograms of RNA per sample derived from F2 progeny of the mapping cross was subjected to alkaline denaturation by the addition of 500 μL of ice-cold 10 mm NaOH and 1 mm EDTA and kept on ice. A total of 94 samples of the F2 progeny were dot blotted onto the nylon membrane together with the AG-5G RNA sample dotted at the top left and bottom right corners as positive controls. Hybridization with the GAPA probe was as described above for northern analyses. The F2 individuals that were homozygous recessive for the uga phenotype (wild type for GAPA mRNA expression) were then matched to the corresponding numbered plant material reserved for DNA isolation. DNA was isolated using the method of Edwards et al. (1991).
PCR of SSLP Markers
About 1 to 10 ng of template DNA was used for PCR using standard reaction conditions provided by Promega (Madison, WI) at 2.5 mm of MgCl2. Three control tubes using DNA isolated from wild-type Ler, wild-type Col, and the AG-5G line were set up and run concurrently with the F2 samples for band size comparison. The primers used for amplifying SSLP markers are as follows: chromosome I, ciw12 and nga111; chromosome II, ciw2, ciw3, and nag168; chromosome III, nag162 and nga6; chromosome IV, ciw5, ciw7, and nga1107; and chromosome V, CTR1, ciw8, ciw9, and ciw10 (Lukowitz et al., 2000). Typically, the annealing temperature was set at 2°C above the higher melting temperature (Tm) of the two primers if they were less than 2°C apart from each other. If the Tms of the two primers were more than 2°C apart, the average between the two Tms was used as the annealing temperature. The PCR cycles are 30 cycles of 95°C, 1 min; 55°C (or other annealing temperature), and 1 min; 72°C, 1 min; followed by 5-min extension at 72°C. About 10 μL of the PCR reaction was resolved on a NuSieve GTG 4% (w/v) agarose gel in 1× Tris-acetate EDTA buffer. Each tier on the DNA agarose gel was run with three control lanes, which carried the respective PCR products of Ler, Col, and AG-5G for band size comparison. F2 wild-type recessive progeny from the mapping cross could then be scored as Ler, heterozygous, or Col ecotypes on the agarose gels for each SSLP marker.
Statistical Analysis
All comparisons between data of mutants versus AG-5G were done by ANOVA one-way analysis with Bonferroni's method, whereas phenotypic ratios of genetic crosses were tested by Chi square analysis. The SigmaStat software (SPSS Sciences, Chicago) was used in each case.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes.
ACKNOWLEDGMENTS
We thank Drs. Erin Irish and Jonathan Poulton for comments on the manuscript. We thank the Arabidopsis Biological Resources Center (Ohio State University, Columbus) for providing cDNA clones of FBA and TIM.
Footnotes
This work was supported by the U.S. Department of Agriculture National Research Initiative Competitive Grants Program (grant no. 9600717 to M.-C.S.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.007849.
