Abstract
In bacteria, the regulatory ACT domains serve as amino acid-binding sites in some feedback-regulated amino acid metabolic enzymes. We have identified a novel type of ACT domain-containing protein family in Arabidopsis whose members contain ACT domain repeats (the “ACR” protein family). There are at least eight ACR genes located on each of the five chromosomes in the Arabidopsis genome. Gene structure comparisons indicate that the ACR gene family may have arisen by gene duplications. Northern-blot analysis indicates that each member of the ACR gene family has a distinct expression pattern in various organs from 6-week-old Arabidopsis. Moreover, analyses of an ACR3 promoter-β-glucuronidase (GUS) fusion in transgenic Arabidopsis revealed that the GUS activity formed a gradient in the developing leaves and sepals, whereas low or no GUS activity was detected in the basal regions. In 2-week-old Arabidopsis seedlings grown in tissue culture, the expression of the ACR gene family is differentially regulated by plant hormones, salt stress, cold stress, and light/dark treatment. The steady-state levels of ACR8 mRNA are dramatically increased by treatment with abscisic acid or salt. Levels of ACR3 and ACR4 mRNA are increased by treatment with benzyladenine. The amino acid sequences of Arabidopsis ACR proteins are most similar in the ACT domains to the bacterial sensor protein GlnD. The ACR proteins may function as novel regulatory or sensor proteins in plants.
Compared with yeast and Escherichia coli, little is known about proteins or regulatory domains involved in amino acid sensing and signaling in plants. The recently identified Arabidopsis PII-like protein and Glu receptor homologs have been proposed to be involved in such processes (Hsieh et al., 1998; Lam et al., 1998; Coruzzi and Bush, 2001). In response to the Gln/α-ketoglutarate status of the cell, the E. coli sensor proteins PII (encoded by the glnB gene) and PII-uridylyl transferase/uridylyl-removing enzyme (GlnD, encoded by the glnD gene) work in concert with the adenylyltransferase and the NtrB/NtrC two-component system to regulate the expression of the Gln synthetase gene and its enzyme activity (Bueno et al., 1985; Rhee et al., 1985; Ninfa and Atkinson, 2000).
The E. coli sensor protein GlnD is composed of a nucleotide transferase domain, a nucleotide hydrolase domain, and two C-terminal ACT domains. The ACT domain, an amino acid-binding domain named after bacterial aspartate kinase, chorismate mutase, and TyrA (prephenate dehydrogenase), is a regulatory domain mostly found in enzymes directly or indirectly involved in amino acid and purine metabolism (Aravind and Koonin, 1999; Chipman and Shaanan, 2001). Because the ACT domain has been found in functionally diverse enzymes, a common origin of allosteric regulation in these enzymes has been proposed (Aravind and Koonin, 1999). In this hypothesis, the regulatory ACT domain, a conserved, evolutionarily mobile module, was independently fused to a variety of enzymes. The acquisition of the ACT domain made these enzymes susceptible to the regulation by their respective ligands. For example, the E. coli 3-phosphoglycerate dehydrogenase (PGDH) is the most well-characterized ACT domain-containing protein. The C-terminal ACT domain of E. coli PGDH is the binding site for its allosteric regulator Ser, and the binding of Ser feedback inhibits the catalytic activity of PGDH (Schuller et al., 1995; Grant et al., 1999, 2001). Besides E. coli PGDH, the ACT domains may also serve as amino acid-binding sites in other feedback-regulated amino acid metabolic enzymes including acetohydroxyacid synthase (AHAS; Mendel et al., 2001), Asp kinase (AK; Kikuchi et al., 1999; Viola, 2001), AK/homo-Ser dehydrogenase (AK/HSD; Omori and Komatsubara, 1993; Omori et al., 1993; Viola, 2001), chorismate mutase (CM; Zhang et al., 1998; Chipman and Shaanan, 2001), and Thr deaminase (TD; Chinchilla et al., 1998; Gallagher et al., 1998; Chipman and Shaanan, 2001).
In addition to the GlnD sensor protein and some amino acid metabolic enzymes, the ACT domain is also found in bacterial RelA/SpoT (GTP pyrophosphokinase/phosphohydrolase) and TyrR (Tyr and phenol metabolism operon regulator). RelA and SpoT proteins are involved in the synthesis of guanosine tetraphosphate (ppGpp) and guanosine pentaphosphate (pppGpp), compounds that play a central role in the bacterial stringent response (Cashel et al., 1996). TyrR is a transcriptional regulator of amino acid metabolism in bacteria (Wilson et al., 1995; Pittard, 1996). In these regulatory or sensor proteins, it is likely that the ACT domain binds to yet unidentified ligands to transduce the signal(s) or to regulate their catalytic activities.
In a survey of the Arabidopsis genome, we have found several ACT domain-containing protein families including amino acid metabolic enzymes AK, AK/HSD, PGDH, the small subunit of AHAS, and a putative prephenate dehydratase (PDT). Here, we report the identification and molecular characterization of a novel Arabidopsis gene family (ACR) encoding proteins with four copies of the ACT domain that extend throughout the entire polypeptide. Other than the ACT domains, the deduced amino acid sequences of the ACR proteins have no homology to any known enzymes or protein motif in the PSI-BLAST conserved domain search. This is in contrast to other known ACT domain-containing proteins where the regulatory ACT domain is usually linked to an enzyme (Aravind and Koonin, 1999). Moreover, the ACT domains of the ACR proteins are highly conserved in the region corresponding to the putative ligand-binding site. Thus far, the only known ligands bound to the ACT domains are amino acids. The possibility that the Arabidopsis ACR proteins may serve as amino acid-binding proteins will be discussed herein.
RESULTS
Molecular Characterization of the Arabidopsis ACR Gene Family
In an attempt to identify plant ACT domain-containing proteins or putative amino acid sensor proteins, we used the ACT consensus sequence (Pfam01842) and bacterial GlnD sequences to search the Arabidopsis expressed sequence tag (EST) database and the recently completed Arabidopsis genome sequence. In our initial search, we found several Arabidopsis EST clones and computer-predicted proteins in the Arabidopsis genome that share homology with the C-terminal ACT domain of bacterial GlnD. These were annotated as uridylyltransferase-like or putative uridylyltransferase in the database (GenBank accession nos. AAD20075, AAF27052, BAB11136, and AAC00631). The genes encoding these “putative uridylyltransferases” and other homologous genes encoding proteins annotated as “unknown” or “hypothetical proteins” in the database form a novel gene family in Arabidopsis. We have named this novel gene family the ACR gene family, because the encoded proteins contain ACT repeats (Fig. 1).
Figure 1.
