Abstract
Mutants defective in the biosynthesis or signaling of brassinosteroids (BRs), plant steroid hormones, display dwarfism. Loss-of-function mutants for the gene encoding the plasma membrane-located BR receptor BRI1 are resistant to exogenous application of BRs, and characterization of this protein has contributed significantly to the understanding of BR signaling. We have isolated two new BR-insensitive mutants (dwarf12-1D and dwf12-2D) after screening Arabidopsis ethyl methanesulfonate mutant populations. dwf12 mutants displayed the characteristic morphology of previously reported BR dwarfs including short stature, short round leaves, infertility, and abnormal de-etiolation. In addition, dwf12 mutants exhibited several unique phenotypes, including severe downward curling of the leaves. Genetic analysis indicates that the two mutations are semidominant in that heterozygous plants show a semidwarf phenotype whose height is intermediate between wild-type and homozygous mutant plants. Unlike BR biosynthetic mutants, dwf12 plants were not rescued by high doses of exogenously applied BRs. Like bri1 mutants, dwf12 plants accumulated castasterone and brassinolide, 43- and 15-fold higher, respectively, providing further evidence that DWF12 is a component of the BR signaling pathway that includes BRI1. Map-based cloning of the DWF12 gene revealed that DWF12 belongs to a member of the glycogen synthase kinase 3β family. Unlike human glycogen synthase kinase 3β, DWF12 lacks the conserved serine-9 residue in the auto-inhibitory N terminus. In addition, dwf12-1D and dwf12-2D encode changes in consecutive glutamate residues in a highly conserved TREE domain. Together with previous reports that both bin2 and ucu1 mutants contain mutations in this TREE domain, this provides evidence that the TREE domain is of critical importance for proper function of DWF12/BIN2/UCU1 in BR signal transduction pathways.
Brassinosteroids (BRs) are poly-hydroxylated plant steroids structurally similar to animal steroid hormones such as ecdysone. Essential roles for BRs in plant growth and development have been demonstrated by the dwarf phenotype displayed in mutants defective in BR biosynthetic or signaling pathways in Arabidopsis, rice (Oryza sativa), tomato (Lycopersicon esculentum), and pea (Pisum sativum). Phenotypes of the light-grown BR dwarf mutants include short stature, dark-green and round leaves, reduced fertility, and a prolonged life cycle, as well as altered skotomorphogenesis in dark-grown plants. Arabidopsis dwarf mutants defective in six genes encoding BR biosynthetic enzymes are rescued by exogenous application of BRs (Li et al., 1996; Szekeres et al., 1996; Choe et al., 1998, 1999a, 1999b, 2000), whereas mutants in signaling components are morphologically similar but insensitive to applied BRs. BR-insensitive mutants in a gene known as bri1 (brassinosteroid insensitive1) were previously isolated based on the phenotype of derepressed root-growth inhibition on media containing BRs (Clouse et al., 1996). Thus far, all BR-insensitive recessive mutants where the affected gene is known are BRI1 alleles (Li and Chory, 1997; Noguchi et al., 1999; Friedrichsen et al., 2000). The BRI1 gene was isolated and shown to encode a Leu-rich repeat receptor protein kinase (Li and Chory, 1997). Evidence from kinase domain swapping experiments with a similar Leu-rich repeat receptor protein kinase encoded by the rice Xa21 gene (He et al., 2000), the cellular localization of BRI1::GFP translational fusion protein to the plasma membrane (Friedrichsen et al., 2000), and in vitro BR-binding assays using an epitope-tagged BRI1 protein (Wang et al., 2001) have shown that BRI1 is a component of a BR receptor located in the plasma membrane.
The isolation and characterization of mutants insensitive or resistant to plant hormones has greatly enhanced our understanding of their signaling pathways (Gray and Estelle, 2000). However, the scarcity of BR-insensitive mutants has delayed unraveling BR-mediated signaling pathways. It has been hypothesized that loss-of-function mutations in additional BR signaling genes may result in lethality because of their essential roles or they may have unnoticeable physiological or morphological phenotypes because of gene redundancy. We have extensively screened both ethyl methanesulfonate (EMS)-induced and T-DNA mutant populations to obtain additional mutants, and recovered two semidominant mutants in a gene we call dwf12 (dwarf12). The semidominant dwf12 mutants are insensitive to exogenously applied BRs and display an extreme dwarf phenotype both in the light and dark. We also show that dwf12-1D plants, like bri1, accumulate significant amounts of BRs. The phenotypic similarity among dwf12 and previously reported BR mutants, the brassinolide (BL) insensitivity of dwf12-1D and dwf12-2D, and the mapping of dwf12 to a chromosomal location distinct from BRI1 indicate that dwf12 mutants define a new gene in the BR signal transduction pathway. Recently, two different groups have independently reported new BR-insensitive mutants. Li et al. (2001) identified two alleles of bin2 (brassinosteroid insensitive 2), and Perez-Perez et al. (2002) found three alleles of ucu1 (ultracurvata 1). The bin2 and ucu1 mutants were shown to have mutations in an Arabidopsis glycogen synthase kinase (GSK) 3β-like kinase gene (Li and Nam, 2002; Perez-Perez et al., 2002), the same gene that is mutant in our dwf12 alleles.
GSKs are a family of cytoplasmic kinases that belong to the mitogen-activated protein kinase superfamily and are found in animals, fungi, and plants. Diverse roles for GSK3s include dorsal/ventral polarity determination in Wnt/Wg signaling in Drosophila melanogaster and Xenopus laevis, endoderm/mesoderm formation in Caenorhabditis elegans, and prespore/prestalk fate determination in Dictyostelium discoideum (Kim and Kimmel, 2000). In mammals, they are involved in insulin-dependent Glc homeostasis, β-catenin-mediated cell signaling, and development of tau-associated Alzheimer's disease (Bienz and Clevers, 2000; Harwood, 2000). In the animal model systems studied thus far, differentially spliced transcripts arise from only one or two GSK3 genes, whereas in plants, GSK3 genes consist of multigene families; currently, the GenBank database contains three genes for alfalfa (Medicago sativa; Pay et al., 1993), five for tobacco (Nicotiana tabacum), three for rice, four for petunia (Petunia hybrida; Decroocq-Ferrant et al., 1995), and 10 for Arabidopsis (Bianchi et al., 1994). Dornelas et al. (1999) reported that the Arabidopsis genes show different temporal and spatial expression patterns. More recently, Dornelas et al. (2000) also showed that antisense down-regulation of the two GSK3-like genes, AtSK11 and AtSK12, results in altered floral development, including increased number of perianth organs and abnormal apical-basal patterning in the gynoecium. Here, we present additional evidence demonstrating that one of the Arabidopsis GSK3-like kinases, previously named ASKη(etha), plays a crucial role in BR signaling.
