Abstract
AtAMT2 is an ammonium transporter that is only distantly related to the five members of the AtAMT1 family of high-affinity ammonium transporters in Arabidopsis. The short-lived radioactive ion 13NH4+ was used to show that AtAMT2, expressed in yeast (Saccharomyces cerevisiae), is a high-affinity transporter with a Km for ammonium of about 20 μm. Changes in external pH between 5.0 and 7.5 had little effect on the Km for ammonium, indicating that NH4+, not NH3, is the substrate for AtAMT2. The AtAMT2 gene was expressed in all organs of Arabidopsis and was subject to nitrogen (N) regulation, at least in roots where expression was partially repressed by high concentrations of ammonium nitrate and derepressed in the absence of external N. Although expression of AtAMT2 in shoots responded little to changes in root N status, transcript levels in leaves declined under high CO2 conditions. Transient expression of an AtAMT2-green fluorescent protein fusion protein in Arabidopsis leaf epidermal cells indicated a plasma membrane location for the AtAMT2 protein. Thus, AtAMT2 is likely to play a significant role in moving ammonium between the apoplast and symplast of cells throughout the plant. However, a dramatic reduction in the level of AtAMT2 transcript brought about by dsRNA interference with gene expression had no obvious effect on plant growth or development, under the conditions tested.
Ammonium and nitrate are thought to be the primary sources of nitrogen (N) for most plants growing in agricultural soils. Acquisition of these inorganic nutrients from the soil solution involves a variety of different transporters, which transport the ions from the apoplast of root epidermal and cortical cells into the symplast. Although ammonium concentrations are often 10 to 1,000 times lower than those of nitrate in well-aerated soil, ammonium nutrition plays an essential role in waterlogged and acid soils (Marschner, 1995). Furthermore, ammonium seems to be a preferred source of N and is taken up more rapidly than nitrate when both ions are presented simultaneously to plants (Gazzarrini et al., 1999).
Physiological studies of ammonium transport into roots have revealed biphasic kinetics in several species (Fried et al., 1965; Vale et al., 1988; Wang et al., 1993). The so-called high-affinity ammonium transport system is predominant at low (submillimolar) concentrations of substrate (NH4+) and exhibits saturation kinetics. A second component of ammonium uptake is the low-affinity transport system, which becomes significant at higher external ammonium concentrations (above 1 mm) and exhibits non-saturation kinetics (Wang et al., 1993; Kronzucker et al., 1996). Although the molecular basis of low-affinity transport system activity remains unknown, there is growing evidence that members of the AMT1 family of transporters are responsible for high-affinity ammonium transport system activity in plants. The first AMT1 gene to be discovered in plants was AtAMT1;1 from Arabidopsis, which was cloned by complementation of a yeast (Saccharomyces cerevisiae) mutant defective in ammonium transport (Ninnemann et al., 1994). Analysis of the Arabidopsis genome sequence reveals four additional closely related genes (AtAMT1;2–5). Several plant AMT1 family members, including three homologs from Arabidopsis and three from tomato (Lycopersicon esculentum) have now been expressed and studied in yeast, and all but one appear to be saturable, high-affinity ammonium transporters that can also transport methylammonium (MA; Gazzarrini et al., 1999; von Wirén et al., 2000). Interestingly, AtAMT1;2 exhibited biphasic transport kinetics when expressed in yeast: In addition to a saturable, high-affinity component, a non-saturable component was manifest at millimolar concentrations of external ammonium (Shelden et al., 2001).
Recently, we reported the isolation and partial characterization of the ammonium transporter AtAMT2 from Arabidopsis, which is only distantly related to plant AMT1 proteins (Howitt and Udvardi, 2000; Sohlenkamp et al., 2000). In fact, AtAMT2 is more closely related to ammonium transporters from prokaryotes than it is to plant AMT1 transporters. Sequencing projects have uncovered novel members of the AMT2 family in several plant species, including Lotus japonicus, Medicago truncatula, rice (Oryza sativa), and Physcomitrella patens. Unlike other ammonium transporters studied to date, AMT2 was unable to transport 14C-MA, which made it difficult to characterize the transport kinetics of this protein (Sohlenkamp et al., 2000).
Plant AMT proteins are presumed to play major roles in the uptake of soil ammonium into roots. For instance, the AtAMT1;1, AtAMT1;2, and AtAMT1;3 genes are all strongly expressed in Arabidopsis roots (Gazzarrini et al., 1999), and regulation of these genes is correlated with changes in ammonium uptake activity of roots (Gazzarrini et al., 1999; Rawat et al., 1999; Shelden et al., 2001). At least two AMT genes from Arabidopsis, AtAMT1;1 and AtAMT2, are also expressed in shoots (Ninnemann et al., 1994; Gazzarrini et al., 1999; Sohlenkamp et al., 2000), where they may play roles in ammonium recycling, especially after photorespiration. The fact that the Arabidopsis genome contains at least six genes encoding ammonium transporters, five from the AMT1 family and one from AMT2, may make it difficult to assign a specific role(s) to any one transporter. Because of the divergence of AtAMT2 from all other AMTs in Arabidopsis, we expected that it might play a role(s) distinct from the other transporters. To uncover this role(s), we have investigated the expression pattern and regulation of AtAMT2, the kinetic properties and intracellular location of the protein, and the physiological and developmental consequences of interfering with expression of the gene. The results of this work are presented here.
RESULTS
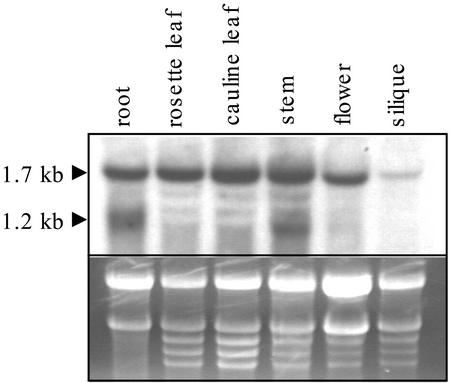
AtAMT2 Is Expressed in All Plant Organs
Previous work showed that AtAMT2 is expressed in both roots and shoots of Arabidopsis (Sohlenkamp et al., 2000). To obtain more information about AtAMT2 expression in different organs, northern-blot analysis was performed using total RNA isolated from stems, rosette leaves, cauline leaves, flowers, and siliques of plants grown in soil for 5 weeks. Root RNA was isolated from plants grown in axenic cultures for 19 d. AMT2 transcripts were detected in all organs examined. Transcript levels were greatest in roots, leaves, and stems, and somewhat lower in flowers and siliques (Fig. 1). In addition to the full-length transcript (1.7 kb), a second shorter transcript of 1.2 kb was detected in both root and stem RNA. As the Arabidopsis genome contains no other sequences that are closely related to AtAMT2, we suspected that the shorter transcript was also derived from AtAMT2. To investigate this further, we hybridized northern blots of total RNA from roots or shoots with antisense riboprobes complementary to either the 5′ end of the AtAMT2 transcript (300 nucleotides) or its 3′ end (400 nucleotides; Fig. 2). The 3′ end probe hybridized to both a 1.7- and a 1.2-kb RNA from roots, whereas the 5′ end probe hybridized to the larger, full-length AMT2 transcript only. Both probes hybridized to only a single, 1.7-kb transcript from leaves. Thus, the 1.2-kb fragment in total RNA from roots appeared to lack the first (5′) 500 bp of the full-length transcript.