LITERATURE CITED
- Ahmad M, Cashmore AR. HY4 gene of Arabidopsis thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature. 1993;366:162–166. doi: 10.1038/366162a0. [DOI] [PubMed] [Google Scholar]
- Ahmad M, Lin C, Cashmore AR. Mutations throughout an Arabidopsis blue-light photoreceptor impair blue-light responsive anthocyanin accumulation and inhibition of hypocotyl elongation. Plant J. 1995;8:653–658. doi: 10.1046/j.1365-313x.1995.08050653.x. [DOI] [PubMed] [Google Scholar]
- Anderson MB, Folta K, Warpeha KM, Gibbons J, Gao, Kaufman LS. Blue light-directed destabilization of the pea Lhcb1*4 transcript depends on sequences within the 5′ untranslated region. Plant Cell. 1999;11:1579–1590. doi: 10.1105/tpc.11.8.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SL, Teakle GR, Martino-Catt SJ, Kay SA. Circadian clock and phytochrome-regulated transcription is conferred by a 78 bp cis-acting domain of the Arabidopsis CAB2 promoter. Plant J. 1994;6:457–470. doi: 10.1046/j.1365-313x.1994.6040457.x. [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J. Selective translation of cytoplasmic mRNAs in plants. Trends Plant Sci. 1999;4:142–148. doi: 10.1016/s1360-1385(99)01386-2. [DOI] [PubMed] [Google Scholar]
- Batschauer A. Photoreceptors of higher plants. Planta. 1998;206:479–492. doi: 10.1007/s004250050425. [DOI] [PubMed] [Google Scholar]
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- Bevan MW. Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 1984;12:8711–8721. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs WR, Beck CF, Cashmore AR, Christie JM, Hughes J, Jarillo JA, Kagawa T, Kanegae H, Liscum E, Nagatani A et al. The phototropin family of photoreceptors. Plant Cell. 2001;13:993–997. doi: 10.1105/tpc.13.5.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs WR, Huala E. Blue-light photoreceptors in higher plants. Annu Rev Dev Biol. 1999;15:33–62. doi: 10.1146/annurev.cellbio.15.1.33. [DOI] [PubMed] [Google Scholar]
- Cerff R. Separation and purification of NAD- and NADP-linked GAPDH from higher plants. In: Edelman ME, Hallick RB, Chua NH, editors. Methods in Chloroplast Molecular Biology. Amsterdam: Elsevier/North Holland; 1982. pp. 683–694. [Google Scholar]
- Chan CS, Guo L, Shih M-C. Promoter analysis of the nuclear gene encoding the chloroplast glyceraldehyde-3-phosphate dehydrogenase B subunit of Arabidopsis thaliana. Plant Mol Biol. 2001;46:131–141. doi: 10.1023/a:1010602031070. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S, Ang LH, Puente P, Deng XW, Wei N. Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell. 1998b;10:673–683. doi: 10.1105/tpc.10.5.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Puente P, Deng XW, Wei N. Combinatorial interaction of light-responsive elements plays a critical role in determining the response characteristics of light-regulated promoters in Arabidopsis. Plant J. 1998a;15:69–77. doi: 10.1046/j.1365-313x.1998.00180.x. [DOI] [PubMed] [Google Scholar]
- Chung HJ, Ferl RJ. Arabidopsis alcohol dehydrogenase expression in both shoots and root is conditioned by root growth environment. Plant Physiol. 1999;121:429–436. doi: 10.1104/pp.121.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clack T, Mathews S, Sharrock RA. The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequences and expression of phyD and phyE. Plant Mol Biol. 1994;25:413–427. doi: 10.1007/BF00043870. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simple method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Conley TR, Park SC, Kwon HB, Peng H-P, Shih M-C. Characterization of cis-acting elements in light regulation of the nuclear gene encoding the A subunit of chloroplast isozymes of glyceraldehyde-3-phosphate dehydrogenase from Arabidopsis thaliana. Mol Cell Biol. 1994;14:2525–2533. doi: 10.1128/mcb.14.4.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley TR, Peng H-P, Shih M-C. Mutations affecting induction of glycolytic and fermentative genes during germination and environmental stresses in Arabidopsis. Plant Physiol. 1999;119:599–607. doi: 10.1104/pp.119.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley TR, Shih M-C. Effects of light and chloroplast functional state on expression of nuclear genes encoding chloroplast glyceraldehyde-3-phosphate dehydrogenase in long hypocotyl (hy) mutants and wild-type Arabidopsis thaliana. Plant Physiol. 1995;108:1013–1022. doi: 10.1104/pp.108.3.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng XW, Quail PH. Signaling in light-controlled development. Semin Cell Dev Biol. 1999;10:121–129. doi: 10.1006/scdb.1999.0287. [DOI] [PubMed] [Google Scholar]
