Abstract
To understand the mechanisms responsible for aluminum (Al) toxicity and tolerance in plants, an expressed sequence tag (EST) approach was used to analyze changes in gene expression in roots of rye (Secale cereale L. cv Blanco) under Al stress. Two cDNA libraries were constructed (Al stressed and unstressed), and a total of 1,194 and 774 ESTs were generated, respectively. The putative proteins encoded by these cDNAs were uncovered by Basic Local Alignment Search Tool searches, and those ESTs showing similarity to proteins of known function were classified according to 13 different functional categories. A total of 671 known function genes were used to analyze the gene expression patterns in rye cv Blanco root tips under Al stress. Many of the previously identified Al-responsive genes showed expression differences between the libraries within 6 h of Al stress. Certain genes were selected, and their expression profiles were studied during a 48-h period using northern analysis. A total of 13 novel genes involved in cell elongation and division (tonoplast aquaporin and ubiquitin-like protein SMT3), oxidative stress (glutathione peroxidase, glucose-6-phosphate-dehydrogenase, and ascorbate peroxidase), iron metabolism (iron deficiency-specific proteins IDS3a, IDS3b, and IDS1; S-adenosyl methionine synthase; and methionine synthase), and other cellular mechanisms (pathogenesis-related protein 1.2, heme oxygenase, and epoxide hydrolase) were demonstrated to be regulated by Al stress. These genes provide new insights about the response of Al-tolerant plants to toxic levels of Al.
Al is one of the most important limiting factors for crop production on acid soils. The most important effect of Al toxicity is a dramatic reduction in root growth, which leads to poor productivity. Severe Al stress threatens the survival of many sensitive crop genotypes. Al has been shown to affect a large number of cellular processes, especially the uptake of K+ (Liu and Luan, 2001), Ca2+ (Huang et al., 1992), and Mg2+ (Keltjens, 1995), cytoskeletal dynamics (Sivaguru et al., 1999), β-(1,3)-glucan (callose) synthesis (Zhang et al., 1994), lipid peroxidation (Yamamoto et al., 2001), and the inositol 1,4,5-triphosphate signal transduction pathway (Jones and Kochian, 1995). Al also induces the secretion of organic acids (e.g. citrate, malate, and oxalate) from roots (Delhaize and Ryan, 1995). These organic acids form a stable complex with Al, preventing the toxic effects of Al and providing the most valuable source of tolerance in the majority of plant species studied. Despite the considerable progress made over the last decade, the rather modest progress in isolating Al-regulated genes has limited our understanding of the molecular mechanisms underlying Al toxicity and tolerance.
Changes in gene expression control normal physiological processes and are also the main effectors of cellular responses to biotic or abiotic stresses (Jiang et al., 2000). Since the cloning of the Wali genes (Snowden and Gardner, 1993; Richards et al., 1994), other genes have been shown to respond to Al stress, such as those identified in rice (Oryza sativa; Yu et al., 1998), tobacco (Nicotiana tabacum; Ezaki et al., 1995; Ezaki et al., 1996), wheat (Triticum aestivum; Richards and Gardner, 1994; Cruz-Ortega et al., 1997; Hamel et al., 1998; Delhaize et al., 1999; Hamilton et al., 2001), Arabidopsis (Richards et al., 1998), and pea (Pisum sativum; Brosché and Strid, 1999; Sävenstrand et al., 2000). These studies provided an initial description of potentially important genes involved in Al stress. Most of them were identified by differential screening of cDNA libraries or suppression-subtractive hybridization. Although these approaches have been invaluable in providing the groundwork for assessing changes in gene-expression profiles during Al stress, more sensitive and efficient techniques are needed to reveal additional genes regulated by Al stress.
An effective method to analyze changes in gene expression under Al stress is to generate expressed sequence tags (ESTs) from normal and stressed tissue. Comprehensively characterizing and contrasting gene expression patterns should provide an alternative strategy to identify candidate genes involved in the Al toxicity and tolerance processes. The generation of high throughput EST sequences is rapid and economical and can be used in identifying differentially regulated mRNAs (Lee et al., 1995; Zhu et al., 2001). However, there are at least two limitations to a comparative EST approach. The first is the elevated number of anonymous ESTs detected (transcripts corresponding to new or previously isolated ESTs of unknown function). In other EST projects, the number of ESTs showing significant similarity to known genes ranged from 25% (Yamamoto and Sasaki, 1997) to 48.4% (Lim et al., 1996). Second, the enormous differences in the number of copies per cell of each mRNA species limit the study of changes in mRNA levels to relatively moderate-to-abundant transcripts (Lee et al., 1995).
In this study, we describe an EST project designed to detect changes in gene expression under Al stress in rye (Secale cereale), one of the most Al-tolerant plant species. Our goals were to identify novel genes regulated by Al stress and to assess the application of a comparative EST approach to the analysis of gene expression in plants. In total, 1,968 sequences were obtained, and ESTs showing significant homology to proteins of known function were functionally classified, providing insights concerning the structure of the mRNA population during the early stages of Al stress in rye. Comparison of two data sets from non-stressed and stressed libraries revealed many previously reported Al-responsive genes and potential genes differentially expressed. Northern analyses of certain genes were performed to confirm their differential expression patterns. Possible functions for these genes during Al stress are discussed.
RESULTS AND DISCUSSION
Al Tolerance Screening
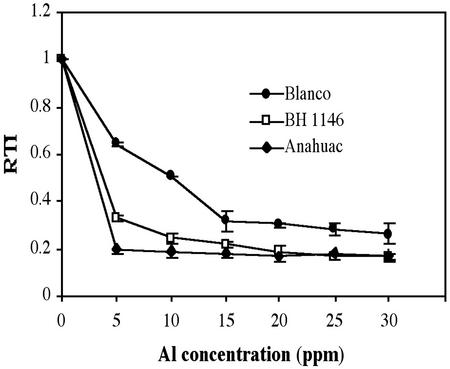
We tested the rye cv Blanco for Al tolerance using the root growth method, because inhibition of root elongation is probably the most reliable symptom of Al toxicity. Wheat cv BH 1146, one of the most tolerant spring wheat cultivars, was included in the screening to compare its level of tolerance against that of rye cv Blanco, and a very sensitive wheat cultivar, Anahuac, was included as a control. After 3 d of growth (Fig. 1), almost complete root growth inhibition was observed for wheat cv Anahuac at every Al concentration tested. Inhibition of root growth in wheat cv BH 1146 was lower from 5 to 20 mg L−1, but then it reached the same level of inhibition as wheat cv Anahuac. Rye cv Blanco showed a 36% reduction in root growth at 5 mg L−1, whereas wheat cvs BH 1146 and Anahuac were already below this level of inhibition (68% and 79%, respectively). At 10 mg L−1, a 50% reduction in root growth was observed in rye cv Blanco, reaching 68% at 15 mg L−1, but increasing Al concentration to 30 mg L−1 (1,110 mm Al) did not lead to the same level of inhibition as observed in the wheat cultivars. Rye cv Blanco was able to grow at Al concentrations up to 70 mg L−1 (data not shown). We chose 5 mg L−1 (185 μm Al) to compare changes in gene expression in rye cv Blanco root apices, because of the inhibitory effects observed at this Al concentration in this cultivar as well as in wheat cvs Anahuac and BH 1146.
Figure 1.
Al dose dependency of rye and wheat root growth at different Al concentrations. The length of the primary root from 10 seedlings (root length approximately 10 mm) was measured after 3 d of growth in control and different Al concentrations (5–30 mg L−1). A “root tolerance index” (RTI) was calculated for each cultivar at every Al concentration as the ratio of the average root length in the presence of Al versus the control at 0 mg L−1 Al. Data represent the means ± se from two independent experiments.
