Abstract
Two MADS box genes, Lily MADS Box Gene 2 (LMADS2) and Eustoma grandiflorum MADS Box Gene 1 (EgMADS1), with an extensive similarity to the petunia (Petunia hybrida) FLORAL BINDING PROTEIN 7/11 and Arabidopsis AGL11, were characterized from the lily (Lilium longiflorum) and lisianthus (Eustoma grandiflorum). The expression of LMADS2 and EgMADS1 mRNA was restricted to the carpel and was absent in the other flower organs or vegetative leaves. LMADS2 mRNA was detected mainly in ovules and weakly in style tissues of the carpel, whereas EgMADS1 mRNA was only expressed in the ovules. Transgenic Arabidopsis plants ectopically expressing LMADS2 or EgMADS1 showed similar novel phenotypes resembling 35S::AGAMOUS plants by significantly reducing plant size, flowering early, and losing inflorescence indeterminacy. Ectopic expression of these two genes also generated similar ap2-like flowers by inducing homeotic conversion of the sepals into carpel-like structures in which stigmatic papillae and ovules were observed. In addition, the petals were converted into stamen-like structures in the second whorl of 35S::LMADS2 and 35S::EgMADS1 transgenic Arabidopsis. Our data indicated that LMADS2 and EgMADS1 are putative D functional MADS box genes in lily and lisianthus with a function similar to C functional genes once ectopically expressed in Arabidopsis.
During development, floral organ identity is specified mainly by a set of flower organ identity genes that contain a conserved MADS box, a DNA-binding domain, in the N terminus of proteins (Purugganan et al., 1995; Rounsley et al., 1995; Theissen and Saedler, 1995; Theissen et al., 2000). On the basis of the sequence and functional similarity, MADS box genes involved in flower development have been classified into five major groups. An ABCDE model was established based on their interactions (Theissen, 2001; Theissen and Saedler, 2001). In this model, A functional genes control the sepal formation; A, B, and E functional genes together regulate petal formation; B, C, and E functional genes control the stamen formation; C and E functional genes regulate carpel formation; and the D functional gene is involved in ovule development. APETALA1 is A functional, APETALA3 and PISTILLATA are B functional, AGAMOUS (AG) is C functional, FLORAL-BINDING PROTEIN (FBP) 7 and FBP11 are D functional, and SEPALLATA1/2/3 (previously described as AGL2, -4, and -9) are E functional flower organ identity genes, respectively (Coen and Meyerowitz, 1991; Drews et al., 1991; Mandel et al., 1992; Jofuku et al., 1994; Weigel and Meyerowitz, 1994; Colombo et al., 1995, 1997a, 1997b; Rounsley et al., 1995; Pelaz et al., 2000; Theissen and Saedler, 2001).
Genes in A, B, and C functional groups have been studied extensively in various plant species (Theissen et al., 2000; Theissen, 2001; Theissen and Saedler, 2001). In contrast, relatively few studies have been reported for the D functional genes (Theissen, 2001). The best known D functional MADS box genes are FBP7 and FBP11 of the petunia (Petunia hybrida), which are expressed specifically in the ovules (Angenent et al., 1995; Colombo et al., 1995, 1997a, 1997b; Angenent and Colombo, 1996). Severe alteration of ovule development was observed in a mutation caused by cosuppression of FBP7 or FBP11 in transgenic petunia plants (Angenent et al., 1995; Colombo et al., 1995, 1997a). Ectopic expression of FBP7 or FBP11 in the petunia induced the formation of ovules on the sepals and petals (Colombo et al., 1995). This data indicated that the FBP7 and FBP11 genes are involved in ovule development. Different from the A or C functional MADS box genes, which form homodimers for regulating flower development, FBP11 has been shown to form heterodimers with E functional genes such as FBP2, FBP5, and FBP9 in the petunia for regulating ovule development (Immink et al., 2002). On the basis of the sequence similarity and the expression pattern, only a few putative orthologs for FBP7/11 were identified in other plant species such as AGL11 of Arabidopsis (Rounsley et al., 1995), ZAG2 and ZMM1 of maize (Zea mays; Schmidt et al., 1993; Theissen et al., 1995), and OsMADS13 of rice (Oryza sativa; Lopez-Dee et al., 1999). However, no corresponding mutants or further functional analysis was performed to indicate their involvement in ovule development.
Several MADS box genes showing sequences similar to D functional genes have also been identified. These genes tend to specify the fourth whorl carpel development (Rounsley et al., 1995). For example, AGL13 of Arabidopsis was also mainly expressed in ovules (Rounsley et al., 1995). SHATTERPROOF (SHP) 1 and SHP2 (previously described as AGL1 and -5) of Arabidopsis were specifically expressed in the carpel (Ma et al., 1991; Rounsley et al., 1995; Savidge et al., 1995; Flanagan et al., 1996; Liljegren et al., 1998, 2000) and are directly regulated by AG during carpel development (Savidge et al., 1995). Interestingly, the D functional genes also share a high sequence identity with the C functional genes that are also involved in carpel formation (Bowman et al., 1989; Yanofsky et al., 1990; Theissen, 2001). For example, FBP7 and FBP11 showed high similarity to FBP6 (AG ortholog) in the petunia, whereas AGL11 looked similar to the AG in Arabidopsis (Angenent et al., 1995; Colombo et al., 1995, 1997a, 1997b; Rounsley et al., 1995). On the basis of their sequence similarity, the C and D functional genes are thought to be possibly produced by duplicated events from an ancestral gene (Theissen et al., 2000). However, different from the D functional mutants in the petunia, the C functional mutants, such as ag of Arabidopsis, produce the perianth organs (petals and sepals) in the inner two whorls normally occupied by reproductive organs (stamens and carpels; Bowman et al., 1989). Moreover, ectopic expression of AG or its orthologs causes the conversion of sepals and petals into carpel- and stamen-like structures and the early-flowering phenotype (Mizukami and Ma, 1992, 1997; Kempin et al., 1993; Pnueli et al., 1994; Kang et al., 1995; Kater et al., 1998; Rutledge et al., 1998; Yu et al., 1999). Therefore, it still remains unclear whether the C and D functional or other related MADS box genes share some similar functions in carpel or ovule development specification. To explore this question, the characterization and functional analyses of more D functional-related or carpel-specific-expressed genes from various plant species is necessary.
Lilies (Lilium longiflorum) and lisianthus (Eustoma grandiflorum) are popular flowers with important economic value in the cut flower market around the world. However, only a few studies regarding flower formation have been reported for these two plant species (Chen and Yang, 2000; Theissen et al., 2000; Tzeng and Yang, 2001). We report here on the isolation and functional analysis of two MADS box genes that may be involved in carpel or ovule development in the lily and lisianthus. The exploration of the relationships between these two genes and their closest counterparts in other plant species is discussed.