Sequence alignment of Arabidopsis ACR proteins. Identical and similar amino acid residues are shaded in black and gray, respectively. The locations of the ACT domains are indicated with solid lines above the sequences. Asterisks shown above the sequences denote the putative nuclear localization sequence of ACR1. The amino acid sequences of ACR1 and ACR3 to ACR8 are deduced from the cDNAs reported here. The amino acid sequence of ACR2 is from accession number NM_122441.
There are at least eight ACR genes in the Arabidopsis genome. Of the eight predicted ACR genes, we have cloned seven ACR cDNAs (ACR1 and ACR3–8) by screening a cDNA library with probes derived from the EST clones or by reverse transcriptase (RT)-PCR using total RNA from 2-week-old Arabidopsis as a template. Two EST clones corresponding to two different genes, ACR3 and ACR7, were used to screen a cDNA library for full-length cDNAs. The amino acid sequence deduced from the full-length ACR3 cDNA is identical to that predicted by computer in the GenBank database (accession no. AAC00631). The ACR7 cDNA sequence is different from the computer-predicted sequence only in the splice site of the second exon. The expected 5′-GU… AG-3′ sequence was found at the splice site; however, a second 5′-GU in the intron following exon 2 was mistakenly picked up as the splice site by the computer prediction. This resulted in the predicted protein having nine extra amino acid residues (VFTLTQPSS) between K26 and I27 (accession no. CAA16563). In addition, the computer-predicted protein is incorrectly annotated as a “translation factor EF-1 α-like protein” in GenBank. A PSI-BLAST search with the deduced ACR7 amino acid sequence revealed that no significant homology was found between this predicted protein and the known translation factor EF-1 α.
On the basis of the predicted open reading frame sequences in the database, we designed 5′ and 3′ primers for RT-PCR and successfully amplified the full-length cDNAs of ACR1, ACR4 to ACR6, and ACR8 using total RNA from 2-week-old Arabidopsis seedlings. We repeatedly failed in obtaining the ACR2 (NM_122441) cDNA by RT-PCR. Together with the results from northern-blot analyses (see below), we conclude that the ACR2 gene is not expressed or is expressed at extremely low levels. The cDNA sequences and the deduced amino acid sequences of ACR1, ACR4, ACR5, ACR6, and ACR8 are identical to the predicted sequences in the database (accession nos. BAB11136, NP_177067, AAD20075, AAF14836, and AAF79655). The deduced ACR amino acid sequences share similarities between 47.4% and 76.5% (Table I).
Table I.
Amino acid similarities among Arabidopsis ACRs
| ACR2 | ACR3 | ACR4 | ACR5 | ACR6 | ACR7 | ACR8 | |
|---|---|---|---|---|---|---|---|
| ACR1 | 53.8 | 54.2 | 53.6 | 52.5 | 48.0 | 47.4 | 49.8 |
| ACR2 | 55.1 | 52.0 | 51.2 | 51.9 | 50.6 | 49.2 | |
| ACR3 | 58.5 | 56.8 | 56.1 | 52.4 | 53.0 | ||
| ACR4 | 76.5 | 68.3 | 56.1 | 58.6 | |||
| ACR5 | 65.3 | 53.8 | 54.5 | ||||
| ACR6 | 53.0 | 55.7 | |||||
| ACR7 | 71.7 |
Alignment of Arabidopsis ACR Proteins
The alignment of the ACR amino acid sequences clearly shows that each member of the ACR protein family contains four copies of the ACT domain (ACT1–4; Fig. 1). Interestingly, these ACT domains are highly conserved and fused together by variable linker sequences (Fig. 1). The first three ACT domains, ACT1 to ACT3, are separated by two different linker sequences. The fourth ACT domain, ACT4, is closely linked to ACT3 and is followed by a C-terminal diverse sequence. In addition to the conserved ACT domains, two highly conserved regions are also found in the ACR protein family. The first conserved region is located in the N terminus before the first ACT domain. The second conserved region is in the middle of the linker sequence between ACT2 and ACT3. No homologous sequences corresponding to these two conserved regions were found in the database.
The deduced amino acid sequences were used with the computer program PSORT to analyze the subcellular localization of the ACR proteins. The ACR1 protein was predicted to be a nuclear protein by the presence of a Robbins and Dingwall consensus sequence (328 KK NGGTLETEGQ RERLR; Fig. 1). The pattern of this consensus sequence (two basic residues, 10 residue spacers, and another basic region consisting of at least three basic residues out of five residues) is based on the nuclear targeting signal in Xenopus nucleoplasmin (Robbins et al., 1991). The other ACR proteins were predicted to be cytosolic proteins by PSORT. A computer program designed to predict the presence of chloroplast transit peptides (http://www.cbs.dtu.dk/services/ChloroP/) was also used to confirm the prediction that none of the ACR proteins are localized in the chloroplast.
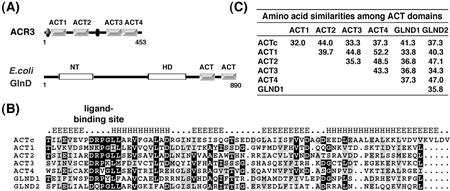
Of the ACT domain-containing proteins in the database, Arabidopsis ACR proteins are most similar to the ACT domains in bacterial GlnD. No significant homology was found between these Arabidopsis proteins and bacterial GlnD outside of the ACT domain. The nucleotide transferase and hydrolase domains of bacterial GlnD were not found in the ACR proteins (Fig. 2A). The sequence alignment of the ACT consensus sequence (Pfam01842), four ACT domains, ACT1 to ACT4, from Arabidopsis ACR3 and two ACT domains (GlnD1 and GlnD2) from E. coli GlnD indicates that these ACT domains are highly conserved in the region corresponding to the ligand-binding site (Fig. 2B).
Figure 2.
A, Schematic diagram of Arabidopsis ACR and E. coli GlnD proteins. The gray boxes indicate the ACT domains. Arabidopsis ACR3 is shown as a representative of the ACR protein family. It contains four ACT domains and two conserved regions (ovals). E. coli GlnD has two ACT domains that show high similarity to those in the Arabidopsis ACR proteins. NT, Nucleotide transferase domain; HD, nucleotide hydrolase domain. B, Sequence alignment of ACT consensus sequence (ACTc) from Pfam01842, four ACT domains (ACT1–ACT4) from Arabidopsis ACR3, and two ACT domains (GlnD1 and GlnD2) from E. coli GlnD. Identical and similar amino acid residues are shaded in black and gray, respectively. The secondary structure of the ACTc predicted by a computer program (http://www.aber.ac.uk/∼phiwww/prof/) is shown above the sequences. The most conserved portion of the ACT domain is the region at the interface between the first strand and the first helix, which corresponds to the ligand-binding site. C, Amino acid sequence similarities among the ACT domains aligned in B.
