Abstract
We have developed a new T-DNA vector, pGA2715, which can be used for promoter trapping and activation tagging of rice (Oryza sativa) genes. The binary vector contains the promoterless β-glucuronidase (GUS) reporter gene next to the right border. In addition, the multimerized transcriptional enhancers from the cauliflower mosaic virus 35S promoter are located next to the left border. A total of 13,450 T-DNA insertional lines have been generated using pGA2715. Histochemical GUS assays have revealed that the GUS-staining frequency from those lines is about twice as high as that from lines transformed with the binary vector pGA2707, which lacks the enhancer element. This result suggests that the enhancer sequence present in the T-DNA improves the GUS-tagging efficiency. Reverse transcriptase-PCR analysis of a subset of randomly selected pGA2715 lines shows that expression of the genes immediately adjacent to the inserted enhancer is increased significantly. Therefore, the large population of T-DNA-tagged lines transformed with pGA2715 could be used to screen for promoter activity using the gus reporter, as well as for creating gain-of-function mutants.
Recent completion of the draft sequence for the rice (Oryza sativa) genome has resulted in an explosion of information on rice genes (Goff et al., 2002; Yu et al., 2002). The challenge for the post-sequencing era is to identify the biological functions for these genes. Of all the approaches used to discover gene function, the most direct is to disrupt the genes and analyze the consequences. Various methods have been developed in plants for this purpose. These include using ethyl methanesulfonate, fast-neutron treatment, or insertion of an element, such as a transposable element or T-DNA (Koornneef et al., 1982; Sundaresan, 1996; Krysan et al., 1999). Insertional mutagenesis has the advantage that the inserted element acts as a tag for gene identification. However, all gene disruption approaches also have some limitations. For example, it is difficult to identify the function of redundant genes, or of genes required in early embryogenesis or gametophyte development.
To overcome those limitations, modified insertional elements have been developed. One of these modified designs is the gene trap system that involves creating fusions between the tagged genes and a reporter gene, such as β-glucuronidase (gus) or green fluorescent protein (gfp; Sundaresan et al., 1995; Springer, 2000). This system provides a way of identifying novel genes based on their expression patterns. Insertion of the promoterless reporter not only destroys normal gene function but also activates expression of the reporter gene. Because expression levels can be monitored in heterozygote plants, the gene trap system is useful for studying the patterns of most plant genes, including essential genes that cause lethal mutations. This system is convenient for observing mutant phenotypes because reporter activation indicates the location, condition, and time of expression for the disrupted gene. In Arabidopsis, activation of reporter genes in a mutated population can be as high as 30% (Sundaresan et al., 1995). Chin et al. (1999) and Jeon et al. (2000a) have shown that at least 5% of the T-DNA and 10% of the transposed Ds elements become activated in various rice tissues, such as roots, leaves, flowers, and seeds. The promoterless gus gene in these gene trap systems contains an intron with multiple splicing acceptor and donor sites in each of three reading frames in front of the gus-coding region. These constructs allow GUS expression when the insertion occurs in either an exon or intron. As a consequence, expression is at high frequencies in plants transformed with such a gene trap system.
Activation tagging is another method for complementing conventional insertional mutagenesis. This system uses T-DNA or a transposable element containing multimerized cauliflower mosaic virus (CaMV) 35S enhancers (Hayashi et al., 1992; Suzuki et al., 2001). Because enhancers can function in either orientation and at a considerable distance from the coding regions, they can cause transcriptional activation of nearby genes, resulting in dominant gain-of-function mutations. Such gene activations may produce novel phenotypes that identify important genes that are either redundant members of a gene family or are essential for survival. This tagging system has been used for cloning several genes in Arabidopsis (Kakimoto, 1996; Kardailsky et al., 1999; Borevitz et al., 2000; Ito and Meyerowitz, 2000; ; van der Graaff et al., 2000; Huang et al., 2001; Zhao et al., 2001), petunia (Petunia hybrida; Zubko et al., 2002), and Madagascar periwinkle (Catharanthes roseus; van der Fits and Memelink, 2000; van der Fits et al., 2001).
Weigel et al. (2000) have characterized over 30 dominant morphological mutants with various phenotypes, at a frequency of approximately one in 1,000. Analysis of a subset of the mutants has shown that the tagging vector causes overexpression of the gene immediately adjacent to the inserted enhancers. With a different type of gain-of-function screen, using a Ds element carrying the CaMV 35S enhancers, Wilson et al. (1996) found four dominant mutants among 1,100 lines analyzed. Recently, an activation tagging method was developed that incorporates the En-I maize (Zea mays) transposon system. With this system, Marsch-Martinez et al. (2002) examined 2,900 insertions and found 31 dominant mutants, a frequency of about 1%. Activation tagging has also been employed as a novel approach for isolating suppressor mutants of a known phenotype (Neff et al., 1999; Lee et al., 2000; Li et al., 2001, 2002).
In Arabidopsis containing an activation-tagging vector, the CaMV 35S enhancers often cause the endogenous expression pattern to be heightened, compared with plants in which the complete CaMV 35S promoter drives a cDNA (Neff et al., 1999; Weigel et al., 2000). This feature would confirm that the phenotype resulting from such enhancement more likely reflects the normal role of the activated gene rather than a situation in which ectopic overexpression might produce the phenotype. An alternative approach could be to use either a tissue-specific promoter to misexpress a gene in a specific tissue, or an inducible promoter to overexpress a gene at a specific time and under certain conditions (Matsuhara et al., 2000; Zuo et al., 2002).
Although the activation-tagging system has been widely used in Arabidopsis, its usefulness has not been demonstrated in monocot plants. In this study, we investigated the possibility of using multimerized 35S enhancer elements for activation tagging of rice genes.