RESULTS
dwf12-1D and dwf12-2D Are New BR-Insensitive Dwarf Mutants
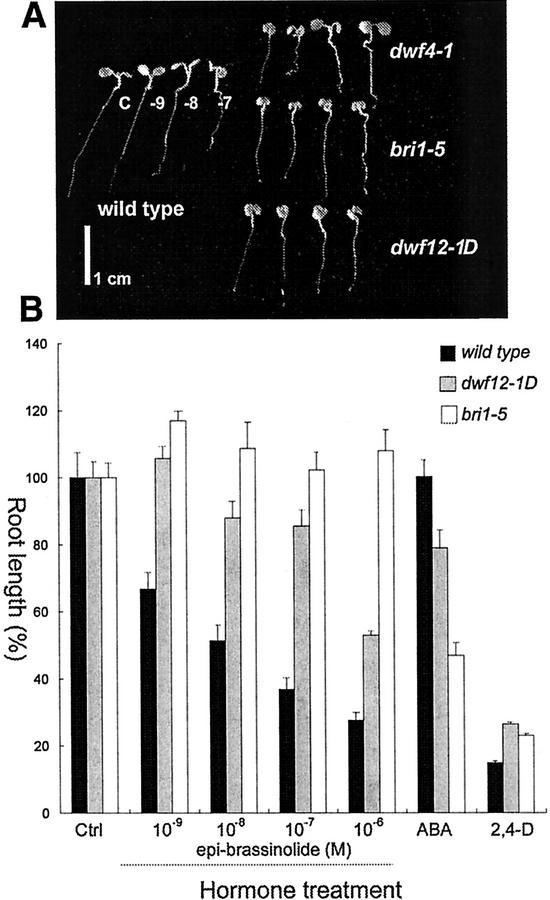
To identify additional players in the BR signaling pathway, we have screened EMS-induced mutant populations (>50,000) for characteristic BR dwarfs (Fig. 1), and identified 43 new dwarf mutants. Most of these 43 new dwarf mutants were rescued to a wild-type phenotype when BRs were topically applied. However, two mutants, wm1-1 and wm5-1, were insensitive to BRs, and phenotypically and genetically distinct from the other mutants. The phenotypes of the two mutants were typical of BR dwarfs, but with slightly twisted inflorescences, and leaves that are severely curled and rolled downward (Fig. 1). Interestingly, the progeny of the two dwarf mutants segregated for discrete groups of wild-type, severe, and weak dwarf plants (Fig. 1), suggesting that the parental dwarf mutants were heterozygous for a semidominant mutation that causes dwarfism. To determine whether dwf12 is allelic to bri1, we chose to map the mutations rather than doing genetic complementation tests because the semidominance of dwf12 may preclude reliable results in a complementation test. We mapped both dwf12 mutations to the middle of chromosome 4 to a position distinct from BRI1, which is linked to the DHS1 marker located on the bottom of chromosome 4 (see “Materials and Methods”). Thus, we designated the two novel BR-insensitive mutants, wm1-1 and wm5-1, dwarf12-1 dominant (dwf12-1D) and dwf12-2D, respectively: The demonstration of allelism comes from sequencing, which is discussed below.
Figure 1.
Phenotypic comparison of the BR biosynthetic mutant dwf4-1, and the insensitive mutants bri1-5, twisted1-60, and dwf12. Plants shown are 25 d old. dwf12 exhibits characteristic BR dwarf phenotypes including short inflorescences, infertility of homozygous mutants, short round leaves, and abnormal de-etiolation. In addition, dwf12 leaves show more severe downward curling than dwf4 and bri1. This phenotype and the mild twisting of inflorescences appear to be weaker version of twd1 (B. Schulz and K. Feldmann, unpublished data). The height of dwf12 heterozygous plants is intermediate between the homozygote and wild type, suggesting that the mutation is semidominant. Unit bar = 2 cm. In the inset, one set each of the adaxial side of a cauline leaf, a rosette leaf, and a silique of Ws-2 wild type, dwf12-1D, dwf12-2D, dwf12-1D/+, and dwf12-2D/+ are shown. Note the extreme downward curling of the leaves. Siliques of the homozygous dwf12 mutants are almost completely sterile and contain few seeds.
The phenotypes of the two alleles were further characterized by morphometric analysis. The total height of aerial parts and the length of pedicels and siliques were all measured and found to be greatly decreased (Table I). Interestingly, the length of these plant organs was noticeably shorter in dwf12-2D mutants, suggesting that dwf12-2D is a more severe allele compared with dwf12-1D. Apical dominance, judging by the number of inflorescences, is increased at 4 weeks of age, but during its prolonged development, dwf12 mutants produced additional inflorescences as a result of decreased apical dominance.
Table I.
Morphometric analysis of the key characteristics of the dwf12 mutants relative to the Ws-2 wild type
| Parameter | Wild Type | dwf12-1D | dwf12-2D | dwf12-2D/DWF12 |
|---|---|---|---|---|
| mm | ||||
| Height | 399.9 ± 20.6 | 88.6 ± 10.4 | 57.5 ± 11.3 | 252.5 ± 27.3 |
| Pedicel | 12.2 ± 1.4 | 3.9 ± 0.6 | 3.7 ± 0.8 | 9.1 ± 1.6 |
| Silique | 12.6 ± 2.6 | 2.9 ± 0.3 | 2.5 ± 0.5 | 6.9 ± 2.8 |
| Petiole | 20.0 ± 1.5 | na | na | 6.0 ± 1.1 |
| Leaf blade length | 10.0 ± 0.9 | na | na | 10.6 ± 2.1 |
| Leaf blade width | 11.6 ± 1.0 | na | na | 11.0 ± 1.1 |
| No. of inflorescences | 3.8 ± 0.8 | 1.6 ± 0.8 | 1.8 ± 0.7 | 3.3 ± 0.9 |
| Branches on a 1° inflorescence | 2.3 ± 0.6 | 2.6 ± 0.5 | 1.9 ± 0.6 | 3.0 ± 0.6 |
All the characteristics were measured at 4 weeks of age. An average (>15) is shown with the sd. na, Not analyzed.