Figure 1.
Northern-blot analysis of AMT2 transcripts in different organs of Arabidopsis. Tissue from different organs was harvested from 5-week-old soil-grown plants, except roots, which were harvested from 19-d-old axenically grown plants. Twenty micrograms of total RNA from each organ was separated, blotted, and probed with an antisense riboprobe specific to the last 400 bp (3′-end) of AtAMT2. Lower, Ethidium bromide-stained gel before blotting.
Figure 2.
Structure of two AMT2 transcripts in roots. A, Schematic representation of the putative structure of the two AMT2 transcripts. Black bars indicate parts to the transcripts that were complementary to the antisense riboprobes. B, Northern blot of 20 μg of total RNA from root and leaf tissue of Arabidopsis C24, using riboprobes specific to the 3′ or 5′ part of the AtAMT2 gene.
Regulation of AMT2 Transcription by Nutrient Supply
To determine whether AtAMT2 expression is subject to N regulation, northern-blot analysis of plants grown under different N regimes was performed. Plants were grown in axenic culture for 19 d in modified Murashige and Skoog medium containing 20.6 mm ammonium nitrate and 18.8 mm potassium nitrate, or for 16 d in this medium followed by transfer to a similar medium lacking N for 3 d. After N deprivation, plants were transferred to fresh medium supplemented with 1 mm nitrate for 30 min or 1 h, to test the influence of nitrate on AMT2 expression. The amount of AtAMT2 transcript increased in roots in response to N deprivation (Fig. 3), as observed before (Sohlenkamp et al., 2000). Addition of nitrate to N-deprived plants resulted in a slight decrease in AtAMT2 transcript levels in roots at 30 and 60 min, although levels did not fall to those observed in ammonium nitrate-grown plants. No differences in AtAMT2 transcript levels were detected in shoots of plants subjected to the various treatments (Fig. 3). Once again, roots contained both 1.7 and 1.2 kb AMT2 transcripts, whereas shoots contained only the larger transcripts.
Figure 3.
N regulation of AtAMT2 expression. Plants were grown in axenic culture in media containing 20.6 mm NH4NO3 and 18.8 mm KNO3 for 19 d (A), for 16 d followed by transfer to a similar medium lacking N for 3 d (B), followed by addition of 1 mm nitrate for 30 min (C) or 1 h (D). Twenty micrograms of total RNA from roots and shoots was loaded in each lane. Hybridization was done with a random-primed full-length AtAMT2 cDNA. Equal loading of RNA was confirmed with a probe to the constitutively expressed gene elF4a.
Photorespiration results in significant ammonium production in photosynthetic tissues in C3 plants, but can be suppressed by increasing the ratio of CO2 to O2 in the atmosphere (Somerville and Ogren, 1979, 1980, 1981). To test whether AtAMT2 may play a role in recycling of photorespiratory ammonium, we performed a northern-blot analysis of plants grown in the presence of high CO2. Plants were grown in soil under ambient atmospheric conditions for 3 weeks before being transferred to high CO2 (800 μL L−1) for a further 2 weeks, whereas control plants were kept under ambient CO2 conditions (approximately 400 μL L−1) for the entire period. The high CO2 treatment resulted in a slight decline in AtAMT2 transcript level in leaves (Fig. 4).
Figure 4.
Effect of high CO2 on AtAMT2 expression. Plants were grown in soil in a normal atmosphere (400 μL L−1 CO2) for 3 weeks before a subset of plants were transferred to high CO2 (800 μL L−1) conditions. Twenty micrograms of total RNA isolated from whole plants was separated, blotted, and probed with an antisense riboprobe specific to the 3′ part of AMT2. Lower, Ethidium bromide-stained gel before blotting.
Promoter β-Glucuronidase (GUS) Studies of AMT2 Expression in Arabidopsis
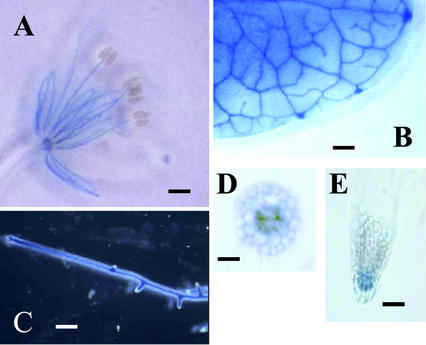
To analyze AtAMT2 expression with greater resolution than was possible with northern blots, transgenic Arabidopsis plants were generated that expressed the GUS reporter gene (Jefferson et al., 1987) under the control of 1.0 kb of the AtAMT2 promoter. T2 individuals of more than 20 transgenic lines were analyzed. Seedlings were grown in vitro in magenta boxes for 4 weeks in Murashige and Skoog medium (Murashige and Skoog, 1962), and all organs were analyzed for GUS activity. GUS activity was detected in the vascular tissues of leaves, stems (not shown), roots, and flowers. GUS activity was also detected in root tips and in the root cortex, but not the root epidermis (Fig. 5). No GUS activity was detected in siliques.
Figure 5.
Localization of AMT2 expression in Arabidopsis with promoter-GUS fusions. Organs from 4-week-old plants were stained for GUS activity: flower (A), rosette leaf (B), root (C), cross section through mature root (D), and root tip (E).