RESULTS
Isolation of D Functional MADS Box Genes from Lily and Lisianthus
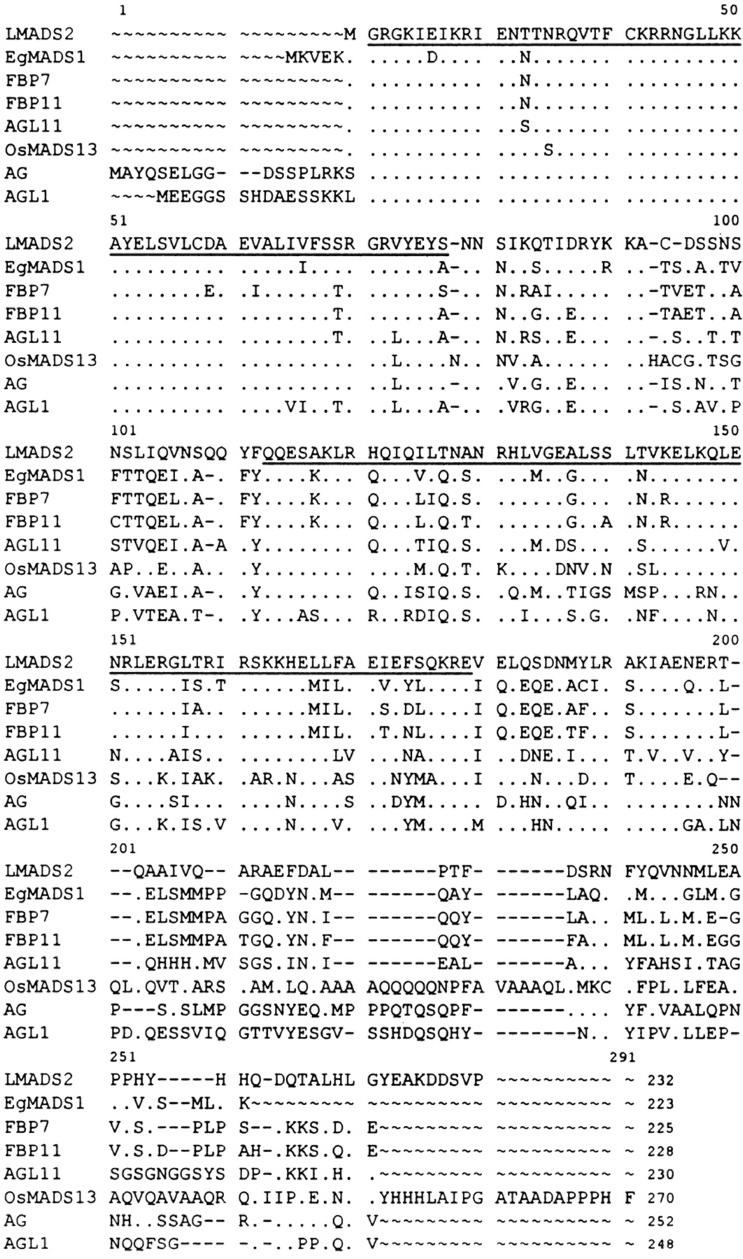
A combined reverse transcriptase-PCR and 5′-RACE strategy was used to isolate the MADS box genes from the lily (Tzeng and Yang, 2001). The cDNA sequence for one gene, Lily MADS Box Gene 2 (LMADS2), showed a high sequence identity (65%) to FBP7/11 and AGL11, the D functional genes of the petunia and Arabidopsis. LMADS2 cDNA was 1,057 bp long and contained an open reading frame (ORF) that encoded a deduced protein with 232 amino acid residues (Fig. 1). Although the full length of the LMADS2 protein showed the highest identity with FBP11 and AGL11, LMADS2 showed a higher identity with the AG of Arabidopsis (98%, 57/58) than with FBP11 (95%, 55/58) or AGL11 (93%, 54/58) in the MADS box domain. In addition to the MADS box domain, a putative protein dimerization K box domain, which showed 76% (51/67), 72% (48/67), and 63% (42/67) identity with FBP11, AGL11, and AG, was found in the middle of the protein (Fig. 1). The high sequence identity between LMADS2 and the D functional genes from various species suggests that LMADS2 is the lily putative D functional MADS box gene ortholog.
Figure 1.
Alignment of amino acid sequence of LMADS2, EgMADS1, and related D and C functional MADS box genes. FBP7 (petunia), FBP11 (petunia), AGL11 (Arabidopsis), and OsMADS13 (rice) are in D functional group; AG (Arabidopsis) and AGL1 (Arabidopsis) are in C functional group. The first underlined region is MADS box domain, whereas the second underlined region is K box domain. Amino acid residues identical to LMADS2 in this alignment are indicated as dots. Dashes were introduced to improve alignment.
The same strategy was used to clone the MADS box genes from the lisianthus. The cDNA sequence for one gene, E. grandiflorum MADS Box Gene 1 (EgMADS1), showing an extremely high sequence identity (80%) to FBP11 was isolated. EgMADS1 cDNA was 1,050 bp long and contained an ORF that encoded a deduced protein with 223 amino acid residues (Fig. 1). The full length of the EgMADS1 protein showed 80%, 64%, and 60% identity with FBP11, LMADS2, and AG, respectively (Fig. 1). In contrast to LMADS2, EgMADS1 showed a higher identity with FBP11 (95%, 55/58) or LMADS2 (93%, 54/58) than with AG of Arabidopsis (91%, 53/58) in the MADS box domain. In the K box domain, 88% (59/67), 73% (49/67), and 61% (41/67) identity were observed for EgMADS1 with FBP11, LMADS2, and AG, respectively (Fig. 1). The high sequence identity between EgMADS1 and FBP11 suggests that EgMADS1 is the putative D functional MADS box gene in lisianthus.
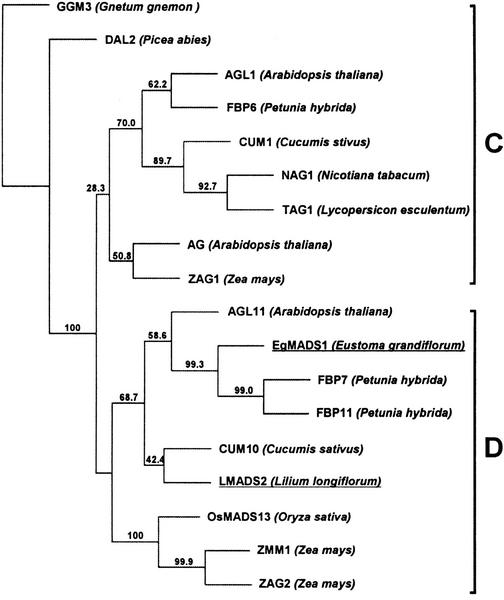
The amino acid sequence alignment shown in Figure 1 was used to construct a phylogenetic tree for the plant D and C functional MADS box genes (Fig. 2). On the basis of the result, LMADS2 was assigned closely related to AGL11 of Arabidopsis, whereas EgMADS1 was assigned closely related to FBP7/11 of the petunia.
Figure 2.
