Abstract
NADPH:cytochrome P450 reductase (CPR) provides reducing equivalents to diverse cytochrome P450 monooxygenases. We isolated cDNAs for three CPR genes (CPR1, CPR2, and CPR3) from hybrid poplar (Populus trichocarpa × Populus deltoides). Deduced CPR2 and CPR3 amino acid sequences were 91% identical, but encoded isoforms divergent from CPR1 (72% identity). CPR1 and CPR2 were co-expressed together with the P450 enzyme cinnamate-4-hydroxylase (C4H) in yeast (Saccharomyces cerevisiae). Microsomes isolated from strains expressing CPR1/C4H or CPR2/C4H enhanced C4H activities approximately 10-fold relative to the C4H-only control strain, and catalyzed NADPH-dependent cytochrome c reduction. The divergent CPR isoforms (CPR1 and CPR2/3) contained entirely different N-terminal sequences, which are conserved in other plant CPRs and are diagnostic for two distinct classes of CPRs within the angiosperms. C-terminal green fluorescent protein fusions to CPR1 and CPR2 were constructed and expressed in both yeast and Arabidopsis. The fusion proteins expressed in yeast retained the ability to support C4H activity and, thus, were catalytically active. Both CPR::green fluorescent protein fusion proteins were strictly localized to the endoplasmic reticulum in transgenic Arabidopsis. The lack of localization of either isoform to chloroplasts, where P450s are known to be present, suggests that an alternative P450 reduction system may be operative in this organelle. Transcripts for the three poplar CPR genes were present ubiquitously in all tissues examined, but CPR2 showed highest expression in young leaf tissue.
Cytochrome P450 monooxygenase enzymes (P450s) play key roles in a wide range of oxidative metabolic reactions in various organisms from microbes to humans. In animals, xenobiotic detoxification and sterol biosynthesis are the major functions of P450s. However, P450-mediated reactions in plants encompass a much broader spectrum, including biosynthesis of plant hormones and signal molecules, defense-related chemicals and structural secondary natural products, and herbicide detoxification (for review, see Werck-Reichhart et al., 2000; Feldmann, 2001). The Arabidopsis genome encodes 273 P450 genes (www.biobase.dk/P450), highlighting the important biochemical roles of P450-mediated reactions that have evolved in plants.
Most eukaryotic P450s are not self-sufficient enzymes, and their catalytic activities strictly rely on an electron donor, NADPH:cytochrome P450 reductase (CPR; Lu et al., 1969). CPR (EC 1.6.2.4) transfers two electrons from NADPH to diverse P450s via two prosthetic groups, FAD and FMN, in sequence as electron entry and exit ports, respectively (Iyanagi and Mason, 1973; Vermilion et al., 1981). Both P450s and CPR are integral membrane-bound proteins, and the reducing equivalents are delivered mainly through transient electrostatic interactions on the endoplasmic reticulum (ER) membrane (Nisimoto, 1986; Shen and Kasper, 1995). CPR is also known to transfer electrons to cytochrome b5, which forms part of electron transport chain for fatty acid and sterol desaturases on the ER (Ilan et al., 1981; Fukuchi-Mizutani et al., 1999), although the physiological significance of this interaction in vivo is yet unclear in plants.
In animals, a single CPR is known to interact with a number of different P450 enzymes in a given species (Simmons et al., 1985). Similarly, single CPR isoforms and corresponding cDNAs were also originally isolated from plant species, such as mung bean (Vigna radiata; Shet et al., 1993), Madagascar periwinkle (Catharanthus roseus; Meijer et al., 1993), and opium poppy (Papaver somniferum; Rosco et al., 1997). However, multiple plant CPR isoforms were inferred from purified CPRs having distinct molecular weights in Jerusalem artichoke (Helianthus tuberosus; Benveniste et al., 1991), and two divergent CPR genes have been characterized in both parsley (Petroselinum crispum) and Arabidopsis (Koopmann and Hahlbrock, 1997; Mizutani and Ohta, 1998). The two CPR genes in these plants are differentially regulated such that one CPR is constitutively expressed, whereas the expression of the other CPR is enhanced by environmental stresses such as UV light and pathogen infection. Therefore, plants appear to deploy distinct CPR isoforms to meet the high reductive demand for the P450-mediated reactions in stressed conditions.
The ER membrane is generally accepted as the primary subcellular target for eukaryotic P450 and CPR enzyme localization, although some P450s are present in animal mitochondria (Okuda, 1994). In plants, subcellular fractionation studies and expression of a green fluorescent protein (GFP) fusion show that a well-characterized plant P450 enzyme, cinnamate-4-hydroxylase (C4H), is exclusively localized to the ER (Benveniste et al., 1978; Ro et al., 2001). However, in recent years, it has become evident that P450-mediated reactions also can occur in the chloroplast. It is apparent from database searches that approximately 20 Arabidopsis P450 genes contain high Ser/Thr content in their N termini, indicative of chloroplast targeting (Watson et al., 2001). In agreement with this data, allene oxide synthase and fatty acid hydroperoxide lyase, members of the CYP74 family and key enzymes for oxylipin (oxygenated fatty acids) metabolism, are known to be localized to the inner and outer membranes of the chloroplast, respectively (Froehlich et al., 2001). Allene oxide synthase and hydroperoxide lyase are unusual, self-sufficient P450 enzymes that do not require a CPR partner for their reactions. However, typical, CPR-requiring P450 enzymes are also chloroplast localized. CYP86B1 (of unknown function) and CYP701A3 (ent-kaurene oxidase involved in GA biosynthesis) are localized to the chloroplast outer membrane (Helliwell et al., 2001; Watson et al., 2001), and CYP79B2/3, proposed to be a key enzyme for indole-3-acetic acid biosynthesis, is predicted to be targeted to the chloroplast where its substrate Trp is synthesized (Hull et al., 2000). Thus, P450 reductase activity competent to transfer reducing equivalents to the chloroplastic P450s is predicted to be present in chloroplast outer membranes.
In vascular plants, lignin is a major natural product and its biosynthesis requires the action of the P450 enzymes CYP73A5 (C4H; Mizutani et al., 1997), CYP98A3 (p-coumaroyl ester-3-hydroxylase; Schoch et al., 2001; Franke et al., 2002), and, in angiosperms, CYP84A1 (ferulate/coniferaldehyde-5-hydroxylase; Meyer et al., 1998) to generate monolignols. Thus, in trees, P450 activity is especially important in wood formation, which typically consists of 20% to 30% lignin (Lewis and Yamamoto, 1990). An understanding of mechanisms by which reductases support P450 activity in trees is important in understanding mechanisms used for partitioning of carbon into monolignol biosynthesis relative to other P450-requiring metabolic pathways.
Poplar (Populus trichocarpa × Populus deltoides) has emerged as a model tree for molecular, genetic, and biochemical studies (Hertzberg et al., 2001; Mellerowicz et al., 2001) and, in this paper, we describe isolation and characterization of cDNAs encoding three CPR isoforms from poplar. When poplar CPR-GFP gene fusions were expressed in Arabidopsis, both CPR isoforms were targeted to the ER, and were not detected in other organelles. To our knowledge, this is the first demonstration of the ER localization of divergent plant CPR isoforms containing entirely different N-terminal sequences.
RESULTS
The Poplar Genome Contains at Least Three CPR Genes
Two divergent Arabidopsis CPR cDNAs (Mizutani and Ohta, 1998) were used to probe a poplar xylem cDNA library for CPR cDNAs. A 1.8-kb 5′-deleted cDNA clone with sequence identity to other plant CPRs (designated CPR181) was isolated, but we were unsuccessful in recovering a full-length CPR cDNA from this library. Subsequently, a poplar young leaf cDNA library was screened for a full-length cDNA using CPR181 as a probe. Sequence analysis of the 5′ portion of the clones obtained showed that two 1.4-kb clones (CPR122 and CPR161) encoded putative ATG start codons and shared a 0.6-kb region with CPR181. However, these and all other clones obtained from this library lacked a 750-bp 3′ portion because of truncation at an internal XhoI site. Sequence comparison revealed that CPR122 and CPR181 were completely identical in their overlapping region (624 bp), whereas CPR161 showed 98.9% (477/482) nucleotide identity to both the CPR122 and CPR181. This suggested that that CPR122 and CPR181 were derived from the same gene, whereas CPR161 might be allelic to CPR122/181. Using two gene-specific primers from 5′- or 3′-untranslated regions of CPR122 and CPR181, respectively, a 2.5-kb CPR cDNA was isolated from a pool of xylem cDNAs by a PCR amplification. This clone showed 100% sequence identity to both CPR122 and CPR181, had a single ATG codon preceded by an in-frame stop codon 9 bp upstream, and contained a 2,076-bp open reading frame (ORF) encoding a polypeptide of 692 amino acids with a predicted molecular mass of 76,726 D. This cDNA clone and the corresponding gene were designated as CPR1 (GenBank accession no. AF302496).