AtAMT2 Encodes a High-Affinity Ammonium Transporter
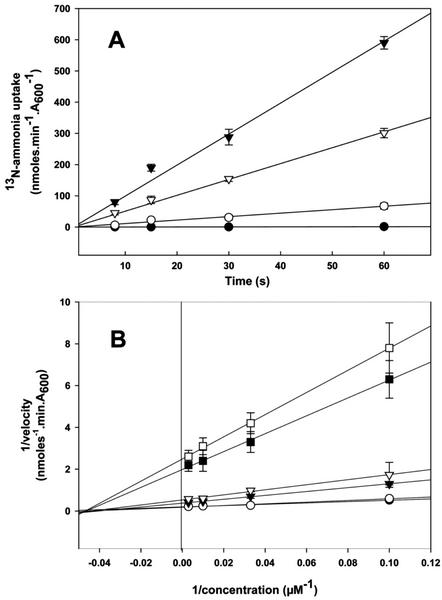
Initial characterization of AtAMT2 showed it to be an ammonium transporter that, unlike AtAMT1 transporters, cannot transport the ammonium analog, MA (Sohlenkamp et al., 2000). The inability of AtAMT2 to transport MA is unfortunate, because it precludes studies of transport kinetics using radioactive 14C-MA. Thus, to determine the transport characteristics of AtAMT2, we resorted to the use of 13N-ammonium, which has a half-life of only 10 min. AtAMT2 was expressed in the yeast ammonium transport mutant, YCW012, which is defective in all three of its known ammonium transporters and does not possess NADPH-dependent Glu dehydrogenase (GDH1), the principle enzyme of ammonia assimilation in yeast. Furthermore, 1 mm l-methionine sulfoximine was added to the cultures before the uptake assays to inhibit Gln synthetase activity. Yeast cells transformed with constructs expressing AtAMT2 were able to take up 13N-ammonium from the external medium (Fig. 6). The same was true for cells expressing the Arabidopsis transporter, AtAMT1;1, which was included as a positive control (Fig. 6). AtAMT1;1 facilitated rapid influx of 13N-ammonium with an apparent Km of 22 μm at pH 6.1 (Table I). Although AtAMT2 also exhibited a high affinity for 13N-ammonium (apparent Km of 21 μm at pH 6.1), its capacity for ammonium transport was at least an order of magnitude lower than that of AtAMT1;1 at pH 5.0 and 6.1 (Fig. 6; Table I). Interestingly, the apparent Vmax of AtAMT2 increased with increasing external pH, whereas that of AtAMT1;1 decreased, so that at pH 7.5 the transport capacity of the two transporters appeared to be similar. The yeast mutant containing the vector pYES3 alone was unable to import 13N-ammonium at either pH 5.0 or 6.1 (Fig. 6). However, significant rates of 13N-ammonium uptake were exhibited by this strain at pH 7.5, which may be attributable to the diffusion of NH3 into the cells.
Figure 6.
13N-ammonium transport by AtAMT1;1 and AtAMT2 expressed in the yeast. A, Time course of 100 μm 13N-ammonium uptake at external pH 6.1 or 7.5. Yeast strain YCW012 was transformed with: pYES3 (●), pAMT2 (○), or pAMT1;1 (▾), and uptake measured at pH 6.1. Uptake was also measured at pH 7.5 for yeast expressing pAMT2 (▿). B, Double reciprocal plots of initial uptake versus ammonium concentration at different external pH. Strain YCW012 was transformed with the following plasmids and assayed at the pH indicated: pAMT1;1 at pH 5.0 (●), pAMT1;1 at pH 6.1 (○), pAMT1;1 at pH 7.5 (▾), pAMT2 at pH 5.0 (□), pAMT2 at pH 6.1 (▪), and pAMT2 at pH 7.5 (▿). The symbols for pAMT1;1 at pH 5.0 and 6.1 partially overlap.
Table I.
Kinetic data for 13N-ammonium uptake by AtAMT1;1 and AtAMT2 expressed in yeast
| Transporter | External pH | Km | Vmaxa |
|---|---|---|---|
| μm | nmol NH4+ min−1 unit A600−1 | ||
| AtAMT1;1 | 5.0 | 16 ± 0.5 | 5.3 ± 1.8 |
| 6.1 | 22 ± 1.1 | 5.6 ± 1.2 | |
| 7.5 | 25 ± 2.5 | 2.7 ± 1.0 | |
| AtAMT2 | 5.0 | 23 ± 5.5 | 0.3 ± 0.2 |
| 6.1 | 21 ± 3 | 0.5 ± 0.3 | |
| 7.5 | 22 ± 1.3 | 1.9 ± 0.5 |
Values are derived from Figure 6B.
The background rate of ammonium uptake measured in the mutant strain YCW012 containing pYES3 was taken into account when calculating Vmax. Estimates of error represent 95% confidence intervals and were calculated from independent samples.
Subcellular Localization of AtAMT2
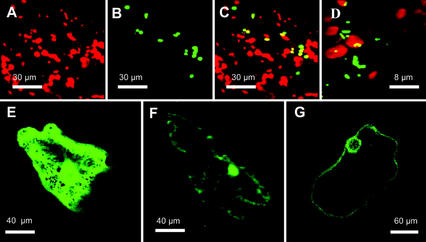
Although circumstantial evidence points toward a plasma membrane location for some AMT1 family members (Lauter et al., 1996; Gazzarrini et al., 1999; Shelden et al., 2001), no direct evidence for this has been published. To determine the intracellular location of AtAMT2 in Arabidopsis, a translational fusion between AMT2 and the fluorescent reporter protein, green fluorescent protein (GFP) was transiently expressed in Arabidopsis leaves under the control of a double cauliflower mosaic virus 35S promoter. To test the efficacy of this approach, a variety of positive controls were included in our experiments. The control constructs were GFP (smGFP without a leader sequence; Davis and Vierstra, 1998), RbcS-GFP (pea [Pisum sativum] RbcS plastid transit peptide fused to GFP; Anderson and Smith, 1986), mGFP5 (which is targeted to the endoplasmic reticulum [ER]; Haseloff et al., 1997), and cox-GFP (transit peptide of mitochondrial cytochrome oxidase fused to GFP). Transient expression of RbcS-GFP in epidermal cells of Arabidopsis led to accumulation of GFP in chloroplasts (Fig. 7, A–C). The cox-GFP fusion protein was found in mitochondria of transformed cells, which were smaller, and clearly distinct from chloroplasts (Fig. 7D). The mGFP5 protein was observed in the ER, whereas GFP without a leader sequence was located in the cytoplasm (Fig. 7, E and F, respectively). Transient expression of the AtAMT2-GFP fusion in leaf epidermal cells invariably led to green fluorescence on the plasma membrane and the nuclear envelope (Fig. 7G). Labeling of the latter may simply reflect the fact that plasma membrane proteins are delivered via the ER, which is contiguous with the nuclear envelope. We have observed this phenomenon for other GFP fusions that are targeted to the plasma membrane (C. Sohlenkamp and M.K. Udvardi, unpublished data).
Figure 7.
Transient expression of AtAMT2-GFP in Arabidopsis leaf epidermal cells. The images shown are representative of numerous transformed cells. Images show chlorophyll fluorescence (red), GFP fluorescence (green), or both. A through C, Epidermal cell expressing RbcS-GFP. A, Chlorophyll fluorescence. B, GFP fluorescence. C, Overlap of both channels, with yellow indicating coincidence of GFP and chlorophyll. D, Overlay of chlorophyll and the GFP fluorescence of an epidermal cell expressing cox-GFP. E, Epidermal cell expressing mGFP5 (Haseloff et al., 1997), which is targeted to the ER. F, Cell expressing GFP without a leader sequence, which distributes between cytoplasm and nucleus. G, Epidermal cell expressing AMT2-GFP. GFP fluorescence can be observed on the border of the cell and on the nuclear envelope.