To rule out the possibility that the lack of rescue of dwf12 mutants in our initial BR application experiment was because of a lower BL dose than required, we tested three different concentrations of epi-BL. The wild-type seedlings responded dramatically to both 10−8 and 10−7 m epi-BL in that the hypocotyls and petioles elongate, whereas root growth was stunted (Fig. 2A). Similarly, seedlings of the BR-deficient mutant dwf4-1 were rescued to wild-type phenotype with 10−8 and 10−7 m epi-BL. In contrast, both dwf12-1D and bri1-5 (a weak bri1 allele) failed to show noticeable responses to epi-BL, even at the highest concentration of 10−7 m. These results indicate that the dwf12 mutants are defective in a step of the BR signal transduction pathway.
Figure 2.
A, Dose response of wild type, dwf4-1, bri1-5, and dwf12-1D to epi-BL. Different doses of epi-BL are C, control; −9, 10−9 m; −8, 10−8 m; and −7, 10−7 m. A wild-type seedling at 10−7 m shows typical BR responses, such as elongated hypocotyls and petioles as well as shortened root length. In addition, the BR biosynthetic mutant dwf4-1 is fully rescued to a wild-type phenotype at a concentration as low as 10−8 m, showing expanded cotyledons and an elongated hypocotyl. However, dwf12-1D seedlings did not respond to epi-BL regardless of the concentrations, confirming that dwf12-1D is defective in sensing or downstream signaling. Similar insensitivity is shown also in the bri1-5 mutant. Unit bar = 1 cm. B, Quantitative analysis of the hormone dose response tests. Root growth inhibition of wild type is decreased linearly and proportionally to the epi-BL concentration. In contrast, both dwf12-1D and bri1-5 are significantly insensitive to epi-BL at all concentrations tested. The two signaling mutants also show altered responses to abscisic acid (ABA) and auxin in that they are more sensitive to ABA but slightly resistant to auxin. Percent root length represents the ratio of the root length grown on BL-supplemented media over the root length grown on the control media containing the same amount of 95% (v/v) ethyl alcohol used to dilute BL from a 4 mm stock solution.
To quantitatively understand the responsiveness of dwf12 mutants to epi-BL and other phytohormones, we measured the root growth after treating the seedlings with epi-BL, ABA, and auxin (2,4-dichlorophenoxyacetic acid [2,4-D]). As shown in Figure 2B, the root growth inhibition in wild type was decreased linearly and proportionally to the epi-BL concentration. In contrast, both dwf12-1D and bri1-5 mutants are significantly less sensitive to different epi-BL concentrations. In response to all the concentrations tested, root growth inhibition in bri1-5 was not noticeable (Fig. 2). However, dwf12-1D was sensitive to this treatment in that root growth was inhibited about 10% in response to 10−8 and 10−7 m epi-BL, and the root length was approximately one-half of the control size at 1 μm. By definition, the control size is the length of roots grown on media that contain the same amount of 95% (v/v) ethyl alcohol used to dilute BL from a 4 mm stock solution. It has been shown previously that root growth of BR mutants is hypersensitive to inhibition by exogenous application of ABA (Ephritikhine et al., 1999). Similarly, the root growth of wild type, bri1-5, and dwf12-1D was unchanged, 23% decreased, and 57% decreased, respectively, compared with their controls in response to 0.5 μm ABA. The inhibition by ABA was more pronounced in bri1-5 than in dwf12-1D. When the two insensitive mutants were treated with a synthetic auxin 2,4-D (0.5 μm), their root growth was greatly inhibited like wild-type seedlings (Fig. 2B); however, it is noteworthy that dwf12-1D and bri1-5 are slightly resistant to the auxin treatment as compared with wild type.
dwf12 Is Defective in Feedback Regulation of BR Biosynthesis
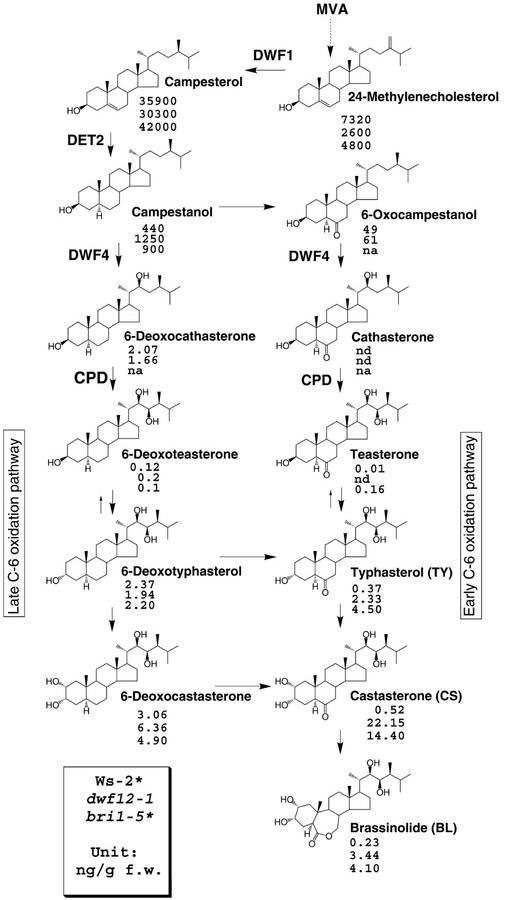
Previously, we have shown that BRs accumulate relative to wild type in direct correlation with the severity of the bri1 allele (Noguchi et al., 1999). The inability to perceive BRs in bri1 mutants also results in increased steady-state levels of BR biosynthetic gene transcripts (Noguchi et al., 1999). These and other data (Mathur et al., 1998) led us to propose a model in which a transcriptional feedback loop downstream of BRI1 regulates BR levels. If DWF12 is a component of the BR signaling pathway, dwf12 mutants might also be defective in feedback regulation and accumulate BRs. We collected tissue from dwf12-1D and analyzed the BR levels and found that dwf12-1D plants accumulate significant quantities of BRs (Fig. 3). The degree of accumulation is more pronounced in downstream compounds in the biosynthetic pathway. The levels of the end product BL and the penultimate compound CS were 15- and 43-fold higher as compared with wild-type levels, respectively. The bri1-5 mutant, which shows similar inflorescence height to dwf12, displayed an 18- and 27-fold increase of BL and CS levels, respectively (Noguchi et al., 1999; Fig. 3).