EST Complexity. Functional Classification of Genes
To identify genes that are regulated by Al, we undertook an analysis of Al-dependent changes in gene expression. We focused on genes induced early in the defense response, because these are most likely to reflect the effects of Al on the plant. Richards et al. (1998) similarly identified several Al-regulated genes in a cDNA library from Arabidopsis seedlings treated with 50 μm Al for 2 h. We used an EST-based approach to detect early changes in gene expression (within 6 h) induced by Al stress in root apices of rye cv Blanco. Two cDNA libraries were constructed, from root tips treated under control and Al-stressed conditions, and a total of 774 and 1,194 ESTs were obtained, respectively. The EST length (after vector and “phred 20” quality screening) ranged from 100 to 703 bp, with an average of 424 and 438 bp for the control and Al-stressed libraries, respectively. To identify the potential biological functions of the 1,968 ESTs, they were translated into all possible open-reading frames and compared with the nonredundant protein database using BLASTX3 (Altschul et al., 1990). A probability cutoff value of 10−3 was used to consider sequence similarities statistically significant. From the two libraries, 1,042 ESTs (53%) showed similarity to proteins of known function, 295 ESTs (15%) showed similarity to proteins of unknown function or predicted from genomic sequences, and 631 ESTs (32%) showed no similarity (Table I).
Table I.
Comparison of EST sequences between control and Al-stressed libraries
| Category | Control
|
Al Stressed
|
||
|---|---|---|---|---|
| No. of ESTs | % | No. of ESTs | % | |
| Summary of EST categories | ||||
| Known function ESTs | 399 | 51 | 643 | 54 |
| Unknown ESTsa | 107 | 14 | 188 | 16 |
| No significant similarityb | 268 | 35 | 363 | 30 |
| Total | 774 | 1,194 | ||
| Functional classification of known genesc | ||||
| Cell division | 6 | 1.6 | 10 | 1.6 |
| Cell structure | 28 | 7.3 | 42 | 6.8 |
| Defense and stress | 58 | 15.1 | 123 | 19.8 |
| Energy | 41 | 10.7 | 54 | 8.7 |
| Intracellular traffic | 6 | 1.6 | 15 | 2.4 |
| Metabolism | 51 | 13.3 | 71 | 11.4 |
| Protein destination | 51 | 13.3 | 88 | 14.2 |
| Protein synthesis | 25 | 6.5 | 40 | 6.4 |
| Secondary metabolism | 41 | 10.7 | 37 | 5.9 |
| Signal transduction | 26 | 6.8 | 60 | 9.6 |
| Transcription | 18 | 4.7 | 32 | 5.1 |
| Transport facilitation | 32 | 8.3 | 46 | 7.4 |
| Transposable elements | 1 | 0.3 | 4 | 0.6 |
| Total | 384 | 622 | ||
ESTs showing similarity to predicted and/or unknown function genes.
A 10−3 BLASTX P value was used to determine significant similarity.
The known function genes were organized in contigs and singletons for a total of 671 unique function genes.
Because the main goal of this research was to study the function of Al-responsive genes, we initially focused on the 1,042 known-function ESTs. Cluster analysis identified 91 redundant genes and 580 “single hits” (522 singletons) yielding 671 unique known genes whose expression patterns were examined in rye cv Blanco root tips under Al stress. Therefore, the transcript profile in rye cv Blanco root apices appeared to be highly heterogeneous, which is consistent with the expected complex gene-expression patterns in root apices. Overall, we found that 78% of the known function genes were singletons, 19% were present in two to three copies, and only 3% were present in more than three copies. The maximum EST frequency was 4.5% for the Wali5 gene (a proteinase inhibitor with five and 25 hits in the control and Al-stressed libraries, respectively). The 671 distinct genes showing similarity to proteins of known function were categorized according to 13 major functional categories (Table I). The complete list of genes is available in Table II, which can be accessed through the on-line version of the manuscript (http://www.plantphysiol.org). Genes representing a wide mix of cellular functions (with the obvious exception of photosynthesis) were present, indicating that our EST database, although certainly not a full representation of root apex transcription, contains tags derived from genes encoding most major cellular functions and should be useful for the discovery of Al-regulated genes involved in several biological processes.
Identification of Previously Reported Al-Responsive Genes
A literature review identified 45 genes previously reported to be regulated by Al, of these 19 showed high identity to genes in our EST database (Table III). One of these genes (encoding a heat shock protein) seemed also to be down-regulated in rye cv Blanco roots, as reported by Richards et al. (1998). Several other genes appeared to be up-regulated in rye cv Blanco root tips within the first 6 h of Al treatment. It is worth mentioning the prevalence of the Wali1, Wali5, and oxalate oxidase genes. Together, these three genes accounted for 7.5% of the ESTs that hit known function genes in the Al-stressed library. One copy of the PEARLI1 and PEARLI4 genes, detected in Arabidopsis during the first hours of exposure to Al (Richards et al., 1998), was also present in the Al-stressed library. In addition to these genes showing high similarity to rye cv Blanco ESTs, other ESTs identified genes that are known to be Al-regulated (e.g. β-1,3-glucanase, Ala aminotransferase, blue copper binding protein, and several glutathione S-transferases and peroxidases), but their similarity with the reported genes was low and probably corresponds to different isoforms. These results indicated that our EST approach was very effective to detect Al-regulated genes in the early stages of Al stress. A further comparison of transcripts in rye cv Blanco roots under control versus stressed conditions should identify more Al-responsive genes.
Table III.
Previously reported Al-regulated genes in plants
| Gene | Accession No.a | Coded Proteinb | Reference | Regulationc | Blanco
Expressiond
|
|
|---|---|---|---|---|---|---|
| Control | Al stressed | |||||
| Wali 1 | L11879 | MT class I | Snowden and Gardner (1993) | Up (24 h) | 8 | 15 |
| Wali 2 | L11880 | Cys-rich protein | Snowden and Gardner (1993) | Up (0.5 h) | 0 | 0 |
| Wali 3 | L11881 | Proteinase inhibitor | Snowden and Gardner (1993) | Up (24 h) | 0 | 0 |
| Wali 4 | L11883 | Phe-ammonia lyase | Snowden and Gardner (1993) | Up (24 h) | 0 | 2 |
| Wali 5 | L11883 | Proteinase inhibitor | Snowden and Gardner (1993) | Up (24 h) | 5 | 25 |
| Wali 6 | L28009 | Proteinase inhibitor | Richards et al. (1994) | Up (24 h) | 2 | 4 |
| Wali 7 | L28008 | Unknown | Richards et al. (1994) | Up (24 h) | 0 | 1 |
| Shh | L11782 | S-adenosyl-l-homo-Cys hydrolase | Richards and Gardner (1994) | Up/down | 0 | 0 |
| H3 | X00937 | Histone H3 | Richards and Gardner (1994) | Up/down | 1 | 5 |
| H4 | X00043 | Histone H4 | Richards and Gardner (1994) | Up/down | 0 | 3 |
| Hsp70 | X13301 | Heat shock protein 70 | Richards and Gardner (1994) | Down (4 h) | 4 | 1 |