Molecular Physiology of AtAMT2
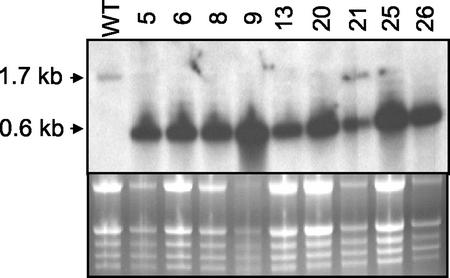
To gain further insight into the possible physiological role(s) of AtAMT2, we used a reverse genetics approach using RNA interference (RNAi; Fire et al., 1998; Kennerdell and Carthew, 1998; Waterhouse et al., 1998; Baulcombe, 1999; Cogoni and Macino, 1999; Smith et al., 2000; Wianny and Zernicka-Goetz, 2000). A binary vector containing the entire AtAMT2 coding sequence (1.5 kb) and a 0.9-kb fragment from the 5′ end of the coding sequence in a head-to-tail configuration was constructed and transformed into Arabidopsis. Seeds from transformed plants were selected on Murashige and Skoog plates containing kanamycin, and resistant seedlings were transferred to soil. Four weeks after germination, rosette leaves from 40 independent lines were harvested for northern-blot analysis of AtAMT2 expression. An antisense riboprobe specific to the 3′ part (500 bp) of the AtAMT2 gene was used for hybridization. The wild-type transcript of AtAMT2 was undetectable in most of the RNAi lines (e.g. Fig. 8). Instead of the 1.7-kb transcript present in the wild type, RNAi lines exhibited an abundant 0.6-kb RNA fragment that was thought to represent the loop connecting the two complementary parts of the RNA hairpin structure (Fig. 8). This was confirmed with two other riboprobes. A sense probe identical to the loop sequence did not hybridize to the 0.6-kb band, as expected, whereas an antisense probe specific to the 5′ part of the wild-type AMT2 transcript hybridized to much smaller, apparently degraded fragments (not shown). Despite the absence of detectable AtAMT2 transcript in many of the RNAi lines, none of them exhibited aberrant growth or development under a variety of different growth conditions (data not shown).
Figure 8.
RNAi inhibition of AMT2 expression in Arabidopsis. Upper, Results of a typical northern-blot analysis of transgenic RNAi lines of Arabidopsis expressing the AMT2 hairpin construct. Randomly chosen leaves were harvested from 4-week-old plants and 10 μg of total RNA was subjected to northern analysis, using an antisense riboprobe specific to the 3′ part (400 bp) of AMT2. Wt, Wild-type C24; numbers refer to different RNAi lines. Lower, Ethidium bromide-stained gel before blotting.
DISCUSSION
The results presented here on the expression pattern and regulation of AtAMT2 confirm and extend previous work on this subject (Sohlenkamp et al., 2000). Transcripts of AtAMT2 were found in all organs tested, with highest levels being detected in roots, leaves, and stems. Lower levels of transcript were found in flowers and siliques (Fig. 1). Promoter-GUS studies showed that the AtAMT2 promoter (1.0 kb) was active in vascular tissues of roots, leaves, and flowers. Lower activity was also detected in other tissue types, including the root cortex and meristematic region (Fig. 5). Expression of AtAMT2 in various tissues indicates that the transporter is likely to play diverse roles in the plant.
Expression of AtAMT2 in roots was subject to regulation by N supply (Fig. 3). Thus, the amount of AtAMT2 transcript increased in roots after 3 d of N deprivation. Subsequent addition of N to the roots, in the form of 1 mm nitrate, resulted in a decrease in AtAMT2 transcript levels toward the level exhibited by plants grown in the presence of both ammonium and nitrate (Fig. 3). In contrast to the situation in roots, AtAMT2 transcript levels remained unchanged in the shoots of plants subjected to the various treatments (Fig. 3). It has been shown for AtAMT1;1 that transcript levels respond to the internal N status of the plant (Rawat et al., 1999). Given the similarities in the responses of AtAMT2 and AtAMT1;1 to altered N supply, it is likely that transcription of AtAMT2 in Arabidopsis roots responds to the same N cues. The lack of response of AtAMT2 expression in shoots of plants exposed to different N regimes may reflect real differences in the regulation of the gene in shoots and roots. However, it is also possible that the shoots were somewhat buffered by the roots against changes in external and internal N pools, which might otherwise have caused similar changes in AtAMT2 expression in shoots.
Unlike AtAMT1 genes, AtAMT2 is expressed at least as strongly in leaves as it is in roots (Figs. 1–3). Photorespiration in shoots leads to production of ammonium, which must be recycled for the plant to maintain its N status. In fact, the flux of ammonium through this cycle can exceed ammonium influx into roots by a factor of 10 (Keys et al., 1978), and photorespiratory mutants that cannot reassimilate this ammonium efficiently are unable to grow under ambient atmospheric conditions (Somerville and Ogren, 1979, 1980, 1981; Wallsgrove et al., 1987). To investigate the possible involvement of AtAMT2 in ammonium recycling after photorespiration, plants were exposed to double the normal concentration of CO2 in the atmosphere to repress photorespiration. Alterations in the CO2 to O2 ratio have been found to alter the expression of GS2 in Phaseolus vulgaris, a gene that is involved in ammonium recycling after photorespiration (Wallsgrove et al., 1987; Cock et al., 1991). In contrast, no such regulation was found for GS2 in Arabidopsis (Beckmann et al., 1997). AtAMT2 transcript levels fell slightly in response to high CO2 (Fig. 4), indicating that the transporter may participate in ammonium recycling during photorespiration.
In addition to the 1.7-kb AtAMT2 transcript that was detected in all organs, a second, 1.2-kb RNA was detected in roots, using an AtAMT2 probe. (Figs. 1–3). Further analysis of the shorter transcript indicated that it probably corresponded to the 3′ end of AtAMT2 because it lacked sequence homologous to the 5′ end of the full-length transcript (Fig. 2). If this is the case, then it will be interesting to determine whether the shorter transcript is derived from the longer by RNA processing, or whether it results from transcription from an alternative promoter in the AtAMT2 gene. Interestingly, the AtAMT2 gene has four introns, one of which is 400 bp downstream of the normal start of transcription. A promoter within this intron could, in theory, produce a 1.2-kb transcript. Multiple transcripts have been found for other genes. For example, cystathione γ-synthase mRNA in Arabidopsis was truncated at the 5′ end in a process that was dependent on Met concentration (Chiba et al., 1999). Mutants not able to process the full-length transcript were isolated during a screen for plants that accumulate Met to high concentrations inside the cells, which highlighted the importance of such processing (Chiba et al., 1999). By analogy, the 1.2-kb AtAMT2 transcript may also play a regulatory role. In this context, it is interesting to note that its synthesis is not only regulated in a developmental manner, but also by N supply to the plant (Figs. 1–3).