- Dewdney J, Conley TR, Shih M-C, Goodman HM. Effects of blue and red light on expression of nuclear genes encoding chloroplast glyceraldehyde-3-phosphate dehydrogenase of Arabidopsis thaliana. Plant Physiol. 1993;103:1115–1121. doi: 10.1104/pp.103.4.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickey LF, Petracek ME, Ngyuen T-T, Hansen ER, Thompson WF. Light regulation of Fed-1 mRNA requires an element in the 5′ untranslated region and correlates with differential polyribosome association. Plant Cell. 1998;10:475–484. doi: 10.1105/tpc.10.3.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolferus R, Jacobs M, Peacock W, Dennis E. Differential interactions of promoter elements in stress responses of the Arabidopsis Adh gene. Plant Physiol. 1994;105:1075–1087. doi: 10.1104/pp.105.4.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald RGK, Cashmore AR. Mutation of either G box or I box sequences profoundly affects expression from the Arabidopsis rbcS-1A promoter. EMBO J. 1990;9:1717–1726. doi: 10.1002/j.1460-2075.1990.tb08295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes BP, Crowell DN. Cytokinin regulates the expression of a soybean beta-expansin gene by a post-transcriptional mechanism. Plant Mol Biol. 1998;37:437–444. doi: 10.1023/a:1005920732211. [DOI] [PubMed] [Google Scholar]
- Edwards K, Hohnstone C, Thompson C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 1991;19:1349. doi: 10.1093/nar/19.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis MH, Dennis ES, Peacock WJ. Arabidopsis roots and shoots have different mechanisms for hypoxic stress tolerance. Plant Physiol. 1999;119:57–64. doi: 10.1104/pp.119.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairchild CD, Schumaker MA, Quail PH. HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev. 2000;14:2377–2391. [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Chory J. Light receptor kinases in plants! Curr Biol. 1999;9:R123–126. doi: 10.1016/s0960-9822(99)80078-5. [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Chory J. RSF1, an Arabidopsis locus implicated in phytochrome A signaling. Plant Physiol. 2000;124:39–45. doi: 10.1104/pp.124.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Yeh K-C, Lagarias JC, Zhang H, Elich TD, Chory J. PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science. 1999;284:1539–1541. doi: 10.1126/science.284.5419.1539. [DOI] [PubMed] [Google Scholar]
- Feinbaum RL, Ausubel FM. Transcriptional regulation of the Arabidopsis thaliana chalcone synthase gene. Mol Cell Biol. 1988;8:1985–1992. doi: 10.1128/mcb.8.5.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR, Bailey-Serres J. Eyes off transcription! The wonderful world of post-transcriptional regulation. Plant Cell. 1997;9:667–673. doi: 10.1105/tpc.9.5.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genoud T, Millar AJ, Nishizawa N, Kay SA, Schafer E, Nagatani A, Chua N-H. An Arabidopsis mutant hypersensitive to red and far-red light signals. Plant Cell. 1998;10:889–904. doi: 10.1105/tpc.10.6.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmartin PM, Sarokin L, Memelink J, Chua N-H. Molecular light switches for plant genes. Plant Cell. 1990;2:369–378. doi: 10.1105/tpc.2.5.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin-Neumann G, Sun L, Tobin EM. Expression of light-harvesting chlorophyll a/b protein genes is phytochrome-regulated in etiolated Arabidopsis thaliana seedlings. Plant Physiol. 1988;88:1323–1331. doi: 10.1104/pp.88.4.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman LS, Thompson WF, Briggs WR. Different red light requirements for phytochrome-induced accumulation of cab and rbcS RNA. Science. 1984;226:1447–1449. doi: 10.1126/science.226.4681.1447. [DOI] [PubMed] [Google Scholar]
