Abstract
Herbicide safeners increase herbicide tolerance in cereals but not in dicotyledenous crops. The reason(s) for this difference in safening is unknown. However, safener-induced protection in cereals is associated with increased expression of herbicide detoxifying enzymes, including glutathione S-transferases (GSTs). Treatment of Arabidopsis seedlings growing in liquid medium with various safeners similarly resulted in enhanced GST activities toward a range of xenobiotics with benoxacor, fenclorim, and fluxofenim being the most effective. Safeners also increased the tripeptide glutathione content of Arabidopsis seedlings. However, treatment of Arabidopsis plants with safeners had no effect on the tolerance of seedlings to chloroacetanilide herbicides. Each safener produced a distinct profile of enhanced GST activity toward different substrates suggesting a differential induction of distinct isoenzymes. This was confirmed by analysis of affinity-purified GST subunits by two-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis. AtGSTU19, a tau class GST, was identified as a dominant polypeptide in all samples. When AtGSTU19 was expressed in Escherichia coli, the recombinant enzyme was highly active toward 1-chloro-2,4-dinitrobenzene, as well as chloroacetanilide herbicides. Immunoblot analysis confirmed that AtGSTU19 was induced in response to several safeners. Differential induction of tau GSTs, as well as members of the phi and theta classes by safeners, was demonstrated by RNA-blot analysis. These results indicate that, although Arabidopsis may not be protected from herbicide injury by safeners, at least one component of their detoxification systems is responsive to these compounds.
Plants actively detoxify both endogenous toxins, such as secondary metabolites and degradation products arising from oxidative stress, and exogenous man-made chemicals, such as herbicides, using a three-phase detoxification system (Neuefeind et al., 1997). In the first phase, oxidation, reduction, or hydrolysis reactions catalyzed by enzymes such as cytochrome P450 monooxygenases result in the exposure, or introduction, of a functional group. Phase two enzymes then catalyze the conjugation of these metabolites with sugars or the tripeptide glutathione (GSH). In the case of GSH, glutathione S-transferases (GSTs) catalyze this conjugation reaction. In the third phase of metabolism, molecules “tagged” with GSH are recognized by ATP-binding cassette transporters in the tonoplast or plasma membrane, which then transfer these conjugates into the vacuole or apoplast (Rea, 1999).
GSTs constitute a family of multifunctional enzymes present in both plants and animals. These dimeric enzymes catalyze the conjugation of GSH to a variety of electrophilic, hydrophobic, and often toxic substrates, thereby reducing their toxicity (Marrs, 1996; Dixon et al., 1998). In addition to GSH conjugation, GSTs may also exhibit glutathione peroxidase (GPOX) or isomerase activities, or function as binding proteins known as ligandins (Edwards et al., 2000). As GPOXs, GSTs have been shown to have major roles in oxidative stress tolerance, reducing organic hydroperoxides to their monohydroxy derivatives with the simultaneous production of oxidized glutathione (Bartling et al., 1993; Roxas et al., 2000).
GSTs were initially studied in plants because of their role in herbicide metabolism and selectivity. Several herbicides are rapidly metabolized via GSH conjugation in crops (Gronwald and Plaisance, 1998). In contrast, many weeds contain lower activities of detoxifying GSTs and are susceptible to herbicides. A notable exception is a biotype of the dicot weed Abutilon theophrasti, which developed resistance to the herbicide atrazine because of increased activity of a specific GST isoenzyme. However, the elevated activity was because of a mutation that increased the catalytic constant of the enzyme toward atrazine, rather than an increase in the abundance of this protein (Plaisance and Gronwald, 1999). The importance of GSTs in herbicide tolerance has been clearly demonstrated in transgenic tobacco (Nicotiana tabacum) plants expressing a maize (Zea mays) GST active in conjugating chloroacetanilide herbicides, such as metolachlor (Jepson et al., 1997). Wild-type tobacco plants were sensitive to metolachlor, whereas plants expressing the maize GST exhibited a high degree of tolerance.
In cereals, but apparently not in dicotyledenous crops, herbicide tolerance can be enhanced using herbicide safeners (Davies and Caseley, 1999). In maize, wheat (Triticum aestivum), rice (Oryza sativa), and sorghum (Sorghum bicolor), much of the protective effect of safeners has been attributed to increases in the detoxification capacity of the respective crops (Davies and Caseley, 1999). For example, in all these cereal crops, safeners enhance the expression of GSTs active in herbicide metabolism. This enhancement is selective, with the specific GST isoenzymes induced depending on the safener used. For example, treatment of wheat with naphthalic anhydride resulted in the induction of the phi class TaGST2-3, which detoxifies the herbicide fluorodifen (Pascal and Scalla, 1999). In contrast, the tau class isoenzymes TaGST1-2, TaGST1-3, and TaGST1-4, which are active in detoxifying fenoxaprop ethyl, were enhanced in response to the safener fenchlorazole ethyl (Cummins et al., 1997). Similarly, GSTs active in detoxifying chloroacetanilide herbicides are selectively induced by fenclorim in rice (Wu et al., 1999; Deng and Hatzios, 2002), by fluxofenim in sorghum (Gronwald and Plaisance, 1998), and by dichlormid in maize (Dixon et al., 1997).
In contrast to cereals, little is known about the effect of safeners on dicotyledenous plants. Many studies have demonstrated that the GSTs of dicots are induced in response to diverse stimuli including infection, exposure to plant hormones, metal ions, and xenobiotics (Edwards et al., 2000), but comprehensive studies on induction of these enzymes by safeners have not been reported. If safener-signaling pathways could be shown to operate in dicots, such as the model plant system Arabidopsis, in a comparable manner to that seen in cereals, this would provide another approach to studying the regulation of safening. A survey of the Arabidopsis genome indicates the presence of approximately 50 candidate GST genes, based on amino acid sequence similarity to known plant and animal GSTs (Wagner et al., 2002; B.P. DeRidder and P.B. Goldsbrough, unpublished data). Many of these Arabidopsis GSTs have amino acid sequence similarity to safener-inducible, herbicide-detoxifying phi (type I) and tau (type III) class GSTs in cereal crops. Therefore, it seems timely to apply the developing tools of genomics and proteomics to study the expression and function of GSTs in Arabidopsis, and to determine if these detoxifying enzymes respond to safeners in a similar way to GSTs in cereal crops.