This paper presents for the first time, to our knowledge, short-term kinetic data for 13N-ammonium transport by an isolated plant AMT (Fig. 6). AtAMT2 and AtAMT1;1 were expressed separately in a yeast mutant defective in all known ammonium transporters. The data indicate that AtAMT2 and AtAMT1;1 are both high-affinity ammonium transporters, with Km for NH4+ of approximately 15 to 25 μm. A similar value was obtained for LeAMT1;1 recently using electrophysiology (Ludewig et al., 2002). Although expression in yeast of both AtAMT2 and AtAMT1;1 were driven from the same Gal-inducible (GAL1) promoter, the Vmax for ammonium transport in AtAMT1;1-containing cells was at least an order of magnitude greater than that exhibited by AtAMT2-containing cells, at pH 5 and 6.1. Although we do not know whether similar numbers of transporter molecules were targeted to and functional in the plasma membrane in both cases, the Vmax data may indicate an important difference between AMT2 and AMT1 transporters. We have consistently observed shorter doubling times for yeast expressing AMT1 proteins from Arabidopsis and L. japonicus (Simon-Rosin et al., 2003) than for those expressing AMT2 transporters from these species, which would be consistent with a higher Vmax for AMT1 transporters.
Apart from the possible difference in Vmax, AtAMT2 and AtAMT1;1 activity in yeast exhibited different responses to changes in external pH. AtAMT1;1 activity was insensitive to pH changes between pH 5.0 and 6.1 and decreased by more than 50% when pH was increased from 6.1 to 7.5. In contrast, AtAMT2 activity increased consistently with increasing pH from 5.0 to 7.5, such that at pH 7.5 the activity of AtAMT2 approached that of AtAMT1;1 (Fig. 6). The pH profiles of AMT activity can help to discern whether NH4+ or NH3 is the transported molecule. Ammonia is a weak base (pKa 9.25), which is more than 99% protonated (NH4+) at neutral pH. As pH increases from pH 5.0 to 7.5, the concentration of NH3 increases 316-fold, whereas that of NH4+ changes by about only 1%. Thus, rates of NH3 transport would be expected to be strongly dependent on pH in this range, whereas those of NH4+ would be independent of pH, other things being equal. Ammonium transport by AtAMT1;1 in yeast was independent of changes of pH between 5.0 and 6.1, and actually decreased between pH 6.1 and 7.5. This, together with the fact that the Km for ammonium was unaffected by external pH, is evidence that NH4+, and not NH3, is the substrate for this transporter (Table I). The same conclusion was reached recently in an elegant electrophysiological study of LeAMT1;1, a related ammonium transporter from tomato (Ludewig et al., 2002). The form of ammonium transported by AtAMT2 is less clear, partly because of the low capacity exhibited by the transporter. Although ammonium transport rates increased with increasing pH, the magnitude of these changes was far less than would be expected if NH3 were transported, especially when the background levels of ammonium uptake exhibited by the control mutant strain were taken into account. On the other hand, the Km for ammonium uptake by AtAMT2 was similar at pH 6.1 and 7.5, which indicates that NH4+ is probably the substrate for this transporter also.
Despite indirect evidence that some plant AMT1 transporters are located on the plasma membrane (Lauter et al., 1996; Gazzarrini et al., 1999; Rawat et al., 1999; Shelden et al., 2001), no direct evidence for this has been published. Our attempts to demonstrate this directly by immunolocalization have failed so far for want of specific antibodies to the AMT proteins. However, using an AtAMT2-GFP fusion construct, we were able to demonstrate that the information inherent in the AtAMT2 protein was sufficient to target the GFP marker to the plasma membrane of Arabidopsis cells (Fig. 7). With the exception of some labeling of the nuclear envelope, which may be accounted for by proteins en route to the plasma membrane via the ER-nuclear membrane continuum, no other membrane systems were labeled in cells expressing the fusion protein. Thus, AtAMT2 is very likely to be a plasma membrane protein in Arabidopsis.
Double-stranded RNA has proved to be highly effective at inducing posttranscriptional gene silencing in plants (Baulcombe, 1999; Cogoni and Macino, 1999; Chen et al., 2000; Smith et al., 2000; Waterhouse et al., 1998). RNAi has been shown to phenocopy a number of well-known mutants, including a set of flower development and meristem identity mutants (Chen and Meyerowitz, 2000). We also found RNAi to be very effective at reducing or eliminating expression of AtAMT2 in Arabidopsis (Fig. 8). Our aim in eliminating AtAMT2 expression was to identify an aberrant phenotype(s) that would provide insight into the role of the transporter in plants. In particular, we were interested in a possible role of AtAMT2 in ammonium recycling after photorespiration. However, despite the dramatic reduction in AtAMT2 transcript levels in many of the transgenic lines, none of the lines showed a photorespiratory phenotype or any other obvious changes compared with wild-type plants. Thus, an essential role for AtAMT2 during photorespiration can be excluded. Detailed molecular analyses of these lines, for instance metabolite profiling, may uncover subtle changes in the RNAi lines. Although residual AtAMT2 activity in the numerous transgenic lines may have been sufficient to fulfill the normal role(s) of the protein, we think that this is unlikely, given the apparent absence of AtAMT2 transcript in many of these lines. What seems more likely is that other ammonium transporters are able to substitute for a lack of AtAMT2 activity. This idea will be tested in the future using AtAMT2 knockout mutants.
MATERIALS AND METHODS
Growth of Arabidopsis
Arabidopsis ecotype C24 was grown either in soil in a phytotron with an 18-/6-h day/night cycle (120 μmol m−2 s−1 light intensity) at 22°C/18°C and 75%/60% humidity, or in a tissue culture room at 22°C with continuous light (120 μmol m−2 s−1). A modified Murashige and Skoog medium was used for growing the plants in axenic culture (Sohlenkamp et al., 2000).
Expression Analysis of AMT2
RNA was isolated from leaves, roots, and flowers using Trizol reagent (Life Technologies, Rockville, MD). For the RNA isolation from stems and siliques, a detergent-based isolation method was used (Jacobsen-Lyon et al., 1995). After separation of the RNA by gel electrophoresis under denaturing conditions (Lehrach et al., 1977), the RNA was transferred to nylon membranes (Porablot NY Amp, Macherey-Nagel, Düren, Germany) and UV cross-linked to the membrane. Blots were probed with either single-strand RNA (a) or a double-stranded DNA (b) probes, as follows: (a) Nylon membranes were prehybridized for at least 1 h at 50°C and hybridized overnight at 50°C in a solution consisting of 50% (v/v) formamide, 0.25 m sodium phosphate buffer (pH 7.2), 0.25 m NaCl, 1 mm EDTA, 7% (w/v) SDS, and α-32P-UTP-labeled RNA probe, which was produced by in vitro transcription from linearized plasmid containing the cDNA of interest (Epicentre Technologies, Madison, WI). Membranes were washed at room temperature for 5 min in 2× SSC, twice in PSE buffer (0.25 m sodium phosphate buffer [pH 7.2], 2% [w/v] SDS, and 1 mm EDTA), twice in PES buffer (0.04 m sodium phosphate buffer [pH 7.2], 1% [w/v] SDS, and 1 mm EDTA) and three times in 2× SSC at room temperature. The filters were treated with 1 μg mL−1 RNase A in 2× SSC for 15 min at room temperature before being washed in 0.1× SSC and 0.1% (w/v) SDS for 30 min at 50°C and subjected to autoradiography; or (b) Nylon membranes were prehybridized for at least 1 h at 65°C and hybridized overnight at 65°C in 250 mm sodium phosphate buffer (pH 7.2) containing 7% (w/v) SDS, 1% (w/v) bovine serum albumin, and 1 mm EDTA. Radioactively labeled probe was prepared using the Rediprime kit (Amersham, Buckinghamshire, UK). After hybridization, membranes were washed at 65°C once in 2× SSC and 1% (w/v) SDS and twice in 0.2× SSC and 0.1% (w/v) SDS, then subjected to autoradiography.