Phylogenetic analysis of represented plant D and C functional MADS box genes. On the basis of similarity of amino acid sequence, LMADS2 was assigned in D group MADS box genes and was closely related to AGL11 of Arabidopsis and FBP7/11 of petunia. EgMADS1 was also assigned in D group and showed the highest similarity to FBP7/11 of petunia. Names of the LMADS2 and EgMADS1 are underlined. Names of the plant species for each MADS box gene are listed behind the protein names. The tree was generated by the neighbor joining (NJ) method, whereas the distance was calculated based on the Dayhoff PAM matrix (Dayhoff et al., 1983) using the PROTDIST program in PHYLIP software package (v3.5c; Kimura, 1980). Numbers on major branches indicate bootstrap estimates for 1,000 replicate analyses.
Flower Structure in Lily and Lisianthus
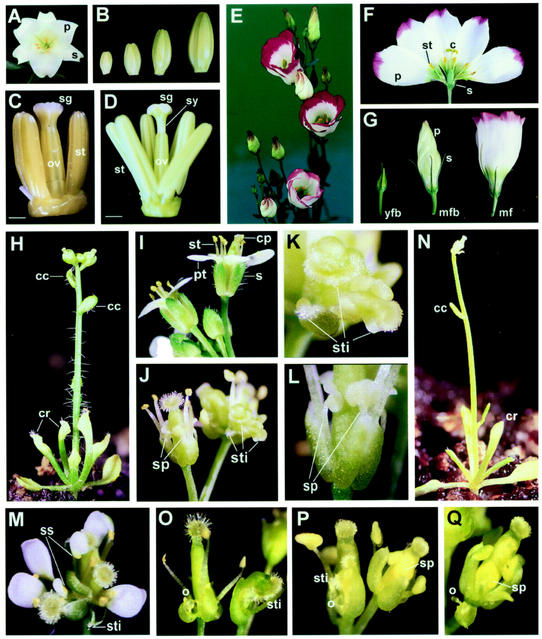
The lily plant produces white flowers that consist of four whorls of organs including three sepals, three petals, six stamens, and three fused carpels (Fig. 3A). The sepals and petals are extremely similar and known as tepals (Fig. 3A). Although the sepals and petals enclose the interior organs (Fig. 3B), these flower organs are well developed in the 10- to 30-mm-long flower buds (Fig. 3, C and D).
Figure 3.
Flowers for lily and lisianthus and phenotypic analysis of transgenic Arabidopsis plants ectopically expressing LMADS2 or EgMADS1. A, Lily flower consists of four whorls of organs including three sepals (s), three petals (p), six stamens, and three fused carpels. B, Lily floral buds at different developmental stages. From left to right, 10-, 15-, 20-, and 30-mm-long floral buds. C, Dissection of a 10-mm-long flower bud. Most part of the stamen (st) was made up of developing anther in this stage. The stigma (sg) and ovary (ov) regions in the carpel are clearly differentiated. The sepals and petals were removed from the flower. Bar = 0.6 mm. D, Dissection of a 30-mm-long flower bud. The stamen (st), stigma (sg), style (sy), and ovary (ov) were clearly distinguishable. The sepals and petals were removed from the flower. Bar = 3 mm. E, Flowers produced in the inflorescence of a lisianthus plant. F, Lisianthus flower consists of four whorls of organs including five sepals (s), five petals (p), five stamens (st), and two fused carpels (c). G, Lisianthus flower at different developmental stages. From left to right, young floral bud (yfb), mature floral bud (mfb), and mature flower (mf). s, Sepals; p, petals. H, Twenty-d-old 35S::LMADS2 Arabidopsis plant grown on soil flowered significantly earlier than wild-type plant after producing only five small, curled rosette leaves (cr) on the base and two small, curled cauline leaves (cc) on inflorescence. This plant was significantly reduced in size to about 3 cm tall, whereas wild-type plant was about 20 cm tall. I, Two mature Arabidopsis wild-type flowers along with several floral buds. In these wild-type flowers, four sepals (s), four petals (pt), six stamens (st), and two fused carpels (cp) were produced. Floral buds were clearly enclosed during early stages. J, Second-whorl organs were transformed into staminoid-petal structures (sp) in a 35S::LMADS2 flower. A flower producing carpelloid sepals with stigmatic papillae (sti) was also observed. K, Close-up of the carpelloid-sepal structures in J. Stigmatic papillae (sti) were clearly observed. L, Close-up of the staminoid-petal structures (sp) in J. M, Terminal flower produced in the end of the inflorescence for a 35S::LMADS2 plant. Carpelloid sepal with stigmatic papillae (sti) and petals containing a staminoid sector (ss) were observed in this terminal flower. N, Similar to 35S::LMADS2 plant in H, a 20-d-old 35S::EgMADS1 Arabidopsis plant also flowered significantly earlier than wild-type plant by producing only four small, curled rosette leaves (cr) on the base and two small, curled cauline leaves (cc) on inflorescence. The size for this plant was also significantly reduced. O, First-whorl organs were transformed into carpel-sepal-like structures in two 35S::EgMADS1 flowers. Stigmatic papillae (sti) and ovules (o) were observed in these flowers. P, 35S::EgMADS1 flowers produced carpelloid sepals with stigmatic papillae (sti) and ovules (o). Stamen-petal-like structures (sp) were also observed. Q, 35S::EgMADS1 flower produced staminoid-petal structures (sp) and carpelloid sepal with ovules (o).
Lisianthus, also called Texas bluebells or the tulip Gentian, is a member of the Gentianaceae family and is known for its many color varieties. Lisianthus produces large and deeply cupped (like a bell) flowers (Fig. 3E) that are red, pink, purple, or violet-blue in color and that consist of four whorls of organs including five sepals, five petals, five stamens, and two fused carpels (Fig. 3F). Various colors are usually seen on the edges of the petals (Fig. 3, E and F).
Gene Expression for LMADS2 and EgMADS1
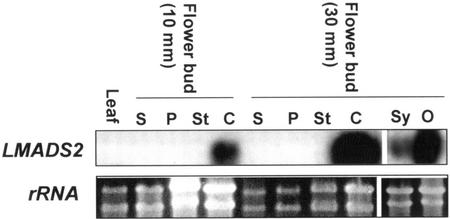
RNA-blot analysis was performed to explore the relationships between sequence similarity and expression pattern for LMADS2 and EgMADS1. As shown in Figure 4, a 1.1-kb fragment corresponding to LMADS2 mRNA was detected in the flower buds of different developmental stages (10 mm and 30 mm in length) and was absent in the vegetative leaves. This indicated that the expression of LMADS2 was flower specific. When floral organs from 10- and 30-mm floral buds were examined (Fig. 3, B–D), LMADS2 was strongly and exclusively expressed in the carpels (Fig. 4). Further analysis indicated that LMADS2 mRNA was mainly expressed in the ovule of the ovary, and a lower level of expression was observed in the style tissue (Fig. 4).
Figure 4.