| PAL111 | NP | Auxin-regulated protein | Ezaki et al. (1995) | Up (10 h) | 0 | 0 |
| PAL142 | D29680 | Glutathione S-transferase | Ezaki et al. (1995) | Up (10 h) | 2 | 5 |
| PAL139 | D29679 | Cys-rich domain protein | Ezaki et al. (1995) | Up (10 h) | 0 | 0 |
| PAL141 | D29681 | Unknown | Ezaki et al. (1995) | Up (10 h) | 0 | 0 |
| PAL 201 | D45891 | Anionic peroxidase | Ezaki et al. (1996) | Up (10 h) | 0 | 0 |
| GDI | AF012823 | GDP-dissociation inhibitor | Ezaki et al. (1997) | Up | 1 | 0 |
| Glc1 | U30323 | β -1,3-Glucanase | Ortega et al. (1997) | Up (6 h) | 0 | 0 |
| Wfim1 | U67717 | Cytoskeletal fimbrin-like | Ortega et al. (1997) | Up (12 h) | 0 | 0 |
| Sali3-2 | U89693 | Auxin down-regulated ADR6 | Ragland and Soliman (1997) | Up | 0 | 0 |
| Sali5-4a | U64866 | Auxin down-regulated ADR6 | Ragland and Soliman (1997) | Up | 0 | 0 |
| RicMT | U77294 | MT class II | Yu et al. (1998) | Down (24 h) | 0 | 0 |
| War 5.2 | AF005088 | Cys proteinase | Hamel et al. (1998) | Up (24 h) | 1 | 1 |
| War 13.2 | AF005084 | Oxalate oxidase | Hamel et al. (1998) | Up (24 h) | 0 | 7 |
| pEARLI8 | NP | Phototropic response transducer | Richards et al. (1998) | Up/down | 0 | 0 |
| pEARLI1 | L43080 | Pro-rich hydrophobic protein | Richards et al. (1998) | Up/down | 0 | 1 |
| pEARLI2 | NP | Unknown | Richards et al. (1998) | Up/down | 0 | 0 |
| ALD | T43001 | Aldolase (chloroplast) | Richards et al. (1998) | Up/down | 0 | 0 |
| pEARLI4 | L43081 | Pro-rich hydrophilic protein | Richards et al. (1998) | Up/down | 0 | 1 |
| pEARLI5 | NP | Berberine bridge enzyme | Richards et al. (1998) | Up (2 h) | 1 | 1 |
| BCB | Z15058 | Blue copper-binding protein | Richards et al. (1998) | Up (2 h) | 0 | 0 |
| PER | X71794 | Peroxidase | Richards et al. (1998) | Up (1 h) | 0 | 0 |
| ALA | T41718 | Ala aminotransferase | Richards et al. (1998) | Down (4 h) | 0 | 0 |
| CAB | X64459 | Chlorophyll a/b-binding protein | Richards et al. (1998) | Down (4 h) | 0 | 0 |
| CZSOD | T04184 | Cu/Zn superoxide dismutase | Richards et al. (1998) | Up (8 h) | 1 | 2 |
| CAT | X64271 | Catalase | Richards et al. (1998) | Down (2 h) | 1 | 0 |
| Sad | AF053638 | Alcohol dehydrogenase (short-chain) | Brosché and Strid (1999) | Up (6 h) | 0 | 0 |
| PU1 | X17020 | Polyubiquitin | Brosché and Strid (1999) | Up (6 h) | 2 | 4 |
| Chs | X63333 | Chalcone synthase | Brosché and Strid (1999) | Up (6 h) | 0 | 0 |
| Psdrr230 | AF139018 | Disease resistance protein 230 | Sävenstrand et al. (2000) | Up (24 h) | 0 | 0 |
| PsLRRP | AF137354 | Leu-rich repeat protein | Sävenstrand et al. (2000) | Up (3 h) | 0 | 1 |
| PsExt | AF155232 | Extensin | Sävenstrand et al. (2000) | Up (3 h) | 0 | 0 |
| VATPase | L11873 | Vacuolar ATPase | Hamilton et al. (2001) | Up (48 h) | 0 | 0 |
| Ef2 | AF005085 | Elongation factor-2 | Hamel et al. (unpublished data) | Down | 0 | 0 |
| Zmal1 | AF031083 | Unknown | Menossi et al. (unpublished data) | Up | 0 | 0 |
Accession nos. of the probes used in each study. NP, The accession no. was not provided in the study.
Coded protein indicates the putative function of the gene product expected by sequence homologies.
Regulation indicates the expression profiles from northern-blot analysis (for more details, see reference).
Blanco expression shows the no. of copies of highly homologous ESTs in Blanco cDNA libraries.
Generation of Abundance Profiles and Northern-Blot Analysis
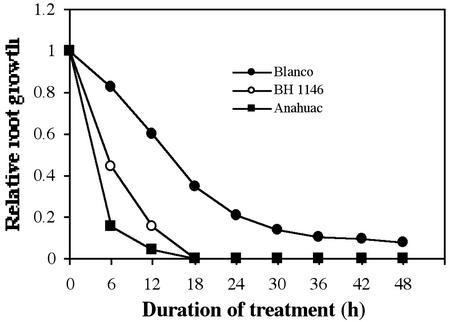
Abundance profiles were obtained for the 91 redundant genes (corresponding to 427 ESTs). Because of the relatively modest sampling size of our EST database, the verification of gene expression profiles using northern analysis was critical to ascertain true differential expression. To confirm and extend the results from EST comparative analysis, we monitored changes in transcript abundance over short- and long-term exposure to Al. A higher Al concentration (30 mg L−1) was used to obtain high levels of stress within a 48-h period. A similar Al concentration (1 mm) was used by Slaski (1994) for a time-course study (up to 24 h) of the effect of Al on the activity of Glc-6-phosphate dehydrogenase (G6PDH) and 6-phosphagluconate dehydrogenase enzymes in an Al-tolerant rye. In the same work, an Al-sensitive wheat was also studied using 0.15 mm Al, with similar results. Because rye cv Blanco survives up to 70 mg L−1 Al, we decided to use 30 mg L−1 (1.11 mm) Al. Figure 2 shows the root growth of wheat cvs Anahuac and BH 1146 and rye cv Blanco in response to Al. Wheat cv Anahuac and BH 1146 showed complete root growth inhibition after 18 h. However, at 18 h, rye cv Blanco roots showed a growth rate that was approximately 40% of control roots, and they continued to grow over the 48-h testing period. We continued to measure rye cv Blanco root growth for 2 more d every 24 h, and it showed a similar rate than that at 48 h (data not shown). The temporal expression of certain genes was monitored using northern analysis at eight time points up to 48 h after the exposure of the roots to Al. Total RNA was isolated at frequent intervals within the first 12-h period to assess early changes in gene expression, and then samples were collected every 12 h to study late response of genes to Al stress. As the first step in the analysis of these libraries, 30 genes (including nine singletons) were selected on the basis of (a) clear differences in copy number between both libraries, and/or (b) potential roles in biological responses to Al stress. Northern-blot analyses revealed 13 genes that clearly responded to Al stress and are described below. Three additional genes (encoding a thioredoxin H, Cys synthase, and kinetochore protein) did not show any change, whereas our analysis was inconclusive for the rest.
Figure 2.
Time course showing rye and wheat root elongation inhibition after exposure to 30 mg L−1 Al. Ten 2-d-old seedlings were treated with or without Al, and the length of the primary root was measured every 6 h over a 48-h period. Relative root growth values were obtained for every interval as the ratio between the root growth at 30 and 0 mg L−1.
Al Effect on Cell Division and Elongation Genes
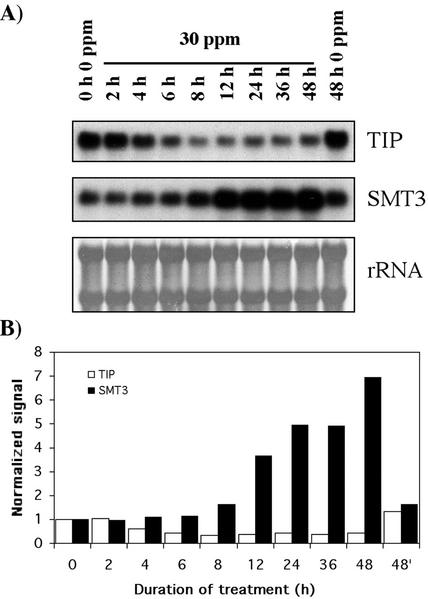
Two different genes coding for tonoplast intrinsic proteins (TIP) were detected in rye cv Blanco libraries, γ-Tip and δ-Tip, both exhibiting the same hybridization profile. That obtained with γ-Tip as a probe is shown in Figure 3. After 6 h of Al stress, the expression of γ-Tip was only 43% of the control, decreasing to 35% at 8 h. Because the γ-Tip and δ-Tip ESTs (GenBank accession nos. BE587109 and BE587310, respectively) show a 73% identity over 409 nt, the use of gene-specific probes (5′- or 3′-untranslated regions) would be necessary to confirm the expression pattern of each gene. Nevertheless, we can conclude that transcripts coding for tonoplast aquaporins decreased in response to Al.
Figure 3.