Location and Gene Structure Comparison of the ACR Gene Family
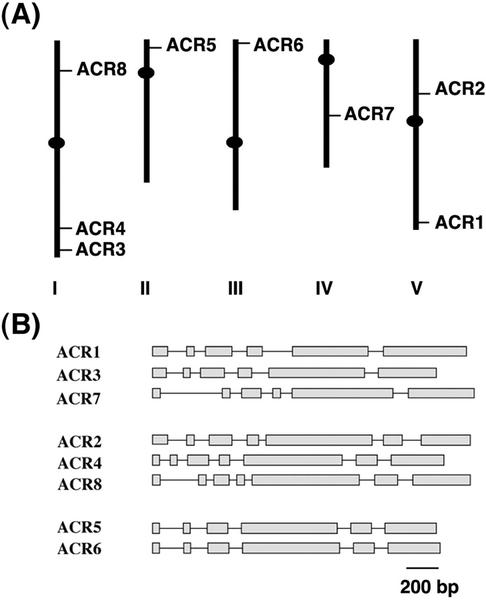
The eight members of the ACR gene family are distributed on all five chromosomes of Arabidopsis (Fig. 3A). Gene structure comparison of the ACR gene family members suggests that this multigene family is the result of gene duplications in the Arabidopsis genome. On the basis of their exon and intron composition, the ACR gene family can be divided into three different groups (Fig. 3B). Similar exon and intron arrangements are found in each group. The first group includes the ACR1, ACR3, and ACR7 genes, and the second group, ACR2, ACR4, and ACR8. Members of the third group, ACR5 and ACR6, have almost identical gene structures with respect to the size and arrangement of their exons and introns.
Figure 3.
A, Chromosomal locations of the Arabidopsis ACR genes. The relative sizes of five Arabidopsis chromosomes are derived from the National Center for Biotechnology Information database. B, Schematic gene structures of the ACR gene family. Exons are shown as gray boxes and solid lines indicate introns.
Expression Patterns of the ACR Gene Family
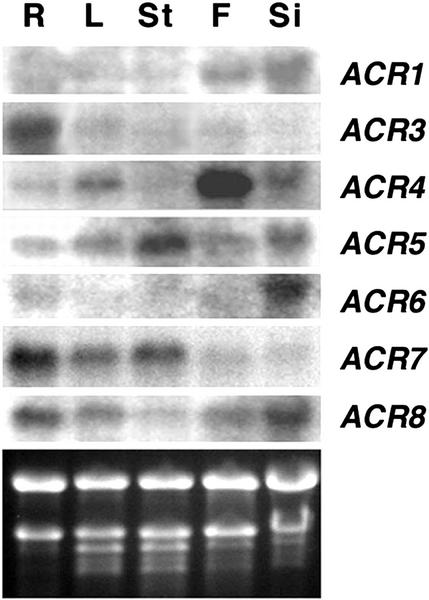
The expression patterns of the ACR gene family was examined by northern-blot analysis using total RNA extracted from roots, leaves, stems, flowers, and siliques of soil-grown 6-week-old Arabidopsis plants (Fig. 4). Distinct expression patterns of the ACR gene family were observed. Higher levels of mRNA were detected in flowers and siliques for ACR1, whereas in the same samples, the steady-state level of ACR2 mRNA was undetectable. Levels of the ACR3 mRNA are high in roots and low or undetectable in the other organs. ACR4 mRNA showed very high levels in flowers but only low to moderate levels in roots, leaves, and siliques. The ACR4 mRNA was not detectable in stems. By contrast, high levels of the ACR5 mRNA were detected in stems whereas the steady-state levels were moderate or low in the other organs. The ACR6 mRNA levels were high in siliques and low or undetectable in roots and other organs. Levels of the ACR7 mRNA were high in roots, moderate in leaves and stems, and low in flowers and siliques. The ACR8 mRNA was detected in all the organs tested with higher levels in roots and siliques, modest levels in leaves and flowers, and low levels in stems. The tissue-specific expression of some ACR genes may reflect their specific roles in a particular organ.
Figure 4.
ACR gene family expression in various organs of 6-week-old Arabidopsis. Total RNA (10 μg) from roots (R), leaves (L), stems (St), flowers (F), and siliques (Si) was used for northern-blot analysis and hybridized with gene-specific probes of ACR1 to ACR8. The ACR2 transcript was not detectable (not shown). The ethidium bromide-stained agarose gel of the same samples is shown at the bottom.
Promoter Activity of the ACR3 Gene
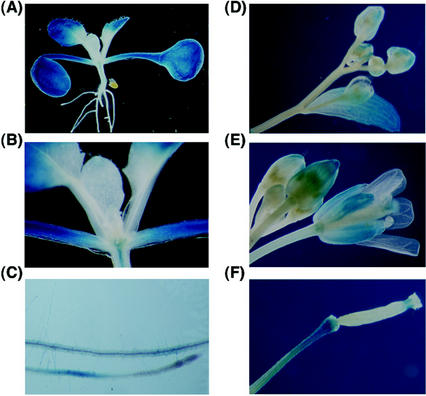
A 0.8-kb putative promoter for ACR3 was used to construct a β-glucuronidase (GUS) reporter gene fusion (ACR3p-GUS) for stable transformation of Arabidopsis. In 2-week-old Arabidopsis seedlings, the GUS activity was detected in the entire cotyledons including the petioles, the distal part of the first leaves, the vascular tissues of the hypocotyls, and the maturation and elongation zones of roots (Fig. 5, A–C). No GUS activity was detected in the emerging young leaves, the basal part of the first leaves, and the root tips of 2-week-old Arabidopsis (Fig. 5, A–C). These regions are mostly composed of dividing and growing young cells (sink). In adult plants, the GUS activity was detected in all mature rosette leaves and mature cauline leaves (data not shown). In contrast, the GUS activity was only detected in the distal part but not the basal part of the young cauline leaves (Fig. 5D). In developing or mature flowers, the GUS activity was detected in the sepal as a gradient from the apical part (high) to the basal part (low to undetectable; Fig. 5, D and E). The GUS activity was also detected in the style of the mature flower (Fig. 5E). In young developing siliques, the GUS activity was detected in the pedicel and the tip region that derived from the style of the mature flower (Fig. 5F). The GUS activity gradient in the young cauline leaves and sepals suggests that the ACR3 gene may be preferentially expressed in the mature cells (source).
Figure 5.
GUS activity in transgenic Arabidopsis containing the ACR3p-GUS fusion. A, Two-week-old seedlings; B, close-up of the emerging leaves and shoot apex in A; C, roots from 2-week-old seedlings; D, young flower buds and a developing cauline leaf; E, flower buds and a mature flower; and F, a developing silique.