RESULTS
Examination of the 35S Enhancer Element in Rice
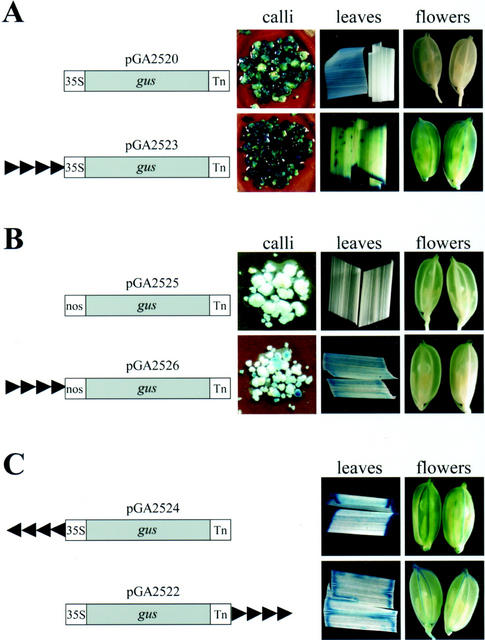
To examine whether the CaMV 35S enhancer elements function in rice, we placed the enhancer sequence upstream of the minimal promoter-gus fusion molecules and introduced this construct into plants. In the transgenic plants transformed with the minimal promoters (pGA2520 and pGA2525), we observed only mild GUS staining from the calli, and no significant GUS staining from the leaves or flowers (Fig. 1). Transformation with vectors pGA2523 and pGA2526, however, revealed that the 35S enhancer sequence was able to increase the level of gene expression over that found with the minimal promoters. Specifically, the presence of the tetramerized 35S enhancers upstream of the CaMV 35S minimal promoter or minimal nos promoter enhanced expression, not only in the calli but also in the leaves and flowers (Fig. 1, A and B).
Figure 1.
Schematic diagrams of the tetramerized CaMV 35S enhancer element constructs and expression patterns of rice calli and plants expressing the gus gene. Left, Plasmid names and structures of the gus fusion constructs. Right, GUS expression patterns of the T1 transgenic calli and plants. A, The minimal 35S promoter (−90) fused to the gus-coding region that was followed by the nos terminator (Tn). B, The minimal nos promoters (−101) fused to the gus-coding region that was followed by the nos terminator (Tn). C, Examination of the inverse orientation of the 35S enhancer elements upstream of the 35S minimal promoter or downstream of the nos terminator. Arrowheads indicate direction of the tetramerized CaMV 35S enhancer elements.
To further evaluate the enhancing activity of the 35S enhancers, we used vector pGA2524 (with the enhancer placed upstream of the minimal 35S promoter, in the reverse direction) and vector pGA2522 (with the enhancer sequences downstream of the nos terminator). In plants transformed with either of those vectors, enhanced GUS activities were detected in the leaves and flowers of the transformants (Fig. 1C). This is consistent with the observations of Fang et al. (1989), who reported that, in dicotyledonous plants, the 35S enhancer element functioned both upstream and downstream of a gene, in either orientation. Our results demonstrate that the CaMV 35S enhancer sequence can be used as an enhancer element for performing activation tagging in rice.
Vector Construction and Production of T-DNA-Tagged Transgenic Rice Plants
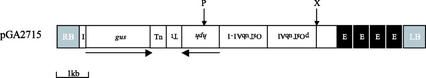
Plasmid pGA2715 (Fig. 2) contained the promoterless gus reporter gene with an intron and multiple splicing donors and acceptors immediately next to the right border. Multimerized transcriptional enhancers from the CaMV 35S promoter were inserted next to the left border. The hygromycin-resistant selectable marker gene, with the rice alpha tubulin (OsTubA1-1) promoter, was placed between the gus gene and the 35S enhancer. Thus, this vector can be used to achieve both gene trapping and activation tagging. The other binary vector, pGA2707, resembles pGA2715 except for its lack of enhancer elements. Using Agrobacterium tumefaciens-mediated rice transformation (Lee et al., 1999), we have now produced 13,450 fertile transgenic plants transformed with pGA2715, and 20,810 fertile transgenic plants with pGA2707.
Figure 2.
Map of the T-DNA region of tagging vector pGA2715. RB and LB in the gray bar represent the right and left borders of T-DNA, respectively. I, OsTubA1 intron 2 carrying three putative splicing acceptor and donor sites; Tn, nos terminator; Tt, OsTubA1 terminator; hph, hygromycin phosphotransferase gene; OsTubA1-1, the first intron of OsTubA1; E, enhancer element of CaMV 35S promoter; P, PstI site; X, XhoI site.
Gene Trapping Using the gus Reporter
To assess the efficiency of gene trapping, we examined GUS expression patterns in various organs of both primary transformants and their progeny. In Table I, a summary of GUS assay results from 6-d-old T2 seedlings is presented. Of 2,990 pGA2707 lines, 194 (6.5%) displayed GUS expression; among these, 158 lines (5.3%) were GUS positive in the mature seeds (including endosperm, scutellum, and seed coat). Likewise, 85 lines (2.8%) were GUS positive in the roots and 115 (3.8%) in the shoots (including the shoot apical meristems, coleoptiles, leaf blades, and sheaths). The frequency of preferential expression in seeds, roots, and shoots was 1.5% (45 lines), 0.2% (six lines), and 0.7% (21 lines), respectively. In contrast, 2.3% (70 lines) expressed the gus gene in all organs at the seedling stage.
Table I.