Effect of Al stress on cell elongation and division-related genes. A, RNA-blot hybridization showing different expression profiles during Al treatment of rye cv Blanco roots over a 48-h period. Total RNA was isolated from the same Al stress experiment. The first (0 h) and last (48 h) lanes correspond to unstressed plants (0 mg L−1). The rRNAs of one representative blot stained with methylene blue show RNA integrity and uniform loading (Herrin and Schmidt, 1988). TIP, aquaporin; SMT3, ubiquitin-like protein SMT3. B, Quantitation of mRNA levels. The hybridization signals obtained from the genes were weighted against those obtained from the scan image of the 25S rRNA stained with methylene blue to correct for minor differences in RNA loading, normalized to that at 0 h (which was set at 1 for each gene) and plotted against time to compare changes in gene expression.
Different studies have shown that these genes are highly expressed in elongating cells (Chaumont et al., 1998; Ferguson et al., 1997). The highest expression of maize (Zea mays) γ-TIP was detected in the apical meristem and the cell-elongation zone, consistent with γ-TIPs permitting the rapid influx of water into the vacuole, generating the turgor pressure that drives cell elongation (Chaumont et al., 1998). Karlsson et al. (2000) suggested that the expression of a δ-TIP gene in spinach (Spinacia oleracea) may be induced during the formation of the large vacuole of elongating cells. Therefore, decreased levels of TIPs under Al stress would generate a lower turgor pressure in the cell elongation zone, resulting in reduced root growth. The factors leading to reduced expression of these genes are unknown, but Smart et al. (2001) reported that some genes homologous to TIPs are down-regulated under drought stress in Nicotiana glauca, and it is possible that Al-induced changes in the cell wall (e.g. binding of Al, lignification, and callose accumulation) may lead to a reduced permeability and, as a result, to water stress.
Al stress up-regulated Smt3 transcripts (accession no. BE587402), which encode an ubiquitin-like protein (Fig. 3). Smt3 showed increased expression after 12 h (3.6-fold), which is when root growth was drastically slowed (Fig. 2). Maximal Smt3 induction (7-fold) was observed after 48 h, when rye cv Blanco root elongation was basal. Unlike ubiquitin, which is also up-regulated by Al (Brosché and Strid, 1999), SMT3-conjugation appears to include regulation of protein subcellular localization and transcription factor activity. Yeast temperature-sensitive SMT3 mutants are blocked at G2/M at the nonpermissive temperature, consistent with a role for SMT3 in mitotic progression (Meluh and Koshland, 1998). In addition, the Saccharomyces pombe SMT3 homolog interacts with PCNA (Tanaka et al., 1999), which is required for DNA replication, DNA repair, and perhaps for the recovery pathway after DNA replication arrest.
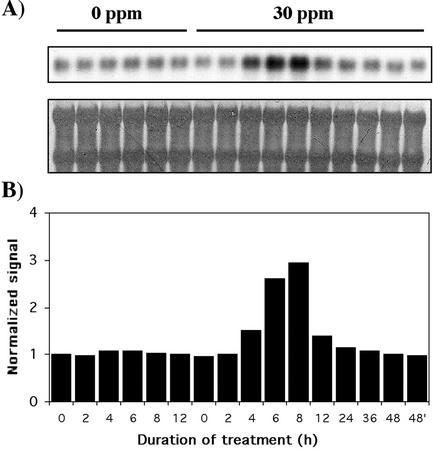
Genes Involved in Oxidative Stress
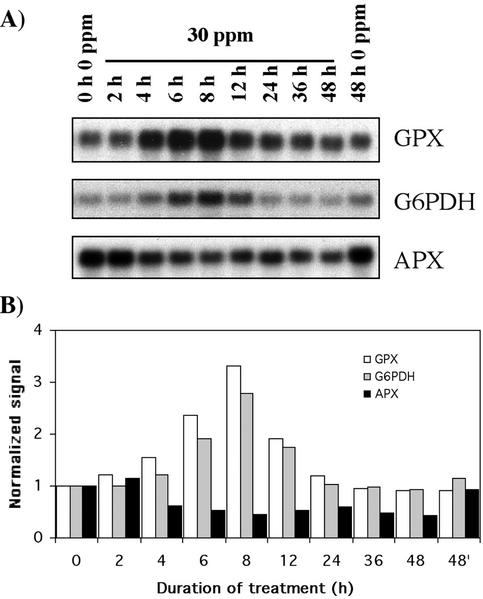
Transcripts encoding a glutathione peroxidase (GPX; accession no. BE587404) were rapidly induced by Al (Fig. 4). The gene showed increased mRNA levels after 4 h, peaking at 8 h (3.3-fold) before decreasing to normal levels at 36 h. The specific substrate of the GPX induced in rye cv Blanco root apices is unknown, but the absence of a predicted transit peptide indicates that the Al-induced gene codes for a cytosolic enzyme. In addition, our BLASTX search found significant similarities with mammalian phospholipid hydroperoxide GPXs. Lipid peroxidation is an early event triggered by Al, as described by Yamamoto et al. (2001) in pea roots. The kinetic patterns of lipid peroxidation in pea roots (reaching a maximum level at 12 h) and the induction of the Gpx gene in rye cv Blanco root apices were similar to each other. Taking into account that pea (a dicot) and rye are distinct plants, it is tempting to speculate that GPX in rye cv Blanco is also involved in the protection of membrane phospholipids, although biochemical characterization of the induced GPX is warranted.
Figure 4.
Time course showing the effect of Al stress on the expression profile of oxidative stress-related genes. Experimental conditions and indications are as in Figure 3.
The expression pattern of the Gpx gene suggested regulation at two different levels. First, it could be argued that the expression of the gene might be related to diurnal and circadian mechanisms rather than to Al stress. In the absence of Al, the expression of the gene did not change significantly compared with expression under Al stress (Fig. 5). However, although this shows that the induction of the gene is attributable to Al stress, it does not preclude a diurnal response of the gene to Al. We then investigated the hypothesis that failure to recycle glutathione could lead to the failure to maintain high levels of Gpx transcripts. Oxidized glutathione is reduced by glutathione reductase by NADPH, and the pentose phosphate pathway is especially important for providing NADPH. The key enzyme controlling the flux of carbon through this pathway is G6PDH. Interestingly, Slaski (1994) observed an increase in G6PDH activity in rye after Al treatment (1 mm, pH 4.5), very similar to the expression pattern of the Gpx gene. The expression profile of a G6pdh gene (accession no. BE587600) present in one copy in the Al-stressed library (Fig. 4) matched almost exactly G6PDH activity reported by Slaski (1994) and Gpx expression. The induction of G6pdh was also Al dependent (data not shown), like Gpx. It is possible that depletion of reduced glutathione might lead to decreased expression of the Gpx gene, although further work is necessary to confirm this hypothesis.
Figure 5.
Time course showing the Al stress-dependent response of the glutathione peroxidase gene. Experimental conditions and indications are as in Figure 3, with the exception that the first six samples (non-stressed) were isolated in a different experiment.
Another gene coding for an enzyme involved in the scavenging of cytosolic hydrogen peroxide, ascorbate peroxidase (APX; accession no. BE587068), was down-regulated by Al stress (Fig. 4). Steady-state Apx transcript levels were found to be only 46% of the control after 6 h. Richards et al. (1998) suggested that both GPX and APX would be induced by Al stress. However, our results indicate that only glutathione-dependent systems (e.g. GPX and glutathione S-transferase) are activated by Al stress. Moreover, Lukaszewski and Blevins (1996) reported that Al stress resulted in a reduction of ascorbate concentration of root apices in squash (Cucurbita pepo).
The role of oxidative stress in Al toxicity is not clear. Richards et al. (1998) showed that Al enhances oxidative stress because Al induces the expression of several genes encoding antioxidant enzymes, although Yamamoto et al. (2001) suggested that lipid peroxidation was not the cause of root elongation inhibition. However, Yamamoto et al. (2002) concluded that Al affects mitochondrial functions, leading to reactive oxygen species (ROS) production, which correlated with inhibition of root elongation. The up-regulation of the Gpx gene supports the hypothesis that Al enhances oxidative stress, but the possible role of ROS production in Al toxicity needs to be clarified.