Effects of Plant Hormones on the Expression of the ACR Gene Family
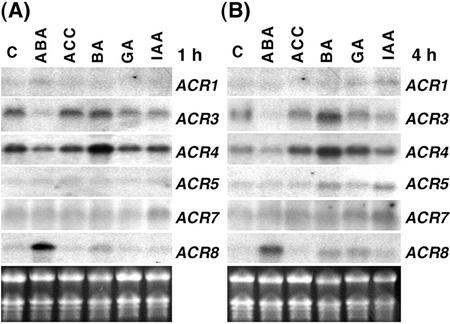
We used 2-week-old Arabidopsis seedlings grown in tissue culture to study the regulation of the ACR gene family. The effects of five classical plant hormones, abscisic acid (ABA), ethylene, cytokinin, gibberellin (GA), and auxin, on the expression of the ACR gene family were examined. The most dramatic response of the ACR gene family to treatment with these plant hormones is the induction of the ACR8 gene by ABA (Fig. 6). The steady-state levels of ACR8 mRNA rapidly increased after 1 h of ABA treatment, whereas treatments with 1-aminocyclopropane-1-carboxylic acid (ACC), benzyladenine (BA), GA, and indole-3-acetic acid (IAA) had little or no significant effects on the levels of ACR8 mRNA (Fig. 6A). Similar results were observed after treatment with plant hormones for 4 h (Fig. 6B).
Figure 6.
Plant hormones differentially regulate steady-state levels of ACR mRNA. Total RNA (10 μg) from 2-week-old Arabidopsis seedlings treated with plant hormones for 1 h (A) or 4 h (B) was used for northern analysis and hybridized with ACR1 to ACR8 gene-specific probes. Levels of ACR8 mRNA were dramatically increased by ABA (compared with C, control). B, Levels of ACR3 and ACR4 mRNA were increased by treatment with BA for 4 h. The transcripts of ACR2 and ACR6 were not detectable. The ethidium bromide-stained gel is shown at the bottom.
In contrast to the positive effect on the expression of the ACR8 gene, ABA negatively regulates the expression of the ACR3 gene. Levels of ACR3 mRNA significantly decreased after 1 or 4 h of ABA treatment and slightly decreased after 1 or 4 h of GA and IAA treatments (Fig. 6). In addition, levels of ACR3 mRNA increased after 4 h of BA treatment, whereas ACC had no significant effect (Fig. 6B). The plant hormones also had various effects on the expression of ACR4. Levels of ACR4 mRNA were increased significantly after treatment with BA for 4 h (Fig. 6B). No significant changes were observed in the levels of ACR1, ACR5, and ACR7 mRNA when the 2-week-old Arabidopsis seedlings were treated with the hormones for 1 or 4 h (Fig. 6). Steady-state levels of ACR2 and ACR6 mRNA in the same samples were not detectable by northern analysis.
Effects of Salt, Cold, and Light/Dark on the Expression of ACR Gene Family
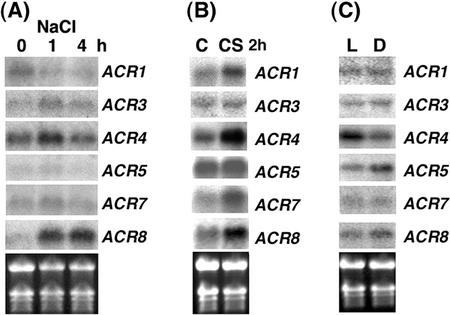
The effects of salt stress, cold stress, and light/dark treatment on the expression of the ACR gene family in 2-week-old Arabidopsis were also tested. Levels of ACR1 mRNA were reduced after treatment with salt for 1 or 4 h (Fig. 7A). A slight increase in levels of ACR3 and ACR4 mRNA was observed after 1 h but not 4 h of salt treatment (Fig. 7A). Little or no change in the steady-state levels of ACR5 and ACR7 mRNA was observed after the salt treatment (Fig. 7A). Interestingly, similar to the ABA treatment, the steady-state levels of ACR8 mRNA were also dramatically increased by the salt treatment (Fig. 7A). The cold treatment at 4°C for 2 h also differentially affected the expression of the ACR gene family. Steady-state levels of ACR1, ACR4, ACR7, and ACR8 mRNA were increased by the cold treatment, whereas levels of ACR3 and ACR5 mRNA were not affected (Fig. 7B). For light/dark treatment, Arabidopsis seedlings grown in a normal 16-h light/8-h dark cycle for 2 weeks were subsequently treated with continuous light or dark for 48 h. Levels of ACR4 mRNA were up-regulated in the light-treated seedlings. By contrast, dark treatment slightly increased levels of ACR5 mRNA (Fig. 7C).
Figure 7.
The levels of ACR mRNA are differentially regulated by salt, cold, and light/dark treatment. Total RNA (10 μg) from 2-week-old Arabidopsis seedlings treated with 250 mm NaCl for 0, 1, or 4 h (A), 4°C for 2 h (B), or light/dark for 48 h (C) was used for northern analysis to detect levels of ACR1 to ACR8 mRNA. C, Control; CS, cold stress; L, light; D, dark. The transcripts of ACR2 and ACR6 were not detectable. The ethidium bromide-stained gel is shown at the bottom.
Other ACT Domain-Containing Protein Families in Arabidopsis
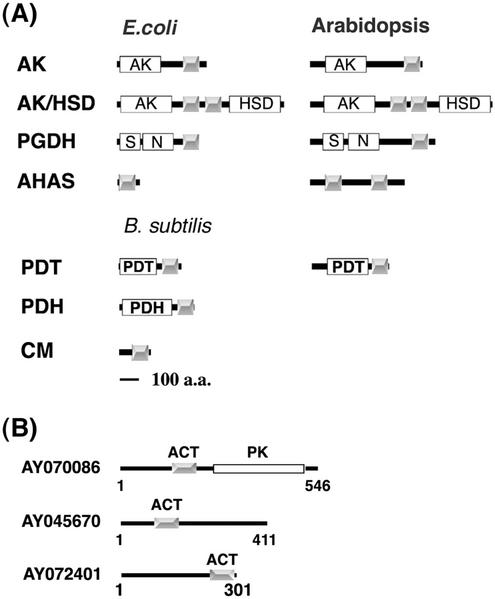
In addition to the ACR protein family, our initial BLAST search of the Arabidopsis genome with the ACT consensus sequence also hit two AHAS-like proteins (NM_121634 and NM_128739; Fig. 8A) and one putative protein kinase (NM_127324; Fig. 8B). Because most of the bacterial ACT domain-containing enzymes have homologs in Arabidopsis, we further analyzed these sequences using the InterProScan computer program (http://www.ebi.ac.uk/interpro/scan.html). The results revealed that some Arabidopsis amino acid metabolic enzymes also contain ACT domains as do their bacterial counterparts (Aravind and Koonin, 1999; Chipman and Shaanan, 2001; Fig. 8A). For example, a C-terminal ACT domain is present in each of the three Arabidopsis monofunctional AK isozymes. In two of the Arabidopsis AK/HSD isozymes that have the AK domain in the N terminus and the HSD domain in the C terminus of the same polypeptide, two ACT domains are located in the linker sequence between the enzymatic domains. Although feedback inhibition by Ser is not strongly supported (Ho et al., 1999), a C-terminal ACT domain also exists in each of the three Arabidopsis PGDH isozymes.