Frequencies of GUS expression in transgenic rice seedlings
| Organ | pGA2707 Vector
|
pGA2715 Vector
|
||||||
|---|---|---|---|---|---|---|---|---|
| Stained lines | Preferential | Stained lines | Preferential | |||||
| n | % | n | % | n | % | n | % | |
| Mature seeds | 158 | 5.3 | 45 | 1.5 | 359 | 9.3 | 91 | 2.4 |
| Roots | 85 | 2.8 | 6 | 0.2 | 241 | 6.3 | 14 | 0.4 |
| Shoots | 115 | 3.8 | 21 | 0.7 | 404 | 10.5 | 78 | 2.0 |
| Total | 194 | 6.5 | 72 | 2.4 | 514 | 13.4 | 183 | 4.8 |
A total of 2,990 lines from pGA2707-tagged plants and 3,842 lines from pGA2715-tagged plants were examined at the T2 seedling stage.
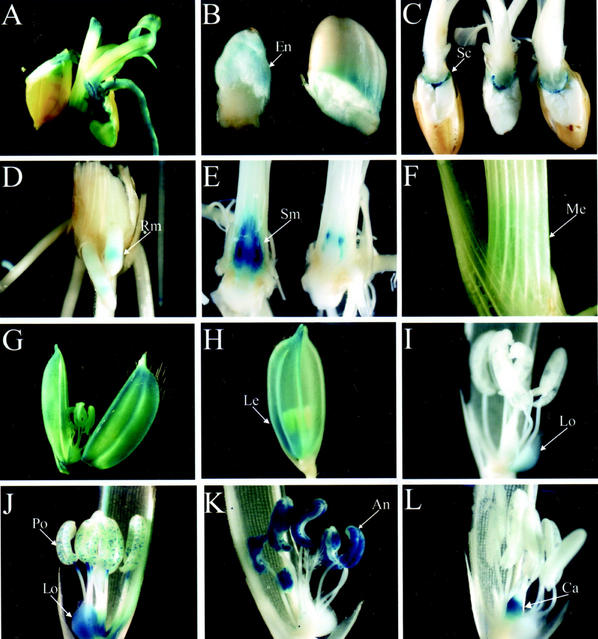
For the pGA2715-transformed lines, the frequency of GUS-positive lines was higher in all organs (Table I). Of the 3,842 lines examined, 514 (13.4%) were GUS positive. Among them, 359 lines (9.3%) were positive in the mature seeds, 241 (6.3%) in the roots, and 404 (10.5%) in their shoots. The frequency of preferential expression in seeds, roots, and shoots was 2.4% (91 lines), 0.4% (14 lines), and 2.0% (78 lines), respectively. A total of 173 lines (4.5%) expressed the reporter in all seedling organs. The frequency of GUS expression in the pGA2715 plants was about 2 times higher than that in the pGA2707 plants, not only overall and at the organ level, but also at the levels for ubiquitous (Fig. 3A) and preferential expression. Figure 3, B through F, provides examples of expression in specific organs at the seedling stage.
Figure 3.
Analysis of GUS activity in transgenic rice plants. A, Line 1B-05504 exhibiting GUS activity in all seedling organs. B, Line 1A-10540 showing GUS activity in the endosperm. C, Line 1A-10620 showing GUS activity in scutellum. D, Line 1A-10919 with root meristem-specific GUS expression. E, Line 1A-10721 displaying preferential GUS activity in the shoot apical meristem. F, Line 1A-10601 exhibiting mesophyll cell-specific GUS staining in the sheath. G, Line 1A-24951 showing GUS activity in all floral organs. H, Line 1B-05625 with GUS activity in lemma. I, Line 1A-25114 with GUS activity in lodicules. J, Line 1B-24512 with GUS activity in pollen and lodicules. K, Line 1A-12905 displaying strong GUS activity in anthers. L, Line 1A-11513 exhibiting carpel-specific GUS expression. An, Anthers; Ca, carpel; En, endosperm; Le, lemma; Lo, lodicules; Me, mesophyll; Po, pollens; Rm, root meristem; Sc, scutellum; Sm, shoot apical meristem.
We also screened the mature flowers of primary transgenic plants to identify lines that were GUS positive, and found expression in 4.7% of the pGA2707 lines (526/11,189) and 9.4% of the pGA2715 lines (515/5,489). The frequencies of GUS expression in various floral organs, e.g. rachillae, pedicels, paleae/lemmae, lodicules, stamens, and carpels, are summarized in Table II. As with the seedlings, the GUS-positive frequency for floral organs in the pGA2715 lines was about twice as high as that in the pGA2707 lines. These results imply that the multimerized CaMV 35S enhancers elevate the expression levels of the gus reporter gene located in the same T-DNA, thereby increasing the efficiency of gene trapping.
Table II.
Frequencies of GUS expression in mature flowers of transgenic rice plants
| Organ | pGA2707 Vector
|
pGA2715 Vector
|
||||||
|---|---|---|---|---|---|---|---|---|
| Stained lines | Preferential | Stained lines | Preferential | |||||
| n | % | n | % | n | % | n | % | |
| Rachilla | 225 | 2.0 | 1 | 0.01 | 186 | 3.4 | 2 | 0.04 |
| Pedicel | 201 | 1.8 | 10 | 0.09 | 248 | 4.5 | 11 | 0.20 |
| Palea/lemma | 329 | 2.9 | 17 | 0.15 | 370 | 6.7 | 30 | 0.55 |
| Lodicules | 224 | 2.0 | 1 | 0.01 | 117 | 2.1 | 2 | 0.04 |
| Stamen | 322 | 2.9 | 58 | 0.52 | 211 | 3.8 | 32 | 0.58 |
| Carpel | 279 | 2.5 | 17 | 0.15 | 156 | 2.8 | 11 | 0.20 |
| Total | 526 | 4.7 | 104 | 0.93 | 515 | 9.4 | 88 | 1.60 |
A total of 11,189 lines from pGA2707-tagged plants and 5,489 lines from pGA2715-tagged plants were examined at the flowering stage in the T1 generation.