The CPR1 sequence was more closely related to the Arabidopsis CPR1 gene than to the Arabidopsis CPR2 gene (Fig. 1), suggesting that other CPR isoforms more closely related to Arabidopsis CPR2 could be present in poplar. Probing the young leaf cDNA library with the Arabidopsis CPR2 cDNA at low stringency allowed us to collect a set of CPR cDNA clones that were classified into two groups by restriction enzyme digestion analysis. Sequencing a few clones from each group revealed two more full-length cDNAs (CPR216 and CPR351) that were 90% identical to each other in their coding regions and 80% identical in their 3′-untranslated regions. Two more partial clones from the CPR216 group were identical to the CPR216 in the sequenced regions. However, as was seen in the CPR1 sequence analysis, the partial nucleotide sequence alignment of five cDNA clones from the CPR351 group revealed that there were also two very similar but distinct subgroups within this group, with 97.8% nucleotide identity. The position and nature of nucleotide changes between these two subgroups were identical (data not shown), indicating that the several nucleotide differences were not caused by cloning or sequencing artifacts but derived from the poplar genome. Thus, these two very similar classes of clones are likely to represent two alleles in the CPR351 group. Assuming the presence of two distinct alleles within the CPR351 group, CPR216 is, therefore, not allelic to CPR351 in the diploid poplar genome.
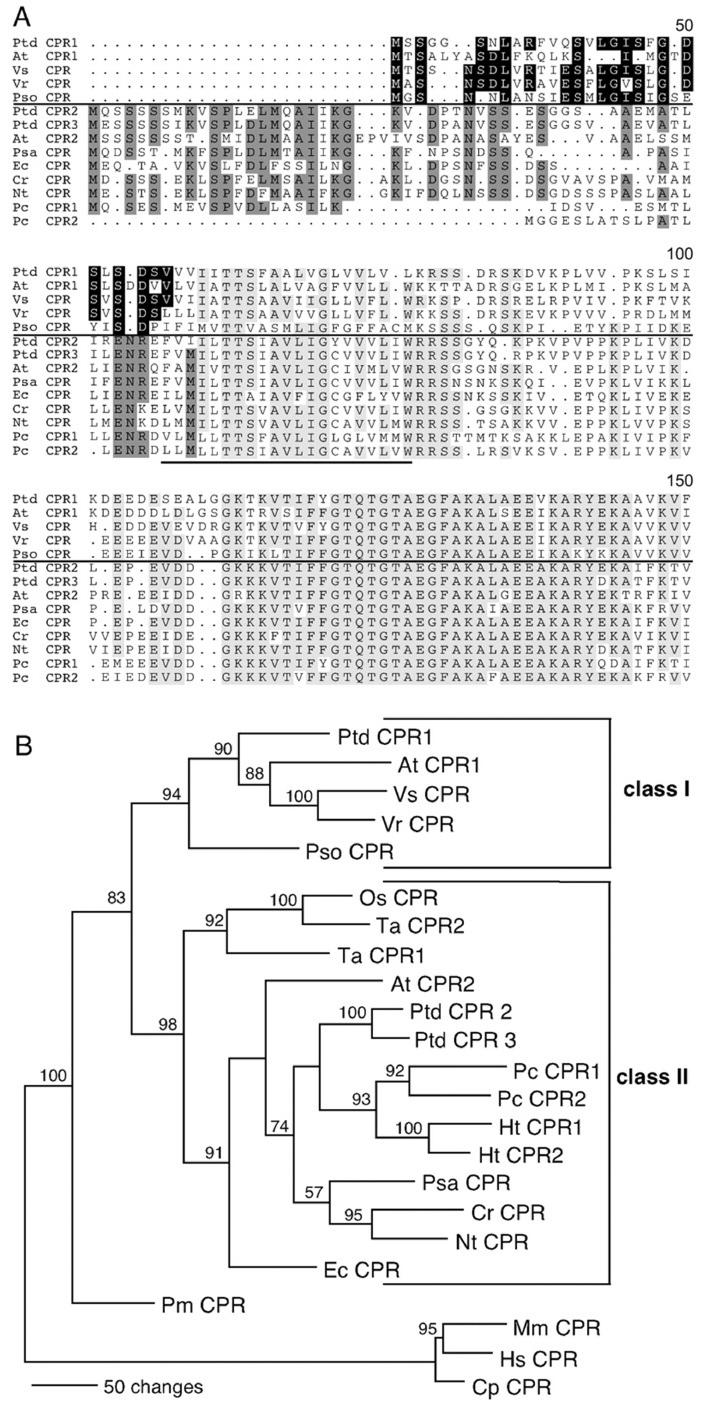
Figure 1.
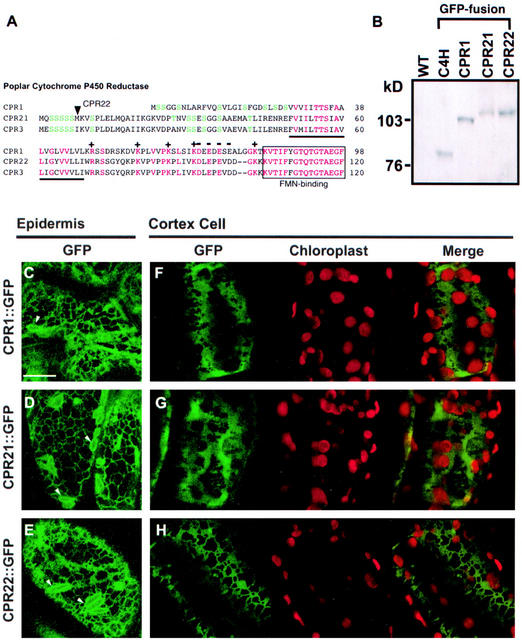
Alignment of multiple N-terminal amino acid sequences and phylogenetic tree reconstruction of plant CPRs. Deduced amino acids from the plants indicated were aligned using ClustalW software and further manually aligned by SeqPup software (Biology Department, Indiana University). A, Alignment of N termini of selected CPRs. Light-gray color indicates conserved amino acids (>60%) in all CPRs after 60th amino acid. Black and dark gray indicate conserved amino acids (>60%) within the N-terminal regions of CPR class I (above the horizontal line) and CPR class II (below the horizontal line), respectively. Underline designates the membrane-anchoring region. B, Maximum parsimony analysis of CPR amino acid sequences, excluding the first 59 positions. Tree reconstruction was performed with the tree-bisection-reconnection heuristic search algorithm using the Phylogenetic Analysis Using Parsimony software, version 4.0 (Sinaur Associates, Sunderland, MA). Branch lengths between nodes are drawn to scale to the number of evolutionary steps. Bootstrap values (percent of 1,000 replicates) for each cluster is shown at the nodes. Three mammalian CPR sequences were used as an out-group to root the tree. At, Arabidopsis; Cp, guinea pig (Cavia porcellus); Cr, Madagascar periwinkle; Ec, California poppy (Eschscholzia californica); Hs, human (Homo sapiens); Ht, Jerusalem artichoke; Mm, mouse (Mus musculus); Nt, tobacco (Nicotiana tabacum); Os, rice (Oryza sativa), Pc, parsley; Pm, Douglas fir (Pseudotsuga menziesii); Psa, pea (Pisum sativum); Pso, opium poppy; Ptd, hybrid poplar; Ta, bread wheat (Triticum aestivum); Vs, spring vetch (Vicia sativa); Vn, mung bean.
The two CPR genes represented by CPR216 and CPR351 were designated as CPR2 (accession no. AF302497) and CPR3 (accession no. AF302498), respectively. The CPR2 and CPR3 cDNAs possess stop codons 9 and 18 bp upstream of the first ATG codons, respectively, suggesting that these are full-length cDNAs. Two more putative ATG codons for CPR2 cDNA and one more ATG codon for CPR3 cDNA are located closely downstream of the first start codon. However, the nucleotides neighboring these ATGs deviated from the dicot start codon consensus, a(a/c) aAUGG, particularly with the occupancy of a pyrimidine at the important −3 position (with A in ATG as +1; Joshi et al., 1997). The flanking sequences of the first ATGs in both CPRs generally matched the consensus for initiation codons with an adenine at −3 positions (aacATGC for CPR2 and aacATGG for CPR3). Thus, the first ATGs were assumed to encode the translation start codons in both CPR2 and CPR3. These two cDNAs both contain 2,136-bp ORFs encoding 712 amino acids with predicted molecular masses of 78,693 D for CPR2 and 78,489 D for CPR3. The deduced amino acid sequences of CPR2 and CPR3 were 91% identical to each other, but they each showed 72% identity to CPR1.
Amino acid sequence alignments showed that all poplar CPR polypeptides share 70% to 77% amino acid identities to CPRs from other plants. Conserved cofactor and substrate-binding domains characteristic for the CPR, such as FMN-, FAD-, NADPH-, cytochrome c-, and P450-binding sites, were identified in all three poplar CPR polypeptides (data not shown), and their N-terminal domains contain an approximately 20-amino acid hydrophobic patch required for membrane anchoring (Fig. 1A). Sequence alignment of N-terminal ends revealed that these could be used to group angiosperm CPRs into two classes, class I and class II, based on their sequences located N terminal to the hydrophobic region. Poplar and Arabidopsis CPR1 and CPRs from spring vetch, mung bean, and opium poppy (class I) contained short N-terminal ends with several conserved amino acids. On the other hand, poplar CPR2/3, Arabidopsis CPR2, parsley CPR1 and CPRs from tobacco, pea, California poppy, and Madagascar periwinkle (class II) contained extended N-terminal ends that showed significant sequence identities. Parsley CPR2 was an exception to these groupings.