Figure 8.
A, Schematic diagram of ACT domains in Arabidopsis amino acid metabolic enzymes. The gray boxes indicate the conserved regulatory ACT domains. The white boxes represent the catalytic domains. S, substrate-binding domain; N, nucleotide-binding domain; PDH, prephenate dehydrogenase. B, Schematic diagram of novel ACT domain-containing proteins in Arabidopsis. The gray boxes indicate the ACT domains. AY070086, AY045670, and AY072401 are representatives of the ACT domain-containing protein kinase family and two unknown protein families, respectively. PK, Protein kinase domain (Pfam00069).
Unlike the E. coli AHAS regulatory subunit that has one ACT domain, the two Arabidopsis AHAS isozymes each have two copies of the ACT domain in the regulatory subunit. In addition, two of the six predicted Arabidopsis proteins annotated as putative PDT also contain ACT domains. These proteins may actually function as arogenate dehydratase, because plants are known to use arogenate, instead of phenylpyruvate, in the Phe biosynthetic pathway (Siehl and Conn, 1988). In E. coli, the CM and PDT are fused together as a bifunctional enzyme with a regulatory ACT domain in the C terminus (Chipman and Shaanan; 2001). In other bacteria, Bacillus subtilis, for instance, monofunctional CM, PDT, and PDH, each have an ACT domain in the C terminus (Fig. 8A). No ACT domains were found in the Arabidopsis CM isozymes (CM1–CM3) or the RelA/SpoT homologs. It is noteworthy that Arabidopsis AK, AK/HSD, and PGDH have very similar domain compositions and arrangements compared with the E. coli enzyme counterparts. The locations in similar positions of the ACT domains in the Arabidopsis amino acid metabolic enzymes and their bacterial counterparts (Fig. 8A) might indicate that the enzyme-linked ACT domain may also function as a regulatory domain in plants.
In addition to the amino acid metabolic enzymes and the ACR protein family reported here, several computer-predicted or expressed Arabidopsis proteins also contained the ACT domain. These proteins include a novel protein kinase family (four members) and two unknown protein families (two members each). Schematic protein domain structures of AY070086, AY045670, and AY072401 representing each of these families are shown in Figure 8B.
DISCUSSION
The ACT domain was recently recognized as a structurally conserved domain involved in the binding of small ligands, such as amino acids, in functionally diverse proteins (Aravind and Koonin, 1999; Chipman and Shaanan, 2001). Of the identified proteins containing the ACT domains, E. coli PGDH is the only protein that has been unambiguously shown to bind its allosteric inhibitor Ser in the ACT domain. The notion that the ACT domain contains the amino acid-binding site is also supported by recent studies on E. coli AHAS and AK. Several lines of evidence derived from biochemical and mutational analyses revealed that the Val-binding region of the AHAS regulatory subunit and the Lys-binding site of AK are both located in the ACT domain (Kikuchi et al., 1999; Mendel et al., 2001). Thus, the ACT domain may represent one type of regulatory domain that evolved independently from the catalytic domain of enzymes involved in amino acid metabolism.
In agreement with this hypothesis, the ACT domain is also present in some Arabidopsis amino acid metabolic enzymes such as AK, AK/HSD, PGDH, AHAS regulatory subunit, and putative PDT (Fig. 8A). Even though the mechanisms of feedback inhibition of plant amino acid metabolic enzymes are not known, the regulatory domains and the amino acid-binding sites in some of the enzymes have been recognized. For instance, the recently identified Arabidopsis AHAS regulatory subunit contains two internal repeats, each repeat starts with an ACT domain, and the first repeat binds Leu whereas the second binds Val or Ile (Chipman and Shaanan, 2001; Lee and Duggleby, 2001). The similarity in protein domain locations and studies on the AHAS regulatory subunit suggest that the conserved ACT domain may also function as a regulatory domain in plants.
We have identified and characterized a novel ACR gene family in Arabidopsis. The proteins encoded by this novel gene family contain four copies of the ACT domain that extend throughout the entire polypeptide. Tissue specificity was observed in some of the ACR genes. For example, in 6-week-old Arabidopsis plants grown in soil, the ACR4 gene is highly expressed in the flower but not in the stem. Levels of ACR6 mRNA are high in siliques and low or undetectable in roots and other organs (Fig. 4). It is conceivable that the ACR4 and ACR6 gene products may have distinct functions in flowers and siliques, respectively. The tissue specificity of ACR3 gene expression has been investigated by studying a GUS reporter fusion. To have a better understanding about the expression and regulation of the entire ACR gene family, similar and more detailed studies should be applied to all members of the ACR gene family. Nevertheless, in the stable Arabidopsis transformants harboring the ACR3p-GUS construct, the GUS activity formed a gradient in the young leaves and sepals, whereas low or no GUS activity was detected in the basal part or younger cells (sink). This suggests that the ACR3 gene may be preferentially expressed in the mature cells (source). It would be interesting to know whether the ACR3 protein plays any role in sensing the source-sink transition inside plant cells.
The effects of exogenous plant hormone treatment, salt stress, cold stress, and light/dark treatment on the expression of the ACR gene family have been tested using 2-week-old Arabidopsis seedlings grown in tissue culture. Interestingly, the steady-state levels of the ACR8 mRNA were significantly increased by ABA and NaCl and moderately increased by cold stress (Figs. 6 and 7). Levels of ACR3 and ACR4 mRNA were increased by BA treatment for 4 h (Fig. 6B). Recent studies have revealed that ABA signaling pathway may interact with sugar signaling pathway (Finkelstein and Gibson, 2002), and cytokinins may be involved in signaling the root-to-shoot nitrogen availability (Sakakibara et al., 1998; Takei et al., 2001, 2002). It will be interesting to test whether the ACR proteins are involved in the networks of metabolic and hormonal signaling pathways.
The existence of the Arabidopsis ACR proteins suggests that the hypothesis that the ACT domain is fused to a variety of enzymes, thereby conveying regulation by their respective ligands, may have further evolved in higher plants. As multiple copies of the ACT domain occupy the entire sequence and no homologs are found in the other organisms to date, the Arabidopsis ACR proteins may be novel regulatory proteins that are specific to plants. Because only amino acids are found to bind ACT domains to date, it is possible that the Arabidopsis ACR proteins also function as amino acid-binding proteins.