Ammonium Uptake Measurements
Yeast (Saccharomyces cerevisiae) strain YCW011 (Δmep1 Δmep2 Δmep3 Δgdh1 MATa) was produced from strain MLY31 (Lorenz and Heitman, 1998), by homologous recombination of the wild-type GDH1 with GDH1::URA3, using pGDH1del (Boles et al., 1993). A uracil auxotroph of strain YCW011 (strain YCW012) was produced by homologous recombination of the functional URA3 gene (within the mutated GDH1) with a dysfunctional form of the gene produced by ApaI digestion, Klenow fill in, and religation of URA3, which generated a frame shift mutation. Strain YCW012 (Δmep1 Δmep2 Δmep3 Δgdh1 ura3 MATa) was selected on media containing 2.5 mg mL−1 5-fluoroorotic acid and 50 μg mL−1 uracil, and later tested on uracil-minus medium to confirm the loss of URA3 gene function.
Yeast plasmid pYES3 (Smith et al., 1995) containing AtAMT1;1 or AtAMT2 was transformed into YCW012 and selected for uracil prototrophy and subsequently the ability to complement growth under ammonium-limiting conditions (Sohlenkamp et al., 2000). Standard molecular methods were used throughout and yeast strains were transformed using the lithium acetate method as described by Gueldener et al. (1996).
For uptake experiments, yeast strains were grown overnight in liquid cultures at 28°C to 30°C in a uracil- and ammonia-free media containing per liter: 0.17% (w/v) yeast minimal salts without amino acids and without ammonia (DIFCO Laboratories, Detroit), 20 g of Gal, 1g of Na-Glu, 1 g of MES buffer, 1.4 g of synthetic amino acid powder lacking uracil (Sigma, St. Louis), and pH adjusted to 6.1 with NaOH. Yeast cells were harvested by centrifugation at 3,000g for 5 min and resuspended in freshly prepared media to a final A600 of 6 to 8. To start reactions, 200 μL of cells was added to 1.5-mL Eppendorf tubes (Eppendorf Scientific, Westbury, NY) containing a fixed amount of 13N-ammonium and a variable amount of 14NH4Cl to give a final ammonium concentration between 10 and 300 μm. After mixing, 200 μL of cells was loaded above a silicon oil layer (100 μL of 3:1 [w/v] AR20:AR200, Fluka, Milwaukee, WI) in a thin-walled 400-μL polyethylene tube (Bio-Rad, Hercules, CA), and reactions were terminated at the appropriate times by centrifugation at 14,000 rpm for 5 s (Beckman MicrofugeE, Beckman Instruments, Fullerton, CA). The cell pellet was then cut from the end of the tube and immediately measured in a gamma counter (Wallac 1480 Wizard 3, PerkinElmer Wallac, Gaithersburg, MD). Uptake was considered stopped at the commencement of centrifugation and the fastest reaction times were typically 8 s. All uptake measurements were independent replicates and were typically repeated three to five times with different batches of cells. In Figure 6, the error bars represent 95% confidence intervals and linear regressions were performed using SigmaPlot software (SPSS, Inc., Chicago).
Production and Purification of 13NH4+
The short-lived radioisotope 13N was produced by proton irradiation of water according to the 16O (p,α) 13N reaction (Meeks, 1993). The cyclotron of Forschungszentrum Jülich GmbH (Japanese Steel BC 1710, Jülich, Germany) provides a 15-μA proton beam. The irradiation procedure generates primarily 13NO3− and only traces of 13NH4+ (Wieneke, 1992). For the chemical conversion of NO3− to ammonia and online clean up of the ammonia generated, a special apparatus was designed to handle small amounts (2–5 mL) of the radioactive solutions. The process was initiated by reduction of 13NO3− using Devarda's alloy under alkaline conditions (in a modification of Kronzucker et al., 1996). The 13NH3 produced was flushed by a stream of N2 gas into a trap containing a solution of 0.01 n H2SO4, adjacent to a radiation detector. The 13N-ammonia-labeled trapping solution was finally adjusted to pH 5. The purity of each sample was checked by gamma spectroscopy and measurements of the gamma emission decay times. Thus, 13N decay (half-life of 9.97 min) can be readily distinguished from the possible contaminant 18F (half-life of 109.7 min).
Construction of Arabidopsis AMT2 Promoter-Reporter Lines
One kilobase pair of sequence upstream (5′) of the start codon of AtAMT2 was PCR amplified using primers that introduced PstI (oAMT2P1, 5′-ACG TCT GCA GAA CAT GAA TCT TAT TGA ATC TCT AA-3′) and SmaI (oAMT2P2, 5′-ACG TCC CGG GTT TGT TAT TCT ATC TTT CCC GGA g-3′) restriction sites into the product. The product was digested with PstI and SmaI and cloned into pUC19 (Yanisch-Perron et al., 1985). After sequence verification, the promoter was subcloned as a PstI/SmaI fragment upstream (5′) of the GUS reporter gene in the binary vector pCAMBIA 1381Z. Arabidopsis C24 plants (T0) were transformed using a floral dip method (Clough and Bent, 1998) and seeds were selected on Murashige and Skoog medium with 1% (w/v) Suc containing 20 μg mL−1 hygromycin A. About 20 T1 transgenic seedlings were grown to maturity in soil and T2 seeds were harvested. T2 plants were then grown in vitro for 4 weeks in magenta boxes on Murashige and Skoog medium. Plants were harvested and the different organs were assayed for GUS activity. Plant organs were transferred to wells of microtiter plates filled with GUS buffer (50 mm sodium phosphate buffer (pH 7.2), 10 mm EDTA, 0.1% (w/v) Triton X-100, 0.1% (w/v) Tween 20, 210 mg L−1 K4Fe(CN)6.3H2O, 166 mg L−1 K3Fe(CN)6, and 0.5 mg mL−1 5-bromo-4-chloro-3-indolyl-β-glucuronic acid), vacuum infiltrated, and then incubated in the dark at 37°C for several hours to overnight. Tissue was then destained in 70% (v/v) ethanol.