Enhancing Gene Expression by Activation Tagging
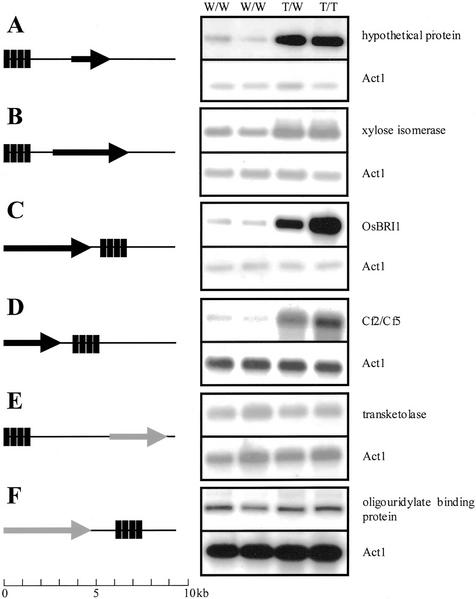
To determine the action of the multimerized CaMV 35S enhancers at the molecular level, we analyzed expression of the genes adjacent to the inserted enhancer elements. Using inverse PCR on 100 lines, we isolated 71 sequences flanking the T-DNA. These isolated sequences were examined by comparing them with entries in publicly available DNA databases, and the sequences were then annotated for their identifying open reading frames. Among them, 28 insertions (39.4%) occurred in the intragenic regions, whereas 43 (60.6%) were located in the intergenic regions. Insertions of enhancer elements within 4.5 kb upstream or downstream of the nearest open reading frame occurred in 12 and nine lines, respectively. We regarded these 21 lines (29.6%) as candidates for activation tagging.
Ten candidate lines were randomly selected and examined for the expression levels of genes near the enhancers. We planted 15 progeny per line and determined their genotypes by PCR analysis, using a gene-specific primer and a T-DNA primer (data not shown). The expression levels for candidate genes in the leaves of 10-d-old seedlings were then examined via reverse transcriptase (RT)-PCR, using gene-specific primers. Although RT-PCR products were obtained from eight lines, the remaining two lines showed no expression in the leaves. Among the lines examined, four showed that the levels of product from the gene near the enhancers were significantly increased in heterozygote and homozygote plants, compared with those in the wild-type tissue (Fig. 4). In those lines, the distance between the enhancers and the overexpressed gene ranged from 1.5 to 4.3 kb and the enhancer sequence was found both upstream and downstream of the target genes. Therefore, we could confirm that the tetramerized 35S enhancer sequence acted as a true enhancer in rice (see also Fang et al., 1989).
Figure 4.
Mapping the insertion position of the 35S enhancer element and RT-PCR analyses of the expression level of tagged genes. Left, Schematic representations of the 35S enhancer elements relative to nearby genes. Black bars indicate the CaMV 35S enhancers. The nearby gene is indicated with an arrow pointing in the direction of transcription. Right, RT-PCR analyses of the expression pattern of genes near the CaMV 35S enhancers. RNAs were isolated from leaves. Rice Act1 transcript was amplified as a control. A, Line 1B-00224 was tagged 2.0 kb upstream of a hypothetical gene. B, Line 1B-03509 was tagged 0.7 kb upstream of a gene homologous to Xyl isomerase. C, Line 1B-00107 was tagged 0.3 kb downstream of the OsBRI1 gene. D, Line 1B-04417 was tagged 0.7 kb downstream of a gene homologous to Cf2/Cf5. E, Line 1B-03714 was tagged 3.5 kb upstream of a gene homologous to transketolase. F, Line 1B-04240 was tagged 1.0 kb downstream of a gene homologous to oligouridylate binding protein. W/W, Wild-type lines; W/T, heterozygote lines for T-DNA insertion; T/T, homozygote lines for T-DNA insertion.
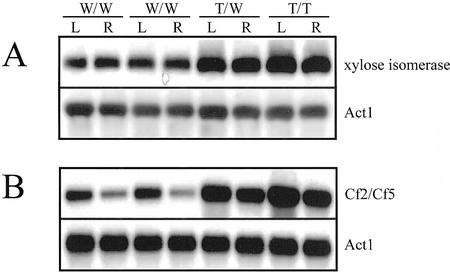
Neff et al. (1999) and Weigel et al. (2000) have reported that, although the CaMV 35S enhancers used with the activation-tagging vector in Arabidopsis studies typically do not lead to constitutive expression of the target genes, they do enhance endogenous expression. Therefore, to investigate whether the same is true for rice, we compared the expression profiles of the Xyl isomerase and Cf2/Cf5 genes in wild-type plants versus activation-tagged lines. In the Xyl isomerase-tagged lines, the gene was expressed at equivalent levels in both roots and leaves, as was also the case in our wild-type plants (Fig. 5A). Furthermore, in the Cf2/Cf5-tagged lines, expression was higher in the leaves than in the roots, which was similar to the pattern observed in wild type (Fig. 5B). These results suggest that the 35S enhancer sequence increases expression of the nearby gene while maintaining the original expression pattern.
Figure 5.
RT-PCR analysis of rice Xyl isomerase and Cf2/Cf5 gene expression. RNAs were extracted from roots (R) and leaves (L) of 10-d-old transgenic seedlings. The rice Act1 gene was used as a control. W/W, Wild-type lines; W/T, heterozygote lines for T-DNA insertion; T/T, homozygote lines for T-DNA insertion.