RESULTS
Effect of Safeners on GST Activity and Herbicide Tolerance in Arabidopsis Seedlings
To examine the effect of safeners on GST activity in Arabidopsis, seedlings growing in liquid medium were exposed to a number of these compounds. The safeners tested are commonly used to increase herbicide tolerance in different cereal crops. The morpholine safener benoxacor and the dichloroacetamide safeners, R-29148 and dichlormid, protect maize from chloroacetanilide injury (Davies and Caseley, 1999). Similarly, the oxime ether safeners (oxabetrinil and fluxofenim) and the safener flurazole are used to enhance tolerance of sorghum to chloroacetanilide herbicides (Gronwald and Plaisance, 1998). Fenclorim, a pyrimidine, enhances chloroacetanilide detoxification in rice (Wu et al., 1999; Deng and Hatzios, 2002), and the safener naphthalic anhydride has been used to increase thiocarbamate herbicide tolerance in maize (Davies and Caseley, 1999).
Protein extracts from safener-treated seedlings were assayed for GST activity with a number of substrates (Table I). GST activity was increased by all safener treatments tested, but the response was dependent on the safener to which the seedlings were exposed and the substrate used for GST activity assays. For example, benoxacor, fenclorim, flurazole, and fluxofenim enhanced GST activity with the model substrate CDNB from 3- to 5-fold, whereas other safeners increased this activity less than 2-fold. However, GST activity with NBC, another model substrate, increased from 8- to 13-fold in response to treatment with benoxacor, fenclorim, and flurazole, but was less affected by oxabetrinil and fluxofenim. BITC was tested as a substrate because isothiocyanates are produced by wounding in plants via the hydrolysis of glucosinolates and may represent natural substrates of GSTs. GST activity against this substrate almost doubled in response to most safeners, with the exception of flurazole. EA, a phenylacetic acid derivative used as a diuretic in mammals, contains an electrophilic group similar to α-alkenals produced in mammals as the result of oxidative stress, and therefore may represent natural GST substrates (Danielson et al., 1987; Berhane et al., 1994). GST activity against EA could not be detected in control tissues but was induced by several safeners, with fluxofenim having the greatest effect. In contrast, GPOX activity with cumene hydroperoxide as substrate was not significantly enhanced above the level seen in controls (0.34 pkat mg−1 protein) by any safener treatment (data not shown).
Table I.
Effect of herbicide safeners on GST-specific activity in Arabidopsis seedlings
| Safener | 1-Chloro-2,4- dinitrobenzene (CDNB) | p-Nitrobenzyl chloride (NBC) | Benzyl Isothiocyanate (BITC) | Ethacrynic Acid (EA) | Metolachlor | Alachlor | Acetochlor |
|---|---|---|---|---|---|---|---|
| nkat mg−1 protein | |||||||
| None | 0.58 | 0.004 | 0.47 | aND | ND | ND | 0.003 |
| Fenclorim | 2.70 | 0.045 | 0.78 | 0.10 | 0.007 | 0.034 | 0.018 |
| Benoxacor | 3.00 | 0.055 | 0.78 | 0.06 | 0.010 | 0.062 | 0.011 |
| Flurazole | 1.93 | 0.032 | 0.31 | ND | ND | ND | 0.006 |
| Dichlormid | 0.92 | 0.012 | 0.88 | ND | ND | ND | 0.008 |
| Oxabetrinil | 1.04 | 0.006 | 0.74 | 0.15 | 0.012 | 0.020 | 0.002 |
| Fluxofenim | 1.53 | 0.007 | 0.89 | 0.35 | 0.012 | 0.016 | 0.006 |
Seven-day-old seedlings were exposed to safeners (100 μm) for 24 h, and specific activity in protein extracts was then determined with a variety of substrates. The results presented are the means of two independent experiments.
ND, None detected.
In cereals, all of the safeners tested in the experiments described above enhance the expression of GSTs that are active in chloroacetanilide herbicide conjugation (Davies and Caseley, 1999). Therefore, we investigated whether treatment of Arabidopsis with the same safeners would result in a similar induction of GSTs capable of conjugating the chloroacetanilide herbicides metolachlor, alachlor, and acetochlor. In the absence of safener treatment, negligible activity toward the chloroacetanilides was detected in Arabidopsis seedlings (Table I). As observed in assays with non-herbicide substrates, fenclorim and benoxacor enhanced GST activity toward the chloroacetanilides, with oxabetrinil and fluxofenim also showing some effect. In contrast, flurazole and dichlormid had little effect. GST activities were also assayed toward the diphenyl ether herbicides fluorodifen, acifluorfen, and fomesafen, the sulfonylurea chlorimuron ethyl, the chloro-s-triazine atrazine, and the aryloxyphenoxypropionate fenoxaprop ethyl. No GST activity was detected with any of these herbicides in untreated seedlings. Modest GST activity toward fluorodifen could be measured after treatment with fenclorim (0.0001 nkat mg−1 protein) and benoxacor (0.029 nkat mg−1 protein). However, the safeners failed to induce measurable activities toward any of the other herbicides (data not shown).
Based on these results, experiments were performed to determine if safeners that enhanced GST activity toward the chloroacetanilides, such as benoxacor and fenclorim, were able to increase tolerance of Arabidopsis to these herbicides. Sterile Arabidopsis seeds were germinated in multiwell plates and after 8 d treated with or without safener before the addition of 10 μm metolachlor at d 10. The plants were then harvested at d 15 and analyzed. Treatment of unsafened seedlings with 10 μm metolachlor resulted in a 24% reduction in fresh weight (0.21 ± 0.01 g mean ± sd, n = 3) compared with untreated control plants that were not exposed to the herbicide. Treatment with 10 mg L−1 benoxacor before addition of metolachlor resulted in a greater reduction in fresh weight (33% ± 4%) as compared with controls, whereas a pretreatment with fenclorim gave no significant protection compared with herbicide treatment alone. Subsequent studies showed that benoxacor significantly inhibited the growth of Arabidopsis seedlings at concentrations above 1 mg L−1; however, sub-toxic doses of the safener still failed to give any significant protection against the growth inhibitory effects of metolachlor. A number of other methods have been used to expose Arabidopsis seedlings to safeners but none of these have been able to increase tolerance to chloroacetanilide herbicides (data not shown).
Effect of Safeners on Glutathione Levels in Arabidopsis Seedlings
Herbicide safeners are known to increase total glutathione content in cereal crops. In maize, for example, total GSH levels were shown to double in shoots and roots after treatment with benoxacor (Farago and Brunold, 1994; Kocsy et al., 2001). One possible explanation for the lack of tolerance to herbicides in Arabidopsis seedlings after safener treatment is that they have insufficient GSH. Therefore, the effects of safeners on total GSH levels (combined GSH and oxidized glutathione) in Arabidopsis seedlings were measured. Seven-day-old seedlings grown in liquid medium were treated with safeners (100 μm), and GSH levels in whole seedlings were then measured (Table II). Compared with untreated seedlings, GSH concentration increased 3-fold in seedlings treated with benoxacor and nearly 2-fold in response to fenclorim and fluxofenim. Therefore, it is unlikely that GSH levels limit the tolerance of Arabidopsis to herbicides in response to the safeners tested.