Genes Involved in the Synthesis of Phytosiderophores and Iron Homeostasis
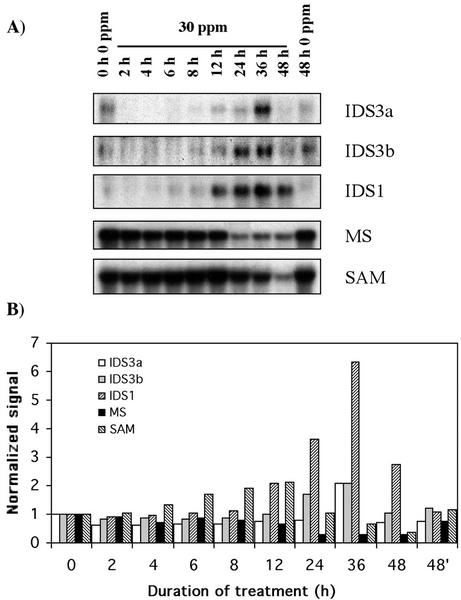
The barley (Hordeum vulgare) Ids3 gene codes for a dioxygenase involved in the synthesis of phytosiderophores, which bind and carry Fe3+ across the plasma membrane in most monocotyledonous (strategy II). Although the barley Ids3 gene is single copy (mapping to the chromosome arm 4HL), several nonallelic isozymes are present in rye (Nakanishi et al., 2000). Our EST comparative analysis detected four different Ids3 homologs in rye cv Blanco roots that seemed to be down-regulated within the first 6 h of Al stress (Table II, secondary metabolism). Rye cv Blanco Ids3a and Ids3b genes transcripts (accession nos. BE587731 and BE587474, respectively) were down-regulated from a low level even further upon Al (Fig. 6). However, longer exposures to Al (24 h for Ids3b and 36 h for Ids3a) led to the up-regulation of the genes, although by 48 h their transcripts were almost undetectable (Fig. 6).
Figure 6.
Time course showing the expression profile of genes involved in the synthesis of phytosiderophores and iron metabolism. IDS3a, Iron deficiency-specific 3a dioxygenase; IDS3b, iron deficiencyspecific 3b dioxygenase; IDS1, iron deficiency-specific 1 MT. Experimental conditions and indications are as in Figure 3.
Minocha et al. (1992) showed that Al treatment for 4 h induced a decrease in cellular levels of Fe in vinca (Catharanthus roseus) a dicot with a completely different Fe-uptake system (strategy I). Chang et al. (1998) more recently demonstrated the early inhibition by Al (within 3 h) of both biosynthesis and secretion of phytosiderophores in wheat. Taken together, these results suggested that both in monocots and dicots, Al induces a very rapid inhibition of Fe uptake. Because Al may directly affect Ids3 transcript levels, it is tempting to speculate that Al-induced oxidative stress is countered by a reduction in Fe uptake. Although it is well known that Al induces oxidative stress, biochemical and physiological studies related to the possible role of the Al ion in this response have not been investigated. Because Al is not a transition metal, it cannot catalyze redox reactions (Yamamoto el al., 2001). Instead, this function is mainly attributed to iron and other metals such as copper. Therefore, as Ranieri et al. (2001) suggested, iron-deficient plants may be protected in some degree against oxidative stress because of a reduction of catalytic iron capable of triggering the Fenton reaction. In other words, a simple idea to reduce radical formation by iron may be the limitation of the iron atom availability. The late response of Ids3a and Ids3b (Fig. 6) agrees with the Fe-uptake pattern in vinca and with Fe-deficiency symptoms observed in several studies after long exposures to Al.
A rye cv Blanco homolog (accession no. BE586403) to barley Ids1, a metallothionein (MT) induced by Fe-deficiency, was strongly activated after 12 h of exposure to Al, peaking at 36 h (Fig. 6), when the Ids3 transcripts were maximally induced. The function of this gene is not clear, but it could be related to the release of phytosiderophores (Okumura et al., 1991). Snowden and Gardner (1993) showed that the wheat Wali1 gene (homologous to MTs) was similar to barley Ids1. However, we detected two similar but clearly different transcripts in rye cv Blanco roots corresponding to the barley Ids1 and wheat Wali1 genes.
Met synthase (MS; accession no. BE587633) transcripts showed no correlation with those of Ids3, as transcripts slowly decreased during the first 12 h, being further reduced after 24 h (Fig. 6). This pattern is consistent with the hypothesis of Yamaguchi et al. (2000) that the Met precursor of phytosiderophores is provided by the Met salvage pathway, in which MS is not involved. In addition, the expression pattern of S-adenosyl-MS (SAM; accession no. BE586457) did not respond to Al stress in the same way as the Ids3 genes (Fig. 6), because its transcripts showed an initial 2-fold induction after 12 h, but decreasing after 24 h. Nevertheless, other members of the Sam gene family may be involved in the pathway. SAM is involved in more than one biosynthesis pathway, and its up-regulation during the first 12 h may be related to an increased demand for adenosyl-Met in lignin biosynthesis. This is in agreement with the activation of Phe-ammonia lyase (Snowden and Gardner, 1993) and S-adenosyl-l-homo-Cys hydrolase (Richards and Gardner, 1994) in response to Al stress in wheat. Finally, other genes related to the synthesis of phytosiderophores (nicotianamine synthase and nicotianamine aminotransferase) were included in the screening, but no hybridization signals were obtained with their EST probes.
Other Genes Regulated by Al Stress
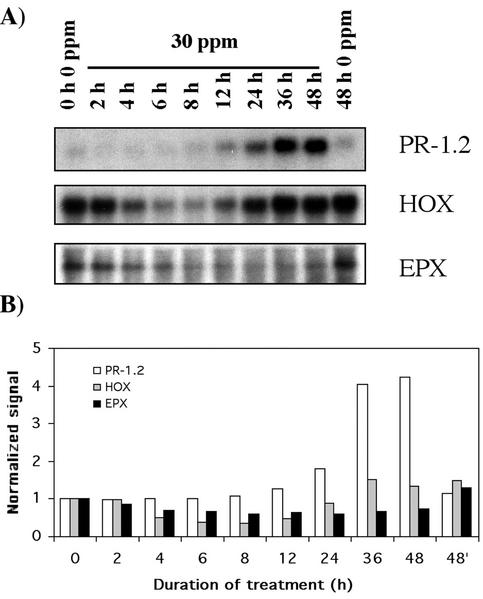
Many plant defense mechanisms that have been associated with the response to pathogens are also associated with symptoms of Al toxicity, which suggests that Al may act as an elicitor of a pathogenesis-related transduction pathway (Hamel et al., 1998). Although induction of a pathogenesis-related protein 1.2 (PR-1.2) started only after 12 h (Fig. 7), it showed the highest up-regulation in our EST comparative analysis (10-fold). The reason for such a difference is unknown, but this pattern was observed using two different ESTs as probes (accession nos. BE586275 and BE587392). The Pr-1.2 gene induced by Al in rye cv Blanco roots belongs to a group that includes mammals, insects, fungi, and plants, although the function of this family of proteins is unclear.
Figure 7.
Time course showing the expression profile of other genes regulated by Al stress. PR-1.2, Pathogenesis-related 1.2; Experimental conditions and indications are as in Figure 3.
A transcript encoding heme oxygenase (HOX; accession no. BE586278) showed a biphasic regulation, transiently decreasing within 4 h of exposure to start a progressive increase after 12 h, returning to unstressed levels at 36 h (Fig. 7). Similar to Gpx, the expression of Hox did not change in the absence of Al (data not shown). Ossola et al. (2000) suggested that human hox-I induction takes place when steady-state levels of the ROS increase and defenses (e.g. GPX) are decreased. If we compare the expression profile of Gpx and Hox, it is clear that Hox and Gpx respond inversely. This is consistent with Gpx and Hox responding to the same signal in different directions. HOX is the rate-limiting enzyme in the degradation of the prosthetic group heme (Platt and Nath, 1998). Among the consequences of heme degradation is the liberation of Fe, a potent oxidant that may exacerbate oxidative stress (Berg et al., 2001). This may explain why the gene is down-regulated in rye cv Blanco Al-stressed roots at the same time that Fe uptake is inhibited and the antioxidant defense (e.g. GPX) is activated.