DISCUSSION
We have reported here the development of a novel T-DNA tagging system that can be used for both gene trapping and activation tagging. Our binary vector pGA2715 is constructed in such a way that it cannot only facilitate identification of genes through GUS-staining patterns, but also activate transcription of neighboring genes by the tetramerized CaMV 35S enhancers. So far, we have produced 13,450 fertile rice lines that are transformed with this activation-tagging vector. Considering that the transgenic plants contain an average of 1.4 loci of T-DNA inserts (Jeon at al., 2000a), we estimate that approximately 18,830 tags have been generated.
We have demonstrated that activation tagging can be applied to a monocot species such as rice. Although one of the main prerequisites for successful activation tagging is a strong enhancer element, until now it had been difficult to isolate such a strong element from monocots because the regulatory elements for gene expression are often found in an intron rather than in the 5′-flanking region of genes. For example, the promoter activity of the rice actin 1 gene, Act1, is dependent on the presence of an intact rice Act1 5′ intron (McElroy et al., 1990). In rice, the CaMV 35S promoter is widely used as a constitutive strong promoter (Terada and Shimamoto, 1990). Likewise, the CaMV 35S enhancer can elevate gene expression in monocotyledonous plants, including maize (Omirulleh et al., 1993) and rice (Mitsuhara et al., 1996). We found in our rice study that the CaMV 35S enhancers were functional in both orientations, either upstream or downstream of nearby genes. Therefore, we developed an activation-tagging vector using the CaMV 35S enhancers, and generated a large number of transgenic plants with this vector.
GUS analysis from the activation-tagging lines has revealed a wide variety of expression patterns—some constitutive and others tissue preferential. The frequency of gene trapping using the gus reporter is 9.4% in flowers and 13.4% in mature seeds and seedlings. Chin et al. (1999) reported a frequency of 10.0% for gene trapping in Ds-tagged rice lines. Screening of the tagging lines after various environmental or chemical treatments is expected to increase GUS-tagging frequency. Considering that approximately 40% of the rice genomic DNA is intragenic, the maximum efficiency should be 20% of the total population if T-DNA is inserted randomly in both orientations. However, if T-DNA inserts preferentially into intragenic regions, gene trap efficiency would be higher. In Arabidopsis, activation of the reporter gene in a gene trap vector is as high as 30% (Sundaresan et al., 1995). However, because the T-DNA- or transposon-tagged lines can contain more than one locus of insertion, actual GUS-positive frequency per insertion would necessarily be lower.
We observed that the GUS trap efficiency for the pGA2715 lines was much higher than with pGA2707, probably because 35S enhancers were present in the vector. Interestingly, the proportion of organ-preferential tags in the GUS-positive lines did not differ between the pGA2715 and pGA2707 lines, which implies that the multimerized CaMV 35S enhancers simply elevated the expression level of the nearby genes without altering the original expression pattern. RT-PCR analysis of the tagged lines also showed that the 35S sequence enhanced expression levels of the endogenous genes without changing their patterns. Our observations are consistent with the previous report that the CaMV 35S enhancer can cause enhanced expression of the endogenous gene without affecting the expression pattern in Arabidopsis (Neff et al., 1999; van der Graaff et al., 2000; Weigel et al., 2000).
The CaMV 35S enhancer did not always increase the expression level of nearby genes. In our study, only four of the 10 genes in which the enhancers were inserted within 4.5 kb displayed elevated expression, with no enhanced expression being found in the remaining lines. This may have resulted from the presence of insulator elements that prevent the mis-regulation of genes by restricting the effects of the regulatory elements to specific domains (Oki and Kamakaka, 2002). Alternatively, elevated gene expression in the remaining lines may occur in other tissue types or under conditions that we did not test. This hypothesis is supported by the fact that the CaMV 35S enhancer can increase nearby gene expression without altering the original expression pattern (Neff et al., 1999; van der Graaff et al., 2000; Weigel et al., 2000).
Although the 35S enhancer element seemed to efficiently activate nearby genes, only a small number of mutants were isolated from our activation-tagging lines. Weigel et al. (2000) and Marsch-Martinez et al. (2002), respectively, have reported frequencies of 0.1% and 1% for dominant mutations caused by activation tagging. These low rates suggest that elevated gene expression by an enhancer may not always generate visible phenotypes. So far, we have isolated nine dominant mutants from 3,290 independent, pGA2715-transformed seedlings (D.H. Jeong and G. An, unpublished data). We are investigating whether any of these mutant phenotypes are the result of activation tagging. It generally has been proposed that the tissue culture process induces somaclonal variants. In the current study, we observed dominant mutants not only from activation-tagging lines but also from untagged lines. Dominant mutant phenotypes that do not cosegregate with T-DNA insertions might be because of somaclonal variation.
To best utilize our activation-tagging lines, we are employing a “reverse activation-tagging” strategy. A PCR method for sequencing the insertion site of a 35S enhancer will help in identifying the activation-tagged gene and for examining the enhancer effects. A DNA pooling system to identify insertion mutants has been widely used in several plant species, including maize, petunia, Arabidopsis, and rice (Krysan et al., 1999; Sato et al., 1999). We are now establishing pools and superpools of DNA prepared from our activation-tagging lines of rice.
The systematic cataloging of tagging mutants by random sequencing has been initiated in Ds-tagged Arabidopsis (Parinov et al., 1999; Ito et al., 2002) and Mutator-tagged maize (Hanley et al., 2000). Flanking sequences have been determined for the Tos17-tagged lines in rice (Hirochika, 2001). We are also generating a flanking sequence database of the activation-tagging lines, using an inverse PCR technique (Spertini et al., 1999). Once this database is established, an insertion in a gene of interest will be searchable in silico. From those selected lines, activation of the tagged genes can then be examined via RT-PCR and RNA gel-blot analyses. A particular mutant phenotype can be carefully investigated by considering the specific location, conditions, or time of gene expression. This reverse activation-tagging strategy will provide a complement to the limitations of forward genetic screening.