Table II.
Effect of safeners on total glutathione content in Arabidopsis seedlings
| Safener | Glutathione Content |
|---|---|
| μmol kg−1 fresh wt | |
| None | 396 ± 51 |
| Benoxacor | 1,251 ± 87 |
| Fenclorim | 702 ± 104 |
| Fluxofenim | 681 ± 74 |
Seven-day-old seedlings were exposed to safeners (100 μm) for 24 h, and total glutathione content was measured. The results presented are the means ± sd of three independent experiments.
Identification of Safener-Induced Arabidopsis GSTs
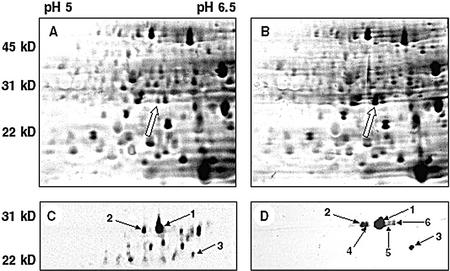
Although safener treatments of Arabidopsis seedlings do not result in tolerance to specific herbicides, they do stimulate GST activity. To further characterize this aspect of the safener response in Arabidopsis, total soluble protein was isolated from untreated and safener-treated seedlings and the polypeptides resolved using two-dimensional SDS-PAGE. One 26-kD polypeptide was noticeably more abundant in the sample from benoxacor-treated cultures compared with untreated controls (Fig. 1, A and B). Significantly, the relative molecular mass of this polypeptide is similar to that of most plant GST subunits.
Figure 1.
Two-dimensional SDS-PAGE gels of Arabidopsis total protein (A and B) and affinity-purified GST fraction (C and D) from seedlings grown 7 d in liquid culture. Total protein extracted from untreated (A) and 100 μm benoxacor-treated (B) tissues were separated on two-dimensional gels. The arrow in B indicates a 26-kD safener-induced protein that is induced by benoxacor. Subsequent experiments identified this as AtGSTU19. GSH-affinity-purified polypeptides from control (C) and safener-treated (D) samples are shown below. Protein spots labeled 1 through 3 are present in each sample. Spots labeled 4 through 6 were induced in response to benoxacor (D). The spot labeled 1 in the benoxacor sample was identified as AtGSTU19 by mass spectrometry.
To determine if specific GSTs were induced by benoxacor treatment, protein extracts from control and safener-treated Arabidopsis cultures were subjected to GSH-affinity chromatography, a method that is known to purify many plant GSTs (Pascal et al., 1998). The affinity-bound proteins were then resolved on two-dimensional gels into a large number of polypeptides (Fig. 1, C and D). The apparent molecular masses of many of these polypeptides were in the range 21 to 29 kD, consistent with that expected for plant GST subunits. When similarly prepared protein samples from control and benoxacor-treated seedlings were compared, a smaller number of intensely stained polypeptides, ranging in mass from 23 to 28 kD, were detected in the sample from benoxacor-treated seedlings. Abundant polypeptides present in samples from both untreated and safener-treated Arabidopsis seedlings were labeled spots 1 through 3, respectively. Polypeptides that were present in the benoxacor-treated seedlings but not detected in the control (spots 4–6) were also identified. Prolonged staining of gels containing proteins from safener-treated plants revealed a similar overall pattern of polypeptides as found in the control sample, suggesting that benoxacor had selectively increased the abundance of polypeptides 1 through 3 without affecting the expression of the other constitutively expressed proteins. Therefore, although polypeptides 1 through 3 are expressed constitutively in seedlings, their abundance increased in response to benoxacor treatment. In contrast, polypeptides 4 through 6 were only observed after benoxacor treatment. Similar changes in the expression of GSH-affinity-purified proteins were observed after treatment with fenclorim and fluxofenim (data not shown).
Polypeptide 1 was chosen for further characterization because the abundance of this protein increased dramatically in response to all three safeners. The protein spot was excised from the gel, digested with trypsin, and the resulting peptides analyzed by reverse-phase HPLC coupled with electrospray ionization time of flight mass spectrometry. The masses of the peptides were then compared with a database of masses predicted for trypsin-digested proteins from Arabidopsis. From this analysis, a gene encoding a 25.6-kD GST was identified in the Arabidopsis genome (GenBank accession no. AAF71809.1). This GST was identified as a member of the tau class of the GST superfamily (Edwards et al., 2000). Comparison of the genomic DNA sequence with an expressed sequence tag for this gene (GenBank accession no. AJ012571) indicated that the protein was encoded within two exons rather than the five indicated in the annotation. This gene corresponds to AtGSTU19 in accordance with the nomenclature system recently suggested for Arabidopsis GSTs (Wagner et al., 2002).
Within the annotated genomic DNA sequence adjacent to AtGSTU19 were two other GST genes, AtGSTU20 and AtGSTU21, whose predicted proteins are approximately 70% similar to AtGSTU19 (Table III). Two additional GST genes, AtGSTU22 and AtGSTU23 (GenBank accession nos. AAF71799 and AAF71800), which are less similar to AtGSTU19, are also present in this region of chromosome one, separated from the others by a mutator-like transposase-coding region. These five GST genes comprise a cluster of closely related sequences contained within 20 kb on chromosome one. From searching the expressed sequence tag databases, at least three of these genes (AtGSTU19, AtGSTU20, and AtGSTU21) are expressed. The AtGSTU19 protein has significant similarity with other Arabidopsis tau class GSTs encoded by genes located outside the cluster on chromosome one (Table III), notably AtGSTU5 (GenBank accession no. D44465), which has been characterized previously (Van der Kop et al., 1996). AtGSTU19 is also very similar to members of the maize tau class of safener-inducible GSTs, ZmGSTU1 and ZmGSTU2 (Table III), which are active in metabolism of atrazine and metolachlor, among others (Dixon et al., 1999).
Table III.
Amino acid sequence similarity of Arabidopsis and maize tau class GSTs
| AtGSTU19 | AtGSTU20 | AtGSTU21 | AtGSTU5 | ZmGSTU1 | ZmGSTU2 | ZmGSTU3 | |
|---|---|---|---|---|---|---|---|
| AtGSTU19 | – | – | – | – | – | – | – |
| AtGSTU20 | 67.9 | – | – | – | – | – | – |
| AtGSTU21 | 72.9 | 56.3 | – | – | – | – | – |
| AtGSTU5 | 34.2 | 32.7 | 33.2 | – | – | – | – |
| ZmGSTU1 | 52.5 | 49.1 | 48.2 | 30.8 | – | – | – |
| ZmGSTU2 | 56.6 | 51.8 | 52.7 | 32.6 | 65.6 | – | – |
| ZmGSTU3 | 35.6 | 36.8 | 36.8 | 36.6 | 35.3 | 33.8 | – |
Protein sequences were aligned using the Clustal method and data are shown as percentage similarity.