Microprojectile Bombardment and GFP Imaging
Using the primers oAMT2;1gfp1 (5′-AGC TCG AGA TGG CCG GAG CTT ACG ATC C-3′) and oAMT2;1gfp2 (5′-AGA CTA GTA CTA GAA CAA TGG TGA CAC CTC-3′), the first 1,200 bp of the coding sequence of AtAMT2 (AF182039) was amplified using Pfu DNA polymerase (Stratagene, La Jolla, CA), then digested with the restriction enzymes cutting at the incorporated sites (XhoI and SalI). The DNA fragment was cloned into the respective restriction sites of the vector pA7-GFP to form a translational fusion between the N terminal of AtAMT2 and GFP. Vectors were then sequenced to confirm the identity of the constructs. pA7-GFP is a pUC19 derivative with the GFP containing cassette from the binary vector pJH-GFP (J. Harper, personal communication).
Microprojectile bombardment of Arabidopsis C24 leaves was performed with a Bio-Rad PDS-1000, as previously described (Hibbert et al., 1998). Imaging of GFP and chlorophyll fluorescence was performed using a confocal scanner TCS SPII together with a DM IRBE microscope (Leica, Heidelberg) 24 to 48 h after bombardment. GFP and chlorophyll were excited with light of wavelength 488 nm.
Construction and Analysis of RNAi Lines
The pBluescript SK− vector containing full-length AMT2 cDNA (Sohlenkamp et al., 2000) was digested with XbaI/BamHI and the AMT2 fragment was subcloned in sense orientation into the binary vector pGPTVkan-35S (Becker et al., 1992) downstream of the CaMV35S promoter to produce pbinAMT2s. Nine hundred base pairs of the 5′ end of the AMT2 cDNA was amplified with primer1 introducing a KpnI site (5′-TGG CCG GAG CTC ACG ATC CAA GCT TGC-3′) and primer2 (5′-ACG TGG TAC CAT GCC TTG AAT TGC TCC-3′). The PCR product was digested with KpnI and EcoRI and cloned into pUC19 (pUC19-AMT2as), before being subcloned into the KpnI and EcoRI sites of pbinAMT2s. The resulting binary vector, pbinAMT2sas, contained the 900-bp fragment downstream (3′) of, and in antisense orientation to, the full-length AtAMT2 cDNA. Arabidopsis ecotype C24 was transformed with this construct using a floral dip method (Clough and Bent, 1998). Transgenic (T1) plants were selected on Murashige and Skoog medium containing 50 μg mL−1 kanamycin before being transferred to soil. Leaves of 4-week-old plants were harvested for RNA analysis. Plants were also monitored daily to detect any aberrant growth or development phenotypes.
ACKNOWLEDGMENTS
We thank Marco Dautzenberg (Forschungszentrum Jülich GmbH, Jülich, Germany) for his excellent technical assistance. We appreciate the generous gifts of haploid strains of yeast MLY31 from Professor Joseph Heitman (Duke University Medical Center, Durham, NC) and the plasmid pGDH1del from Professor Eckhard Boles (University of Düsseldorf, Germany).
Footnotes
This work was supported by the Max Planck Society for the Advancement of Science and by the Alexander von Humboldt Foundation.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.008599.
LITERATURE CITED
- Anderson S, Smith SM. Synthesis of the small subunit of ribulose-bisphosphate carboxylase from genes cloned into plasmids containing the SP6 promoter. Biochem J. 1986;240:709–715. doi: 10.1042/bj2400709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe DC. Fast forward genetics based on virus-induced gene silencing. Curr Opin Plant Biol. 1999;2:109–113. doi: 10.1016/S1369-5266(99)80022-3. [DOI] [PubMed] [Google Scholar]
- Becker D, Kemper E, Schell J, Masterson R. New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol. 1992;20:1195–1197. doi: 10.1007/BF00028908. [DOI] [PubMed] [Google Scholar]
- Beckmann K, Dzuibany C, Biehler K, Fock H, Hell R, Migge A, Becker TW. Photosynthesis and fluorescence quenching, and the mRNA levels of plastidic glutamine synthetase or of mitochondrial serine hydroxymethyltransferase (SHMT) in the leaves of the wild-type and of the SHMT-deficient stm mutant of Arabidopsis thaliana in relation to the rate of photorespiration. Planta. 1997;202:379–386. doi: 10.1007/s004250050140. [DOI] [PubMed] [Google Scholar]
- Boles E, Lehnert W, Zimmermann FK. The role of the Nad-dependent glutamate-dehydrogenase in restoring growth on glucose of a Saccharomyces cerevisiae phosphoglucose isomerase mutant. Eur J Biochem. 1993;217:469–477. doi: 10.1111/j.1432-1033.1993.tb18266.x. [DOI] [PubMed] [Google Scholar]
- Chen CF, Meyerowitz EM. Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2000;97:4985–4990. doi: 10.1073/pnas.060034297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba Y, Ishikawa M, Kijima F, Tyson RW, Kim J, Yamamoto A, Nambara E, Leustek T, Wallsgrove RM, Naito S. Evidence for autoregulation of cystathionine γ-synthase mRNA stability in Arabidopsis. Science. 1999;286:1371–1374. doi: 10.1126/science.286.5443.1371. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–753. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cock JM, Brock IW, Watson AT, Swarup R, Morby AP, Cullimore JV. Regulation of glutamine synthetase genes in leaves of Phaseolus vulgaris. Plant Mol Biol. 1991;17:761–772. doi: 10.1007/BF00037059. [DOI] [PubMed] [Google Scholar]
- Cogoni C, Macino G. Homology-dependent gene silencing in plants and fungi: a number of variations on the same theme. Curr Opin Microbiol. 1999;2:657–662. doi: 10.1016/s1369-5274(99)00041-7. [DOI] [PubMed] [Google Scholar]
- Davis SJ, Vierstra RD. Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol Biol. 1998;36:321–328. doi: 10.1023/a:1005991617182. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SA, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fried MF, Zsoldos F, Vose PB, Shatokin IL. Characterizing the NO3− and NH4+ uptake of rice roots by use of 15N labelled NH4NO3−. Plant Physiol. 1965;18:313–320. [Google Scholar]
- Gazzarrini S, Lejay L, Gojon A, Ninnemann O, Frommer WB, von Wirén N. Three functional transporters for constitutive, diurnally regulated, and starvation induced uptake of ammonium into Arabidopsis roots. Plant Cell. 1999;11:937–947. doi: 10.1105/tpc.11.5.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueldener U, Heck S, Fiedler T, Beinhauer J, Hegemann JH. A new efficient disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci USA. 1997;94:2122–2127. doi: 10.1073/pnas.94.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbert JM, Linley PJ, Khan MS, Gray JC. Transient expression of green fluorescent protein in various plastid types following microprojectile bombardment. Plant J. 1998;16:627–632. [Google Scholar]