Figure 3.
Endogenous levels of BRs in wild-type (Ws-2), dwf12-1D, and bri1-5 plants. Similar to Arabidopsis bri1 plants, dwf12-1D accumulates significant amounts of BRs, especially typhasterol (TY), castasterone (CS), and BL. BR biosynthetic pathways with major intermediates and enzyme names are shown with their chemical structures. nd, Not detected; na, not analyzed; unit = ng g fresh weight−1.
DWF12 Encodes a Member of the GSK3 Gene Family
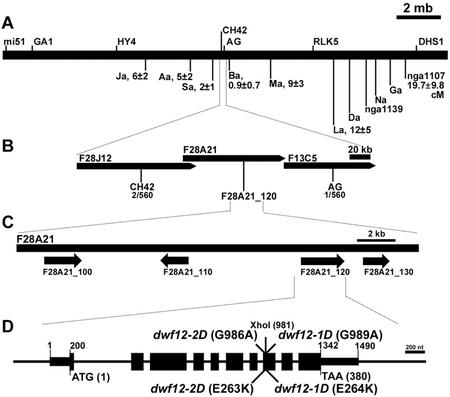
To understand the role of DWF12 in BR signaling, we isolated the DWF12 gene using conventional map-based cloning techniques. First, we found that dwf12 mutations are located between the AG and CH42 markers in the middle of chromosome 4 (Fig. 4, A–D). The Arabidopsis Genome Initiative has sequenced and annotated this region; thus, we took advantage of this information in selecting candidate genes for dwf12. The gene in which we found the G to A transition mutations, typical of EMS-induced changes, was a GSK/shaggy-like kinase (F28A21.120). Both dwf12-1D and dwf12-2D carried base changes in this gene (Fig. 4D). Interestingly, the two mutations were only 3 bp apart: dwf12-2D possessed a G to A transition at nucleotide position 986, whereas dwf12-1D had a G to A change at nucleotide position 989 (Fig. 4D). These two mutations resulted in substitution of an acidic Glu (GAA) to a basic Lys (AAA) residue in two adjacent amino acids (263 [dwf12-2D] and 264 [dwf12-1D], respectively; Fig. 4D).
Figure 4.
Map-based cloning of DWF12. A, Chromosome 4 with major genetic markers. In addition to previously available markers, we have developed novel simple sequence length polymorphism (SSLP) markers, including Ga, Na, Da, La, Ma, Ba, Sa, Aa, and Ja (Table III). B, Localization of the mutation between CH42 and AG markers. The number of recombinants found with CH42 and AG and dwf12-1D is two and one of 560 examined chromatids, respectively. C, Geography around the DWF12 gene annotated as F28A21.120 by the Arabidopsis Genome Initiative. D, The schematic of DWF12 with mutations. The gene consists of 10 exons and nine introns. The two dwf12 mutations are in exon 8, changing consecutive Glu residues to Lys. The dwf12-2D mutation abolishes an XhoI site (CTCGAG to CTCGAA). Schematics are drawn to the scales that are depicted on the top right of each diagram.
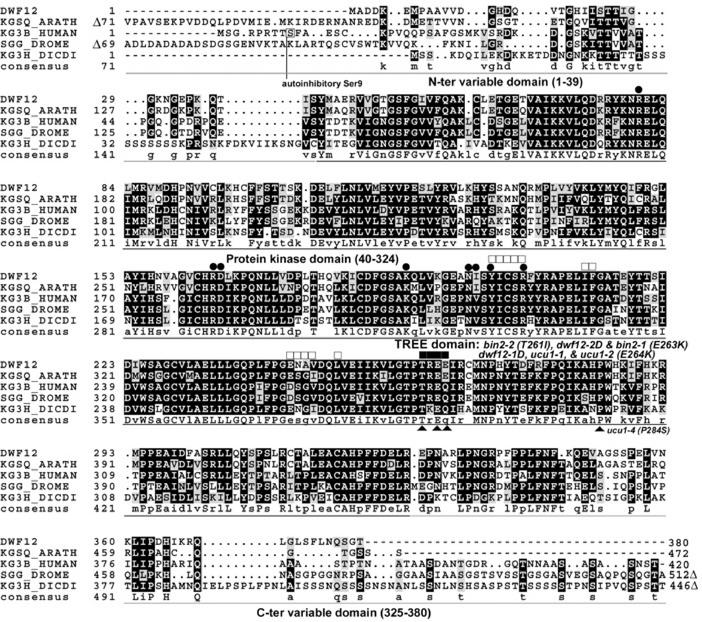
To learn more about the role of DWF12 as well as the importance of the changes in these two amino acids, we conducted protein sequence analysis. First, we searched the GenBank and SwissProt databases, and found a group of kinases belonging to the GSK3 family as the most similar sequences to DWF12. Multiple sequence alignments revealed that the DWF12 protein sequence could be subdivided into three discrete domains: (a) N-terminal variable domain (1–39), (b) conserved kinase domain (40–324), and (c) C-terminal domain (325–380). When these domains were individually compared with the corresponding domains of human GSK3β, the sequence identity was 44%, 71%, and 49%, respectively. The lower sequence identity at the N-terminal domain was attributable partly to the short length of this domain in DWF12, only 39 amino acids, but 53 in the human GSK3β (Fig. 5). Arabidopsis has 10 copies of the GSK3-like genes, named AtSKs (Arabidopsis shaggy-like kinases) after their similarity to D. melanogaster shaggy-like kinase (Dornelas et al., 1998). The Arabidopsis AtSK sequences deviate substantially in the length of their N-terminal domains. The total number of amino acids ranged from 380 (DWF12) to as long as 472 (KGSQ_ARATH). The length of the DWF12 N-terminal domain is the shortest when compared with GSK3 from other organisms including human, D. melanogaster, and D. discoideum (Fig. 5).
Figure 5.