Characterization of Arabidopsis GSTU19
To determine the substrate specificity of AtGSTU19, the open reading frame encoding this GST was cloned and expressed in Escherichia coli. The resulting recombinant dimer, AtGSTU19-19, was purified by GSH-affinity chromatography and assayed for GST activity. The enzyme was highly active in conjugating CDNB and BITC, and showed limited activity with NBC and EA (Table IV). AtGSTU19-19 was also active as a GPOX, reducing cumene hydroperoxide. With herbicide substrates, recombinant AtGSTU19-19 was able to catalyze the conjugation of the chloroacetanilide herbicides, alachlor, acetochlor, and metolachlor (Table IV). The specific activities of AtGSTU19-19 toward these herbicides were comparable with those reported for purified tau class GSTs from maize (Dixon et al., 1999) and wheat (Cummins et al., 1997) and for phi class GSTs from sorghum (Gronwald and Plaisance, 1998). However, AtGSTU19-19 showed no detectable activity toward other herbicides including acifluorfen, fomesafen, fluorodifen, chlorimuron-ethyl, fenoxaprop ethyl, or atrazine (data not shown). AtGSTU19-19 catalyzed the conjugation of GSH to chloroacetanilide herbicides with the same order of efficiency (i.e. alachlor > acetochlor > metolachlor) as observed in protein extracts from safener-treated Arabidopsis seedlings (compare Tables I and IV), consistent with the suggestion that AtGSTU19 makes a significant contribution to the herbicide-conjugating GST activity induced in response to safener treatment.
Table IV.
Specific activities of affinity-purified recombinant AtGSTU19 towards different substrates
| Substrate | Specific Activity |
|---|---|
| nkat mg−1 protein | |
| CDNB | 295.0 |
| NBC | 3.0 |
| EA | 2.6 |
| BITC | 35.7 |
| Cumene hydroperoxidea | 17.6 |
| Alachlor | 2.8 |
| Acetochlor | 1.9 |
| Metolachlor | 0.2 |
Data are means of three independent experiments for model substrates and two independent experiments for herbicide substrates.
GPOX activity determined with cumene hydroperoxide.
Expression of AtGSTU19 and Other GST Genes in Arabidopsis in Response to Safeners
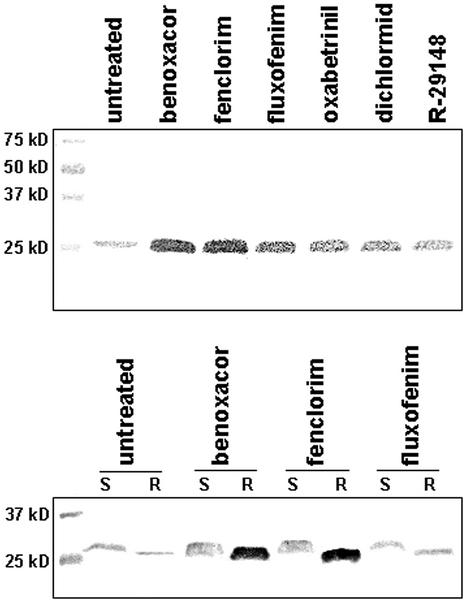
Polyclonal antibodies raised against purified AtGSTU19-19 were used in immunoblot assays to determine the abundance and tissue specificity of this protein in control and safener-treated Arabidopsis seedlings. The antiserum recognized a 26-kD polypeptide in proteins extracted from untreated plants (Fig. 2A), whereas the pre-immune antiserum did not react with this polypeptide (data not shown). Although this protein was present in untreated seedlings, its abundance increased noticeably in response to all safeners tested (Fig. 2A). The magnitude of induction by the various safeners was in the order: benoxacor > fenclorim > fluxofenim > oxabetrinil > dichlormid > R-29148. Notably, this order is the same as that observed for these safeners to induce GST activity toward CDNB (Table I). In addition, this protein was shown to be highly induced in roots, and to a lesser degree in shoots, after benoxacor and fenclorim treatment (Fig. 2B). There is an apparent difference in mobility of the protein detected by the AtGSTU19 antiserum in root and shoot tissue. Whether this reflects a modification of the protein or an artifact of the immunoblot procedure is under investigation.
Figure 2.
Immunoblot assay of Arabidopsis total proteins using antisera raised against AtGSTU19. A, Seven-day old seedlings grown in liquid medium were treated with various safeners (100 μm) and extracted total soluble proteins were separated using SDS-PAGE. Blots were then probed with antiserum raised against recombinant AtGSTU19 and a 26.8-kD polypeptide was detected. B, Three-week old seedlings grown hydroponically were treated with three safeners (100 μm) and total soluble protein from root (R) and shoot (S) tissues were resolved using SDS-PAGE. Blots were probed with AtGSTU19 antiserum.
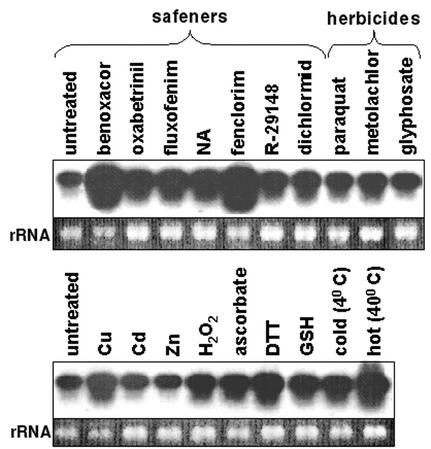
To characterize the expression of AtGSTU19 in response to various compounds, a cDNA for AtGSTU19 was used as a probe to examine the mRNA expression of this gene in Arabidopsis seedlings treated with a range of chemicals, including several safeners (Fig. 3). AtGSTU19 RNA was readily detected under control conditions. All of the safeners tested increased the level of AtGSTU19 RNA, with benoxacor and fenclorim giving the greatest induction. Treatment with the herbicides metolachlor and paraquat also gave a modest increase in expression of AtGSTU19 RNA, whereas glyphosate did not. The effects of a number of other chemical treatments and environmental conditions were also examined. AtGSTU19 RNA was modestly induced by exposure to Cu2+ ions; hydrogen peroxide; the reducing agents ascorbic acid, DTT, and GSH; and high temperature (Fig. 3). Treatment at 4°C and exposure to Cd2+ or Zn2+ ions had little or no effect. None of these nonspecific chemical treatments gave the level of AtGSTU19 RNA induction seen with benoxacor or fenclorim.
Figure 3.