- Kenigsbuch D, Tobin EM. A region of the Arabidopsis Lhcb1*3 promoter that binds to CA-1 activity is essential for high expression and phytochrome regulation. Plant Physiol. 1995;108:1023–1027. doi: 10.1104/pp.108.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BC, Soh MS, Hong SH, Furuya M, Nam HG. Two dominant photomorphogenic mutations of Arabidopsis thaliana identified as suppressor mutations of hy2. Plant J. 1996;9:441–456. doi: 10.1046/j.1365-313x.1996.09040441.x. [DOI] [PubMed] [Google Scholar]
- Kim BC, Soh MS, Hong SH, Furuya M, Nam HG. Photomorphogenic development of the Arabidopsis shy2-1D mutation and its interaction with phytochromes in darkness. Plant J. 1998;15:61–68. doi: 10.1046/j.1365-313x.1998.00179.x. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Doi M, Suetsugu N, Kagawa T, Wada M, Shimazaki K. Phot1 and phot2 mediate blue light regulation of stomatal opening. Nature. 2001;414:656–660. doi: 10.1038/414656a. [DOI] [PubMed] [Google Scholar]
- Kwon HB, Park SC, Peng HP, Goodman HM, Dewdney J, Shih M-C. Identification of a light-responsive region of the nuclear gene encoding the B subunit of chloroplast glyceraldehyde-3-phosphate dehydrogenase from Arabidopsis thaliana. Plant Physiol. 1994;105:357–367. doi: 10.1104/pp.105.1.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Culligan K, Dixon RA, Chory J. CUE1: a mesophyll cell-specific positive regulator of light-controlled gene expression in Arabidopsis. Plant Cell. 1995;7:1599–1610. doi: 10.1105/tpc.7.10.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. Plant blue-light receptors. Trends Plant Sci. 2000;5:337–342. doi: 10.1016/s1360-1385(00)01687-3. [DOI] [PubMed] [Google Scholar]
- Liscum E, Briggs WR. Mutations in the nph1 locus of Arabidopsis disrupt the perception of phototropic stimuli. Plant Cell. 1995;7:473–485. doi: 10.1105/tpc.7.4.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Juez E, Jarvis RP, Takeuchi A, Page AM, Chory J. New Arabidopsis cue mutants suggest a close connection between plastid- and phytochrome regulation of nuclear gene expression. Plant Physiol. 1998;118:803–815. doi: 10.1104/pp.118.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W, Gilmor CS, Scheible W-R. Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol. 2000;123:795–805. doi: 10.1104/pp.123.3.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia JF, Huq E, Quail PH. Direct targeting of light signals to a promoter element-bound transcription factor. Science. 2000;288:859–863. doi: 10.1126/science.288.5467.859. [DOI] [PubMed] [Google Scholar]
- Mazzella MA, Cerdán PD, Staneloni RJ, Casal JJ. Hierarchical coupling of phytochromes and cryptochromes reconciles stability and light modulation of Arabidopsis development. Development. 2001;128:2291–2299. doi: 10.1242/dev.128.12.2291. [DOI] [PubMed] [Google Scholar]
- Michelet L, Lukaszewicz M, Dupriez V, Boutry M. A plant plasma membrane proton-ATPase gene is regulated by development and environment and shows signs of a translational regulation. Plant Cell. 1994;6:1375–1389. doi: 10.1105/tpc.6.10.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH. Binding of phytochrome B to its nuclear signaling partner PIF3 is reversibly induced by light. Nature. 1999;400:781–784. doi: 10.1038/23500. [DOI] [PubMed] [Google Scholar]
- Palomares R, Herrmann RG, Oelmuller R. Post-transcriptional and post-translational regulatory steps are crucial in controlling the appearance and stability of thylakoid polypeptides during the transition of etiolated tobacco seedlings to white light. Eur J Biochem. 1993;217:345–352. doi: 10.1111/j.1432-1033.1993.tb18252.x. [DOI] [PubMed] [Google Scholar]