In bacterial amino acid metabolic enzymes, most amino acids are bound to the ACT domains as feedback inhibitors. However, Phe is an activating ligand for rat Phe hydroxylase (Kobe et al., 1999). In some enzymes with ACT repeats, for example, E. coli AK/HSD and TD and Arabidopsis AHAS regulatory subunit and TD, nonequivalent ligand binding was observed (Gallagher et al., 1998; Wessel et al., 2000; Chipman and Shaanan, 2001; Lee and Duggleby, 2001). These findings complicated the functional prediction of proteins containing ACT domain repeats. For example, in an Arabidopsis ACR protein, the ACT repeats may bind different amino acids or the same amino acid with different affinity. Upon amino acid binding, the ACR protein may function as a positive or negative regulator in plants. Because none of the ACR proteins are localized in the chloroplast, and most of the amino acid biosynthetic pathways are compartmentalized in the chloroplast, it is unlikely that the ACR proteins directly regulate the catalytic activities of the amino acid metabolic enzymes in plants. This argument is also supported by the presence of the ACT domain in several plant amino acid metabolic enzymes (Fig. 8A). In these enzymes, the enzyme linked-ACT domains may regulate their own catalytic activities. Nevertheless, it is critical to test whether these ACR proteins bind amino acids.
Although amino acids are the only ligands known to bind the ACT domain to date, we cannot exclude the possibility that small ligands other than amino acids may be bound to the ACT domains of the ACR proteins. The composition of Arabidopsis ACR proteins indicates that the ACT domain may duplicate and further evolve as an independent regulatory protein in plants. The evolution of the non-enzyme-linked ACT repeat proteins in plants may reflect the special needs for binding to yet unidentified plant-specific small ligands. By binding to specific ligands, or sensing the environmental conditions, the ACR proteins may exert their regulatory functions on their respective target proteins or enzymes. Therefore, to unravel the functions of Arabidopsis ACR proteins, it is important to identify the ligands or environmental signals and their interacting target proteins.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis ecotype Columbia-0 was used in all experiments. Plants grown in soil were placed in the greenhouse on a 16-h light/8-h dark cycle at 23°C. Roots, leaves, stems, flowers, and siliques from the same batch of 6-week-old soil-grown Arabidopsis plants were used for total RNA extraction. Two-week-old Arabidopsis seedlings grown in tissue culture were used for ACR gene regulation studies in Figures 6 and 7. Arabidopsis seeds were sterilized and plated on one-half-strength Murashige and Skoog salt phytoagar plates (Murashige and Skoog salts [Invitrogen, Carlsbad, CA], pH adjusted to 5.7 with 1 n KOH and 0.8% [w/v] agar) containing 2% (w/v) Suc. After cold treatment at 4°C for 48 h, plates were wrapped with parafilm, vertically oriented, placed in a 23°C chamber on a 16-h light/8-h dark cycle for 2 weeks. For cold stress treatment, 2-week-old plants were transferred to 4°C for 2 h. Plants for hormone and salt treatments were also grown in the same tissue culture conditions for 2 weeks, except the sterilized seeds were sown on nylon nets placed on the media (Lam et al., 1998). For transfer to a new media, the nylon nets were lifted, and the plants were transferred to 15- × 90- × 90-mm square plates with 15 mL of H2O (control), 250 mm NaCl, or 50 μm plant hormones (ABA, ACC for ethylene, BA, GA, and IAA) for 1 or 4 h.
cDNA Library Screening and RT-PCR
The [32P]dATP-labeled probe consisting of a mix of ACR3 and ACR7 EST clones (109F21T7 and ATTS0354) was used to screen an Arabidopsis cDNA library from the Arabidopsis Stock Center (CD4-14 and CD4-15). Full-length cDNA clones of ACR3 and ACR7 were obtained and subcloned in pBluescript KS. Total RNA extracted from 2-week-old Arabidopsis seedlings was used for RT-PCR to generate the computer-predicted full-length cDNA clones of ACR1, ACR4-6, and ACR8. First-strand cDNA was synthesized using the SuperScript II RNase H− RT Kit (Invitrogen). The following forward and reverse primer pairs were used to amplify full-length ACR cDNA clones. ACR1: MH355, 5′-AACACATGTTGGTACGTAGG-3′; MH190, 5′-GTTCATCTATATATACAACG-3′. ACR2: MH455, 5′-CCGTAGAGACGATGCAGAAA; MH456, 5′-AACTTGAAGCTTACGTAAGG-3′. ACR4: MH182, 5′-ACCTCACAGCTAAGTGAAGT-3′; MH352, 5′-TGGGAATTTTTTGATTGCAG-3′. ACR5: MH194, 5′-ACCCTGTAAAAATGAATCTA-3′; MH354, 5′-GGACAGATGTTTGCTTGAGC-3′. ACR6: MH184, 5′-CTACTCTGTCTAACCTATAC-3′; MH356, 5′-ATCCTCCTTCAACTTCCCTC-3′. ACR8: MH188, 5′-TCCGCGTGCCTCTCAAAGGA-3′; MH353, 5′-AGAGGTTGAAAACGACGGCG-3′. TOPO TA Cloning Kit (Invitrogen) was used to clone the ACR cDNAs amplified by RT-PCR.
Sequence Analysis and Nomenclature
The consensus sequence of the ACT domain (Pfam01842) was from the Pfam database site (http://www.sanger.ac.uk/Software/Pfam/). The deduced amino acid sequences of Arabidopsis ACR proteins were aligned with the program ClustalW with the default setting (http://npsa-pbil.ibcp.fr) and ACR numbers 1 to 8 were named according to the output alignment from top to bottom. The amino acid sequences of ACR1 and ACR3 to -8 were deduced from the isolated cDNAs reported here and ACR2 was from the GenBank database (accession no. NM_122441). The National Center for Biotechnology Information PSI-BLAST conserved domain search detected four ACT domains (ACT1–4) in ACR3, three (ACT2–4) in ACR5, and two (ACT2 and ACT4) in the other ACRs. Multiple sequence alignment revealed that sequences corresponding to the four ACT domains of ACR3 were conserved in all ACR proteins (Fig. 1). Moreover, InterProScan (http://www.ebi.ac.uk/interpro/scan.html) detected four amino acid-binding ACT domains in all ACR proteins. We thus propose that all the ACR proteins reported here contain four copies of the ACT domain.