Expression of AtGSTU19 mRNA in liquid cultures of Arabidopsis seedlings treated for 24 h with safeners (100 μm), herbicides (100 μm), 50 μm CuSO4, 90 μm CdCl2, 400 μm ZnCl2, 3 mm H2O2, 1 mm ascorbic acid, 1 mm dithiothreitol (DTT), and 1 mm GSH. In addition, cultures were exposed to low temperature (4°C) and high temperature (40°C) for 24 h. NA, Naphthalic anhydride.
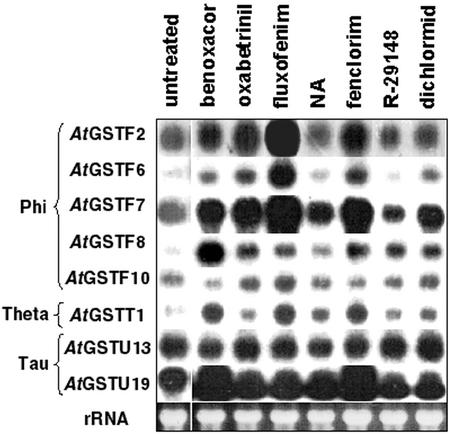
The effect of safeners on RNA expression of several other Arabidopsis GST genes was also examined (Fig. 4). Among the genes chosen were five from the phi class (AtGSTF2, AtGSTF6, AtGSTF7, AtGSTF8, and AtGSTF10), two from the tau class (AtGSTU13 and AtGSTU19), and one from the theta class (AtGSTT1; Table V). cDNAs encoding these GSTs were used as probes to examine the mRNA expression of these GST genes in response to a number of herbicide safeners. The abundance of several GST mRNAs increased in response to safeners. As shown above, AtGSTU19 RNA was markedly induced by treatment with benoxacor or fenclorim. In contrast, expression of the other tau class gene examined, AtGSTU13, was not induced by any of the safeners tested. Expression of the theta class gene, AtGSTT1, was enhanced by benoxacor, fluxofenim, and fenclorim. Among the phi class GSTs, expression of AtGSTF10 was not altered by any of the treatments. AtGSTF2, AtGSTF6, and AtGSTF7 showed similar patterns of RNA induction, with fluxofenim having the greatest effect and more modest responses to fenclorim, oxabetrinil, and benoxacor. AtGSTF6 and AtGSTF7 were also modestly induced by dichlormid. AtGSTF8 differed from all other GST genes by showing the greatest induction in response to treatment with benoxacor.
Figure 4.
RNA expression of various Arabidopsis GSTs after treatment with herbicide safeners. Total RNA was isolated from Arabidopsis seedlings grown for 7 d in liquid culture followed by treatment with safeners (100 μm for 24 h). RNA was separated on a gel, transferred to a nitrocellulose membrane, and hybridized with cDNAs encoding various Arabidopsis GSTs. Equal RNA loading was confirmed by the ethidium bromide staining of rRNA present as shown at the bottom. NA, Naphthalic anhydride.
Table V.
Summary of Arabidopsis GST genes discussed in the text
| New Name | Old Name | Reference(s) |
|---|---|---|
| AtGSTF1 | PM239x14, GST3 | Bartling et al. (1993) |
| AtGSTF2 | GST2 | Zhou and Goldsbrough (1993) |
| Atpm24.1 | Zettl et al. (1994) | |
| AtGSTF6 | ERD11 | Kiyosue et al. (1993) |
| GST1 | Greenberg et al. (1994); Yang et al. (1998) | |
| AtGSTF7 | GST11 | Wagner et al. (2002) |
| AtGSTF8 | GST6 | Chen et al. (1996) |
| AtGSTF10 | ERD13, GST4 | Kiyosue et al. (1993) |
| AtGSTU5 | GST5 | Watshiki et al. (1995) |
| 103-1a | Van der Kop et al. (1996) | |
| AtGSTU19 | AtGST8 | Wagner et al. (2002) |
| AtGSTU13 | T7N9.15 | Wagner et al. (2002) |
| AtGSTT1 | AtGST10 | Dixon et al. (1999) |
Where possible, the nomenclature proposed by Wagner et al. (2002) is used.
DISCUSSION
A number of mechanisms have been proposed for safener activity. As herbicide antagonists, safeners may compete at the site(s) of action, preventing plant injury. Alternatively, safeners may lower the amount of herbicide that reaches the site of action by reducing uptake or translocation. Safeners may also enhance the metabolism of herbicides to inactive forms. Most studies favor the last hypothesis, which is thought to involve the induction of detoxification systems by safeners (Davies and Caseley, 1999).
Here, we show that safeners are also able to induce the expression of herbicide detoxifying enzymes in Arabidopsis. After safener treatment of seedlings, GST activity against model substrates and herbicides was elevated, as is the case in wheat (Cummins et al., 1997) and maize (Dixon et al., 1998). Increased GST activity was the result of increased levels of RNAs encoding GSTs and the accumulation of specific GST proteins. The observation that some Arabidopsis GSTs are induced by herbicide safeners is significant because it demonstrates that the recognition, signaling, and gene activation processes required for this facet of safener activity are present in dicotyledenous plants as well as in cereals. Additional evidence that a safener-signaling pathway operates in Arabidopsis comes from the observation that the promoter of the safener-inducible In2-2 gene from maize is activated in Arabidopsis by several benzene sulfonamide safeners in a tissue-specific manner (DeVeylder et al., 1997). Another similarity in how cereals and Arabidopsis respond to safeners is an increase in the total GSH pool. The significance of this response is unknown but suggests there would be sufficient GSH available for herbicide conjugation in Arabidopsis seedlings after safener treatment.
Very little has been published on the effects of safeners on dicots, largely because of the lack of any discernible protective effect of these compounds toward herbicides in broad leaf crops (Davies and Caseley, 1999). Similarly, we have been unable to demonstrate any safening of Arabidopsis seedlings toward chloroacetanilide herbicides despite testing numerous combinations of safener and herbicide treatments. One possible explanation for this is that safener treatment of dicots does not induce expression of GSTs in tissues where this activity is required to protect plants from herbicide damage. Immunoblot experiments indicated that at least two safeners induced the expression of AtGSTU19 preferentially in roots. It has also been shown that the maize safener dichlormid enhanced GST activity toward atrazine in pea (Pisum sativum) seedlings, but only in root tissues (Edwards, 1996). These results suggest that safener-induced GSTs must not only have activity toward the target herbicide but also be expressed in the appropriate tissue(s) to provide tolerance. However, there may be additional factors that contribute to the failure of safeners to protect Arabidopsis from chloroacetanilide herbicides. These might include inadequate expression of genes encoding other components of the detoxification system, such as conjugate transporters.