- Park SC, Kwon HB, Shih MC. Cis-acting elements essential for light regulation of the nuclear gene encoding the A subunit of chloroplast glyceraldehyde-3-phosphate dehydrogenase in Arabidopsis thaliana. Plant Physiol. 1996;112:1563–1571. doi: 10.1104/pp.112.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks BM, Quail PH. hy8, a new class of Arabidopsis long hypocotyl mutants deficient in functional phytochrome A. Plant Cell. 1993;5:39–48. doi: 10.1105/tpc.5.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente P, Wei N, Deng XW. Combinatorial interplay of promoter elements constitutes the minimal determinants for light and developmental control of gene expression in Arabidopsis. EMBO J. 1996;15:3732–3743. [PMC free article] [PubMed] [Google Scholar]
- Quail P. The phytochromes: a biochemical mechanism of signaling in sight? BioEssays. 1997;19:571–579. doi: 10.1002/bies.950190708. [DOI] [PubMed] [Google Scholar]
- Quail PH, Boylan MT, Parks BM, Short TW, Xu Y, Wagner D. Phytochromes: photosensory perception and signal transduction. Science. 1995;268:675–680. doi: 10.1126/science.7732376. [DOI] [PubMed] [Google Scholar]
- Sakai T, Kagawa T, Kasahara M, Swartz TE, Christie JM, Briggs WR, Wada M, Okada K. Arabidopsis nph1 and npl1: blue light receptors that mediate both phototropism and chloroplast relocation. Proc Natl Acad Sci USA. 2001;98:6969–6974. doi: 10.1073/pnas.101137598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih M-C, Goodman HM. Differential light regulated expression of nuclear genes encoding chloroplast and cytosolic glyceraldehyde-3-phosphate dehydrogenase in Nicotiana tabacum. EMBO J. 1988;7:893–898. doi: 10.1002/j.1460-2075.1988.tb02893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh MS, Kim YM, Han SJ, Song PS. REP1, a basic helix-loop-helix protein, is required for a branch pathway of phytochrome A signaling in Arabidopsis. Plant Cell. 2000;12:2061–2074. doi: 10.1105/tpc.12.11.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville CR, Ogren WL. Isolation of photorespiration mutants in Arabidopsis thaliana. In: Edelman M, Hallick RB, Chua N-H, editors. Methods in Chloroplast Molecular Biology. Amsterdam: Elsevier Biomedical Press; 1982. pp. 129–138. [Google Scholar]
- Terzaghi WB, Cashmore AR. Light-regulated transcription. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:445–474. [Google Scholar]
- Wagner D, Hoecker U, Quail PH. RED1 is necessary for phytochrome B-mediated red light-specific signal transduction in Arabidopsis. Plant Cell. 1997;9:731–743. doi: 10.1105/tpc.9.5.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Guo L, Sjolund R, Shih MC. Immunolocalization of GAPDH in Arabidopsis thaliana. Protoplasma. 1997a;198:155–162. [Google Scholar]
- Wang ZY, Kenigsbuch D, Sun L, Harel E, Ong MS, Tobin EM. A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell. 1997b;9:491–507. doi: 10.1105/tpc.9.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam GC, Johnson E, Peng J, Carol P, Anderson ML, Cowl JS, Harbed NP. Phytochrome A null mutants display a wild-type phenotype in white light. Plant Cell. 1993;7:2013–2022. doi: 10.1105/tpc.5.7.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woeste KE, Ye C, Kieber JJ. Two Arabidopsis mutants that overproduce ethylene are affected in the posttranscriptional regulation of 1-aminocyclopropane-1-carboxylic acid synthase. Plant Physiol. 1999;119:521–529. doi: 10.1104/pp.119.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Wu R. Rice alcohol dehydrogenase genes: anaerobic induction, organ specific expression and characterization of cDNA clones. Plant Mol Biol. 1989;13:53–68. doi: 10.1007/BF00027335. [DOI] [PubMed] [Google Scholar]
- Yang Y, Kwon HB, Peng H-P, Shih M-C. Stress responses and metabolic regulation of GAPDH genes in Arabidopsis. Plant Physiol. 1993;101:209–216. doi: 10.1104/pp.101.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh K-C, Lagarias JC. Eukaryotic phytochromes: light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc Natl Acad Sci USA. 1998;95:13976–13981. doi: 10.1073/pnas.95.23.13976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh K-C, Wu SH, Murphy JT, Lagarias JC. A cyanobacterial phytochrome two-component light sensory system. Science. 1997;277:1505–1508. doi: 10.1126/science.277.5331.1505. [DOI] [PubMed] [Google Scholar]