MATERIALS AND METHODS
Construction of Vectors
Plasmid pGA2520 was made by inserting the DNA fragment containing the CaMV 35S minimal promoter (−90 to −1), the gus-coding region derived from pBI101.1, and the nopaline synthase (nos) terminator into the HindIII and HpaI sites of pGA1605 (Lee et al., 1999). Via the same method, pGA2525 was constructed to contain the minimal nos promoter (−101 to −1; Kim et al., 1993). To construct the enhancer fusion vectors, we amplified the tetramerized CaMV 35S enhancer by PCR, using pSKI15 as template (see Weigel et al., 2000). The PCR primers were 5′-GGAATTCCTAGAACTAGTGGATCC-3′ and 5′-GGAATTCACTGATAGTTTCGGATC-3′. Underlined sequences corresponded to the EcoRI sites. The pGA2523 and pGA2526 vectors were constructed by inserting the EcoRI fragment, which carried the tetramerized CaMV 35S enhancers (nucleotide −417 to −86 relative to the transcription start; Weigel et al., 2000) upstream of the 35S minimal promoter or the nos minimal promoter, respectively. Plasmid pGA2524 was made by inserting the EcoRI fragment that carried the enhancer sequence, in the inverse orientation, to the 35S minimal promoter of pGA2520. The pGA2522 plasmid was constructed by inserting the tetramerized CaMV 35S enhancers downstream of the nos terminator of pGA2525.
The pGA2707 vector was constructed by altering our previous tagging vector pGA2144 (Jeon et al., 2000a). The modified OsTubA1 intron2 (Jeon et al., 2000b) was placed in front of the gus gene. This modification was achieved via PCR, using the OsTubA1 intron2 as template and primers that contained triple splicing donors or acceptors. The PCR primer sequences were 5′-GGGGATCCGAGGTACCAGGTACCAGGTGAGTTCCATT-3′ and 5′-CGCCCGGGACCTGCATATAACCTGCATATAACCTGTAAGATTTAG-3′, with the underlined sequences corresponding to the BamHI and SmaI sites. The second modification was to replace the transcription terminator for Gene 7 of pTiA6 with the OsTubA1 terminator for the chimeric hph gene. In addition, the direction of the chimeric hph gene was inverted.
The tetramerized 35S enhancer sequence was cloned into the BamHI and EcoRI sites of pGA617, a derivative of pUC19. We named the resulting plasmid pGA2702. Finally, the pGA2715 plasmid was constructed by inserting the tetramerized 35S enhancers between XhoI and SalI of pGA2702 in the XhoI site of pGA2707.
Production and Growth of Transgenic Rice Plants
Rice (Oryza sativa var. japonica) was transformed using the Agrobacterium-mediated cocultivation method described by Lee et al. (1999). We included the following modifications: (a) cocultivation temperature reduced to 20°C, (b) exclusion of hygromycin in the regeneration media, and (c) direct transfer of the regenerated plantlets to soil from the shoot induction medium, thereby omitting the rooting procedure in Murashige and Skoog medium. Plants were grown in the greenhouse at a minimum night temperature of 20°C, with a day length of at least 14 h, supplemented with artificial light.
GUS Assay
Histochemical GUS-staining was performed according to the method of Jeon et al. (2000a). Mature flowers of the primary transformants were sampled and stained, and three or four seedlings were dissected and stained as well. Each tissue type was incubated at 37°C for 12 h; then, the staining solution was replaced with 70% (w/v) ethanol to remove the chlorophyll. Afterward, we examined the tissues under a dissecting microscope and analyzed their staining patterns.
Isolation of the Sequence-Flanking T-DNA
We extracted genomic DNA from immature leaves according to the procedure of Chen and Ronald (1999), except that we ground the samples with an MM300 Mixer Mill (Retsch, Haan, Germany). The sequence-flanking T-DNA was isolated by inverse PCR, as previously described by Spertini et al. (1999). Modifications to their technique included digesting 1 μg of the genomic DNA with PstI or XhoI. After overnight digestion, the samples were ethanol precipitated and dissolved in water. Self-ligation of the cut DNA was performed in a 50-μL volume containing 1 unit of ligase (Boehringer Mannheim/Roche, Basel) and 5 μL of 10× buffer at 8°C for 12 h. About 20 ng of the ligated DNA was used for template. PCR was performed with an initial 5-min denaturation at 94°C, followed by 35 cycles (each cycle: 94°C, 1 min; 62°C, 1 min; and 72°C, 3 min), then a final 10 min at 72°C.
RT-PCR Analysis
We used Tri-reagent (MRC Inc., Cincinnati) to isolate total RNA from immature leaves and roots. For the first strand cDNA synthesis, 2 μg of total RNA was reverse transcribed in a total volume of 25 μL that contained 10 ng of oligo(dT)12-18 primer, 2.5 mm dNTPs, and 200 units of Moloney murine leukemia virus RT (New England Biolabs, Beverly, MA) in a reaction buffer. PCR was performed in a 50-μL solution containing a 1-μL aliquot of the cDNA reaction, 0.2 μm gene-specific primers, 10 mm dNTPs, 1 unit of rTaq DNA polymerase (TakaRa Shuzo, Shiga, Japan), and 10× reaction buffer. The reaction included an initial 5-min denaturation at 94°C, followed by 18 to 24 cycles of PCR (94°C, 1 min; 56°C, 1 min; and 72°C, 1 min), and a final 10 min at 72°C. Afterward, we analyzed 10 μL of the reaction mixture on a 1.2% (w/v) agarose gel, and transferred it to a nylon membrane, Hybond N+ (Amersham, Buckinghamshire, UK). The blot was hybridized at 60°C for 12 h in Church buffer (Church and Gilbert, 1984), using a P32-labeled probe. Membranes were washed once for 10 min with 0.2× SSC and 0.1% (w/v) SDS at 25°C, and twice, for 10 min each, with 0.2× SSC and 0.1% (w/v) SDS at 58°C. These washed membranes were then exposed to RT for 30 min on x-ray film with intensifying screens.