Expression of several Arabidopsis GST genes increased in seedlings in response to safeners. Given the large number of GST genes in Arabidopsis, it is possible that the RNA expression profiles of some of these genes might be complicated by cross hybridization between related sequences. Microarray analysis of RNA expression of maize and soybean (Glycine max) GSTs indicates there is no significant cross hybridization among GST cDNAs if sequence similarity is less than 80% (McGonigle et al., 2000). Among the Arabidopsis GST genes examined in these experiments, AtGSTF6 and AtGSTF7 have the highest similarity (93% in the coding region) and these genes exhibit similar responses to various safeners (Fig. 4). AtGSTF2 shows 94% similarity to AtGSTF3, but the latter was not studied in these experiments. Further experiments will be required to determine which of these closely related genes is induced in response to these safeners. Because the promoter sequences in these pairs of genes are also very similar, it is possible they may be coordinately regulated by safeners. Nevertheless, RNA hybridization and two-dimensional SDS-PAGE studies have clearly demonstrated that treatment of Arabidopsis seedlings with various safeners resulted in specific changes in the expression of different GST genes derived from several classes, rather than invoking a similar response to all safeners. This suggests that multiple signaling pathways must be involved in regulating GST expression in response to these compounds. It will now be of interest to identify other safener-inducible genes in Arabidopsis, and use a molecular genetic approach to identify the signaling pathways involved in safener-inducible gene expression. Further studies using Arabidopsis may also shed light on the basis for differences in safener-induced herbicide tolerance between monocots and dicots.
MATERIALS AND METHODS
Chemicals
Substrates for GST assays were purchased from the Aldrich Chemical Company (Dorset, UK). Analytical grade (95%–99% pure) safeners and herbicides were provided by the following companies: benoxacor, oxabetrinil, fluxofenim, fenclorim, R-29148, dichlormid, metolachlor, fomesafen, and fluorodifen (Syngenta, Greensboro, NC); flurazole, alachlor, and acetochlor (Monsanto, St. Louis); acifluorfen and atrazine (BASF Corporation, Mount Olive, NJ); chlorimuron-ethyl (DuPont, Wilmington, DE); and fenoxaprop-ethyl (Greyhound Chemicals, Merseyside, UK). Stocks of 100 mm herbicide safeners and herbicides were prepared in acetone and stored at −20°C.
Plant Material and Safener and Herbicide Treatments
Seedlings of Arabidopsis ecotype Columbia were grown for 7 d in liquid culture containing one-half-strength Murashige and Skoog balanced salt solution and Gamborg's vitamin solution under sterile conditions (Murashige and Skoog, 1962). Plants were grown under continuous soft-white fluorescent lighting with gentle shaking on a rotary shaker at 25°C. Seedlings were treated with safeners for 24 h at a final concentration in liquid culture of 100 μm. An equal volume of acetone, which has been shown to have no effect on GST RNA levels or enzyme activity (data not shown), was used in control treatments. Plant tissue was then frozen in liquid nitrogen and stored at −70°C until use. For herbicide phytotoxicity trials, sterile Arabidopsis seeds were sown into 25-well plastic plates (seven seeds per well) containing one-half-strength Murashige and Skoog medium (3.5 mL per well). The plants were then grown in sterile conditions with a 16-h photoperiod (light intensity) for 8 d at 25°C before the addition of safeners, which were added in acetone to give a final concentration of 10 mg L−1, with acetone alone serving as the control treatment. At d 10, metolachlor was added to a final concentration of 10 μm and the plants then harvested at d 15, carefully blotted dry, and weighed. Treatments were carried out in triplicate.
Protein Extraction and Purification
All extraction and purification steps were carried out at 4°C or on ice. Arabidopsis seedlings (25–30 g) were ground to a fine powder in liquid nitrogen using a mortar and pestle and then homogenized in 20 mm Tris-HCl (pH 7.8), 1 mm EDTA, and 5 mm DTT with a Polytron. The slurry was filtered through eight layers of cheesecloth and centrifuged (12,000g for 30 min). The resulting supernatant was applied to GSH-agarose (G-4510, Sigma, St. Louis) packed in a 10-mL column equilibrated with buffer A (20 mm Tris-HCl [pH 7.8], 1 mm EDTA, and 5 mm DTT). After washing with three-column volumes of buffer A, the GSTs were eluted as a single peak with 10 mm GSH in buffer B (20 mm Tris-HCl [pH 7.8] and 1 mm EDTA). The affinity-purified GSTs were immediately concentrated and desalted in buffer A using a Centricon YM-10 spin column (10-kD cutoff, Millipore Corp., Bedford, MA) before storage at −70°C.
Glutathione Content and GST Enzyme Assays
The spectrophotometric assay used to determine total GSH content in Arabidopsis seedlings was as described by Scheller et al. (1987). For GST enzyme assays, total protein extracts were prepared as described above from safener-treated seedlings (5 g). (NH4)2SO4 was added to the resulting supernatant to 80% saturation and the protein precipitate was recovered by centrifugation (12,000g for 20 min) and stored at −70°C. Before assay, the protein was desalted in buffer A using a Sephadex G-25 column (PD10, Pharmacia Biotech, Piscataway, NJ). All protein samples were quantified by the method of Bradford (1976) using bovine gamma globulin as a standard. Spectrophotometric assays described by Edwards (1996) were used to determine GST-specific activity toward CDNB, NBC, and EA. GST activity toward BITC and herbicide substrates, and GPOX activity toward cumene hydroperoxide, were conducted as described by Dixon et al. (1998).
Two-Dimensional Gel Electrophoresis
For separation of total protein by two-dimensional SDS-PAGE, samples were prepared for isoelectric focusing (IEF) as described by Tsugita and Masaharu (1999). Precast Immobiline Drystrips (7 cm, pH 4–7 linear gradient, Amersham-Pharmacia Biotech, Uppsala) were rehydrated for 12 h with buffer containing 250 μg of protein. IEF was performed on an IPGphor instrument for a total of 8,750 volt hours (Amersham-Pharmacia Biotech). IEF gels were then applied to the surface of an SDS-PAGE gel (15% [w/v] acrylamide and 0.6% [v/v] N,N′-methylene bisacrylamide). After electrophoresis, proteins were visualized by silver staining (Wray et al., 1981). For GSH-affinity-purified samples, 15 μg of protein was incubated in IEF buffer (8 m urea, 2% [w/v] CHAPS, 0.5% [w/v] Immobilized pH gradient buffer [Amersham-Pharmacia Biotech]), at room temperature for 1 h, before application and electrophoresis as described above.