The overall sequence relatedness of CPRs within each class was further confirmed by phylogenetic analysis. The same grouping pattern as depicted in Figure 1A was evident in the parsimony analysis of CPR sequences, either using full-length amino acid sequences or using N-terminally (first 59 residues) deleted sequences (Fig. 1B). Therefore, two classes of CPR isoforms with distinct, conserved N-terminal ends are present in angiosperms, which appear to have evolved at least two CPR isoforms after an ancient gene duplication.
Functional Expression of CPRs in Yeast (Saccharomyces cerevisiae)
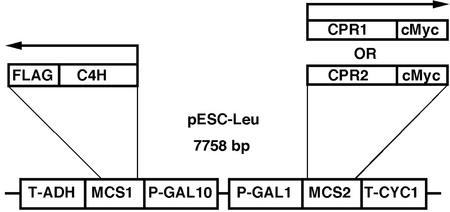
To confirm the bona fide CPR activity encoded by the poplar cDNAs, we expressed CPR1 and CPR2 in a yeast system together with C4H. It is known that endogenous CPR activity is a limiting factor in yeast when foreign P450s are overexpressed (Urban et al., 1994). Therefore, enhanced P450 activity by CPR co-expression in yeast is solid evidence for the biochemical identities of cloned CPR cDNAs. A Gal-inducible yeast dual expression vector pESC-Leu (Stratagene, La Jolla, CA) was employed for simultaneous expression of C4H and CPR in yeast (Fig. 2). The C4H coding sequence was inserted into the multiple cloning site behind the Gal-10 promoter in-frame with the FLAG epitope. Within the same vector, CPR1 or CPR2 were inserted behind the Gal-1 promoter in-frame with the c-Myc epitope. As a result, three pESC expression vectors were generated that harbor C4H alone, C4H/CPR1, or C4H/CPR2, and three corresponding yeast strains were generated by independent transformation of these constructs.
Figure 2.
Representation of the cloning and expression strategy for C4H and CPR1/2 in the yeast pESC dual expression vector. T-ADH and T-CYC1, Transcription termination sequences for alcohol dehydrogenase and cyclin 1 gene, respectively; MCS, multiple cloning site; P-GAL1 and P-GAL10, promoters for GAL1 and GAL10 gene, respectively.
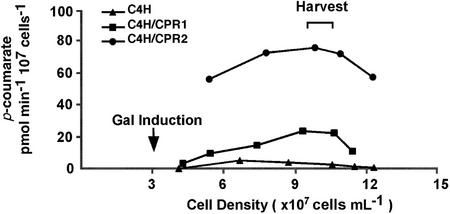
We used a convenient in vivo enzyme assay to evaluate C4H enzyme activity in these strains to establish optimal cell densities for harvesting cells and culture filtrates. Figure 3 shows that the catalytic activity of C4H was significantly higher when co-expressed with poplar CPR1 or CPR2 than when C4H was expressed alone, indicative of supportive role of the CPRs for C4H activity in yeast cells. Strains with a combination of C4H with CPR2 showed 3 to 4 times higher conversion rate of cinnamate to p-coumarate than those with C4H and CPR1. The maximum level of p-coumarate was produced at a cell density of approximately 109 cells mL−1.
Figure 3.
In vivo production of p-coumarate from yeast strains expressing C4H only, C4H and CPR1, and C4H and CPR2. Each yeast strain was precultured in Glc for 12 h, and the trans-genes were induced by 2% (w/v) Gal for 12 h. Formation of p-coumarate from cinnamate was quantified in aliquots harvested at 2- to 3-h intervals after induction. The same patterns for p-coumarate accumulation were repeated in two independent experiments.
Yeast microsomal fractions were prepared from yeast cells harvested at optimal cell densities (Fig. 3) to verify the CPR activities in vitro. Using microsomal preparations, recombinant CPR activities were evaluated either by measuring the formation of p-coumarate from cinnamate (C4H activity) or by measuring the reduction of cytochrome c, an artificial substrate for CPR. Patterns for the formation of p-coumarate from cinnamate in the microsomes prepared from various yeast strains (Table I) were very similar to those from in vivo estimations of C4H activity. In addition, when reduction of cytochrome c was measured in vitro, the microsomal fractions from CPR1- and CPR2-expressing yeast showed approximately 6- and 3-fold higher cytochrome c reduction activities, respectively, than yeast transformed with C4H alone. The reduction of cytochrome c by recombinant CPR1 and CPR2 protein was dependent on NADPH, but not on NADH, as is known for other CPRs (Koopmann and Hahlbrock, 1997; Mizutani and Ohta, 1998). Based on the in vivo and in vitro enzyme assays, therefore, we concluded that the CPR1 and CPR2 cDNAs encode functional, authentic CPR enzymes that properly interact with C4H and reduce cytochrome c in an NADPH-dependent manner.
Table I.
Catalytic activities of C4H and CPR in microsomal fractions prepared from C4H- or C4H/CPR-expressing yeast strains
| CPR Acta
|
||||
|---|---|---|---|---|
| Construct | C4H Acta | (Relative Act)b | NADH | NADPH |
| C4H/− | 0.9 ± 0.1 | (1.0 ± 0.1) | 528 ± 28 | 344 ± 25 |
| C4H/CPR1 | 2.8 ± 0.2 | (9.7 ± 1.3) | 504 ± 39 | 1,848 ± 95 |
| C4H/CPR2 | 7.4 ± 0.3 | (14.3 ± 1.3) | 437 ± 30 | 1,096 ± 60 |
Specific activities are given as nmol p-coumarate min−1 microsomal protein mg−1 for the C4H assay and nmol cyt c reduction min−1 microsomal protein mg−1 for the CPR assay. Each value is a mean ± se from four independent microsomal preparations.
For relative activities, the C4H-specific activities were normalized as arbitrary units derived from the densitometric quantification of the C4H signals detected by enhanced chemiluminescence (ECL) immunoblot analysis. The C4H activity from the C4H-only-expressing yeast was used as a reference to calculate relative activities in the other strains.
Characterization of C4H/CPR Dual Expression in Yeast
It was evident from the in vivo and in vitro assays that both CPR1 and CPR2 enhance C4H activity. However, in both cases, C4H activity was consistently higher when associated with CPR2 than with CPR1. These results led us to further investigate whether the higher C4H activity is related to a relatively higher affinity of C4H toward the CPR2 isoform or simply a higher level of C4H protein in the C4H/CPR2 strains.
Total P450 content in microsomes can be quantified by CO differential absorption spectroscopy (Omura and Sato, 1964). However, we were unable to detect this P450 signature indicative of C4H in yeast microsomes from C4H- or C4H/CPR-expressing strains. Apparently, the amount of recombinant C4H in these strains was below the detection limit of the spectroscopic method (<10 pmol mg−1 microsomal protein). In agreement with this conclusion, the presence of C4H was not discernible by SDS-PAGE analysis after Coomassie Blue staining, whereas new intense bands for CPR1 and CPR2 recombinant proteins with predicted molecular masses (78.4 and 80.4 kD after cMyc epitope fusion) were detected on the gel (data not shown). Thus, the absolute amount of CPR appeared to be much higher than that of C4H in this expression system, and C4H may be a limiting component in this dual expression system where CPR is highly expressed.
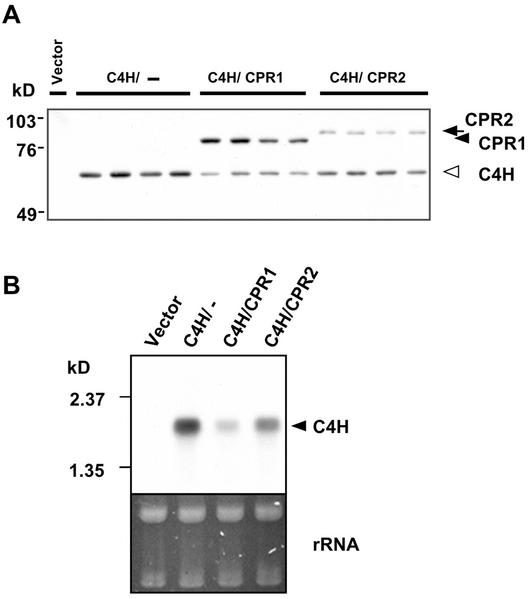
Subsequently, the expression level of C4H in differently transformed yeast strains was compared by semiquantitative immunoblot analysis and by northern-blot analysis. For immunoblot analysis, the same microsomal proteins, independently prepared and previously used for the enzyme assays, were fractionated by SDS-PAGE, and FLAG and c-Myc monoclonal antibodies used to detect specific recombinant proteins of predicted molecular weights on blots (Fig. 4A). Unexpectedly, C4H protein levels varied significantly in the different strains, although the same Gal-10 promoter in an identical vector background drove the expression of C4H in all cases. C4H levels were consistently lower in strains co-expressing CPRs than strains expressing C4H alone, and were lower in CPR1-co-expressing strains than in CPR2 co-expressing strains. C4H transcript levels in the different yeast strains showed a similar pattern, with highest amount of C4H expression in strains expressing C4H alone, and lower amounts in strains co-expressing CPR1 and CPR2 (Fig. 4B).
Figure 4.