ACKNOWLEDGMENTS
We thank all the members in Gynheung An's laboratory for help with the GUS assays of the primary transgenic plants, Shi-In Kim for growing the plants, and Priscilla Licht for critical reading of the manuscript.
Footnotes
This work was supported in part by the Korea Institute of Science and Technology Evaluation and Planning (National Research Laboratory program grant), and by the Crop Functional Genomic Center (21st Century Frontier Program grant).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.014357.
LITERATURE CITED
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell. 2000;12:2383–2394. doi: 10.1105/tpc.12.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DH, Ronald PC. A rapid DNA minipreparation method suitable for AFLP and other PCR applications. Plant Mol Biol Rep. 1999;17:53–57. [Google Scholar]
- Chin HG, Choe MS, Lee SH, Park SH, Koo JC, Kim NY, Lee JJ, Oh BG, Yi GH, Kim SC et al. Molecular analysis of rice plants harboring an Ac/Ds transposable element-mediated gene trapping system. Plant J. 1999;19:615–623. doi: 10.1046/j.1365-313x.1999.00561.x. [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1191–1195. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang RX, Nagy F, Sivasubramaniam S, Chua NH. Multiple cis regulatory elements for maximal expression of the cauliflower mosaic virus 35S promoter in transgenic plants. Plant Cell. 1989;1:141–150. doi: 10.1105/tpc.1.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- Hanley S, Edwards D, Stevenson D, Haines S, Hegarty M, Schuch W, Edwards KJ. Identification of transposon-tagged genes by the random sequencing of Mutator-tagged DNA fragments from Zea mays. Plant J. 2000;23:557–566. doi: 10.1046/j.1365-313x.2000.00830.x. [DOI] [PubMed] [Google Scholar]
- Hayashi H, Czaja I, Lubenow H, Schell J, Walden R. Activation of a plant gene by T-DNA tagging: auxin-independent growth in vitro. Science. 1992;258:1350–1353. doi: 10.1126/science.1455228. [DOI] [PubMed] [Google Scholar]
- Hirochika H. Contribution of the Tos17 retrotransposon to rice functional genomics. Curr Opin Plant Biol. 2001;4:118–122. doi: 10.1016/s1369-5266(00)00146-1. [DOI] [PubMed] [Google Scholar]
- Huang S, Cerny RE, Bhat DS, Brown SM. Cloning of an Arabidopsis patatin-like gene, STURDY, by activation T-DNA tagging. Plant Physiol. 2001;125:573–584. doi: 10.1104/pp.125.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Meyerowitz EM. Overexpression of a gene encoding a cytochrome P450, CYP78A9, induces large and seedless fruit in Arabidopsis. Plant Cell. 2000;12:1541–1550. doi: 10.1105/tpc.12.9.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Motohashi R, Kuromori T, Mizukado S, Sakurai T, Kanahara H, Seki M, Shinozaki K. A new resource of locally transposed Dissociation elements for screening gene-knockout lines in silico on the Arabidopsis genome. Plant Physiol. 2002;129:1695–1699. doi: 10.1104/pp.002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon JS, Lee S, Jung KH, Jun SH, Jeong DH, Lee J, Kim C, Jang S, Yang K, Nam J et al. T-DNA insertional mutagenesis for functional genomics in rice. Plant J. 2000a;22:561–570. doi: 10.1046/j.1365-313x.2000.00767.x. [DOI] [PubMed] [Google Scholar]
- Jeon JS, Lee S, Jung KH, Jun SH, Kim C, An G. Tissue-preferential expression of a rice alpha-tubulin gene, OsTubA1, mediated by the first intron. Plant Physiol. 2000b;123:1005–1014. doi: 10.1104/pp.123.3.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. Activation tagging of the floral inducer FT. Science. 1999;286:1962–1965. doi: 10.1126/science.286.5446.1962. [DOI] [PubMed] [Google Scholar]
- Kakimoto T. CKI1, a histidine kinase homolog implicated in cytokinin signal transduction. Science. 1996;274:982–985. doi: 10.1126/science.274.5289.982. [DOI] [PubMed] [Google Scholar]
- Kim SR, Kim Y, An G. Identification of methyl jasmonate and salicylic acid response elements from the nopaline synthase (nos) promoter. Plant Physiol. 1993;103:97–103. doi: 10.1104/pp.103.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Dellaert LW, van der Veen JH. EMS- and radiation-induced mutation frequencies at individual loci in Arabidopsis thaliana (L.) Heynh. Mutant Res. 1982;93:109–123. doi: 10.1016/0027-5107(82)90129-4. [DOI] [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Sussman MR. T-DNA as an insertional mutagen in Arabidopsis. Plant Cell. 1999;11:2283–2290. doi: 10.1105/tpc.11.12.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Suh SS, Park E, Cho E, Ahn JH, Kim SG, Lee JS, Kwon YM, Lee I. The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 2000;14:2366–2376. doi: 10.1101/gad.813600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Jeon JS, Jung KH, An G. Binary vectors for efficient transformation of rice. J Plant Biol. 1999;42:310–316. [Google Scholar]
- Li J, Lease KA, Tax FE, Walker JC. BRS1, a serine carboxypeptidase, regulates BRI1 signaling in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2001;98:5916–5921. doi: 10.1073/pnas.091065998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002;110:213–222. doi: 10.1016/s0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- Marsch-Martinez N, Greco R, van Arkel G, Herrera-Estrella L, Pereira A. Activation tagging using the En-I maize transposon system in Arabidopsis. Plant Physiol. 2002;129:1544–1556. doi: 10.1104/pp.003327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuhara S, Jingu F, Takahashi T, Komeda Y. Heat-shock tagging: a simple method for expression and isolation of plant genome DNA flanked by T-DNA insertions. Plant J. 2000;22:79–86. doi: 10.1046/j.1365-313x.2000.00716.x. [DOI] [PubMed] [Google Scholar]