GenBank Accession Numbers
The accession numbers for the Arabidopsis ACR sequences are ACR1, AF528058; and ACR3 to ACR8, AF528059 to AF528064, respectively. The accession numbers for the Escherichia coli sequences in Figures 2 and 8 are AK, P08660; AK/HSD, P00561; PGDH, 1127236; AHAS, P08143; and GlnD, M96431. The accession numbers for the Bacillus subtilis sequences in Figure 8 are PDT, AAA22507; PDH, AAA20868; and CM, D32804. The accession numbers for the Arabidopsis protein sequences in Figure 8A are: monofunctional AK, NP_186851, NP_196910 and NP_196832; bifunctional AK/HSDs, NP_174408 and NP_193706; PGDH, NP_564034, NP_195146, and NP_566637; AHAS, NP_180740 and NP_197133; and PDT, NP_172644 and NP_187420. The accession numbers for the Arabidopsis sequences in Figure 8B are putative protein kinases AY070086, T04683, T05675, and T08864; Unknown proteins, AY045670 and AAD31570; unknown proteins, AY072401 and NP_564010.
RNA-Blot Analysis
Arabidopsis total RNA was isolated using a phenol extraction protocol (Jackson and Larkins, 1976). Total RNA (10 μg) was separated in standard formaldehyde gel by electrophoresis and blotted onto a nylon membrane. For detection of mRNA, gene-specific DIG-labeled single-stranded DNA probes were generated by PCR (Myerson, 1991). The following primer pairs (located in the last exon and the 3′-untranslated region of each ACR gene) were used to make probes for RNA-blot analysis. ACR1: MH189, 5′-CTTTTCGACACGGTGTGTGC-3′; MH190, 5′-GTTCATCTATATATACAACG-3′. ACR2: MH456, 5′-AACTTGAAGCTTACGTAAGG-3′; MH457, 5′-GAGGTCAAGAATGAAGACAC-3′. ACR3: MH32, 5′-GTTGGAGTTTGGAGCTCTGC; MH162, 5′-CGGTATCGATAAGCTTGATA. ACR4: MH181, 5′-GGATTGAAGTTGGAACTGTG-3′; MH182, 5′-ACCTCACAGCTAAGTGAAGT-3′. ACR5: MH193, 5′-GCACTTCAGATAGAGTCGGG-3′; MH194, 5′-ACCCTGTAAAAATGAATCTA-3′. ACR6: MH183, 5′-ATCAGCAGAAGACCGAGTTG-3′; MH184, 5′-CTACTCTGTCTAACCTATAC-3′. ACR7: MH35, 5′-TCAGAACCGGAGAGACAACG-3′; MH195, 5′-GTTGTTGACAAACACGATTG-3′. ACR8: MH187, 5′-TGAGATTAGAGCTGAGGCAT-3′; MH188, 5′-TCCGCGTGCCTCTCAAAGGA-3′. Probe labeling, prehybridization, hybridization, wash conditions, and detection were performed according to the Boehringer Mannheim Genius System User's Guide (v3.0).
ACR3 Promoter-GUS Fusion
A 0.8-kb region upstream of the ACR3 (AC002291) initial Met was amplified from the Arabidopsis genomic DNA by PCR using the primer pair MH28 5′-AATTTCGATGTCGACGCACC-3′ and MH29 5′-GTCTTGTGGATATTAAGCTC-3′. The PCR product was cloned into the pCR2.1-TOPO vector (TOPO TA Cloning Kit, Invitrogen) and the sequence was confirmed. A HindIII/XbaI fragment containing the entire 0.8-kb ACR3 promoter region was subcloned into the pBI101 binary vector to create a ACR3p-GUS fusion construct that was transformed into the Agrobacterium tumefaciens strain GV3101. The floral dip method was used for Arabidopsis transformation (Clough and Bent, 1998). Several independent ACR3p-GUS Arabidopsis transgenic lines were carried to T3 homozygous and stained for GUS activity (Jefferson et al., 1987).
Footnotes
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.007484.
LITERATURE CITED
- Aravind L, Koonin EV. Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database search. J Mol Biol. 1999;287:1023–1040. doi: 10.1006/jmbi.1999.2653. [DOI] [PubMed] [Google Scholar]
- Bueno R, Pahel G, Magasanik B. Role of glnB and glnD gene products in regulation of the glnALG operon of Escherichia coli. J Bacteriol. 1985;164:816–822. doi: 10.1128/jb.164.2.816-822.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashel M, Gentry DR, Hernandez VJ, Binella D. The stringent response. In: Neidhardt FC, Ingraham JL, Lin ECC, Low KB, Magasanik B, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Washington, DC: ASM Press; 1996. pp. 1458–1496. [Google Scholar]
- Chinchilla D, Schwarz FP, Eisenstein E. Amino acid substitutions in the C-terminal regulatory domain disrupt allosteric effector binding to biosynthetic threonine deaminase from Escherichia coli. J Biol Chem. 1998;273:23219–23224. doi: 10.1074/jbc.273.36.23219. [DOI] [PubMed] [Google Scholar]
- Chipman DM, Shaanan B. The ACT domain family. Curr Opin Struct Biol. 2001;11:694–700. doi: 10.1016/s0959-440x(01)00272-x. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Coruzzi G, Bush DR. Nitrogen and carbon nutrient and metabolite signaling in plants. Plant Physiol. 2001;125:61–64. doi: 10.1104/pp.125.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Gibson SI. ABA and sugar interactions regulating development: cross-talk or voices in a crowd? Curr Opin Plant Biol. 2002;5:26–32. doi: 10.1016/s1369-5266(01)00225-4. [DOI] [PubMed] [Google Scholar]
- Gallagher DT, Gilliland GL, Xiao G, Zondlo J, Fisher KE, Chinchilla D, Eisenstein E. Structure and control of pyridoxal phosphate dependent allosteric threonine deaminase. Structure. 1998;6:465–475. doi: 10.1016/s0969-2126(98)00048-3. [DOI] [PubMed] [Google Scholar]
- Grant GA, Hu Z, Xu XL. Specific interactions at the regulatory domain-substrate binding domain interface influence the cooperativity of inhibition and effector binding in Escherichia colid-3-phosphoglycerate dehydrogenase. J Biol Chem. 2001;276:1078–1083. doi: 10.1074/jbc.M007512200. [DOI] [PubMed] [Google Scholar]
- Grant GA, Kim SJ, Xu XL, Hu Z. The contribution of adjacent subunits to the active sites of d-3-phosphoglycerate dehydrogenase. J Biol Chem. 1999;274:5357–5361. doi: 10.1074/jbc.274.9.5357. [DOI] [PubMed] [Google Scholar]
- Ho CL, Noji M, Saito M, Saito K. Regulation of serine biosynthesis in Arabidopsis: crucial role of plastidic 3-phosphoglycerate dehydrogenase in non-photosynthetic tissues. J Biol Chem. 1999;274:397–402. doi: 10.1074/jbc.274.1.397. [DOI] [PubMed] [Google Scholar]