Immunoblot and RNA-blot analysis of the transgenic yeast strains expressing C4H and CPR. A, Immunoblot of vector-, C4H-, C4H/CPR1-, and C4H/CPR2-transformed yeast strains. Two micrograms of microsomal proteins were fractionated by the 10% (w/v) polyacrylamide gel and transferred to the polyvinylidene difluoride (PVDF) membrane. CPR1/2 and C4H recombinant proteins were immunoreacted on the same blot by anti-cMyc antibody and anti-FLAG antibody. Four protein samples were prepared from independently transformed yeast strains. Specificities of antibodies were verified individually on separate blots before this immunoblot. B, RNA blot of vector-, C4H-, C4H/CPR1-, and C4H/CPR2-transformed yeast strains. Ten micrograms of total RNA was fractionated on a 1.5% (w/v) formaldehyde agarose gel, transferred to a nylon membrane, and hybridized to a poplar C4H probe.
Based on quantitation of immunodetectable protein levels, the relative C4H activity in yeast expressing C4H alone was approximately 10% or less than that in the C4H/CPR dual expressers (Table I), and within the dual expressers, the amount of C4H transcript and protein was generally proportional to the C4H activities previously measured by in vivo and in vitro enzyme assays. Thus, considering differences in C4H protein amount in different strains, the difference in the relative C4H activities between yeast strains expressing C4H/CPR1 and C4H/CPR2 was small (less than 2-fold; Table I). The enhanced C4H activity in the yeast strain co-expressing CPR2 was largely the result of elevated C4H transcript and protein levels in this strain relative to the CPR1-expressing strain, and not the result of enhanced ability of CPR2 to support C4H activity in yeast.
Two Distinct CPR Isoforms Are Localized to the ER
The three poplar CPR isoforms contain highly conserved amino acid sequences in the FMN-, FAD-, cytochrome c-, and NADPH-binding sites (data not shown). However, the CPR1 and CPR2/3 cDNAs encode two entirely different N termini, each of which has higher identity to its apparent isoform ortholog in Arabidopsis than to the other poplar isoform (Fig. 1). This could suggest important roles of the evolutionarily conserved N-terminal sequences in directing CPR1 and CPR2/3 to distinct subcellular locations, as similarly suggested for Arabidopsis CPRs by Urban et al. (1997) and Mizutani and Ohta (1998). Figure 5A illustrates the amino acid sequences of the poplar CPR N termini, highlighting non-conserved Ser/Thr-rich regions, a 20-amino acid hydrophobic region, a partially conserved (35% identity between CPR1 and CPR2/3) charged amino acid-rich region, and the highly conserved FMN-binding site. The N termini of CPR1 and CPR2 contain 30% (8/27 residues) and 25% (12/49 residues) Ser/Thr, respectively. Although there is no strictly conserved amino acid motif for chloroplast targeting of proteins, high-Ser/Thr composition in the N terminus is considered as an indication for the chloroplast localization (von Heijne et al., 1989). Analyzing the CPR amino acid sequence with the program ChloroP (www.cbs.dtu.dk/services/ChloroP) for the in silico prediction of chloroplast localization did not provide concrete answers. Scores for potential chloroplast targeting of CPR isoforms were 0.513 for CPR1, 0.484 for CPR2, and 0.479 for CPR3, where a value >0.5 is considered as indicative of chloroplast localization.
Figure 5.
N-terminal amino acid sequences of poplar CPRs, immunoblot analysis of the CPR::GFP proteins from transformed Arabidopsis, and localization of CPR::GFP in Arabidopsis seedlings by confocal microscopy. A, Comparison of predicted N termini of CPR1, CPR2, and CPR3. Green letters indicate Ser or Thr, and red letters indicate conserved amino acids in all three CPRs. Hydrophobic domains are shown by an underline, and conserved charged amino acids are shown as + or −. An arrowhead points to the start Met of the CPR22 version of CPR2-GFP. B, Immunoblot of microsomal proteins from transformed or non-transformed control (WT) Arabidopsis plants. Proteins were resolved in a 7.5% (w/v) polyacrylamide gel, transferred to a PVDF membrane, and immunoreacted with anti-GFP-antibody. The ECL-plus detection system was used to visualize signals. C through H, Confocal microscopy in living Arabidopsis seedlings transgenic for CPR1::GFP, CPR21::GFP, and CP22::GFP. GFP signals were detected in epidermal cells from cotyledon (C) or hypocotyl (D and E), and cortex cells from hypocotyl (F–H). Autofluorescence from chlorophyll in chloroplasts and GFP signals in hypocotyl cortex cells were separately collected through red and green channels, respectively, and merged using PhotoShop 5.0 software (Adobe Systems, Mountain View, CA; F–H). White arrows indicate elongated organelles associated with the ER. Bar = 10 μm.
To definitely determine the subcellular localization of the poplar CPRs, we fused GFP to the C termini of the predicted full-length CPR1 polypeptide and to two versions of CPR2, and expressed them in Arabidopsis under the control of the cauliflower mosaic virus 35S promoter. As shown in Figure 5A, CPR2 and CPR3 contain very similar N termini; therefore, CPR3 was not used in these studies. CPR21 is a full-length version of CPR2, whereas CPR22 was truncated such that its second ATG is the translation start codon, resulting in deletion of five Ser (Fig. 5A). To meet the nucleotide preference for the translation initiation (Joshi et al., 1997), the cytosine at the −3 position in CPR22 was replaced with an adenine during PCR amplification.
A total of >80 independently transformed T1 and T2 Arabidopsis seedlings for each construct were prescreened for the presence of green fluorescence by fluorescence microscopy. Most of the kanamycin-resistant plants presented weak GFP signals associated with the perinuclear membrane in the guard cells but showed no fluorescence in the epidermal cells (data not shown). Several Arabidopsis lines containing each construct showed detectable green fluorescence in the epidermis and some cortex cells. Immunoblot analysis of the microsomal proteins from these Arabidopsis seedlings demonstrated that intact fusion proteins were expressed in these Arabidopsis lines (Fig. 5B).
Confocal microscopy showed that all three different GFP-fused CPRs were strictly localized to the ER membrane in the epidermal cells of transgenic plants (Fig. 5, C–E). The GFP localization patterns observed here were identical to those observed for poplar C4H::GFP (Ro et al., 2001). As observed in the Arabidopsis lines expressing the C4H::GFP and other ER-targeted GFPs (Hawes et al., 2001; Hayashi et al., 2001; Ro et al., 2001), mobile, elongated organelles were also detected in the epidermal cells from all three CPR::GFP-expressing Arabidopsis (Fig. 5, C–E, arrowheads). Some cortex cells from hypocotyls had detectable GFP signals, and reticulated ER-like patterns were observed along with red autofluorescence from the chloroplasts (Fig. 5, F–H). These two signals did not overlap with each other, showing that the CPRs were not localized to the chloroplast. Therefore, we conclude that two divergent poplar CPR isoforms are targeted to ER, that their distinct N termini do not specify differential subcellular localization, and that neither CPR1 nor CPR2 is targeted to the chloroplast.
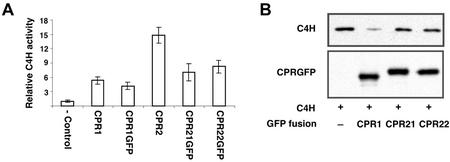
Functional verification of GFP fusion protein activity can be used to exclude potential localization artifacts caused by nonfunctional GFP fusions. For this purpose, the CPR::GFP constructs were recloned into a pESC yeast expression vector already containing the C4H, and co-expressed with C4H in yeast. In vivo analysis of C4H activities in the absence or presence of the CPR::GFP-fusion proteins in these strains was used to assay CPR::GFP function. Figure 6A shows that yeast transformed with the CPR::GFP constructs supported the C4H reaction at significantly higher rates relative to C4H activity in control cells expressing only C4H. To validate this observation, the presence and relative amounts of the CPR::GFP and C4H proteins were estimated by immunoblot analysis using anti-GFP or anti-FLAG antibodies to detect CPR::GFPs and C4H in yeast microsomal proteins (Fig. 6B). As previously observed, the highest amount of C4H was detected in the yeast strain expressing C4H alone, although this strain had the lowest C4H activity. These data demonstrate that CPR::GFP enzymes are active and that the C-terminal GFP fusions do not significantly disrupt interaction with C4H in vivo.
Figure 6.
CPR::GFP activity and protein levels in transgenic yeast strains. A, Relative in vivo C4H activities in the presence of GFP-fused CPRs. The C4H-only expressed yeast was used as a negative control (−control) and set as a relative value 1 (5.4 ± 1.1 pmol p-coumarate min−1 107 cell−1). B, Immunoblot analysis of C4H and GFP-fused CPR recombinant proteins expressed in yeast. Two micrograms of microsomal proteins from the yeast strains expressing the indicated proteins was fractionated on 7.5% (w/v) SDS-PAGE, and protein blots were prepared on PVDF membranes. C4H or CPR::GFP recombinant proteins were detected by the ECL method using anti-FLAG or anti-GFP antibodies, respectively.