Northern analysis for LMADS2. Total RNA was isolated from four flower organs of 10- and 30-mm-long floral buds; from style (Sy) and ovules (O) of carpel; and from vegetative leaves. For northern hybridization, LMADS2-specific DNA probes (without MADS box domain) were used. The results indicated that the RNA for LMADS2 was detected specifically in carpel (C) during different stages of flower development. LMADS2 expression was mainly detected in ovules (O) of carpel and weakly in style (Sy). No signals could be detected in sepal (S), petal (P), stamen (St), and vegetative leaves (Leaf) for LMADS2. An ethidium bromide-stained gel before blotting and hybridization was shown as control.
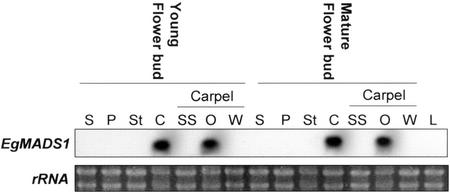
Similar to LMADS2, the expression of EgMADS1 was also floral specific (Fig. 5). EgMADS1 mRNA was detected in both young and mature flower buds but was undetectable in the vegetative leaves. When the floral organs were examined in young and mature flower buds (Fig. 3G), EgMADS1 was highly expressed only in the carpel (Fig. 5). When the carpel tissues were examined, EgMADS1 was strongly and exclusively expressed in the ovules (Fig. 5). The expression pattern for both LMADS2 and EgMADS1 was similar to other D functional MADS box genes such as FBP7/11 in the petunia, AGL11 in Arabidopsis, and OsMADS13 in rice (Angenent et al., 1995; Rounsley et al., 1995; Lopez-Dee et al., 1999). This expression pattern strongly suggested that LMADS2 and EgMADS1 are putative D functional genes that regulate ovule development in plants.
Figure 5.
Northern analysis for EgMADS1. Total RNA was isolated from four flower organs of young and mature floral buds; from stigma plus style (SS), ovules (O), and ovary wall (W) of carpel; and from vegetative leaves (L). For northern hybridization, EgMADS1-specific DNA probes (without MADS box domain) were used. The results indicated that the RNA for EgMADS1 was detected specifically in carpel (C) during different stages of flower development. EgMADS1 expression was exclusively detected in ovules (O) and was absent in other tissues of carpel. No signals could be detected in sepal (S), petal (P), stamen (St), and vegetative leaves for EgMADS1. An ethidium bromide-stained gel before blotting and hybridization was shown as control.
Ectopic Expression of LMADS2 Causes Early Flowering and the Conversion of Sepals and Petals to Carpel- and Stamen-Like Structures in Transgenic Arabidopsis Plants
To further investigate whether the sequence and structure similarity is coupled to the functional similarity between LMADS2 and D functional genes, functional analysis of LMADS2 through transgenic plants is necessary. LMADS2 cDNA driven by cauliflower mosaic virus 35S promoter was therefore transformed into Arabidopsis plants for functional analysis.
Eight independent transgenic Arabidopsis T1 plants were obtained. Three plants were indistinguishable from untransformed wild-type plants, whereas five other plants produced nearly identical and severe alterations in both the vegetative and reproductive development (Fig. 3H). The alterations observed in these five plants included extreme reduction in plant size (about 2 cm tall when mature), early flowering, production of only five to six small, curled filamentous leaves with yellowed tips, loss of inflorescence indeterminacy, and production of less than 10 flowers (Fig. 3H). These alterations were similar to those described in Arabidopsis plants transformed with AG or AG orthologs from heterologous plant species (Mizukami and Ma, 1992; Kater et al., 1998; Rutledge et al., 1998).
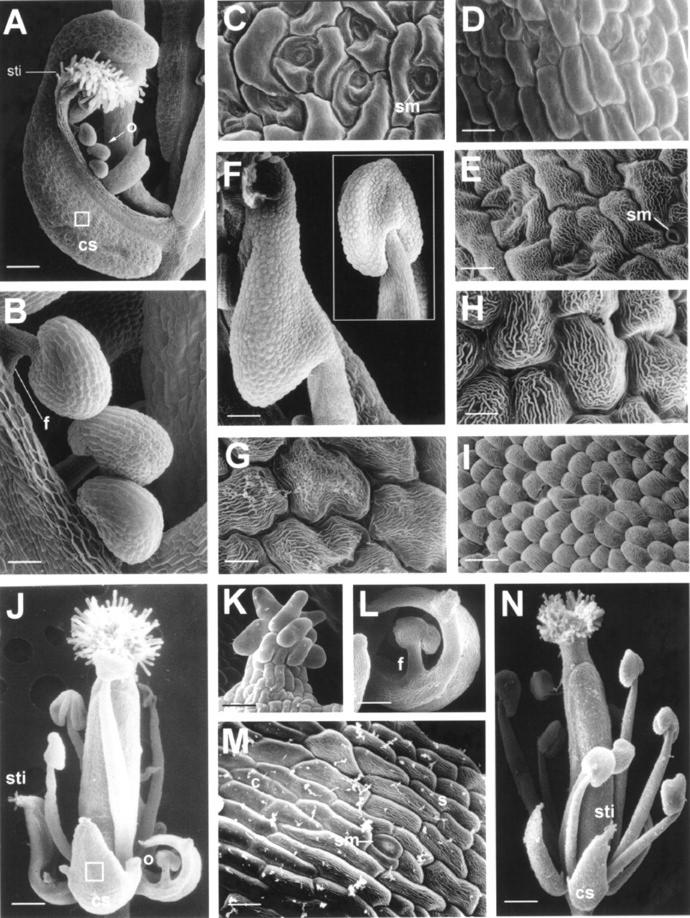
When the flowers in these five plants were analyzed, they clearly opened prematurely (Fig. 3H) and showed the typical homeotic conversion of sepals and petals similar to that observed in ap2 mutants or AG overexpressed transgenic Arabidopsis plants (Mizukami and Ma, 1992, 1997). The first whorl presented a carpel-like organ (Figs. 3, J and K, and 6A) that was small and abnormal in shape compared with wild-type sepals (Fig. 3I). The stigmatic papillae (Figs. 3, J and K, and 6A) and ovules (Fig. 6, A and B) were often observed at the tip of these structures. When the epidermal cells (Fig. 6C) in these first-whorl carpel-like structures were examined, they were morphologically similar to wild-type carpel epidermis (Fig. 6D) and distinct from wild-type sepal epidermis (Fig. 6E). The organs in second whorl were usually absent. Stamen-like structures (Figs. 3, J and L, and 6F) similar to those observed in ap2 mutants were sometimes produced in the second whorl of the mutant flowers. When the epidermal cells (Fig. 6G) in these second-whorl stamen-like structures were examined, they were morphologically similar to wild-type anther epidermis (Fig. 6H) and distinct from wild-type petal epidermis (Fig. 6I). All five plants produced terminal flowers (Fig. 3M) similar to those observed in Arabidopsis plants, ectopically expressing the AG or AG orthologs (Mizukami and Ma, 1992; Rutledge et al., 1998). Different from the terminal flowers produced in Arabidopsis terminal flower 1 mutants, petals with clear stamen-like structures or staminoid sectors (Fig. 3M) were observed in the second-whorl organs of these 35S::LMADS2 terminal flowers. The stigmatic papillae were also observed at the tip of the sepals in these terminal flowers (Fig. 3M).