- Hsieh MH, Lam HM, van de Loo FJ, Coruzzi G. A PII-like protein in Arabidopsis: putative role in nitrogen sensing. Proc Natl Acad Sci USA. 1998;95:13965–13970. doi: 10.1073/pnas.95.23.13965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AD, Larkins BA. Influence of ionic strength, pH and chelation of divalent metals on isolation of polyribosomes from tobacco leaves. Plant Physiol. 1976;57:5–10. doi: 10.1104/pp.57.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y, Kojima H, Tanaka T. Mutational analysis of the feedback sites of lysine-sensitive aspartokinase of Escherichia coli. FEMS Microbiol Lett. 1999;173:211–215. doi: 10.1111/j.1574-6968.1999.tb13504.x. [DOI] [PubMed] [Google Scholar]
- Kobe B, Jennings IG, House CM, Michell BJ, Goodwill KE, Santarsiero BD, Stevens RC, Cotton RGH, Kemp BE. Structural basis of autoregulation of phenylalanine hydrogenase. Nat Struct Biol. 1999;6:442–448. doi: 10.1038/8247. [DOI] [PubMed] [Google Scholar]
- Lam HM, Chiu J, Hsieh MH, Meisel L, Oliveira IC, Shin M, Coruzzi G. Glutamate-receptor genes in plants. Nature. 1998a;396:125–126. doi: 10.1038/24066. [DOI] [PubMed] [Google Scholar]
- Lam HM, Hsieh MH, Coruzzi G. Reciprocal regulation of distinct asparagine synthetase genes by light and metabolites in Arabidopsis thaliana. Plant J. 1998b;16:345–353. doi: 10.1046/j.1365-313x.1998.00302.x. [DOI] [PubMed] [Google Scholar]
- Lee YT, Duggleby RG. Identification of the regulatory subunit of Arabidopsis thalianaacetohydroxyacid synthase and reconstitution with its catalytic subunit. Biochemistry. 2001;40:6836–6844. doi: 10.1021/bi002775q. [DOI] [PubMed] [Google Scholar]
- Mendel S, Elkayam T, Sella C, Vinogradov V, Vyazmensky M, Chipman DM, Barak Z. Acetohydroxyacid synthase: a proposed structure for regulatory subunits supported by evidence from mutagenesis. J Mol Biol. 2001;307:465–477. doi: 10.1006/jmbi.2000.4413. [DOI] [PubMed] [Google Scholar]
- Myerson D. Producing single-stranded DNA probes with the Taq DNA polymerase: a high yield protocol. Biotechniques. 1991;10:35–38. [PubMed] [Google Scholar]
- Ninfa AJ, Atkinson MR. PII signal transduction proteins. Trends Microbiol. 2000;8:172–179. doi: 10.1016/s0966-842x(00)01709-1. [DOI] [PubMed] [Google Scholar]
- Omori K, Imai Y, Suzuki S, Komatsubara S. Nucleotide sequence of the Serratia marcescensthreonine operon and analysis of the threonine operon mutations which alter feedback inhibition of both aspartokinase I and homoserine dehydrogenase I. J Bacteriol. 1993;175:785–794. doi: 10.1128/jb.175.3.785-794.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omori K, Komatsubara S. Role of serine 352 in the allosteric response of Serratia marcescensaspartokinase I-homoserine dehydrogenase I analyzed by using site-directed mutagenesis. J Bacteriol. 1993;175:959–965. doi: 10.1128/jb.175.4.959-965.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittard J. The various strategies within the TyrR regulation of Escherichia colito modulate gene expression. Genes Cells. 1996;1:717–725. doi: 10.1111/j.1365-2443.1996.tb00012.x. [DOI] [PubMed] [Google Scholar]
- Rhee SG, Park SC, Koo JH. The role of adenylyltransferase and uridylyltransferase in the regulation of glutamine synthetase in Escherichia coli. Curr Top Cell Reg. 1985;27:221–232. doi: 10.1016/b978-0-12-152827-0.50026-8. [DOI] [PubMed] [Google Scholar]
- Robbins J, Dilworth SM, Laskey RA, Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Suzuki M, Takei K, Deji A, Taniguchi M, Sugiyama T. A response-regulator homologue possibly involved in nitrogen signal transduction mediated by cytokinin in maize. Plant J. 1998;14:337–344. doi: 10.1046/j.1365-313x.1998.00134.x. [DOI] [PubMed] [Google Scholar]
- Schuller DJ, Grant GA, Banaszak LJ. The allosteric ligand site in the V-max-type cooperative enzyme phosphoglycerate dehydrogenase. Nat Struct Biol. 1995;2:69–76. doi: 10.1038/nsb0195-69. [DOI] [PubMed] [Google Scholar]
- Siehl DL, Conn EE. Kinetic and regulatory properties of arogenate dehydratase in seedlings of Sorghum bicolor(L.) Moench. Arch Biochem Biophys. 1988;260:822–829. doi: 10.1016/0003-9861(88)90513-9. [DOI] [PubMed] [Google Scholar]
- Takei K, Sakakibara H, Taniguchi M, Sugiyama T. Nitrogen-dependent accumulation of cytokinins in root and the translocation to leaf: implication of cytokinin species that induces gene expression of maize response regulator. Plant Cell Physiol. 2001;42:85–93. doi: 10.1093/pcp/pce009. [DOI] [PubMed] [Google Scholar]
- Takei K, Takahashi T, Sugiyama T, Yamaya T, Sakakibara H. Multiple routes communicating nitrogen availability from roots to shoots: a signal transduction pathway mediated by cytokinin. J Exp Bot. 2002;53:971–977. doi: 10.1093/jexbot/53.370.971. [DOI] [PubMed] [Google Scholar]
- Viola RE. The central enzymes of the aspartate family of amino acid biosynthesis. Acc Chem Res. 2001;34:339–349. doi: 10.1021/ar000057q. [DOI] [PubMed] [Google Scholar]
- Wessel PM, Graciet E, Douce R, Dumas R. Evidence for two distinct effector-binding sites in threonine deaminase by site-directed mutagenesis, kinetic, and binding experiments. Biochemistry. 2000;39:15136–15143. doi: 10.1021/bi001625c. [DOI] [PubMed] [Google Scholar]
- Wilson TJ, Argaet VP, Howlett GJ, Davidson BE. Evidence for two aromatic amino acid-binding sites, one ATP-dependent and the other ATP-independent, in the Escherichia coliregulatory protein TyrR. Mol Microbiol. 1995;17:483–492. doi: 10.1111/j.1365-2958.1995.mmi_17030483.x. [DOI] [PubMed] [Google Scholar]
- Zhang S, Pohnert G, Kongsaeree P, Wilson DB, Clardy J, Ganem B. Chorismate mutase-prephenate dehydratase from Escherichia coli: study of catalytic and regulatory domains using genetically engineered proteins. J Biol Chem. 1998;273:6248–6253. doi: 10.1074/jbc.273.11.6248. [DOI] [PubMed] [Google Scholar]