CPR Expression Patterns in Poplar
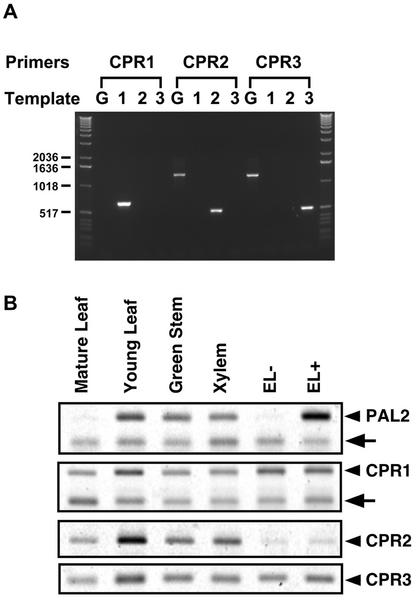
The mRNA accumulation patterns of poplar CPRs in various tissues and in poplar cultured cells with or without elicitor treatment were examined by semiquantitative reverse transcriptase (RT)-PCR analysis. This RT-PCR analysis employed CPR gene-specific primers and 18S-specific primers as internal controls for RNA amount. PCR cycle number was optimized for each gene to ensure that amplification endpoints were in the logarithmic phase. CPR1-specific primers were designed in the coding region of CPR1. For CPR2 and CPR3, a common forward primer in the 5′-coding region of CPR2/3 and distinct reverse primers each for CPR2 or CPR3 in the 3′-untranslated regions were designed. Figure 7A shows that these gene-specific primer pairs were able to distinguish different CPR cDNAs used as templates, and, thus, are gene specific. However, using poplar genomic DNA as templates, the CPR1-specific primers did not amplify any PCR product, whereas the CPR2 and CPR3 primers showed about 800 bp longer PCR products than those calculated from the corresponding CPR cDNAs. These data suggest that the CPR1 primers span an exon-intron junction or flank a very long intron in genomic DNA (resulting in no PCR products). The longer genomic fragments amplified by CPPR2/3 primers relative to the cDNAs likely arose because of the presence of introns in poplar CPRs as shown in Arabidopsis CPR (17 introns for each Arabidopsis isoform). Therefore, the primer sets for each CPR isoform were not only gene specific, but also distinguished CPR cDNA amplicons from those of genomic DNA.
Figure 7.
RT-PCR expression analysis of CPR1, CPR2, and CPR3 in poplar. A, Specificities of PCR primers specific to the three CPR genes were tested using 100 ng of poplar genomic DNA (G) or 1 ng of CPR1 (1), CPR2 (2), and CPR3 (3) cDNAs. PCR products were resolved on agarose gels stained with ethidium bromide. B, RT-PCR amplification of specific CPR isoforms from poplar tissues and cell culture. Total RNA (100 ng) isolated from poplar tissues or control (EL−) and 2-h elicitor-treated (EL+) cell cultures was used for RT-PCR after the optimization of PCR cycle number (20 cycles for PAL2 and CPR3; 25 cycles for CPR1 and CPR2). Arrowheads indicate specific RT-PCR products, and arrows indicate the 18S internal control amplified by 18S primers mixed with “competimer” (Ambion, Austin, TX). All four RT-PCR products were amplified from the same RNA batch, and similar expression patterns were obtained in an independent experiment.
We have documented previously the expression pattern of the poplar PAL2 gene (PAL7 cDNA) by northern-blot analysis (Subramaniam et al., 1993). Thus, the reliability of the overall RT-PCR analysis was first evaluated using this gene as a control. Figure 7B (top) shows that the relative levels of the predicted PAL2 RT-PCR products using PAL2-specific primers and RNA templates from different sources closely matched the expression pattern of this gene estimated by northern-blot analysis (i.e. highest expression in young leaf and strong induction by elicitor; Subramaniam et al., 1993). RT-PCR analyses of CPR transcript levels using the same total RNA preparations were then carried out and showed that CPR1 transcripts were ubiquitously present in all tissues and cells with lowest expression in mature leaves when normalized to 18S amplification. (Fig. 7B).
Amplified 18S ribosomal RNA fragments interfered with RT-PCR using CPR2 and CPR3 primers, necessitating amplification of transcripts from these two genes without this internal control. Figure 7B shows that the highest amount of CPR2 transcript was detected in young leaf, and that its expression in cultured poplar cells was barely detectable. The CPR3 transcripts were also ubiquitously present in tissues and cultured cells as observed in CPR1. The CPR3 gene appeared to have the strongest expression level among the CPR genes because, with the same amount of total RNA, it required about five fewer PCR cycles than CPR1 and CPR2. We have shown previously a strong induction of genes involved in phenylpropanoid metabolism, such as PAL, C4H, and 4CL, from the poplar cell cultures treated with an elicitor derived from cell walls of Fusarium oxysporum (Moniz de Sa et al., 1992; Ro et al., 2001). However, we did not find a significant induction of any CPR isoform by the same elicitor treatment, whereas the transcripts of control PAL2 were dramatically induced. CPR enzyme assays based on cytochrome c reduction prepared from elicitor-treated cells resulted in about 4 times higher activity than those from non-treated control cells (data not shown). Therefore, although we could not detect transcriptional up-regulation of the three poplar CPR genes, CPR enzyme activity was induced by elicitor in cultured poplar cells.
DISCUSSION
Multiple Isoforms of CPR in Poplar
As is the case for other poplar phenylpropanoid genes such as PAL, 4CL, and C4H (Subramaniam et al., 1993; Allina et al., 1998; Cukovic et al., 2001; Ro et al., 2001), CPR is encoded by a small gene family in poplar. Among the three CPR cDNAs studied here, CPR1 and CPR2/3 displayed highly divergent sequences, whereas the CPR2 and CPR3 showed 91% amino acid identity to each other. In addition, partial sequencing of several additional CPR cDNA clones suggested the presence of two putative alleles each for CPR1 and CPR3. The cDNA libraries in this study were constructed from a poplar F1 hybrid; thus, complete heterozygosity for genes is expected in its genome and in cDNA libraries. Considering that two probable CPR3 alleles were found in the cDNA population, CPR2 and CPR3 appear to be two different genes, likely to have arisen from a recent gene duplication. CPR2 and CPR3 show similar but not identical expression patterns in the RT-PCR analysis (Fig. 7B), further supporting the idea that they are two different genes.
The identification of two divergent classes of poplar CPR genes (CPR1 and CPR2/3) shows that this plant contains two distantly related CPR genes very similar to those in Arabidopsis (Urban et al., 1997; Mizutani and Ohta, 1998). Furthermore, analysis of N-terminal sequence identities and phylogenetic analysis of complete amino acid sequences of CPRs from several plant species revealed that all angiosperm CPRs fell into two classes that closely resembled either the poplar CPR1- or CPR2/3-type CPR. Although parsley contains two differentially regulated CPR isoforms with 80% amino acid identity (Koopmann and Hahlbrock, 1997), these two CPRs are both more closely related to poplar CPR2/3 and Arabidopsis CPR2 in the phylogenetic analysis (Fig. 1B). Therefore, it is probable that other plants (including parsley) contain other divergent CPR isoforms equivalent of CPR1 or CPR2/3. A potential third CPR isoform was inferred from immunoblot analysis in parsley (Koopmann and Hahlbrock, 1997). The phylogenetic position of the Douglas fir CPR (PmCPR) at the base of all angiosperm sequences found in the parsimony analysis (Fig. 1B) remains uncertain because conflicting results were obtained with neighbor-joining analyses, which placed this sequence either within the class I or class II cluster depending on whether full-length sequences or N-terminal deleted sequences were used (data not shown). Therefore, it cannot be concluded if the ancient gene duplication leading to class I and II CPR occurred before or after the divergence of angiosperms and gymnosperms. Additional CPR sequences from gymnosperms may help in resolving this question.
The occurrence of multiple CPRs in plants may reflect the diversity of P450s that catalyze reactions for the synthesis of a wide range of primary and secondary metabolites essential for plant growth and development (Werck-Reichhart et al., 2000; Feldmann, 2001). Despite the diversity of P450-mediated reactions, 62% of Arabidopsis P450s (153/246) subjected to phylogenetic analysis clustered within the same lineage group of so-called plant-specific “A-type P450s” (Durst and Nelson, 1995; www.biobase.dk/P450). All the P450s belonging to this A type are presumed to have evolved after divergence of plant and animal/fungal lineages. Thus, it is reasonable that plants have evolved different CPR isoforms to cope with demands for reduction of diverse plant P450s.
The authentic activities of recombinant poplar CPR isoforms were verified by expression in yeast. The dual expression vector employed allowed convenient co-expression of C4H and CPR in a single yeast strain. The Gal-1 and Gal-10 promoters controlling C4H and CPR expression are strictly regulated by Glc/Gal at the level of transcription and the promoters comparably induce reporter genes (information supplied by Stratagene). Thus, we expected that similar levels of both recombinant proteins would accumulate. This was expected to simplify in vivo evaluation of differential ability of CPR to support C4H activity without membrane reconstitution in vitro using individually prepared recombinant proteins. However, we unexpectedly found unequal amounts of recombinant C4H and CPR in different strains (Fig. 4).