- Howitt SM, Udvardi MK. Structure, function and regulation of ammonium transporters in plants. Biochim Biophys Acta. 2000;1465:152–170. doi: 10.1016/s0005-2736(00)00136-x. [DOI] [PubMed] [Google Scholar]
- Jacobsen-Lyon K, Jensen EO, Jorgensen JE, Marcker KA, Peacock WJ, Dennis ES. Symbiotic and nonsymbiotic hemoglobin genes of Casuarina glauca. Plant Cell. 1995;7:213–223. doi: 10.1105/tpc.7.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions, β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennerdell JR, Carthew RW. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled2 act in the same pathway. Cell. 1998;95:1017–1026. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- Keys AJ, Bird IF, Cornelius MJ, Lea PJ, Wallsgrove RM, Miflin BJ. Photorespiratory nitrogen cycle. Nature. 1978;275:741–743. [Google Scholar]
- Kronzucker HJ, Siddiqi MY, Glass ADM. Kinetics of NH4+ influx in spruce. Plant Physiol. 1996;110:773–779. doi: 10.1104/pp.110.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauter FR, Ninnemann O, Bucher M, Riesmeier JW, Frommer WB. Preferential expression of an ammonium transporter and two putative nitrate transporters in root hairs of tomato. Proc Natl Acad Sci USA. 1996;93:8139–8144. doi: 10.1073/pnas.93.15.8139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrach H, Diamond D, Wozney JM, Boetcker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical re-examination. Biochemistry. 1977;16:4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- Lorenz MC, Heitman J. The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 1998;17:1236–1247. doi: 10.1093/emboj/17.5.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludewig U, von Wiren N, Frommer WB. Uniport of NH4+ by the root hair plasma membrane ammonium transporter LeAMT1;1. J Biol Chem. 2002;277:13548–13555. doi: 10.1074/jbc.M200739200. [DOI] [PubMed] [Google Scholar]
- Marschner H. Mineral Nutrition of Higher Plants. Ed 2. London: Academic Press; 1995. [Google Scholar]
- Meeks JC. 13N techniques. In: Knowles R, Blackburn TH, editors. Nitrogen Isotope Techniques. San Diego, CA: Academic Press; 1993. pp. 273–303. [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue. Physiol Plant. 1962;15:473–495. [Google Scholar]
- Ninnemann O, Jauniaux JC, Froomer WB. Identification of a high-affinity transporter from plants. EMBO J. 1994;13:3463–3471. doi: 10.1002/j.1460-2075.1994.tb06652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat SR, Silim SN, Kronzucker HJ, Siddiqi MY, Glass ADM. AtAMT1 gene expression and NH4+ uptake in roots of Arabidopsis thaliana: evidence for regulation by root glutamine levels. Plant J. 1999;19:143–152. doi: 10.1046/j.1365-313x.1999.00505.x. [DOI] [PubMed] [Google Scholar]
- Shelden MC, Dong B, De Bruxelles G, Trevaskis B, Whelan J, Ryan PR, Howitt SM, Udvardi MK. Arabidopsis ammonium transporters, AtAMT1;1 and AtAMT1;2 have different biochemical properties and functional roles. Plant Soil. 2001;231:151–160. [Google Scholar]
- Simon-Rosin U, Wood CC, Udvardi MK. Molecular and cellular characterisation of LjAMT2;1, an ammonium transporter from the model legume Lotus japonicus. Plant Mol Biol. 2003;51:99–108. doi: 10.1023/a:1020710222298. [DOI] [PubMed] [Google Scholar]
- Smith F, Ealing P, Hawkesford M, Clarkson D. Plant members of a family of sulfate transporters reveal functional subtypes. Proc Natl Acad Sci USA. 1995;92:9373–9377. doi: 10.1073/pnas.92.20.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith NA, Singh SP, Wang MB, Stoutjesdijk PA, Green AG, Waterhouse PM. Gene expression: Total silencing by intron-spliced hairpin-RNAs. Nature. 2000;407:319–320. doi: 10.1038/35030305. [DOI] [PubMed] [Google Scholar]
- Sohlenkamp C, Shelden M, Howitt S, Udvardi M. Characterization of Arabidopsis AtAMT2, a novel ammonium transporter in plants. FEBS Lett. 2000;476:273–278. doi: 10.1016/s0014-5793(00)01153-4. [DOI] [PubMed] [Google Scholar]
- Somerville CR, Ogren WL. A phosphoglycolate phosphatase-deficient mutant of Arabidopsis. Nature. 1979;280:833–836. [Google Scholar]
- Somerville CR, Ogren WL. Inhibition of photosynthesis in Arabidopsis mutants lacking leaf glutamate synthase activity. Nature. 1980;286:257–259. [Google Scholar]
- Somerville CR, Ogren WL. Photorespiration-deficient mutants of Arabidopsis thaliana lacking mitochondrial serine transhydroxymethylase activity. Plant Physiol. 1981;67:666–671. doi: 10.1104/pp.67.4.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale FR, Volk RJ, Jackson WA. Simultaneous influx of ammonium and potassium into maize roots: kinetics and interactions. Planta. 1988;173:424–431. doi: 10.1007/BF00401031. [DOI] [PubMed] [Google Scholar]
- von Wirén N, Lauter FR, Ninnemann O, Gillisen B, Walch-Liu P, Engels C, Jost W, Frommer WB. Differential regulation of three functional ammonium transporter genes by nitrogen in root hairs and by light in leaves of tomato. Plant J. 2000;21:167–175. doi: 10.1046/j.1365-313x.2000.00665.x. [DOI] [PubMed] [Google Scholar]
- Wallsgrove RM, Turner JC, Hall NP, Kendall AC, Bright SWJ. Barley mutants lacking chloroplast glutamine synthetase: biochemical and genetic analysis. Plant Physiol. 1987;83:155–158. doi: 10.1104/pp.83.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MY, Siddiqi MY, Ruth TJ, Glass ADM. Ammonium uptake by rice roots: II. Kinetics of 13NH4+ influx across the plasmalemma. Plant Physiol. 1993;103:1259–1267. doi: 10.1104/pp.103.4.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse PM, Graham MW, Wang MB. Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc Natl Acad Sci USA. 1998;95:13959–13964. doi: 10.1073/pnas.95.23.13959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wianny F, Zernicka-Goetz M. Specific interference with gene-function by double-stranded RNA in early mouse development. Nat Cell Biol. 2000;2:70–75. doi: 10.1038/35000016. [DOI] [PubMed] [Google Scholar]
- Wieneke J. Nitrate fluxes in squash seedlings measured with 13N. J Plant Nutr. 1992;15:99–124. [Google Scholar]
- Yanisch-Perron C, Viera J, Messing J. Improved M13 phage cloning vector and host strains: nucleotide sequences of the M13 mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]