Identification of AtGSTU19
Polypeptide spots from two-dimensional gels were identified by in-gel enzymatic digestion followed by mass spectrometry (Arnott et al., 1998). In brief, the spots were excised and placed in 25 μL of 50:50 (v/v) acetonitrile:100 mm ammonium bicarbonate and shaken at room temperature for 15 min. The solution was removed and replaced with enough 10 mm DTT in 100 mm ammonium bicarbonate to cover the spots. After incubating at 55°C for 1 h, the solution was discarded and the samples were alkylated with 50 mm iodoacetamide in 100 mm ammonium bicarbonate in the dark at room temperature for 1 h. After washing the gel pieces twice with 50 μL of 50:50 (v/v) acetonitrile:100 mm ammonium bicarbonate, the gel pieces were dried in a vacuum evaporator (Centrivap, Labconco, Kansas City, MO). The gel pieces were rehydrated for 10 min with 6 μL of 0.01% (w/v) SDS in 50 mm ammonium bicarbonate containing 0.05 μg μL−1 sequencing grade-modified trypsin (Promega, Madison, WI). The gel pieces were covered with 50 mm ammonium bicarbonate and incubated overnight at 45°C. The supernatant was removed and the gel pieces washed two times, each for 20 min, with 50:50 (v/v) acetonitrile:0.5% (v/v) trifluoroacetic acid. The enzyme digest solution and the washes were then pooled and lyophilized to a final volume of 5 to 10 μL. The sample (2.5 μL) was loaded onto a C18 microbore HPLC column. The column was connected to the electrospray ionization source of an ion-trap mass spectrometer (Esquire-LC, Bruker Daltonics, Billerica, MA) and a gradient (5%–95% [v/v] acetonitrile in 30 min) was used to elute the digested peptides into the mass spectrometer. The instrument was set to analyze positive ions with mass-to-charge ratio between 300 and 2,000. The resulting peptide ion signals were entered into the MS-TAG protein database searching program (http://prospector.ucsf.edu/ucsfhtml4.0/mstagfd.htm). The best protein match was found to be AtGSTU19 from Arabidopsis (GenBank accession no. AJ012571). Peptide fragments that accounted for approximately 70% of AtGSTU19 were identified in this analysis.
Expression and Purification of AtGSTU19
A cDNA clone for AtGSTU19 was obtained from the Arabidopsis Biological Resource Center (clone no. 35A3T7, GenBank accession no. T04270). For PCR amplification, the 5′ primer (5′-TCGTAACCATGGCGAACGAGGTGATTC-3′) included an NcoI site (underlined) at the translation start site. The 3′ primer (5′-CGCGCAGGATCCGAACCATATGACAAGAG-3′) included the translation stop site and a BamHI site (underlined) for cloning into an expression vector. After PCR amplification of the coding sequence, the DNA fragment was digested with NcoI and BamHI and ligated into the pET11d vector (New England Biolabs, Beverly, MA) and subsequently transformed into competent BL21(DE3) cells. Cultures were induced with 1 mm isopropyl β-d-thiogalactopyranoside for 4 to 6 h. Cells were collected by centrifugation and disrupted by sonication in buffer A. The crude protein extract was then applied directly to a GSH-agarose column and the GST recovered as a single peak as described above. The purified sample was precipitated by the addition of solid (NH4)2SO4 to 80% saturation and stored at −70°C until assayed. Polyclonal antibodies against AtGSTU19 were raised in rabbits according to the standard protocols of Cocalico Biologicals Inc. (Reamstown, PA).
ACKNOWLEDGMENTS
The authors acknowledge the technical assistance of Malcolm Slater in carrying out the herbicide phytotoxicity trials and Jen Victor in optimizing the two-dimensional SDS-PAGE methods.
Footnotes
This work was supported in part by the U.S. Department of Agriculture (National Needs fellowship grant).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010066.
LITERATURE CITED
- Arnott D, O'Connell KL, King KL, Stults JT. An integrated approach to proteome analysis: identification of proteins associated with cardiac hypertrophy. Anal Biochem. 1998;258:1–18. doi: 10.1006/abio.1998.2566. [DOI] [PubMed] [Google Scholar]
- Bartling D, Radzio R, Steiner U, Weiler EW. A glutathione S-transferase with glutathione-peroxidase activity from Arabidopsis thaliana: molecular cloning and functional characterization. Eur J Biochem. 1993;216:579–586. doi: 10.1111/j.1432-1033.1993.tb18177.x. [DOI] [PubMed] [Google Scholar]
- Berhane K, Widersten M, Engström Å, Kozarich JW, Mannervik B. Detoxification of base propenals and other unsaturated aldehyde products of radical reactions and lipid peroxidation by human glutathione transferases. Proc Natl Acad Sci USA. 1994;91:1480–1484. doi: 10.1073/pnas.91.4.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chen WQ, Chao G, Singh KB. The promoter of a H2O2-inducible, Arabidopsis glutathione S-transferase gene contains closely linked OBF- and OBP1-binding sites. Plant J. 1996;10:955–966. doi: 10.1046/j.1365-313x.1996.10060955.x. [DOI] [PubMed] [Google Scholar]
- Cummins I, Cole DJ, Edwards R. Purification of multiple glutathione transferases involved in herbicide detoxification from wheat (Triticum aestivumL.) treated with the safener fenchlorazole-ethyl. Pestic Biochem Phys. 1997;59:35–49. [Google Scholar]
- Danielson UH, Esterbauer H, Mannervik B. Structure-activity relationships of 4-hydroxyalkenals in the conjugation catalysed by mammalian glutathione transferases. Biochem J. 1987;247:707–713. doi: 10.1042/bj2470707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J, Caseley JC. Herbicide safeners: a review. Pestic Sci. 1999;55:1043–1058. [Google Scholar]
- Deng F, Hatzios KK. Characterization and safener induction of multiple glutathione S-transferases in three genetic lines of rice. Pestic Biochem Phys. 2002;72:24–39. [Google Scholar]
- DeVeylder L, VanMontagu M, Inzé D. Herbicide safener-inducible gene expression in Arabidopsis thaliana. Plant Cell Physiol. 1997;38:568–577. doi: 10.1093/oxfordjournals.pcp.a029206. [DOI] [PubMed] [Google Scholar]
- Dixon DP, Cole DJ, Edwards R. Purification, regulation and cloning of a glutathione transferase (GST) from maize resembling the auxin-inducible type-III GSTs. Plant Mol Biol. 1997;36:75–87. doi: 10.1023/a:1005958711207. [DOI] [PubMed] [Google Scholar]
- Dixon DP, Cole DJ, Edwards R. Dimerization of maize glutathione transferases in recombinant bacteria. Plant Mol Biol. 1999;40:997–1008. doi: 10.1023/a:1006257305725. [DOI] [PubMed] [Google Scholar]