The significantly decreased C4H recombinant protein levels relative to CPR in C4H/CPR dual-expressing strains could result from the presence of C4H codons unfavorable for use in yeast. Codon use in both CPR1 and CPR2 mRNAs does not severely deviate from that in yeast, but the predicted C4H mRNA contains a few unfavorable codons for yeast expression such as CTC (Leu) and AGG (Arg). We recovered >100 pmol C4H mg−1 microsomal protein from yeast grown in rich medium (Ro et al., 2001). However, in the minimal medium conditions used for dual expression in this study, rare tRNAs could be limiting factors for efficient translation of C4H, especially in combination with CPR. It has been reported that codon usage is a serious problem for yeast expression of plant P450s that contain a high proportion of rare yeast codons (Batard et al., 2000). In addition, however, expression of different combinations of C4H and CPR resulted in different levels of C4H transcripts. We do not have a clear explanation for this variation, but other unknown variations in transcription seem to be introduced by dual expression of C4H and CPR.
Although different levels of recombinant proteins that accumulated complicated analysis, the data here in general show that both CPR isoforms could nearly equally support the activity of C4H, the best characterized A-type P450 enzyme. This is in agreement with findings reported from other plants (Urban et al., 1997; Cabello-Hurtado et al., 1998; Mizutani and Ohta, 1998), and suggests that there may be no specificity in vivo in the interactions between P450s and individual reductases.
Two Divergent CPR Isoforms Are Localized to the ER
Sequence comparison of the CPR isoforms from poplar and Arabidopsis revealed striking cross species identities in the N termini within the same CPR group. Other angiosperm species also contain CPRs that group either with class I or II CPR isoform from poplar and Arabidopsis not only in their N-terminal sequences but by phylogenetic analysis of complete coding amino acid sequences (Fig. 1). The overall length of N termini and even many amino acids were conserved in each class of CPR in an isoform-specific manner. This conservation of this primary sequence suggests functional relevance of the N-terminal domain such that it has been maintained through evolution in distantly related species. One plausible explanation for this is that the different N termini contain information for the different subcellular destination of each CPR isoform. In particular, poplar CPR2/3 and its ortholog Arabidopsis CPR2 possess an extended N terminus with high Ser/Thr content, generally believed to be required for chloroplast targeting (von Heijne et al., 1989). However, the GFP localization experiments for poplar CPRs (Fig. 5) showed that both classes of CPR isoforms are localized and retained on the ER in the same pattern as C4H::GFP (Ro et al., 2001), and that neither is localized to chloroplasts in transgenic Arabidopsis. In addition, the CPR2 isoform engineered to utilize the second Met, in which the Ser-rich region was deleted, was also localized to the ER, suggesting that the Ser-rich region does not function significantly in subcellular targeting, and that minor N-terminal modification does not interfere with localization of CPR. Therefore, the conserved N-terminal regions do not direct differential subcellular localization of CPRs in plants.
As was shown in C4H::GFP localization studies (Ro et al., 2001), fluorescent, elongated, mobile organelles along the ER were observed in the epidermal layers of Arabidopsis expressing the various CPR::GFP constructs (Fig. 5, C–E, arrowheads). Based on the accumulation of Cys proteinases and a vacuolar-processing enzyme in these organelles, Hayashi et al. (2001) have recently proposed that these organelles are proteinase-storing ER bodies. However, Golgi-targeted secretory forms of GFP are also transiently retained in these elongated organelles before being sorted to the destined organelles (Hawes et al., 2001). In addition, it is obvious from our work that these organelles can harbor both CPR and the P450 enzyme C4H. Thus, the organelles are more likely to be an integral part of the ER where many ER proteins are localized, rather than a specialized organelle only for a specific subset of enzymes.
The CPR::GFP localization results were further substantiated by the ability of CPR::GFP to support C4H activity in yeast, indicating that the GFP domain of the fusion enzyme does not significantly interfere with CPR-C4H interaction in living cells, and that correct protein folding and orientation of CPR on the ER is maintained in the fusions. In contrast, the CPR::GFP fusion enzymes were incapable of supporting reduction of cytochrome c in microsomes in an in vitro assay (data not shown). The observed preferential reduction of C4H by CPR::GFP compared with cytochrome c may be because of the fact that cytochrome c interacts with CPR from the aqueous phase in the in vitro assay, whereas C4H interacts with CPR in a tight membrane-bound form through the ER. The membrane anchoring domains of P450s and CPR may stabilize the P450-CPR association either by direct N-terminal hydrophobic interactions or by conferring the favorable structural conformations for P450-CPR interactions. Alternatively, the GFP fusion may result in steric hindrance that prevents CPR interaction with cytochrome c in the presence of C4H.
CPR-Independent Electron Transport Path to P450s in Chloroplasts?
Arabidopsis P450 enzymes have been identified that are likely to be chloroplast localized based on the high Ser/Thr content of their N termini (Watson et al., 2001). Some P450s have been shown experimentally to be chloroplast outer membrane localized (Helliwell et al., 2001; Watson et al., 2001). Thus, electron donors for these P450s should be associated with chloroplasts, but the absence of CPR::GFP targeting of either poplar CPR isoform to the chloroplast observed is not consistent with roles of CPRs as electron donors for chloroplast-localized P450s. Although it is possible that unidentified divergent genes encoding a chloroplast-targeted CPR isoform exist, this is unlikely because the Arabidopsis genome appears to contain only two CPRs, CPR1 and CPR2. One additional gene (At3g02280) in the Arabidopsis genome shows limited sequence identity (approximately 28%) to Arabidopsis CPR1 and CPR2. However, the predicted At3g02280 ORF does not contain a transit peptide for chloroplast targeting, and the sequences for strictly conserved FMN- and cytochrome P450-binding sites deviate from those of all other CPRs, making its identity as a CPR questionable.
In the apparent absence of a chloroplast-targeted CPR species, it is likely that a non-homologous electron donor system is present in chloroplast to support P450 activity in this organelle. In animals, a specific mitochondrial electron donor system composed of two proteins, adrenodoxin and adrenodoxin reductase, supplies electrons from NADPH to mitochondrial P450s (Omura et al., 1966; Atsuta and Okuda, 1978). Chloroplast thylakoid membranes contain ferredoxin (Fd) and ferredoxin:NADP+ reductase (FNR), which serve as electron carriers in photosynthetic electron transport chain to produce NADPH. By a reverse reaction, these proteins can also serve as electron donors from NADPH for various physiological enzymes (for review, see Knaff and Hirasawa, 1991; Neuhaus and Emes, 2000), for chloroplast-targeted bacterial P450 in vivo (O'Keefe et al., 1994), and for the reconstituted in vitro P450 system (Yamazaki et al., 1995; Dong et al., 1996). However, the NADPH-FNR-Fd electron transport chain is operative in the stroma and is not likely to support P450s in the chloroplast outer membrane. There are several Fd- and FNR-like genes in Arabidopsis genome, and it would be interesting to see if these homologs are functional and interact with P450s in the outer membrane.
CPR Expression Patterns in Poplar
RT-PCR analysis detected all three CPR transcripts in several poplar tissues, but young leaves always showed higher levels of CPR transcript accumulation. CPR2 showed similar but slightly different expression patterns from CPR1 and CPR3. Accumulation of CPR2 transcripts was more predominant in young leaf, and CPR2 expression was low in both elicitor-treated and non-treated poplar cell cultures. Although speculative, this suggests a specific role for CPR2 in poplar young leaves in which other phenylpropanoid genes are also highly expressed (Subramaniam et al., 1993; Hu et al., 1998; Cukovic et al., 2001). The expression of poplar genes encoding specific PAL and 4CL isoforms in the epidermal or subepidermal cells of young leaf has been shown by in situ hybridization or promoter-GUS analysis (Subramaniam et al., 1993; Hu et al., 1998; Gray-Mitsumune et al., 1999). Poplar buds and young leaves are rich in phenolic compounds, including flavonoids and hydroxycinnamic acid conjugates and esters (Pearl and Darling, 1971; English et al., 1991), whose biosynthesis requires P450 enzymes, suggesting that the epidermal and subepidermal cells of these organs may require high levels of CPR activity. In situ hybridization and promoter-GUS fusions could determine the precise cell type-specific expression of CPR2 relative to CPR1 and CPR3 in poplar leaves.
There was no significant induction of any of the three CPR genes in an elicitor-treated cell culture harvested 2 h after elicitor treatment, when PAL and 4CL mRNAs are maximally expressed (Moniz de Sa et al., 1992). However, a significant increase in CPR enzyme activity was observed in the same elicitor-treated cell cultures (data not shown). The increased CPR enzyme activity in cells in which CPR transcripts were not induced suggests that posttranslational regulation of CPR activity may play an important role in coordinating CPR activity levels with those of other enzymes in phenylpropanoid metabolism.
MATERIALS AND METHODS
Isolation of CPR cDNA Clones
A cDNA library from hybrid poplar (Populus trichocarpa × Populus deltoides) H11-11 young leaf or xylem mRNAs was previously constructed in λZAPII (Subramaniam et al., 1993; Yuji Tsutsumi, Shizuoka University, Japan; C. Douglas, unpublished data) according to the manufacturer's instructions (Stratagene). A mixture of two Arabidopsis CPR cDNA clones (AR1 and AR2; Mizutani and Ohta, 1998) was radiolabeled by a random priming kit (Life Technologies/Gibco-BRL, Cleveland), and these probes were used to screen for CPR cDNA clones in the xylem cDNA library (105 plaque-forming unit total). Hybridization of the probes was performed for 16 h at 45°C in buffer containing 1% (w/v) bovine serum albumin (BSA), 7% (w/v) SDS, 50 mm sodium phosphate (pH 7.5), and 1 mm EDTA. A final wash was performed for 20 min at 45°C in 2× SSC with 0.1% (w/v) SDS. Three 1.8-kb clones isolated were confirmed to encode an identical 5′-deleted partial CPR sequence that was very similar to known plant CPRs. One of these clones (CPR181) was used to rescreen the H11-11 young leaf cDNA library for a full-length clone, using the following high-stringency conditions: hybridization, 16 h at 65°C in the same buffer as above; and final wash, 20 min at 65°C in 0.2× SSC with 0.1% (w/v) SDS. Among 30 positive clones, the longest clone isolated and sequenced was a 1.4-kb 3′-deleted clone (CPR122).