Multiple sequence alignment of two Arabidopsis GSK3-like protein and GSK3s from human, D. melanogaster, and D. discoideum. DWF12 (GenBank accession no. AY157149) and KGSQ_ARATH (Q96287), possessing the longest amino acid sequence among 10 Arabidopsis GSK3-like proteins, were compared with GSK3β proteins from human (P49841), D. melanogaster (SGG_DROME, P18431), and D. discoideum (KG3H_DICDI, P51136). Alignment was performed using the PileUp program of the Genetics Computer Group software with a gap creation penalty of 3 and a gap extension penalty of 1. The aligned sequences were further annotated using BOXSHADE and Photoshop programs. Annotations are based on structural characterization described by Dajani et al. (2001). Black circles indicate the residues involved in active site formation. White squares identify the residues that are involved in homodimerization. The TREE domain was identified to be a putative Thr phosphorylation site by caseine kinase II, and the two dwf12 mutations were located in this domain. Thick lighter bars delimit an N- or C-terminal variable domain, and a darker bar spans the protein kinase domain. Numbers in parentheses are based on the DWF12 protein sequence. Dashes and dots indicate gaps introduced to maximize alignment. Δ, Truncation of amino acid sequences for better comparison.
DISCUSSION
The phenotypes of the dwf12 mutants can be summarized by the statement that all organs are reduced in size. The specific characteristics examined in this research, such as plant height and the length of pedicels, siliques, petioles, hypocotyls, and roots, all are significantly shorter than wild type (Table I; Figs. 1 and 2). These morphological alterations are typical of BR biosynthetic or signaling mutants (Figs. 1 and 2). Thus, it is likely that the dwf12 mutants are defective in BR biosynthesis or signaling. One interesting exception in dwf12 is a typical leaf curling in the abaxial direction. Downward curling leaves are often found in mutants that are defective in auxin signaling, such as axr1 and axr2 (Lincoln et al., 1990; Timpte et al., 1992). These shared phenotypes between dwf12 and auxin mutants suggest that DWF12 also plays a role in auxin signaling in specific cell types.
Two lines of evidence strongly suggest that the dwf12 mutants are BR insensitive. First, a BR biosynthetic mutant dwf4-1 was rescued to wild-type phenotype at 10−8 and 10−7 m epi-BL (Fig. 2A), but these concentrations did not induce noticeable responses such as elongating hypocotyls and petioles from dwf12-1D and bri1-5. In addition, root growth inhibition by concentrated epi-BL in the dwf12-1D mutants was significantly less sensitive compared with wild type. Second, previously we have shown that bri1 mutants accumulate significant amounts of BRs (Noguchi et al., 1999). Similarly, here we reported that BRs also accumulate in dwf12-1D plants. Because increased BR signaling often accompanies decreased levels of BL through negative feedback regulation (Choe et al., 2001), it is likely that the mechanisms controlling endogenous BR levels act downstream of the two signaling components BRI1 and DWF12.
Recent structural determination of the human GSK3β revealed that the N-terminal domain (55 amino acids) plays a key role as an intramolecular inhibitory domain: Once Ser at position 9 from the N terminus (Ser-9) is phosphorylated, this domain occupies the active site and prevents access to substrates (Dajani et al., 2001; ter Haar et al., 2001). However, in DWF12, the first 15 amino acids, corresponding to the self-inhibitory domain of human GSK3β, are missing, suggesting that the auto-inhibitory role of the N-terminal region may be lacking in DWF12. Protein kinase B, which inactivates GSK3 by phosphorylation at Ser-9, has also not been found in plants. In addition, human GSK3β was found to be in an active kinase conformation if Ser-9 is not phosphorylated: The structure of the human GSK3β activation segment is completely superimposed with that of the activated kinase ERK2-P2 (Dajani et al., 2001), and the active site is buried when two monomers form a dimer. Thus, dissociation into a monomer is required for activity of this protein. We found that the residues participating in dimerization are well conserved in DWF12 (white squares in Fig. 5). Based on these findings, we postulate that DWF12, which is lacking the inhibitory N-terminal domain, is a permanently active kinase that does not require prior activation, and dimerization and dissociation may affect the activity of this protein.
A search for possible protein modification sites in DWF12 using ScanProsite utility (http://www.expasy.ch/prosite/) revealed that the two acidic Glu residues at 263 and 264 could help the phosphorylation of the adjacent Thr residue at 261 (Thr-261), possibly by casein kinase II, because these amino acids are most similar to the CKII consensus phosphorylation site (http://www.expasy.org/cgi-bin/get-prodoc-entry?PDOC00006). Because this region, including Thr-261 to Glu-264 (TREE domain), is highly conserved in GSK3s, it likely plays a critical role. Interestingly, structural determination of the human GSK3β protein revealed that the TREE domain is exposed at dimerization, implying that access by another modifier like CKII is possible. Assuming that phosphorylation of Thr-261 is essential in the regulation of DWF12, the dwf12 alleles may have negative effects on phosphorylation at Thr-261, leading to the dominant nature of DWF12 mutant protein. Thus, phosphorylation of the TREE domain in DWF12 could replace the protein kinase B phosphorylation at Ser-9 as a negative regulatory event. Alternatively, the TREE domain may define an interaction domain with itself or with another protein, with the dwf12 mutations causing either stronger or reduced binding, resulting in a dominant phenotype. Our mapping and sequence data indicate that our dwf12 mutants are allelic with the recently identified BR-insensitive mutants bin2 (Li and Nam, 2002) and ucu1 (Perez-Perez et al., 2002). Remarkably, the six semidominant alleles are all missense mutations in the TREE domain (Table II), further emphasizing the importance of this domain in the regulation of BIN2/UCU1/DWF12.
Table II.