- McElroy D, Zhang W, Cao J, Wu R. Isolation of an efficient actin promoter for use in rice transformation. Plant Cell. 1990;2:163–171. doi: 10.1105/tpc.2.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuhara I, Ugaki M, Hirochika H, Ohshima M, Murakami T, Gotoh Y, Katayose Y, Nakamura S, Honkura R, Nishimiya S et al. Efficient promoter cassettes for enhanced expression of foreign genes in dicotyledonous and monocotyledonous plants. Plant Cell Physiol. 1996;37:49–59. doi: 10.1093/oxfordjournals.pcp.a028913. [DOI] [PubMed] [Google Scholar]
- Neff MM, Nguyen SM, Malancharuvil EJ, Fujioka S, Noguchi T, Seto H, Tsubuki M, Honda T, Takatsuto S, Yoshida S et al. BAS1: A gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc Natl Acad Sci USA. 1999;96:15316–15323. doi: 10.1073/pnas.96.26.15316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oki M, Kamakaka RT. Blockers and barriers to transcription: competing activities? Curr Opin Cell Biol. 2002;14:299–304. doi: 10.1016/s0955-0674(02)00327-7. [DOI] [PubMed] [Google Scholar]
- Omirulleh S, Abraham M, Golovkin M, Stefanov I, Karabaev MK, Mustardy L, Morocz S, Dudits D. Activity of a chimeric promoter with the doubled CaMV 35S enhancer element in protoplast-derived cells and transgenic plants in maize. Plant Mol Biol. 1993;21:415–428. doi: 10.1007/BF00028800. [DOI] [PubMed] [Google Scholar]
- Parinov S, Sevugan M, De Y, Yang WC, Kumaran M, Sundaresan V. Analysis of flanking sequences from dissociation insertion lines: a database for reverse genetics in Arabidopsis. Plant Cell. 1999;11:2263–2270. doi: 10.1105/tpc.11.12.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Sentoku N, Miura Y, Hirochika H, Kitano H, Matsuoka M. Loss-of-function mutations in the rice homeobox gene OSH15 affect the architecture of internodes resulting in dwarf plants. EMBO J. 1999;18:992–1002. doi: 10.1093/emboj/18.4.992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spertini D, Beliveau C, Bellemare G. Screening of transgenic plants by amplification of unknown genomic DNA flanking T-DNA. Biotechniques. 1999;27:308–314. doi: 10.2144/99272st01. [DOI] [PubMed] [Google Scholar]
- Springer PS. Gene traps: tools for plant development and genomics. Plant Cell. 2000;12:1007–1020. doi: 10.1105/tpc.12.7.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan V. Horizontal spread of transposon mutagenesis: new uses of old elements. Trends Plant Sci. 1996;1:184–191. [Google Scholar]
- Sundaresan V, Springer P, Volpe T, Haward S, Jones JD, Dean C, Ma H, Martienssen R. Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 1995;9:1797–1810. doi: 10.1101/gad.9.14.1797. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Uemura S, Saito Y, Murofushi N, Schmitz G, Theres K, Yamaguchi I. A novel transposon tagging element for obtaining gain-of-function mutants based on a self-stabilizing Ac derivative. Plant Mol Biol. 2001;45:123–131. doi: 10.1023/a:1006455130098. [DOI] [PubMed] [Google Scholar]
- Terada R, Shimamoto K. Expression of CaMV 35S-Gus gene in transgenic rice plants. Mol Gen Genet. 1990;220:389–392. [Google Scholar]
- van der Fits L, Hilliou F, Memelink J. T-DNA activation tagging as a tool to isolate regulators of a metabolic pathway from a genetically non-tractable plant species. Transgenic Res. 2001;10:513–521. doi: 10.1023/a:1013087011562. [DOI] [PubMed] [Google Scholar]
- van der Fits L, Memelink J. ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science. 2000;289:295–297. doi: 10.1126/science.289.5477.295. [DOI] [PubMed] [Google Scholar]
- van der Graaff E, Dulk-Ras AD, Hooykaas PJ, Keller B. Activation tagging of the LEAFY PETIOLE gene affects leaf petiole development in Arabidopsis thaliana. Development. 2000;127:4971–4980. doi: 10.1242/dev.127.22.4971. [DOI] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blazquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrandiz C, Kardailsky I, Malancharuvil EJ, Neff MM et al. Activation tagging in Arabidopsis. Plant Physiol. 2000;122:1003–1013. doi: 10.1104/pp.122.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K, Long D, Swinburne J, Coupland G. A Dissociation insertion causes a semidominant mutation that increases expression of TINY, an Arabidopsis gene related to APETALA2. Plant Cell. 1996;8:659–671. doi: 10.1105/tpc.8.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science. 2001;291:306–309. doi: 10.1126/science.291.5502.306. [DOI] [PubMed] [Google Scholar]
- Zubko E, Adams CJ, Machaekova I, Malbeck J, Scollan C, Meyer P. Activation tagging identifies a gene from Petunia hybrida responsible for the production of active cytokinins in plants. Plant J. 2002;29:797–808. doi: 10.1046/j.1365-313x.2002.01256.x. [DOI] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Frugis G, Chua NH. The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. Plant J. 2002;30:349–359. doi: 10.1046/j.1365-313x.2002.01289.x. [DOI] [PubMed] [Google Scholar]