To obtain a full-length CPR cDNA, forward (5′-GCTAGTGACCACGTTTCAAGACAAC-3′) and reverse (5′-AGCTTTAATTCTACCTTGACACCTGG-3′) primers specific to the 3′- and 5′-untranslated regions of CPR122 and CPR181 partial cDNA clones were used to PCR amplify a full-length CPR cDNA from a cDNA pool of H11-11 xylem in a 50-μL reaction mixture containing 20 pmol each primer, 2.5 units of Pyrococcus furiosus polymerase (Stratagene), 100 ng of xylem cDNA template, 0.2 mm dNTP, and 2 mm Mg2+. The PCR conditions used were: one cycle of 3 min at 94°C, 25 cycles of 1 min at 94°C, 1 min at 62°C, and 3 min at 72°C. The PCR product was incubated with 5 units of Taq polymerase (Life Technologies/Gibco-BRL) at 72°C for 10 min to add overhang adenine nucleotides. The 2.5-kb PCR products were cloned into a pCR2.1 vector using a PCR TA cloning kit (Invitrogen, Carlsbad, CA), according to the manufacturer's instructions. This CPR clone was named CPR1. Other CPR isoforms were recovered from the poplar young leaf cDNA library, using an AR2 coding region as a hybridization probe under low-stringency conditions, as described above. As a result, 38 positive clones were identified, which were classified into two groups based on restriction fragment analyses using HindIII and PvuII. The longest clones from each group were completely sequenced, and named CPR2 and CPR3.
Construction of C4H, CPR, and CPR::GFP in the pESC-Leu Vector
Coding regions for C4H, CPR1, and CPR2 were amplified by PCR using the Pwo DNA polymerase (Roche Applied Science, Indianapolis). Gene-specific primers with NotI, SalI, or ApaI restriction enzyme sites (underlined) used for PCR were: for the C4H coding region, a forward primer, 5′-ATAAGACTGCGGCCGCATCATGGATCTCCTTCTCCTG-3′ and a reverse primer, 5′-AAGTAGTAGCGGCCGCAAAGGACCTTGGCTTTGCAACAATAG-3′; for the CPR1 coding region, a forward primer, 5′-TACAGCGGGCCCAGGATGAGTTCAGGTGGTTCAAAT-3′ and a reverse primer, 5′-TACAGCGTCGACCCAGACATCTCTTAGATACCGTCC-3′; and for the CPR2, a forward primer, 5′-TACAGCGGGCCCAACATGCAATCATCAAGCAGCTCG-3′ and a reverse primer, 5′-TACAGCGTCGACCCATACACGCAGATACCTGC-3′. The pESC-Leu yeast dual expression vector (Stratagene) was used to clone and express these genes. The PCR fragment for C4H was digested with NotI and inserted into the corresponding site behind the Gal-10 promoter in-frame with the FLAG epitope in the correct orientation. The PCR fragments for CPR1 or CPR2 were digested by SalI and ApaI and inserted into the corresponding sites behind the Gal-1 promoter in-frame with the c-Myc epitope. Thus, three different forms of the pESC-Leu expression vector were prepared to contain C4H alone, C4H and CPR1, or C4H and CPR2.
A series of CPR::GFP genes in the pBin19 vector (see below) were recloned into a pESC dual expression vector along with C4H. The CPR1::GFP fusion gene constructed in the pBin19 vector was used as a template in a PCR amplification. The coding sequence for CPR1::GFP was amplified using Pwo polymerase, a forward primer 5′-TACAGCGGGCCCAGGATGAGTTCAGGTGGTTCAAAT-3′, and a reverse primer, 5′-TACAGCGTCGACTTATTTGTATAGTTCATCCATGCC-3′. The PCR products were digested by ApaI and SalI, and inserted into the ApaI and XhoI sites of the pESC-Leu vector in which the C4H had previously been cloned. For yeast expression of the GFP-fusion of CPR21 and CPR22, DNA fragments containing a CPR21::GFP or CPR22::GFP coding sequence were isolated by ApaI/XhoI digestion from the pSL1180-based CPR::GFP constructs. These fragments were directionally cloned into the ApaI/XhoI sites of a pESC-Leu vector in which the C4H had been cloned previously.
Yeast Culture Media, Transformation, and Gene Induction
Untransformed YPH499 strain (MATa, ura3-52, lys2-801, ade2-101, trp1-Δ63, his3-Δ200, and leu2-Δ1) was maintained in yeast extract-peptone-dextrose plus adenine medium containing 0.08 g L−1 adenine hemisulfate salt, 10 g L−1 yeast extract, 10 g L−1 bactopeptone, and 20 g L−1 dextrose. For solid medium, 15 g of agar was supplemented. Transformed YPH499 strains with pESC-Leu vector were screened and maintained in Leu dropout minimal medium. One liter of Leu dropout synthetic minimal medium contains 6.7 g of yeast nitrogen base without amino acids (DIFCO Laboratories, Detroit), 1.3 g of Leu-dropout amino acid powder (Sigma), and 20 g of dextrose (for synthetic dextrose [SD]-Leu dropout medium) or Gal (for synthetic galactose-Leu dropout medium; DIFCO Laboratories). All the yeast expression vectors previously prepared were independently transformed into yeast strain YPH499 by the polyethylene glycol-LiOAc method (Gietz et al., 1992), and transformants were screened on the solid SD-Leu dropout medium.
For C4H and CPR gene induction in the YPH499 strain, the transformed yeast strains were subcultured in 3 mL of SD-Leu dropout medium overnight and 250 mL of SD-Leu dropout medium was subsequently inoculated by a 1:100 (v/v) dilution. Yeast cells were cultured at 28°C on a shaker at 150 rpm for 12 to 15 h until cultures reached mid-log phase (approximately 3–5 × 107 cells mL−1). The culture medium was changed to fresh synthetic galactose-Leu dropout medium to initiate gene induction, and the yeast cells were cultured for an additional 15 to 20 h until cell batches reached a cell density of nine, approximately 12 × 107 cells mL−1.
Enzyme Assays
C4H enzyme assays were performed as described previously (Ro et al., 2001). For in vitro CPR enzyme assay, cytochrome c (bovine heart; Sigma) was used as an artificial electron acceptor to measure cytochrome c reduction activity of CPR. The rate of reduction was calculated by differential absorption coefficiency of 21 mm−1 cm−1 at 550 nm. The reaction was initiated by addition of 0.1 mm NADPH or NADH to a final volume of 1 mL of reaction solution containing 100 mm sodium phosphate buffer (pH 7.4), 0.05 mm cytochrome c, 1 mm KCN, and 3 to 6 μg of microsomal protein. For the reference sample, buffer solution was added instead of NADPH. Formation of reduced cytochrome c was measured for 90 s using a 1601 Biospec spectrophotometer (Shimadzu, Columbia, MD) at room temperature.
For in vivo enzyme assays in yeast, the number of yeast cells, estimated by the spectrophotometric optical density at 600 nm (OD600), was used to normalize the enzyme activity data obtained from in vivo enzyme assays. An OD600 of 1 was determined to be equivalent to 3.0 × 107 cells mL−1, when OD600 value from the Shimadzu Biospec 1601 was calibrated by comparing hemocytometeric cell counting with the OD600 value, using YPH499 yeast strain (2.94 × 107 and 3.15 × 107 cells mL−1 per 1 OD600 in two independent countings). Ten milliliters of late-log phase yeast cells was harvested at 1,000g after Gal induction (in 2–3-h time intervals for Fig. 3) and resuspended in 5 mL of Tris-EDTA buffer (pH 7.5) with 0.2 mm cinnamate. Transformed yeast strains were incubated with cinnamate at 28°C for 20 min in a 150-rpm shaker. Cells were centrifuged at 4,000g and the supernatant was analyzed by a HPLC using the same program as described (Ro et al., 2001).
Microsomal Protein Preparation
Microsome preparation from yeast was performed as described (Pompon et al., 1996), except that ultracentrifugation at 100,000g was performed for 60 min. For Arabidopsis microsomal protein preparation, approximately 200 2-week-old Arabidopsis seedlings transformed with different GFP-fusion constructs, or wild-type control seedlings, were ground in liquid nitrogen. The resulting fine powders were resuspended in a buffer containing 50 mm Tris (pH 7.5), 1 mm phenylmethylsulfonyl fluoride, 5 mm MgCl2, one tablet of protease inhibitor mixture (Roche Molecular Biochemicals), and 14 mm β-mercaptoethanol. The suspension was centrifuged at 13,000g for 5 min at 4°C and the supernatant was recentrifuged at 100,000g for 30 min at 4°C in a bench-top ultracentrifuge (Beckman Instruments, Fullerton, CA). The pellets were redissolved in 50 μL of Tris buffer (pH 7.5) containing 20% (w/v) glycerol.