Finally, transcripts of a gene encoding epoxide hydrolase (EPX; accession no. BE587663) decreased slowly after the Al treatment (59% of the control level at 8 h; Fig. 7), confirming the predictions from the EST comparative analysis. In comparison with animals, characterization of plant EPXs is very limited, although they are involved in the metabolism of epoxy fatty acids and show a higher expression in meristematic tissue than in mature leaves (Stapleton et al., 1994). Further research is necessary to understand the effects of Al on Epx expression.
CONCLUDING REMARKS
The EST comparative analysis described here was successful in the identification of Al-regulated genes. In general, various patterns of gene expression were observed, and for most genes, we could see a gradual change over time. However, the magnitude of change in mRNA levels predicted by EST analysis did not correlate as well as the direction of change. Lee et al. (1995) reported similar results, suggesting that at these sampling depths, EST approaches are suitable for qualitative rather than quantitative comparisons. For instance, induction of the Wali5 gene started after 8 h of Al stress (data not shown), which is consistent with the results obtained by Richards et al. (1998) in Arabidopsis. Overall, the correlation between the EST copy number and northern analysis can be considered as acceptable at the sampling depths of the project. Increasing the number of ESTs will undoubtedly lead to a better estimation of the real transcript profiles, although additional verification will always be necessary. To date, our biochemical understanding of the molecular basis of Al toxicity and tolerance is still in its infancy, and the expression profiles obtained in rye cv Blanco root apices can provide new insights into the molecular mechanisms underlying the root response of an Al-tolerant plant (rye cv Blanco) to toxic levels of Al. The 1,194 ESTs from the Al-stressed library provide an overall picture about the structure of the mRNA population of root cells during the early stages of Al stress. Significant findings from this study were the rapid down-regulation by Al stress of tonoplast aquaporins possibly affecting cell elongation, and the late induction of the ubiquitin-like protein SMT3 gene, involved in the control and recovery of the cell cycle. In addition, our results suggest that glutathione-dependent systems (e.g. GPX and glutathione S-transferase) provide the most important antioxidant defense in root apices under Al stress, whereas ascorbate-related systems seem to be negatively affected. The strong and complex effects of Al stress on several genes involved in the control of Fe uptake and homeostasis suggest an important role of this mechanism during the response of plants to Al, probably connected to oxidative stress. Finally, our results support the further use of rye (the most Al-tolerant cereal) for the discovery of Al-responsive genes that might be related to Al tolerance or used to obtain some degree of tolerance (e.g. overexpressing oxidative stress-related genes). It is very likely that other genome-scale gene expression approaches (e.g. microarrays, SAGE) will provide additional information about changes in mRNA levels during Al stress, especially if future studies focus on the comparison of expression profiles between Al-tolerant and -sensitive genotypes.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Seeds of uniform size of rye (Secale cereale L. cv Blanco) and wheat (Triticum aestivum L. cvs BH 1146 and Anahuac) were sterilized with 25% (v/v) sodium hypochlorite for 5 min, rinsed with deionized water, and incubated on filter paper in the dark. Rye seeds were incubated first at 10°C for 24 h and then at 25°C, whereas wheat seeds were germinated directly at 25°C. Seedlings with a primary root of about 10 mm were placed on a styrofoam sheet with a nylon net bottom. The styrofoam sheets were floated on a nutrient solution (2 L) in a plastic tray. For treatments longer than 3 d, the sheets were transferred to 4-L plastic trays. The nutrient solution was prepared with ultra pure water and contained all macronutrients except Pi, to prevent Al precipitation: 0.4 mm CaCl2, 0.01 mm (NH4)2 SO4, 0.65 mm KNO3, 0.25 mm MgCl2, and 0.04 mm NH4NO3. For Al treatments, AlCl3 was added to final concentrations from 0 to 30 mg L−1 (1 mg L−1 = 37 μm). The pH of the solution was adjusted to 4.0 with 0.1 n HCl or 1 n NaOH after the addition of Al. Nutrient solutions were changed every 24 h. Seedlings were grown in aerated medium under controlled environmental conditions (26 ± 1°C and a 16-h photoperiod).
Root Growth Measurements
Germplasm was screened for Al tolerance by using the root growth method. The length of the primary root from 10 seedlings was measured after 3 d of growth in control or different Al concentrations (5–30 mg L−1 Al). A root tolerance index was calculated for each cultivar at every Al concentration as the ratio of the average root length in the presence of Al versus the control at 0 mg L−1 Al. Time course root elongation inhibition during a 48-h period of Al treatment was also determined. For each cultivar, two sets of 10 seedlings each were grown in nutrient solutions without Al for 2 d, and then one set received 30 mg L−1 Al. The control set was simultaneously moved to fresh medium. The length of the primary root (from control and Al-stressed seedlings) was measured every 6 h to examine the temporal response of the cultivars to toxic Al levels. Relative root growth values were obtained for every 6-h interval as the ratio between the root growth at 30 and 0 mg L−1.
cDNA Libraries Construction
The ESTs used in this study were obtained from two different libraries (control and Al-stressed). Rye cv Blanco seedlings were grown in an Al-free solution, at pH 4.0 for 2 d. After this period, seedlings for the Al-stressed library were transferred to a solution containing 5 mg L−1 Al (185 μm Al). Plant-to-plant variation of gene expression was reduced by bulk harvesting of a total of approximately 600 plants. Root tips were harvested from seedlings from both treatments at 2, 4, and 6 h after the addition of Al to the treated set. Time points were used to detect genes whose expression in response to Al is transient. The seedlings were removed from the medium, root tips were cut off, and tissue was frozen and ground in liquid nitrogen before extraction of RNA. Total RNA was isolated from pooled samples (2, 4, and 6 h) by an adaptation of the guanidine isothiocyanate/CsCl method (Chirgwin et al., 1979). Poly(A+) RNA was isolated using the PolyATtract system (Promega, Madison, WI) and cDNA synthesized. Double-stranded cDNA was ligated with SalI-NotI oligonucleotide adapters and cloned in the pSPORT vector (Invitrogen, Carlsbad, CA) following manufacturer's instructions, and recombinants were propagated in Escherichia coli strain DH12S.
EST Generation and Data Analysis
Plasmid DNA from randomly selected clones was extracted and sequenced from the 5′ end. Single-pass partial sequences were determined with an automated DNA sequencer (model ABI Prism 3700, Applied Biosystems, Foster City, CA, or CEQ2000, Beckman Coulter, Fullerton, CA) as part of the National Science Foundation Wheat Genome Project. Chromatogram traces were evaluated using PHRED (Ewing and Green, 1998). Vector sequences were removed and bases having “phred” scores lower than 20 were trimmed off. Sequences of less than 100 bases were removed from the analysis. A total of 1,968 high-quality ESTs were obtained from the two libraries. A list of the nucleotide sequences is available at http://wheat.pw.usda.gov/NSF/data.html.
The ESTs generated were compared with the nonredundant protein databases using the BLASTX program provided by the National Center for Biotechnology Information. A minimum P value cutoff of 10−3 (the probability that alignment would be generated randomly is <1 in 1,000) was used to determine homology of ESTs to known proteins. The 10−3 cutoff was chosen for empirical reasons. For example, some ESTs with long 5′ or 3′ non-coding regions generated perfect matches of only a few amino acids and, as a result, had low BLASTX P values. Therefore, a few false-positives were likely attributable to gene family members. Each sequence in the EST database was classified as single hit (represented by a single EST in either library [i.e. singletons] or in both) or redundant (two or more copies in one or both libraries). Redundant ESTs were assembled in overlapping contigs and nonoverlapping sequences that corresponded to different parts of the same gene. ESTs were identified as the protein showing the highest score among the candidate proteins and classified according to 13 functional categories. For each gene, abundance profiles (relative levels of gene expression) were inferred from the common gene transcript frequency, computed by summing the number of ESTs matching that particular gene and dividing the sum by the total of ESTs for each library.