Figure 6.
Scanning electron micrographs of various floral organs observed in 35S::LMADS2 and 35S::EgMADS1 transgenic Arabidopsis plants. A, Carpel-like sepal (cs) with stigmatic papillae (sti) and ovules (o) in a 35S::LMADS2 flower. Bar = 125 μm. B, Close-up of the ovules (o) in A. f, Funiculus. Bar = 25 μm. C, Surface cells from carpel-like structure (boxed in A) contained irregularly shaped cells with smooth surface along with the interspersed stomata (sm). Bar = 10 μm. D, Surface cells of a mature wild-type ovary that was similar to that observed in C. Bar = 10 μm. E, Irregularly shaped cells with cuticular thickenings along with the interspersed stomata (sm) in epidermis of wild-type sepals. Bar = 10 μm. F, Stamen-like structure in second whorl of a 35S::LMADS2 flower. A wild-type stamen in same magnification was boxed in right. Bar = 70 μm. G, Irregularly shaped and uniformly sized cells with rugose surface in epidermis of a stamen-like structure in F. Bar = 5 μm. H, Surface cells of a mature wild-type anther that was similar to that observed in G. Bar = 5 μm. I, Surface cells of a mature wild-type petal. Bar = 10 μm. J, 35S::EgMADS1 flower produced carpel-sepal-like (cs) structures in the first-whorl organs. Stigmatic papillae (sti) and ovules (o) were observed in these flowers. Bar = 200 μm. K, Close-up of the stigmatic papillae (sti) in J. Bar = 25 μm. L, Close-up of the ovules (o) in J. f, Funiculus. Bar = 80 μm. M, Surface cells from carpel-sepal structure (boxed in J) containing both carpel (c) and sepal (s) cells. Wild-type sepals in E contained irregularly shaped cells with cuticular thickenings in surface, whereas wild-type carpels in D contained irregularly shaped cells with smooth surface along with the interspersed stomata (sm). Bar = 10 μm. N, A 35S::EgMADS1 flower produced carpel-sepal-like (cs) structures with stigmatic papillae (sti) in the first-whorl organs. Second-whorl organs were completely transformed into stamen-like structures in this flower. Bar = 200 μm.
Ectopic Expression of EgMADS1 Causes Abnormal Phenotypes Similar to Those Observed in 35S::LMADS2 Transgenic Arabidopsis Plants
To further investigate the function of EgMADS1, transgenic plants ectopically expressing EgMADS1 cDNA were generated. Eleven independent transgenic Arabidopsis T1 plants were obtained. Four plants were phenotypically indistinguishable from the wild-type plants, whereas seven plants showed identical novel phenotypes similar to that observed in 35S::LMADS2 transgenic plants. These seven plants were also smaller and flowered significantly earlier than the wild-type plants by producing only four to six small, curled rosette leaves and two small, curled cauline leaves on inflorescence (Fig. 3N). In the same stage, the wild-type plants produced only round rosette leaves with long petioles and did not show elongated inflorescence. Similar to those observed in 35S::LMADS2 transgenic plants, floral buds produced in the inflorescence of these plants also opened prematurely (Fig. 3O).
When the flowers in these seven plants were analyzed, they showed the homeotic conversion of sepals and petals similar to that observed in 35S::LMADS2 transgenic Arabidopsis plants. The first-whorl sepals were converted into sepal-carpel-like organs (Figs. 3, O–Q, and 6J) containing stigmatic papillae (Fig. 6K) and ovules (Fig. 6L). When the epidermal cells in these first-whorl sepal-carpel-like structures were examined, they were morphologically similar to the wild-type carpel epidermis or a mixture of both sepal and carpel epidermis (Fig. 6M). In addition to the conversion of the sepal into a carpel, the second-whorl organs of these 35S::EgMADS1 flowers were usually absent (Fig. 3, O and P) or converted into stamen-like structures (Figs. 3Q and 6N).
DISCUSSION
On the basis of DNA sequence conservation in MADS box genes, we were able to clone and characterize MADS box genes from the lily and lisianthus to initiate a molecular investigation into flower development for these two important species. In this study, MADS box genes specifically expressed in the female reproductive organ carpels were characterized for these two species.
The protein sequence and phylogenetic analysis indicated that LMADS2 of lily and EgMADS1 of lisianthus are in the D functional group and are closely related to AGL11 of Arabidopsis and FBP7/11 in the petunia (Figs. 1 and 2). Therefore, LMADS2 and EgMADS1 are likely putative D functional genes in these two species. This assumption was supported by their expression pattern. Similar to FBP11 (Angenent et al., 1995; Colombo et al., 1995), EgMADS1 was also exclusively expressed in the ovules within the fourth whorl carpel (Fig. 5). The high degree of similarity in protein sequence (about 80% identity) and in expression pattern strongly indicates that EgMADS1 is an FBP11 ortholog and is also involved in ovule development in lisianthus.
Similar to EgMADS1, LMADS2 mRNA also accumulated specifically in the carpel (Fig. 4). However, the expression pattern for LMADS2 was slightly different from that observed for EgMADS1, FBP11, or AGL11 detected uniformly in ovules of the ovary (Angenent et al., 1995; Colombo et al., 1995; Rounsley et al., 1995). Although LMADS2 mRNA was also strongly detected in the ovules, a weak expression was observed in the style tissue (Fig. 4). One possible explanation is that the slight difference between LMADS2 and other D functional genes may reflect the diversity in the D group genes in various plant species during evolution. As an alternative, LMADS2 may possibly be an ortholog for other MADS box genes specifically expressed in the carpel. Because LMADS2 also showed a high sequence similarity to the C functional gene AG, especially in the MADS box domain (Fig. 1), the AG gene is thus a good candidate. However, AG or its orthologs reported to date were not only expressed in the fourth whorl carpel but also in the third whorl stamen of the flower (Kempin et al., 1993; Schmidt et al., 1993; Pnueli et al., 1994; Kang et al., 1995; Kater et al., 1998; Rutledge et al., 1998; Yu et al., 1999; Winter et al., 1999). This result distinguished LMADS2 from AG orthologs and indicated that LMADS2 is not likely the ortholog for AG. Two other candidates for LMADS2 orthologs are SHP1 and SHP2 (AGL1 and AGL5) because both genes were also specifically expressed in the carpel (Ma et al., 1991; Savidge et al., 1995; Flanagan et al., 1996). However, mRNA for SHP1 and SHP2 were detected in particular regions of the gynoecium and ovule and were absent in other parts of the carpel (Savidge et al., 1995; Flanagan et al., 1996), indicating their difference from LMADS2. In addition, these two genes have been thought to encode functionally redundant proteins in regulating the development of the fruit dehiscence zone (Liljegren et al., 1998, 2000). Moreover, protein sequence and phylogenetic analysis clearly separated LMADS2 from these two genes (Figs. 1 and 2). Therefore, we favor the assumption that LMADS2 is the lily putative D functional gene that regulates ovule formation.