- Dixon DP, Cummins I, Cole DJ, Edwards R. Glutathione-mediated detoxification systems in plants. Curr Opin Plant Biol. 1998;1:258–266. doi: 10.1016/s1369-5266(98)80114-3. [DOI] [PubMed] [Google Scholar]
- Edwards R. Characterization of glutathione transferases and glutathione peroxidases in pea (Pisum sativum) Physiol Plant. 1996;98:594–604. [Google Scholar]
- Edwards R, Dixon DP, Walbot V. Plant glutathione S-transferases enzymes with multiple functions in sickness and in health. Trends Plant Sci. 2000;5:193–198. doi: 10.1016/s1360-1385(00)01601-0. [DOI] [PubMed] [Google Scholar]
- Farago S, Brunold C. Regulation of thiol contents in maize roots by intermediates and effectors of glutathione synthesis. J Plant Physiol. 1994;144:433–437. [Google Scholar]
- Greenberg JT, Guo AL, Klessig DF, Ausubel FM. Programmed cell-death in plants—a pathogen-triggered response activated coordinately with multiple defense functions. Cell. 1994;77:551–563. doi: 10.1016/0092-8674(94)90217-8. [DOI] [PubMed] [Google Scholar]
- Gronwald JW, Plaisance KL. Isolation and characterization of glutathione S-transferase isozymes from sorghum. Plant Physiol. 1998;117:877–892. doi: 10.1104/pp.117.3.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jepson I, Holt DC, Roussel V, Wright SY, Greenland AJ. Transgenic plant analysis as a tool for the study of maize glutathione S-transferases. In: Hatzios KK, editor. Regulation of Enzymatic Systems Detoxifying Xenobiotics in Plants. Ed 1, No 3. Vol. 37. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1997. pp. 313–323. [Google Scholar]
- Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K. Characterization of two cDNAs (ERD11 and ERD13) for dehydration-inducible genes that encode putative glutathione S-transferases in Arabidopsis thaliana L. FEBS Lett. 1993;335:189–192. doi: 10.1016/0014-5793(93)80727-c. [DOI] [PubMed] [Google Scholar]
- Kocsy G, von Ballmoos P, Ruegsegger A, Szalai G, Galiba G, Brunold C. Increasing the glutathione content in a chilling-sensitive maize genotype using safeners increased protection against chilling-induced injury. Plant Physiol. 2001;127:1147–1156. [PMC free article] [PubMed] [Google Scholar]
- Marrs KA. The functions and regulation of glutathione S-transferases in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:127–158. doi: 10.1146/annurev.arplant.47.1.127. [DOI] [PubMed] [Google Scholar]
- McGonigle B, Keeler SJ, Lan SMC, Koeppe MK, O'Keefe DP. A genomics approach to the comprehensive analysis of the glutathione S-transferase gene family in soybean and maize. Plant Physiol. 2000;124:1105–1120. doi: 10.1104/pp.124.3.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio-assays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Neuefeind T, Reinemer P, Bieseler B. Plant glutathione S-transferases and herbicide detoxification. Biol Chem. 1997;378:199–205. [PubMed] [Google Scholar]
- Pascal S, Debrauwer L, Ferte MP, Anglade P, Rouimi P, Scalla R. Analysis and characterization of glutathione S-transferase subunits from wheat (Triticum aestivumL.) Plant Sci. 1998;134:217–226. [Google Scholar]
- Pascal S, Scalla R. Purification and characterization of a safener-induced glutathione S-transferase from wheat (Triticum aestivum) Physiol Plant. 1999;106:17–27. [Google Scholar]
- Plaisance KL, Gronwald JW. Enhanced catalytic constant for glutathione S-transferase (atrazine) activity in an atrazine-resistant Abutilon theophrastibiotype. Pest Biochem Physiol. 1999;63:34–49. [Google Scholar]
- Rea PA. MRP subfamily ABC transporters from plants and yeast. J Exp Bot. 1999;50:895–913. [Google Scholar]
- Roxas VP, Lodhi SA, Garrett DK, Mahan JR, Allen RD. Stress tolerance in transgenic tobacco seedlings that overexpress glutathione S-transferase/glutathione peroxidase. Plant Cell Physiol. 2000;41:1229–1234. doi: 10.1093/pcp/pcd051. [DOI] [PubMed] [Google Scholar]
- Scheller HV, Huang B, Hatch E, Goldsbrough PB. Phytochelatin synthesis and glutathione levels in response to heavy-metals in tomato cells. Plant Physiol. 1987;85:1031–1035. doi: 10.1104/pp.85.4.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugita A, Masaharu K. 2-D electrophoresis of plant proteins. In: Link A, editor. 2-D Proteome Analysis Protocols (Methods in Molecular Biology Series, Vol 112). Totowa, NJ: Humana Press Inc.; 1999. pp. 513–530. [DOI] [PubMed] [Google Scholar]
- Van der Kop DAM, Schuyer M, Sheres B, Van der Zaal BJ, Hooykaas PJJ. Isolation and characterization of an auxin-inducible glutathione S-transferase gene of Arabidopsis thaliana. Plant Mol Biol. 1996;30:839–844. doi: 10.1007/BF00019016. [DOI] [PubMed] [Google Scholar]
- Wagner U, Edwards R, Dixon DP, Mauch F. Probing the diversity of the Arabidopsis glutathione S-transferase gene family. Plant Mol Biol. 2002;49:515–532. doi: 10.1023/a:1015557300450. [DOI] [PubMed] [Google Scholar]
- Watahiki MK, Mori H, Yamamoto KT. Inhibitory effects of auxins and related substances on the activity of an Arabidopsis glutathione S-transferase isozyme expressed in Escherichia coli. Physiol Plant. 1995;94:566–574. [Google Scholar]
- Wray W, Boulikas T, Wray VP, Hancock R. Silver staining of proteins in polyacrylamide gels. Anal Biochem. 1981;118:197–203. doi: 10.1016/0003-2697(81)90179-2. [DOI] [PubMed] [Google Scholar]
- Wu JR, Cramer CL, Hatzios KK. Characterization of two cDNAs encoding glutathione S-transferases in rice and induction of their transcripts by the herbicide safener fenclorim. Physiol Plant. 1999;105:102–108. [Google Scholar]
- Yang KY, Kim EY, Kim CS, Guh JO, Kim KC, Cho BH. Characterization of a glutathione S-transferase gene ATGST 1 in Arabidopsis thaliana. Plant Cell Rep. 1998;17:700–704. doi: 10.1007/s002990050468. [DOI] [PubMed] [Google Scholar]
- Zettl R, Schell J, Palme K. Photoaffinity-labeling of Arabidopsis thalianaplasma-membrane vesicles by 5-Azido-[7-H-3]indole-3-acetic acid: identification of a glutathione S-transferase. Proc Natl Acad Sci. 1994;91:689–693. doi: 10.1073/pnas.91.2.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JM, Goldsbrough PB. An Arabidopsis gene with homology to glutathione S-transferases is regulated by ethylene. Plant Mol Biol. 1993;22:517–523. doi: 10.1007/BF00015980. [DOI] [PubMed] [Google Scholar]