Northern Hybridization and Densitometry
For time-course experiments of gene expression, approximately 60 seedlings were used for RNA isolation from each sample. To minimize plant-to-plant variation, seedlings were grown for 5 d in nutrient solutions with no Al, and only roots of similar length were selected. Total RNA from rye cv Blanco root tips non-stressed (0 mg L−1, 0 and 48 h) or treated with Al (30 mg L−1) for 2, 4, 6, 8, 12, 24, 36, and 48 h was isolated using TRIZOL reagent (Invitrogen), following manufacturer's protocol. Equal amounts of total RNA (10 μg) were electrophoresed in a 1.5% (w/v) agarose-formaldehyde gel, transferred to nylon membranes (Hybond-N, Amersham Biosciences, Piscataway, NJ), and then fixed by UV cross-linking (Fisher Scientific, Pittsburgh). Blots were stained with methylene blue (Herrin and Schmidt, 1988). They were immersed briefly in a methylene blue staining solution (0.3 m sodium acetate, pH 5.2, and 0.06% [w/v] methylene blue) and rinsed in deionized distilled water. The moist blots were wrapped in clear plastic and scanned. Probes were obtained by PCR amplification or restriction digestion with appropriate enzymes of plasmid inserts. The DNA was purified using the Geneclean kit (Bio-101, Carlsbad, CA), and 50 to 100 ng was labeled with [α-32P]dCTP using DNA Polymerase I Klenow (Promega). Probes were denatured at 95°C before adding to the hybridization solution. Prehybridization (8 h) and hybridization (36 h) were carried out at 65°C in 7% (w/v) SDS, 0.191 m Na2HPO4, 0.058 m NaH2PO4, 1% (w/v) bovine serum albumin, and 100 μg mL−1 denatured salmon sperm DNA. Washes were done at room temperature with 2× SSC, 0.5% (w/v) SDS (probes with accession nos. BE587600, BE587633, BE586275, BE587392, and BE586278) and with 0.1× SSC and 0.1% (w/v) SDS at 65°C for the rest of the probes. Membranes were wrapped in plastic sheets and exposed to x-ray films at −80°C. Autoradiographs were scanned, and the hybridization signals were quantitated using the Molecular Analyst software (Macintosh v2.1, Apple Computer, Cupertino, CA). The methylene blue images were quantitated using the same software. The signals obtained from the genes were weighted against those obtained from the 25S rRNA methylene blue scan images (Auger et al., 2001) to correct for minor differences in RNA loading and normalized to that at 0 h, which was set at 1 for each gene.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. R.C. Gardner (University of Auckland) for providing the wali1, wali3, and wali5 wheat clones used as a control in the early stages of this study. We thank Dr. D. Blevins, Dr. L. Darrah, Dr. H. Krishnan, Dr. M. McMullen, Dr. J. Polacco, and Miftahudin (University of Missouri-Columbia) for critical reading of the manuscript.
Footnotes
This work was supported in part by the National Science Foundation (grant no. 9975989) and by the Instituto Nacional de Investigacion y Tecnologia Agraria y Alimentaria, Spain (scholarship to M.A.R.M.). This is a contribution from the U.S. Department of Agriculture, Agricultural Research Service, Plant Genetics Research Unit, and the University of Missouri Agricultural Experiment Station. Mention of a proprietary product does not constitute an endorsement or a recommendation for its use by the U.S. Department of Agriculture-Agricultural Research Service or the University of Missouri.
The online version of this article contains Web-only data. The supplemental material is available at www.plantphysiol.org.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.009969.
LITERATURE CITED
- Altschul SF, Gish W, Miller W, Myers EW, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Auger DL, Newton KJ, Birchler JA. Nuclear gene dosage effects upon the expression of maize mitochondrial genes. Genetics. 2001;157:1711–1721. doi: 10.1093/genetics/157.4.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg D, Gerlach M, Youdim MBH, Double KL, Zecca L, Riederer P, Becker G. Brain iron pathways and their relevance to Parkinson's disease. J Neurochem. 2001;79:225–236. doi: 10.1046/j.1471-4159.2001.00608.x. [DOI] [PubMed] [Google Scholar]
- Brosché M, Strid A. Cloning, expression, and molecular characterization of a small pea gene family regulated by low levels of ultraviolet b radiation and other stresses. Plant Physiol. 1999;121:479–487. doi: 10.1104/pp.121.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YC, Ma JF, Matsumoto H. Mechanisms of Al-induced iron chlorosis in wheat (Triticum aestivum): Al-inhibited biosynthesis and secretion of phytosiderophore. Physiol Plant. 1998;102:9–15. doi: 10.1034/j.1399-3054.1998.1020102.x. [DOI] [PubMed] [Google Scholar]
- Chaumont F, Barrieu F, Herman EM, Chrispeels MJ. Characterization of a maize tonoplast aquaporin expressed in zones of cell division and elongation. Plant Physiol. 1998;117:1143–1152. doi: 10.1104/pp.117.4.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin JM, Przybyla AE, MacDonald RJ, Rutter WJ. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5300. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Cruz-Ortega R, Cushman JC, Ownby J. cDNA clones encoding 1,3-β-glucanase and a fimbrin-like cytoskeletal protein are induced by Al toxicity in wheat roots. Plant Physiol. 1997;114:1453–1460. doi: 10.1104/pp.114.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Hebb DM, Richards KD, Lin J-M, Ryan PR, Gardner RC. Cloning and expression of a wheat (Triticum aestivumL.) phosphatidylserine synthase cDNA. J Biol Chem. 1999;274:7082–7088. doi: 10.1074/jbc.274.11.7082. [DOI] [PubMed] [Google Scholar]
- Delhaize E, Ryan PR. Aluminum toxicity and tolerance in plants. Plant Physiol. 1995;107:315–321. doi: 10.1104/pp.107.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using PHRED: II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Ezaki B, Tsugita S, Matsumoto H. Expression of a moderately anionic peroxidase is induced by aluminum treatment in tobacco cells: possible involvement of peroxidase isozymes in aluminum ion stress. Physiol Plant. 1996;96:21–28. [Google Scholar]
- Ezaki B, Yamamoto Y, Matsumoto H. Cloning and sequencing of the cDNAs induced by aluminum treatment and Pi starvation in cultured tobacco cells. Physiol Plant. 1995;93:11–18. [Google Scholar]
- Ferguson DL, Turley RB, Kloth RH. Identification of a δ-TIP cDNA clone and determination of related A and D genome subfamilies in Gossypiumspecies. Plant Mol Biol. 1997;34:111–118. doi: 10.1023/a:1005844016688. [DOI] [PubMed] [Google Scholar]
- Hamel F, Breton C, Houde M. Isolation and characterization of wheat aluminum-regulated genes: possible involvement of aluminum as a pathogenesis response elicitor. Planta. 1998;205:531–538. doi: 10.1007/s004250050352. [DOI] [PubMed] [Google Scholar]
- Hamilton CA, Good AG, Taylor GJ. Induction of vacuolar ATPase and mitochondrial ATP synthase in an aluminum-resistant cultivar of wheat. Plant Physiol. 2001;125:2068–2077. doi: 10.1104/pp.125.4.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrin DL, Schmidt GW. Rapid, reversible staining of northern blots prior to hybridization. Biotechniques. 1988;6:196–197. [PubMed] [Google Scholar]
- Huang JW, Grunes DL, Kochian LV. Aluminum effects on the kinetics of calcium uptake into cells of the wheat root apex. Planta. 1992;188:414–421. doi: 10.1007/BF00192809. [DOI] [PubMed] [Google Scholar]