Further evidence supported that LMADS2 and EgMADS1 are orthologs from the phenotypic analyses of transgenic plants. Both 35S::LMADS2 and 35S::EgMADS1 transgenic Arabidopsis plants produced very similar phenotypes by flowering early and generating ap2-like flowers in which sepal-carpel and stamen-petal structures were observed in the first and second whorls of the flowers (Figs. 3 and 6). This result suggested that LMADS2 and EgMADS1 have the same effect on flower development. These transgenic phenotypes were quite different from those observed in the transgenic petunia ectopically expressing FBP11 in which ovule-like structures were produced on the sepal and petal, whereas the leaf morphology and flowering time were normal (Colombo et al., 1995). This difference may be because of the diversity in the D gene functions. As an alternative, this might also be the difference between backgrounds of these transgenic plants. Because no functional analysis was available for FBP7/11 in Arabidopsis or for other D functional genes in the petunia, both assumptions require further investigation.
Interestingly, the phenotypes produced in 35S::LMADS2 and 35S::EgMADS1 transgenic Arabidopsis were clearly similar to Arabidopsis plants that ectopically expressed AG or its orthologs from heterologous plants (Mizukami and Ma, 1992, 1997; Rutledge et al., 1998). This result indicated that the D and C functional genes should have a similar effect on floral induction and formation once ectopically expressed in Arabidopsis. This assumption can be supported by the high sequence identity in the MADS box domain among LMADS2, EgMADS2, and AG. Because the MADS box sequence was the DNA-binding domain recognized and bound specifically in different CC-A rich-GG DNA sequence elements in regulating gene expression (Huang et al., 1993; Shiraishi et al., 1993; Tilly et al., 1998; Egea-Cortines et al., 1999), it is reasonable to believe that these MADS box genes share a higher degree of similarity in the MADS box domain and should present greater function similarity by targeting similar downstream genes. As shown in Figure 1, LMADS2 showed the highest identity to AG in the MADS box domain. Only one amino acid was found to differ between LMADS2 and AG in this domain. In contrast 3 to 5 amino acid difference exists between LMADS2 and the other D functional genes. Interestingly, similar to LMADS2, there is also only one amino acid difference in the MADS box domain between AG and OsMADS13, a putative D functional gene from rice (Fig. 1). Because both rice and lily are monocots, this relatively similar characteristic was not surprising. EgMADS1 also showed a high identity to AG in the MADS box domain with a 5-amino acid difference. A 3- to 6-amino acid difference was observed between EgMADS1 and the other D functional genes in this domain (Fig. 1). Therefore, a high sequence identity in the MADS box domain for LMADS2, EgMADS1, and AG supported the production of similar phenotypes in transgenic Arabidopsis as seen in our result. This assumption was further supported by the result that the same homeotic conversion (carpelloid sepals and staminoid petals) was produced in transgenic Arabidopsis plants ectopically expressing SHP1 or SHP2 (Liljegren et al., 2000). On the basis of our result, the C and D functional genes and their closely related MADS box genes specifically expressed in the carpel should have overlapping functions in regulating female reproductive organ development. This data also supported the notion that the C and D functional genes were possibly raised from gene duplication and diversification events (Theissen et al., 2000). It is reasonable to believe that significant changes in the expression patterns occurred for these genes during evolution. These changes separated their roles from one another and restricted each gene's function to a particular timing and/or region during flower development. Ectopic expression of these genes by 35S promoter will disrupt this restriction and alter the expression of the similar downstream genes, resulting in the production of the same phenotypes in transgenic plants.
In summary, two MADS box genes, LMADS2 and EgMADS1, specifically expressed in the carpel female reproductive organ were characterized in the lily and lisianthus. Sequence comparison and phylogenetic analysis indicated that they are putative D functional genes in these two plant species. Ectopic expression of these two genes in heterologous Arabidopsis plants produced similar phenotypes by flowering early and generating ap2-like mutant flowers. The characteristics of these two genes provide useful information in the understanding of the relationships between the C and D functional MADS box genes in regulating flower development. Efforts are under way to clone and analyze more carpel-specific MADS box genes from the lily and lisianthus. The results should lead to a deeper understanding of the diverse roles played by these closely related MADS box genes during evolution.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Plants of lily (Lilium longiflorum Thunb. cv Snow Queen) and lisianthus (Eustoma grandiflorum) used in this study were grown in the field in Tein Wei County, Chang Haw, Taiwan. Seeds for Arabidopsis were sterilized and placed on agar plates containing 0.5× Murashige and Skoog (1962) medium at 4°C for 2 d. The seedlings were then grown in growth chambers under long-day conditions (16-h light/8-h dark) at 22°C for 10 d before being transplanted to soil. The light intensity of the growth chambers was 150 μE m−2 s−1.
Cloning of cDNA for LMADS2 and EgMADS1
Total RNA was isolated from floral buds of lily or lisianthus using ULTRASPEC RNA Isolation System (BIOTECX Company, Houston). cDNA was synthesized from 500 μg total RNA using a cDNA synthesis kit (no. 200401, Stratagene, La Jolla, CA). Synthesized cDNA was size fractionated, and the fractions containing 1- to 1.5-kb cDNA fragments were collected and used as templates in following PCR experiments. PCR amplification was performed by touchdown program and by using MADS box degenerate primer M7 (5′-GCTCTCTGTNCTITGYGAYGC-3′) and K box degenerate primer K1 (5′-GGAATTCTCAGC(A/G/T) AT(C/T) TTNGC(C/T) CT-3′) or M7 and poly(T) primer as described by Tzeng and Yang (2001). PCR products about 800 bp [M7 + poly(T)] or 400 bp (M7 + K1) long were cloned and sequenced. Partial sequence for LMADS2 (800 bp) and EgMADS1 (400 bp), which showed similarity to D functional MADS box genes, were identified. Internal gene-specific primers were designed for LMADS2 for 5′-RACE and for EgMADS1 for both 5′- and 3′-RACE by using the SMART RACE cDNA Amplification Kit (BD Biosciences Clontech, Palo Alto, CA). Gene-specific primer for 5′-RACE of LMADS2: 5′-GTACATGTTGTCATTCTGAAGCT-3′. Gene-specific primers for 5′-RACE of EgMADS1: 5′-ATGCTTCTTTGATCTGATTCTTG-3′; for 3′-RACE of EgMADS1: 5′-AGTCAACAATTGACCGTTACAGG-3′. The full-length cDNA for LMADS2 and EgMADS1 were obtained by PCR amplification using the following 5′ primers: LMADS2, 5′-CACTTGGGATCCAGTGGTGACTGTCCT-3′; EgMADS1, 5′-ACGCGGGGGGATCCCAAAAGTGTT-3′; and the 3′ primers: LMADS2, 5′-CACTTGGGATCC(T)18-3′; EgMADS1, 5′-CCCGGGCATGGTAAACACAGATTACC-3′. Both the specific 5′ and 3′ primers for LMADS2 contained the generated BamHI recognition site (5′-GGATCC-3′, underlined) to facilitate the cloning of this cDNA. Full-length cDNA for EgMADS1 was cloned into PGEM-T Easy Vector (Promega, Madison, WI). Because a BamHI site was present in the end of 3′-untranslated region of EgMADS1 cDNA, a BamHI fragment-contained ORF of EgMADS1 was obtained by BamHI digestion.