Alleles of Arabidopsis BIN2/UCU1/DWE12
| Allele | DNA Sequence Change | Amino Acid Change | Ecotype | Reference |
|---|---|---|---|---|
| bin2-1 | G989A (GAA→AAA) | E263K (=dwf12-2D) | Columbia (Col-0) | Li and Nam (2002) |
| bin2-2 | C981T (ACT→ATT) | T261I | Col-0 | Li and Nam (2002) |
| ucu1-1 | G986A (GAA→AAA) | E264K (=ucu1-2, dwf12-1D) | Landsburg erecta (Ler) | Perez-Perez et al. (2002) |
| ucu1-2 | G986A (GAA→AAA) | E264K (=ucu1-1, dwf12-1D) | Ler | – |
| ucu1-3 | C1049T (CCC→TCC) | P248S | Ler | – |
| dwf12-1D | G986A (GAA→AAA) | E264K (=ucu1-1, ucu1-2) | Wassilewskija-2 (Ws-2) | This paper |
| dwf12-2D | G989A (GAA→AAA) | E263K (=bin2-1) | Ws-2 | This paper |
To address the role of BIN2/UCU1/DWF12 in BR signaling, Li and Nam (2002) purified both a wild-type kinase and a kinase with the bin2-1 mutation, and found that the mutant kinase phosphorylated a substrate approximately 30% more than the wild-type kinase. These authors provide further support for the model that the BR insensitivity of the bin2/ucu1/dwf12 mutants is caused by an overly active kinase by generating plants expressing the bin2-1 mutant kinase under the control of its own promoter in addition to the two endogenous BIN2(+) copies. In these plants, there was a good correlation between RNA levels and the severity of the dwarf phenotype (Li and Nam, 2002). Reducing the levels of the BIN2/UCU1/DWF12 RNA can suppress the phenotype of a weak bri1 allele, providing further evidence that this gene encodes a negative regulator that acts downstream of BRI1 (Li and Nam, 2002).
These results indicate that BIN2/UCU1/DWF12 is a kinase of the GSK3 family with a role in BR signaling. Our evidence includes the observations that dwf12 mutants share characteristic BR dwarf phenotypes, including short stature, insensitivity to BRs, and the accumulation of BRs. Characterization of the DWF12 gene indicates that the encoded protein shows >70% sequence identity to the human GSK3β protein, except that DWF12 lacks the N-terminal auto-inhibitory domain containing the Ser-9 that is phosphorylated in GSK3β. Our model is that DWF12 is a naturally active kinase that functions as a repressor of BR signaling pathways. Because BIN2/UCU1/DWF12 has no targeting sequences, it is most likely located in the cytoplasm. This location predicts that BIN2/UCU1/DWF12 operates downstream of BRI1, but there is no evidence yet that BRI1 interacts with or phosphorylates BIN2/UCU1/DWF12. The TREE domain does not match a phosphorylation site consensus derived from in vitro studies of BRI1 (Oh et al., 2000).
Recently, Yin et al. (2002) provided evidence that BIN2 physically interacts with a novel type of nuclear protein named BES1 (bri1-EMS-suppressor). Furthermore, Wang et al. (2002) also showed that a BES1-like nuclear protein BZR1 (brassinazole resistant) acts downstream of BIN2 and plays a role as a transcriptional inhibitor of BR biosynthetic genes. In the future, it will be important to elucidate the signaling components that regulate BIN2, and to identify additional components downstream of BIN2 that lead to the transcription of BR biosynthetic and BR response genes.
MATERIALS AND METHODS
Plant Materials and Endogenous BR Analysis
More than 50,000 M2 seeds of Arabidopsis ecotype Ws-2 ecotype were planted on presoaked Metromix (350, Grace Sierra, Milpitas, CA). The seeds were cold treated (4°C) for 3 d and transferred to a 16-:8-h-light (200 μmol m−2 s−1):dark cycle (22°C and 21°C, and 70%–90% humidity) until the plants reached maturity. Forty-three mutants were selected based on their characteristic dwarf phenotype. BR treatment and genetic mapping of these dwarfs showed that most of these were found to be alleles of existing loci, and have been reported previously (Choe et al., 1999a). However, among these mutants, five were not rescued to the wild-type phenotype after topical application of BR, and were designated as insensitive mutants. Three of the five were alleles of bri1 (Noguchi et al., 1999). Two additional mutants were out-crossed to Ws-2 wild type several times to dilute out background mutations, and also crossed to the Col-0 ecotype to obtain a mapping population. The mapping population consisted of 280 homozygous F2 dwarf mutants.
BR dose response tests were performed according to the method described in Choe et al. (1998). In brief, cold-treated seeds (3 d) of Ws-2 wild type, dwf4-1, bri1-5, and dwf12-1D were cocultured in 1× liquid Murashige and Skoog media supplemented with designated concentrations of epi-BL:0 (control), 10−9, 10−8, and 10−7 m. After incubation for 3 d, the seedlings were recovered from the culture media, their root and hypocotyl lengths were measured, and photographs were taken using representative seedlings for each concentration.
In the analysis of root growth inhibition in response to epi-BL, ABA (Gibco-BRL, Cleveland), and 2.4-d, seeds of wild type, dwf12-1D, and bri1-5 were germinated on Murashige and Skoog solid media for 4 d, and transferred to plates supplemented with 10−9, 10−8, 10−7, and 10−6 molar concentrations of epi-BL, 0.5 μm ABA, and 2,4-D. Seedlings on the agar plates were grown vertically for 7 more d in the light and their root length was measured.
Endogenous levels of BRs in 4-week-old dwf12-1D mutant plants were determined using gas chromatography/mass spectrometry. Procedures for gas chromatography/mass spectrometry were described previously (Choe et al., 1999b), and the endogenous BR levels of bri1-5 were taken for comparison from Noguchi et al. (1999).
Map-Based Cloning
The approximate location of dwf12 in the Arabidopsis genome was determined by testing genomic DNAs from 24 F2 homozygous dwf12 plants from a mapping population with SSLP markers distributed on the five Arabidopsis chromosomes. Markers included nga59, nga280, and nga111 from chromosome 1, nga1145 and nga168 from chromosome 2, nga172, nga162, and nga6 from chromosome 3, nga8, nga1139, and nga1107 from chromosome 4, and nga151, nga76, and nga129 from chromosome 5. PCR was performed as described by Bell and Ecker (1994). In the course of fine mapping dwf12, we developed new sets of SSLP markers. The novel SSLP markers were developed based on the prediction available from the Arabidopsis sequence table (http://www.Arabidopsis.org/cgi-bin/maps/Seqtable.pl?chr=4). The new SSLP markers are shown in Table III. The names of the markers, orientation, sequences, corresponding bacterial artificial chromosome clones, and the size of the PCR products amplified using Col-0 DNA are included in the table. PCR products were run on 4% (w/v) agarose gels for >2 h for maximum separation of the polymorphic fragments.
Table III.