- Jiang H, Kang D, Alexandre D, Fisher PB. RaSH, a rapid subtraction hybridization approach for identifying and cloning differentially expressed genes. Proc Natl Acad Sci USA. 2000;97:12684–12689. doi: 10.1073/pnas.220431297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DL, Kochian VL. Aluminum inhibition of the inositol 1,4,5-triphosphate signal transduction pathway in wheat roots: a role in aluminum toxicity? Plant Cell. 1995;7:1913–1922. doi: 10.1105/tpc.7.11.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson M, Johansson I, Bush M, McCann MC, Maurel C, Larsson C, Kjellbom P. An abundant TIP expressed in mature highly vacuolated cells. Plant J. 2000;21:83–90. doi: 10.1046/j.1365-313x.2000.00648.x. [DOI] [PubMed] [Google Scholar]
- Keltjens WG. Magnesium uptake by Al-stressed maize plants with special emphasis on cation interactions at root exchange sites. Plant Soil. 1995;171:141–146. [Google Scholar]
- Lee NH, Weinstock KG, Kirkness EF, Earle-Hughes JA, Fuldner RA, Marmaros S, Glodek A, Gocayne JD, Adams MD, Kerlavage AR et al. Comparative expressed-sequence-tag analysis of differential gene expression profiles in PC-12 cells before and after nerve growth factor treatment. Proc Natl Acad Sci USA. 1995;92:8303–8307. doi: 10.1073/pnas.92.18.8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CO, Kim HY, Kim MG, Lee SI, Chung WS, Park SH, Hwang I, Cho MJ. Expressed sequence tags of Chinese cabbage flower bud cDNA. Plant Physiol. 1996;111:577–588. doi: 10.1104/pp.111.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Luan S. Internal aluminum block of plant inward K+channels. Plant Cell. 2001;13:1453–1465. doi: 10.1105/tpc.13.6.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukaszewski KM, Blevins DG. Root growth inhibition in boron-deficient or aluminum-stressed squash may be a result of impaired ascorbate metabolism. Plant Physiol. 1996;112:1135–1140. doi: 10.1104/pp.112.3.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh P, Koshland D. Abstracts of the 1998 Yeast Genetics and Molecular Biology Meeting. University of Maryland, College Park. 1998. Ubiquitin-related Smt3p, required for cell cycle progression in budding yeast, is attached to a subset of nuclear proteins and to a component of the mother-bud junction; p. 300. [Google Scholar]
- Minocha R, Minocha SC, Long SL, Shortle WC. Effects of aluminum on DNA synthesis, cellular polyamines, polyamine biosynthetic enzymes and inorganic ions in cell suspension cultures of a woody plant, Catharanthus roseus. Physiol Plant. 1992;85:417–424. [Google Scholar]
- Nakanishi H, Yamaguchi H, Sasakuma T, Nishizawa NK, Mori S. Two dioxygenase genes, Ids3 and Ids2, from Hordeum vulgareare involved in the biosynthesis of mugeneic acid family phytosiderophores. Plant Mol Biol. 2000;44:199–207. doi: 10.1023/a:1006491521586. [DOI] [PubMed] [Google Scholar]
- Okumura N, Nishizawa NK, Umehara Y, Mori S. An iron deficiency-specific cDNA from barley roots having two homologous cysteine-rich MT domains. Plant Mol Biol. 1991;17:531–533. doi: 10.1007/BF00040651. [DOI] [PubMed] [Google Scholar]
- Ossola JO, Kristoff G, Tomaro ML. Heme oxygenase induction by menadione bisulfite adduct-generated oxidative stress in rat liver. Comp Biochem Physiol Part C. 2000;127:91–99. doi: 10.1016/s0742-8413(00)00133-x. [DOI] [PubMed] [Google Scholar]
- Platt JL, Nath KA. Heme oxygenase: protective gene or Trojan horse. Nat Med. 1998;4:1364–1365. doi: 10.1038/3947. [DOI] [PubMed] [Google Scholar]
- Ragland M, Soliman KM. Sali5-4a (accession no. U64866) and Sali 3-2 (accession no. U89693): two genes induced by aluminum in soybean roots (PGR 97-071) Plant Physiol. 1997;114:395. [Google Scholar]
- Ranieri A, Castagna A, Baldan B, Soldatini GF. Iron deficiency differently affects peroxidase isoforms in sunflower. J Exp Bot. 2001;52:25–35. [PubMed] [Google Scholar]
- Richards KD, Gardner RC. The effect of aluminum treatment on wheat roots: expression of heat shock, histone and SHH genes. Plant Sci. 1994;98:37–45. [Google Scholar]
- Richards KD, Schott EJ, Sharma YK, Davis KR, Gardner RC. Aluminum induces oxidative stress genes in Arabidopsis thaliana. Plant Physiol. 1998;116:409–418. doi: 10.1104/pp.116.1.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards KD, Snowden KC, Gardner RC. Wali6 and wali7: genes induced by aluminum in wheat (Triticum aestivumL.) roots. Plant Physiol. 1994;105:1455–1456. doi: 10.1104/pp.105.4.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sävenstrand H, Brosché M, Ängehagen M, Strid Å. Molecular markers for ozone stress isolated by suppression subtractive hybridization: specificity of gene expression and identification of a novel stress-regulated gene. Plant Cell Environ. 2000;23:689–700. [Google Scholar]
- Sivaguru M, Baluska F, Volkmann D, Felle HH, Horst WJ. Impacts of aluminum on the cytoskeleton of the maize root apex: short-term effects on the distal part of the transition zone. Plant Physiol. 1999;119:1073–1082. doi: 10.1104/pp.119.3.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaski JJ. Differences in the metabolic responses of root tips of wheat and rye to aluminum stress. Plant Soil. 1994;167:165–171. [Google Scholar]
- Smart LB, Moskal WA, Cameron KD, Bennett AB. MIP genes are down-regulated under drought stress in Nicotiana glauca. Plant Cell Physiol. 2001;42:686–693. doi: 10.1093/pcp/pce085. [DOI] [PubMed] [Google Scholar]
- Snowden KC, Gardner RC. Five genes induced by aluminum in wheat (Triticum aestivumL.) roots. Plant Physiol. 1993;103:855–861. doi: 10.1104/pp.103.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton A, Beetham JK, Pinot F, Garbarino JE, Rockhold DR, Friedman M, Hammock BD, Belknap WR. Cloning and expression of soluble epoxide hydrolase from potato. Plant J. 1994;6:251–258. doi: 10.1046/j.1365-313x.1994.6020251.x. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Nishide J, Okazaki K, Kato H, Niwa O, Nakagawa T, Matsuda H, Kawamukai M, Murakami Y. Characterization of a fission yeast SUMO-1 homologue, Pmt3p, required for multiple nuclear events, including the control of telomere length and chromosome segregation. Mol Cell Biol. 1999;19:8660–8672. doi: 10.1128/mcb.19.12.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Nakanishi H, Nishizawa NK, Mori S. Induction of the IDI1 gene in Fe-deficient barley roots: a gene encoding a putative enzyme that catalyses the methionine salvage pathway for phytosiderophore production. Soil Sci Plant Nutr. 2000;46:1–9. [Google Scholar]
- Yamamoto Y, Kobayashi Y, Devi SR, Rikiishi S, Matsumoto H. Aluminum toxicity is associated with mitochondrial dysfunction and the production of reactive oxygen species in plant cells. Plant Physiol. 2002;128:63–72. [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Kobayashi Y, Matsumoto H. Lipid peroxidation is an early symptom triggered by aluminum, but not the primary cause of elongation inhibition in pea roots. Plant Physiol. 2001;125:199–208. doi: 10.1104/pp.125.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Sasaki T. Large-scale EST sequencing in rice. Plant Mol Biol. 1997;35:135–144. [PubMed] [Google Scholar]
- Yu L-H, Umeda M, Liu J-Y, Zhao N-M, Uchimiya H. A novel MT gene in rice plants is strongly expressed in the node portion of the stem. Gene. 1998;206:29–35. doi: 10.1016/s0378-1119(97)00577-5. [DOI] [PubMed] [Google Scholar]
- Zhang G, Hoddinott J, Taylor GJ. Characterization of 1,3-β-glucan (callose) synthesis in roots of Triticum aestivumin response to aluminum toxicity. J Plant Physiol. 1994;144:229–234. [Google Scholar]
- Zhu H, Nowrousian M, Kupfer D, Colot HV, Berrocal-Tito G, Lai H, Bell-Pedersen D, Roe BA, Loros JJ, Dunlap JC. Analysis of expressed sequence tags from two starvation, time-of-day-specific libraries of Neurospora crassareveals novel clock-controlled genes. Genetics. 2001;157:1057–1065. doi: 10.1093/genetics/157.3.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.