RNA Gel-Blot Analysis
Total RNA was isolated from various organs and tissues of plants. For northern hybridization, 10 μg total RNA was electrophoresed in formaldehyde-agarose gels and transferred to Hybond N+ membranes (Amersham Biosciences UK, Ltd., Buckinghamshire, UK). The membranes were prehybridized for 30 min and hybridized with a 32P-labeled DNA probes overnight at 65°C in the same solution (0.25 m Na2HPO4, pH 7.2, and 7% [w/v] SDS) and then washed twice each in solution 1 (20 mm Na2HPO4, pH 7.2, and 5% [w/v] SDS) and solution 2 (20 mm Na2HPO4, pH 7.2, and 1% [w/v] SDS) for 30 min per wash. The blots were then air dried, covered with plastic wrap, and autoradiographed. The DNA probes specific for LMADS2 or EgMADS1 genes were partial cDNA fragments (without MADS box) amplified by PCR, respectively.
Plant Transformation and Transgenic Plants Analysis
A BamHI fragment containing the full-length cDNA for LMADS2 or EgMADS1 gene was cloned into binary vector PBI121 (BD Biosciences Clontech) under the control of cauliflower mosaic virus 35S promoter. The sense construct for each MADS box gene was an orientation determinant using PCR and was used for further plant transformation. Arabidopsis plants were transformed using vacuum infiltration method as described elsewhere (Bechtold et al., 1993). Transformants that survived in the medium containing kanamycin (50 μg mL−1) were further verified by PCR and reverse transcriptase-PCR analyses.
Scanning Electron Microscopy
Scanning electron microscopy was performed according to the method of Haung and Yang (1998) and Tzeng and Yang (2001). Various floral organs were fixed in 2% (w/v) glutaraldehyde in 25 mm sodium phosphate buffer (pH 6.8) at 4°C overnight. After dehydration in a graded ethanol series, specimens were critical-point dried in liquid CO2. The dried materials were mounted and coated with gold-palladium in a sputter-coater (model 5150, JBS, Watford, UK). Specimens were examined in a Topcon scanning electron microscope (model ABT-150S) with an accelerating voltage of 15 kV.
ACKNOWLEDGMENTS
We thank Sum-Wen Chen and Sum-Li Wang for helping to grow lily and lisianthus plants used in this research in the field.
Footnotes
This work was supported by the Council of Agriculture and National Science Council (Taiwan, Republic of China; grant nos. 90AS–2.1.1–FD–Z1 and NSC89–2311–B–005–050 to C.-H.Y.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.007948.
LITERATURE CITED
- Angenent GC, Colombo L. Molecular control of ovule development. Trends Plant Sci. 1996;1:228–232. [Google Scholar]
- Angenent GC, Franken J, Busscher M, van Dijken A, van Went JL, Dons HJM, van Tunen AJ. A novel class of MADS box genes is involved in ovule development in petunia. Plant Cell. 1995;7:1569–1582. doi: 10.1105/tpc.7.10.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Ser III Sci Vie. 1993;316:1194–1199. [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. Genes directing flower development in Arabidopsis. Plant Cell. 1989;1:37–52. doi: 10.1105/tpc.1.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H-Y, Yang CH. Molecular characterization of MADS box genes regulating ovule development and floral organ formation from Eustoma grandiflorum (abstract no. 82). 2000 Annual Meeting of The American Society of Plant Physiologists. California: San Diego; 2000. . July 15–19, 2000, p 39. [Google Scholar]
- Coen ES, Meyerowitz EM. The war of the whorls: genetic interactions controlling flower development. Nature. 1991;353:31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- Colombo L, Franken J, Alexander R, van der Krol R, Wittich PE, Dons HJM, Angenent GC. Downregulation of ovule-specific MADS box genes from petunia results in maternally controlled defects in seed development. Plant Cell. 1997a;9:703–715. doi: 10.1105/tpc.9.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo L, Franken J, Koetje E, van Went J, Dons HJM, Angenent GC, van Tunen AJ. The petunia MADS box gene FBP11 determines ovule identity. Plant Cell. 1995;7:1859–1868. doi: 10.1105/tpc.7.11.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo L, van Tunen AJ, Dons HJM, Angenent GC. Molecular control of flower development in Petunia hybrida. Adv Bot Res. 1997b;26:229–250. [Google Scholar]
- Dayhoff MO, Barker WC, Hunt LT. Establishing homology in protein sequences. Methods Enzymol. 1983;91:524–545. doi: 10.1016/s0076-6879(83)91049-2. [DOI] [PubMed] [Google Scholar]
- Drews GN, Bowman JL, Meyerowitz EM. Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell. 1991;65:991–1002. doi: 10.1016/0092-8674(91)90551-9. [DOI] [PubMed] [Google Scholar]
- Egea-Cortines M, Saedler H, Sommer H. Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 1999;18:5370–5379. doi: 10.1093/emboj/18.19.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan CA, Hu Y, Ma H. Specific expression of the AGL1 MADS-box gene suggests regulatory functions in Arabidopsis gynoecium and ovule development. Plant J. 1996;10:343–353. doi: 10.1046/j.1365-313x.1996.10020343.x. [DOI] [PubMed] [Google Scholar]
- Haung M-D, Yang C-H. EMF genes interact with late-flowering genes to regulate Arabidopsis shoot development. Plant Cell Physiol. 1998;39:382–393. doi: 10.1093/oxfordjournals.pcp.a029381. [DOI] [PubMed] [Google Scholar]
- Huang H, Mizukami Y, Hu Y, Ma H. Isolation and characterization of the binding sequences for the product of the Arabidopsis floral homeotic gene AGAMOUS. Nucleic Acids Res. 1993;21:4769–4776. doi: 10.1093/nar/21.20.4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immink RGH, Gadella TWJ, Ferrario S, Busscher M, Angenent GC. Analysis of MADS box protein-protein interactions in living plant cells. Proc Natl Acad Sci USA. 2002;99:2416–2421. doi: 10.1073/pnas.042677699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofuku KD, den Boer BGW, van Montagu M, Okamuro JK. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell. 1994;6:1211–1225. doi: 10.1105/tpc.6.9.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H-G, Noh Y-S, Chung Y-Y, Costa MA, An K, An G. Phenotypic alterations of petal and sepal by ectopic expression of a rice MADS box gene in tobacco. Plant Mol Biol. 1995;29:1–10. doi: 10.1007/BF00019114. [DOI] [PubMed] [Google Scholar]