SSLP markers used in cloning of DWF12
| Name | Orientation | Sequence (5′ to 3′) | Bacterial Artificial Chromosomea | Sizeb |
|---|---|---|---|---|
| bp | ||||
| Ga | GaF | ctccaacttgttgaaacccgt | AP22 | 226 |
| GaR | aacaatctaactattgatggtcttt | |||
| Na | NaF | gggattttcacgatgtatttaattggt | F23E12 | 304 |
| NaR | aggtgttttaggtggcttacac | |||
| Da | DaF | gcaagtaattaatacatttatcaactc | F8B4 | 107 |
| DaR | gcgttaatagtgtgtatgttgtaa | |||
| La | LaF | aagaagctcaagcggtggaaat | T10C21 | 287 |
| LaR | agaatgtccagggcagccat | |||
| Ma | MaF | ggttcagtgttactatgtaccaag | F16G20 | 270 |
| MaR | gagaccagtggcctaagtagat | |||
| Ba | BaF | caaaatcgtggatgcacttacag | T18B16 | 295 |
| BaR | gtcttaccaataaatggttctctcat | |||
| Sa | SaF | gtgccttattctgactatagtca | FCA9 | 461 |
| SaR | gttacaggtacataatgtataggta | |||
| Aa | AaF | cgtcgctcttatatttgatggatt | FCA6 | 426 |
| AsR | tccatttgtagcatttccattaggt | |||
| Ja | JaF | caccacttttaagggcactaca | F8L21 | 358 |
| JaR | gatacatcaacatgctgtagaatccat |
The BAC clone name where markers are located.
PCR product size of the Col-0 ecotype.
The mutations in the two dwf12 alleles were detected and confirmed by sequencing genomic DNA from dwf12 homozygous mutants amplified using PCR. Primers used in the amplification of the genomic DNA and sequencing are, from 5′ to 3′, D12F2, gagggttttgagttctgagc; D12F3, gccaacatttcttacatctgct; D12F4, tttttcttgcctttgtttct; D12F5, tggctacaaaatcctcactg; D12R2, ggaagatctaacataacaaaggaagtaa; D12R3, gttacatggcggagcgagtt; D12R4, caagatagaagatacaagaaccgagaact; D12-OVF1, gtcgaattcgccatggctgatgataag; and D12-OVR1, gtctctagacccttttaagttccagattgattc.
The structure of the DWF12 gene was confirmed by comparing the cDNA sequence with that of the genomic DNA. cDNA was synthesized using D12-OVR1 as a primer for reverse transcriptase-PCR. Ten exons and nine introns were delimited by local alignment of the two sequences, and the schematic diagrams shown in Figure 4 were prepared using BestFit (Genetics Computer Group, Madison, WI), Vector NTI (InforMax, Bethesda, MD), and Photoshop (Adobe, San Jose, CA) software. Multiple sequence alignment was performed using the protein sequences available in Swiss-Prot protein knowledge bases. The His at 350 of the KG3B_HUMAN sequence was corrected to Leu according to Dajani et al. (2001) before comparison. The KGSQ_ARATH was chosen from 10 Arabidopsis GSK-like sequences because it has the longest total amino acid sequence. The five sequences were subjected to multiple sequence alignment using the PileUp program of GCG package. The conserved sequences were shaded using the BOXSHADE program (http://www.ch.embnet.org/software/BOX_form.html), and further annotation was carried out using Photoshop.
ACKNOWLEDGMENTS
We thank Amanda Ross, Brian Gregory, and Masayo Sekimoto for technical assistance, Laurence H. Pearl for comments on the structural importance of dwf12 mutations, and the sequencing facilities in the Arizona Research Laboratory at the University of Arizona (Tucson).
Footnotes
This work was supported by Ceres, Inc. (to S.C.), by the U.S. Department of Agriculture (grant no. 97–35304–4708), and by the Honors Undergraduate Research Grant Program and the Undergraduate Biology Research Program at the University of Arizona (grant to R.J.S.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010496.
LITERATURE CITED
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- Bianchi MW, Guivarc'h D, Thomas M, Woodgett JR, Kreis M. Arabidopsis homologs of the shaggy and GSK-3 protein kinases: molecular cloning and functional expression in Escherichia coli. Mol Gen Genet. 1994;242:337–345. doi: 10.1007/BF00280424. [DOI] [PubMed] [Google Scholar]
- Bienz M, Clevers H. Linking colorectal cancer to Wnt signaling. Cell. 2000;103:311–320. doi: 10.1016/s0092-8674(00)00122-7. [DOI] [PubMed] [Google Scholar]
- Choe S, Dilkes BP, Fujioka S, Takatsuto S, Sakurai A, Feldmann KA. The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22α-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell. 1998;10:231–243. doi: 10.1105/tpc.10.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Dilkes BP, Gregory BD, Ross AS, Yuan H, Noguchi T, Fujioka S, Takatsuto S, Tanaka A, Yoshida S et al. The Arabidopsis dwarf1 mutant is defective in the conversion of 24-methylenecholesterol to campesterol in brassinosteroid biosynthesis. Plant Physiol. 1999a;119:897–907. doi: 10.1104/pp.119.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Fujioka S, Noguchi T, Takatsuto S, Yoshida S, Feldmann KA. Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J. 2001;26:573–582. doi: 10.1046/j.1365-313x.2001.01055.x. [DOI] [PubMed] [Google Scholar]
- Choe S, Noguchi T, Fujioka S, Takatsuto S, Tissier CP, Gregory BD, Ross AS, Tanaka A, Yoshida S, Tax FE et al. The Arabidopsis dwf7/ste1 mutant is defective in the Δ7 sterol C-5 desaturation step leading to brassinosteroid biosynthesis. Plant Cell. 1999b;11:207–221. [PMC free article] [PubMed] [Google Scholar]
- Choe S, Tanaka A, Noguchi T, Fujioka S, Takatsuto S, Ross AS, Tax FE, Yoshida S, Feldmann KA. Lesions in the sterol Δ7 reductase gene of Arabidopsis cause dwarfism due to a block in brassinosteroid biosynthesis. Plant J. 2000;21:431–443. doi: 10.1046/j.1365-313x.2000.00693.x. [DOI] [PubMed] [Google Scholar]
- Clouse SD, Langford M, McMorris TC. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111:671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dajani R, Fraser E, Roe SM, Young N, Good V, Dale T, Pearl LH. Crystal structure of glycogen synthase kinase 3 beta: structural basis for phosphate-primed substrate specificity and autoinhibition. Cell. 2001;105:721–732. doi: 10.1016/s0092-8674(01)00374-9. [DOI] [PubMed] [Google Scholar]
- Decroocq-Ferrant V, van Weng J, Bianchi MW, de Vries SC, Kreis M. Petunia hybrida homologues of shaggy/zeste-white 3 expressed in female and male reproductive organs. Plant J. 1995;7:897–911. doi: 10.1046/j.1365-313x.1995.07060897.x. [DOI] [PubMed] [Google Scholar]
- Dornelas MC, Lejeune B, Dron M, Kreis M. The Arabidopsis SHAGGY-related protein kinase (ASK) gene family: structure, organization and evolution. Gene. 1998;212:249–257. doi: 10.1016/s0378-1119(98)00147-4. [DOI] [PubMed] [Google Scholar]
- Dornelas MC, van Lammeren AA, Kreis M. Arabidopsis thaliana SHAGGY-related protein kinases (AtSK11 and 12) function in perianth and gynoecium development. Plant J. 2000;21:419–429. doi: 10.1046/j.1365-313x.2000.00691.x. [DOI] [PubMed] [Google Scholar]
- Dornelas MC, Wittich P, von Recklinghausen I, van Lammeren A, Kreis M. Characterization of three novel members of the Arabidopsis SHAGGY-related protein kinase (ASK) multigene family. Plant Mol Biol. 1999;39:137–147. doi: 10.1023/a:1006102812280. [DOI] [PubMed] [Google Scholar]
- Ephritikhine G, Fellner M, Vannini C, Lapous D, Barbier-Brygoo H. The sax1 dwarf mutant of Arabidopsis thaliana shows altered sensitivity of growth responses to abscisic acid, auxin, gibberellins and ethylene and is partially rescued by exogenous brassinosteroid. Plant J. 1999;18:303–314. doi: 10.1046/j.1365-313x.1999.00454.x. [DOI] [PubMed] [Google Scholar]
- Friedrichsen DM, Joazeiro CA, Li J, Hunter T, Chory J. Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 2000;123:1247–1256. doi: 10.1104/pp.123.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Estelle M. Function of the ubiquitin-proteasome pathway in auxin response. Trends Plant Sci. 2000;25:133–138. doi: 10.1016/s0968-0004(00)01544-9. [DOI] [PubMed] [Google Scholar]
- Harwood AJ. Signal transduction: life, the universe and development. Curr Biol. 2000;10:R116–119. doi: 10.1016/s0960-9822(00)00307-9. [DOI] [PubMed] [Google Scholar]
- He Z, Wang ZY, Li J, Zhu Q, Lamb C, Ronald P, Chory J. Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science. 2000;288:2360–2363. doi: 10.1126/science.288.5475.2360. [DOI] [PubMed] [Google Scholar]
- Kim L, Kimmel AR. GSK3, a master switch regulating cell-fate specification and tumorigenesis. Curr Opin Genet Dev. 2000;10:508–514. doi: 10.1016/s0959-437x(00)00120-9. [DOI] [PubMed] [Google Scholar]
- Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- Li J, Nagpal P, Vatart V, McMorris TC, Chory J. A role for brassinosteroids in light-dependent development of Arabidopsis. Science. 1996;272:398–401. doi: 10.1126/science.272.5260.398. [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science. 2002;295:1299–1301. doi: 10.1126/science.1065769. [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH, Vafeados D, Chory J. BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 2001;127:14–22. doi: 10.1104/pp.127.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln C, Britton JH, Estelle M. Growth and development of the axr1 mutants of Arabidopsis. Plant Cell. 1990;2:1071–1080. doi: 10.1105/tpc.2.11.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Molnar G, Fujioka S, Takatsuto S, Sakurai A, Yokota T, Adam G, Voigt B, Nagy F, Maas C et al. Transcription of the Arabidopsis CPD gene, encoding a steroidogenic cytochrome P450, is negatively controlled by brassinosteroids. Plant J. 1998;14:593–602. doi: 10.1046/j.1365-313x.1998.00158.x. [DOI] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Yoshida S, Yuan H, Feldmann KA, Tax FE. Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 1999;121:743–752. doi: 10.1104/pp.121.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh MH, Ray WK, Huber SC, Asara JM, Gage DA, Clouse SD. Recombinant brassinosteroid insensitive 1 receptor-like kinase autophosphorylates on serine and threonine residues and phosphorylates a conserved peptide motif in vitro. Plant Physiol. 2000;124:751–766. doi: 10.1104/pp.124.2.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pay A, Jonak C, Bogre L, Meskiene I, Mairinger T, Szalay A, Heberle-Bors E, Hirt H. The MsK family of alfalfa protein kinase genes encodes homologues of shaggy/glycogen synthase kinase-3 and shows differential expression patterns in plant organs and development. Plant J. 1993;3:847–856. doi: 10.1111/j.1365-313x.1993.00847.x. [DOI] [PubMed] [Google Scholar]
- Perez-Perez JM, Ponce MR, Micol JL. The UCU1 Arabidopsis gene encodes a SHAGGY/GSK3-like kinase required for cell expansion along the proximodistal axis. Dev Biol. 2002;242:161–173. doi: 10.1006/dbio.2001.0543. [DOI] [PubMed] [Google Scholar]
- Szekeres M, Nemeth K, Koncz-Kalman Z, Mathur J, Kauschmann A, Altmann T, Redei GP, Nagy F, Schell J, Koncz C. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450 controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85:171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- ter Haar E, Coll JT, Austen DA, Hsiao HM, Swenson L, Jain J. Structure of GSK3β reveals a primed phosphorylation mechanism. Nat Struct Biol. 2001;8:593–596. doi: 10.1038/89624. [DOI] [PubMed] [Google Scholar]
- Timpte CS, Wilson AK, Estelle M. Effects of the axr2 mutation of Arabidopsis on cell shape in hypocotyl and inflorescence. Planta. 1992;188:271–278. doi: 10.1007/BF00216824. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature. 2001;410:380–383. doi: 10.1038/35066597. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Nakano T, Gendron J, He J, Chen M, Vafeados D, Yang Y, Fujioka S, Yoshida S, Asami T et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell. 2002;2:505–513. doi: 10.1016/s1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]