- Kater MM, Colombo L, Franken J, Busscher M, Masiero S, Van Lookeren Campagne MM, Angenent GC. Multiple AGAMOUS homologs from cucumber and petunia differ in their ability to induce reproductive organ fate. Plant Cell. 1998;10:171–182. doi: 10.1105/tpc.10.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempin SA, Mandel MA, Yanofsky MF. Conversion of perianth into reproductive organs by ectopic expression of the tobacco floral homeotic gene NAG1. Plant Physiol. 1993;103:1041–1046. doi: 10.1104/pp.103.4.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Liljegren SJ, Ditta GS, Eshed Y, Savidge B, Bowman JL, Yanofsky MF. SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature. 2000;404:766–770. doi: 10.1038/35008089. [DOI] [PubMed] [Google Scholar]
- Liljegren SJ, Ferrándiz C, Alvarez-Buylla ER, Pelaz S, Yanofsky MF. Arabidopsis MADS-box genes involved in fruit dehiscence. Flowering Newslett. 1998;25:9–19. [Google Scholar]
- Lopez-Dee ZP, Wittich P, Enrico Pe M, Rigola D, Del Buono I, Gorla MS, Kater MM, Columbo L. OsMADS13, a novel rice MADS-box gene expressed during ovule development. Dev Genet. 1999;25:237–244. doi: 10.1002/(SICI)1520-6408(1999)25:3<237::AID-DVG6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Ma H, Yanofsky MF, Meyerowitz EM. AGL1-AGL6, an Arabidopsis gene family with similarity to floral homeotic and transcription factor genes. Genes Dev. 1991;5:484–495. doi: 10.1101/gad.5.3.484. [DOI] [PubMed] [Google Scholar]
- Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature. 1992;360:273–277. doi: 10.1038/360273a0. [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Ma H. Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell. 1992;71:119–131. doi: 10.1016/0092-8674(92)90271-d. [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Ma H. Determination of Arabidopsis floral meristem identity by AGAMOUS. Plant Cell. 1997;9:393–408. doi: 10.1105/tpc.9.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–479. [Google Scholar]
- Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature. 2000;405:200–203. doi: 10.1038/35012103. [DOI] [PubMed] [Google Scholar]
- Pnueli L, Hareven D, Rounsley SD, Yanofsky MF, Lifschitz E. Isolation of tomato AGAMOUS gene TAG1 and analysis of its homeotic role in transgenic plants. Plant Cell. 1994;6:163–173. doi: 10.1105/tpc.6.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purugganan MD, Rounsley SD, Schmidt RJ, Yanofsky MF. Molecular evolution of flower development: diversification of the plant MADS-box regulatory gene family. Genetics. 1995;140:345–356. doi: 10.1093/genetics/140.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounsley SD, Ditta GS, Yanofsky MF. Diverse roles for MADS box genes in Arabidopsis development. Plant Cell. 1995;7:1259–1269. doi: 10.1105/tpc.7.8.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutledge R, Regan S, Nicolas O, Fobert P, Cote C, Bosnich W, Kauffeldt C, Sunohara G, Seguin A, Stewart D. Characterization of an AGAMOUS homologue from the conifer black spruce (Picea mariana) that produces floral homeotic conversions when expressed in Arabidopsis. Plant J. 1998;15:625–634. doi: 10.1046/j.1365-313x.1998.00250.x. [DOI] [PubMed] [Google Scholar]
- Savidge B, Rounsley SD, Yanofsky MF. Temporal relationship between the transcription of two Arabidopsis MADS box genes and the floral organ identity genes. Plant Cell. 1995;7:721–733. doi: 10.1105/tpc.7.6.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt RJ, Veit B, Mandel MA, Mena M, Hake S, Yanofsky MF. Identification and molecular characterization of ZAG1, the maize homolog of Arabidopsis floral homeotic gene AGAMOUS. Plant Cell. 1993;5:729–737. doi: 10.1105/tpc.5.7.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi H, Okada K, Shimura Y. Nucleotide sequences recognized by the AGAMOUS MADS domain of Arabidopsis thaliana in vitro. Plant J. 1993;4:385–398. doi: 10.1046/j.1365-313x.1993.04020385.x. [DOI] [PubMed] [Google Scholar]
- Theissen G. Development of floral organ identity: stories from the MADS house. Curr Opin Biol. 2001;4:75–85. doi: 10.1016/s1369-5266(00)00139-4. [DOI] [PubMed] [Google Scholar]
- Theissen G, Becker A, Di Rosa A, Kanno A, Kim JT, Münster T, Winter K-U, Saedler H. A short history of MADS-box genes in plants. Plant Mol Biol. 2000;42:115–149. [PubMed] [Google Scholar]
- Theissen G, Saedler H. MADS-box genes in plant ontogeny and phylogeny: Haeckel's “biogenetic law” revisited. Curr Opin Genet Dev. 1995;5:628–639. doi: 10.1016/0959-437x(95)80032-8. [DOI] [PubMed] [Google Scholar]
- Theissen G, Saedler H. Floral quartets. Nature. 2001;409:469–471. doi: 10.1038/35054172. [DOI] [PubMed] [Google Scholar]
- Theissen G, Strater T, Fischer A, Saedler H. Structural characterization, chromosomal location and phylogenetic evaluation of two pairs of AGAMOUS-like MADS box genes from maize. Gene. 1995;156:155–166. doi: 10.1016/0378-1119(95)00020-7. [DOI] [PubMed] [Google Scholar]
- Tilly JJ, Allen DW, Jack T. The CArG boxes in the promoter of the Arabidopsis floral organ identity gene APETALA3 mediate diverse regulatory effects. Development. 1998;125:1647–1657. doi: 10.1242/dev.125.9.1647. [DOI] [PubMed] [Google Scholar]
- Tzeng T-Y, Yang C-H. A MADS box gene from lily (Lilium longiflorum) is sufficient to generate dominant negative mutation by interacting with PISTILLATA (PI) in Arabidopsis thaliana. Plant Cell Physiol. 2001;42:1156–1168. doi: 10.1093/pcp/pce151. [DOI] [PubMed] [Google Scholar]
- Weigel D, Meyerowitz EM. The ABCs of floral homeotic genes. Cell. 1994;78:203–209. doi: 10.1016/0092-8674(94)90291-7. [DOI] [PubMed] [Google Scholar]
- Winter KU, Becker A, Münster T, Kim JT, Saedler H, Theissen G. MADS-box genes reveal that gnetophytes are more closely related to conifers than to flowering plants. Proc Natl Acad Sci USA. 1999;96:7342–7347. doi: 10.1073/pnas.96.13.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature. 1990;346:35–39. doi: 10.1038/346035a0. [DOI] [PubMed] [Google Scholar]
- Yu D, Kotilainen M, Pollanen E, Mehto M, Elomaa P, Helariutta Y, Albert VA, Teeri TH. Organ identity genes and modified patterns of flower development in Gerbera hybrida (Asteraceae) Plant J. 1999;17:51–62. doi: 10.1046/j.1365-313x.1999.00351.x. [DOI] [PubMed] [Google Scholar]