Construction of CPR::GFP Fusions in the pBin19 Vector
A C4H::GFP fusion construct had been generated previously in the pSL1180 vector (Ro et al., 2001). To make a series of CPR::GFP fusion constructs, the C4H portion was removed from C4H::GFP in the pSL1180 vector by ApaI and XbaI digestion. The CPR coding fragments for CPR1 and two versions of CPR2 (CPR21 and CPR22) were amplified by Pwo polymerase. Forward and reverse primers with ApaI and XbaI restriction sites used in the PCR were: for CPR1, 5′-TACAGCGGGCCCAGGATGAGTTCAGGTGGTTCAAAT-3′ and, 5′-CAGCTCTAGACCAGACATCTCTTAGATACC-3′; for CPR21, 5′-TACAGCGGGCCCAACATGCAATCATCAAGCAGCTCG-3′ and 5′-CAGCTCTAGACCATACATCACGCAGATACCT-3′; for CPR22, 5′-TACAGCGGGCCCAACATGAAAGTGTCACCACTTGAACTT-3′ and the same reverse primer used for the CPR21 amplification. The amplified PCR fragments were digested by ApaI and XbaI, followed by the ligation into the corresponding ApaI and XbaI sites of pSL1180-GFP. These ligations resulted in the construction of in-frame CPR::GFP gene fusions in the pSL1180 vector. All three constructs, CPR1::GFP, CPR21::GFP, and CPR22::GFP, were sequenced to verify the fidelity of the PCR products. The three constructs were digested by SalI and SpeI. These fragments were inserted into the XhoI and XbaI site of the previously generated Bin19/pRT101 vector (Ro et al., 2001), which placed the fusion constructs appropriately behind the cauliflower mosaic virus 35S promoter in the pBin19 vector background. These three constructs were independently transformed into Agrobacterium tumefaciens GV3101 strains by electroporation.
Plant Growth Conditions and Transformation
Arabidopsis ecotype Columbia was germinated and grown on medium containing a 0.5× Murashige and Skoog basal salt mixture (Sigma) for 1 week at 20°C under constant light, and the seedlings were then transferred to soil (Redi-Earth, W.R. Grace and Co., Ontario, Canada) under the same growth conditions. Plants were grown until abundant immature floral clusters had formed, and then transformed by the floral dip method (Clough and Bent, 1998) using 0.05% (w/v) Silwet L-77 (Lehle Seeds, Round Rock, TX). Primary transformants (T1) were selected by screening on 0.5× Murashige and Skoog medium containing 50 mg L−1 kanamycin and 0.7% (w/v) phytoagar (Life Technologies/Gibco-BRL).
Fluorescence and Confocal Microscopy Analysis
To detect fluorescent signals in CPR::GFP-transformed Arabidopsis lines, at least 80 independently transformed seedlings (3–5 d old) were prescreened by fluorescence microscopy using a light microscope equipped with a GFP filter set, and the strongest fluorescent lines were further analyzed by a Radiance confocal microscope (Bio-Rad Laboratories, Hercules, CA). Seedling samples were gently squeezed between cover and slide glass with a drop of distilled water, and a line of a 488-nm argon laser (10%–20%) was used as an excitation light source. Green and red fluorescent signals from GFP and chloroplasts were separately collected through the HQ515/30 or E600LP barrier filter, respectively. NIH Image software (National Institutes of Health, Bethesda, MD) was used to process the confocal microscopy data. Five to 10 sections at 0.2-μm intervals were overlapped to enhance the fluorescence signals. Collected images were further processed by the NIH Image or PhotoShop 5.0 software.
SDS-PAGE and Immunoblots
Protein concentrations were quantified by the Bradford method using BSA as a standard. Microsomal protein samples (2 μg for yeast and 20 μg for Arabidopsis) were separated on 7.5% (w/v; for GFP-fused proteins) or 10% (w/v; for non-GFP-fused proteins) polyacrylamide gels, and either stained with Coomassie Blue or transferred to PVDF membrane (Amersham-Pharmacia Biotech, Uppsala) for immunoblot analysis. Primary antibodies used, depending on the target proteins, were anti-FLAG (Stratagene), anti-cMyc (a gift from Dr. Michael Gold, Department of Microbiology and Immunology, University of British Columbia), or anti-GFP (Sigma) monoclonal antibodies. Immunodetection was performed using the ECL system (Amersham-Pharmacia Biotech). For simultaneous detection of C4H and CPR, sequential immunodetection was employed. Duplicate blots were prepared and reacted with FLAG or cMyc primary antibodies independently for 30 min, followed by secondary antibody binding and ECL-based signal detection in standard conditions. Each primary antibody identified a protein band of the predicted size, and no cross-reaction to other proteins was detected. The two blots were then washed with 1× Tris-buffered saline buffer supplemented with 0.05% (v/v) Tween 20 for 1 h, and reacted again with the reciprocal second primary antibody for 1 h. After incubation with secondary antibody at a 1:5,000 (v/v) dilution for 1 h, signals were detected as before. X-ray films were exposed to a level of under-saturation and the intensities of signals on the film were quantified by a densitometer (Bio-Rad Laboratories). The dual (FLAG and cMyc) and single signals (FLAG or cMyc) detected in the duplicated blots during this experiment showed very similar patterns.
Nucleic Acid Isolation and Northern Blots
For isolation of poplar genomic DNA, 1 g of frozen poplar (clone H11-11) young leaf was ground to a fine powder in liquid nitrogen, and the Nucleon PhytoPure beads (Amersham-Pharmacia Biotech) were used to retrieve DNA, according to the manufacturer's instructions. For RNA isolation from poplar cell culture and organs, the Nucleon PhytoPure method (Kiefer et al., 2000) was used, except that approximately 300 mg of starting material was used with a repeated Nucleon PhytoPure (50 μL) wash step. For total RNA isolation from yeast, cell pellets from 50-mL cultures of YPH499 yeast strain after 15 h of Gal induction were ground in liquid nitrogen, and extracted with the Trizol reagent (Life Technologies/Gibco-BRL). For northern blots, 10 μg of total RNA was resolved on 1.5% (w/v) formaldehyde agarose gels. RNA was transferred onto Hybond XL membrane (Amersham-Pharmacia Biotech), according to standard methods (Sambrook et al., 1989). Radioactive probes were prepared from the 1.5-kb-sized 5′-C4H coding region by use of a random priming kit (Life Technologies/Gibco-BRL). Probe hybridization was performed overnight at 65°C in a buffer containing 1% (w/v) BSA, 7% (w/v) SDS, 50 mm sodium phosphate (pH 7.5), and 1 mm EDTA. Membranes were washed twice for 20 min at 65°C in 2× SSC with 0.1% (w/v) SDS. The final wash was performed at 65°C in 0.2× SSC and 0.1% (w/v) SDS for 1 h.
RT-PCR Expression Analysis
Gene-specific primers used to amplify a cDNA fragment of PAL1/2 were: a forward primer, 5′-GTTGCATCCATTGCTGGTCATGATAC-3′ and a reverse primer, 5′-GAATCCAGATTCAATGCCAGCTGCTT-3′; for CPR1, a forward primer, 5′-CCTAGCGAGGCAGATAGACTCAAGT-3′ and a reverse primer, 5′-TAGTTCATATCTCTGCTGCTCTATC-3′; for CPR 2, a forward primer, 5′-TCATTATGATTGGCCCTGGAACTGGT-3′ and a reverse primer, 5′-CAAGGCTTCAACGGAGTTAACTTTTG-3′; and for CPR3, the same forward primer as for the CPR2 and a reverse primer, 5′-GGCTTCGGTATTTATAGAGTAAACTTT-3′. The specificities of the CPR primers were verified by PCR using 1 ng of PAL and CPR cDNA or 100 ng of poplar genomic DNA as templates. Taq polymerase (Life Technologies/Gibco-BRL) was used with 30 cycles of PCR under the following conditions: 30 s of denaturation at 94°C, 30 s of annealing at 62°C, and 1 min of polymerization at 72°C. OneStep RT-PCR reagent in a 25-μL final volume was used for optimization and experiments according to the manufacturer's protocol (Qiagen USA, Valencia, CA). Using young leaf total RNA, PCR cycle numbers were optimized for each gene such that product accumulation was in the exponential range. In the optimized cycle-range (20–25 cycles), a 7:3 ratio of 18S ribosomal RNA primers and competimers (QuantumRNA universal 18S internal standard, Ambion) was determined to be optimal, and 1 μL of this mixture was added to the final 25-μL reaction volume. Using gene-specific primers and 100 ng of total RNA from various tissues and cell cultures, 543-bp PAL2, 669-bp CPR1, 516-bp CPR2, and 513-bp CPR3 fragments were amplified as estimated by agarose gel electrophoresis with 100-bp DNA molecular markers (Life Technologies/Gibco-BRL). The specific PCR conditions used were as follows: 30 s of denaturation at 94°C, 30 s of annealing at 62°C, and 30 s of polymerization at 72°C for 20 cycles (PAL and CPR3) or 25 cycles (CPR1 and CPR2). The RT-PCR products were resolved on a 1.5% (w/v) agarose gel (4-mm thickness), and stained for 30 min by Sybr Green I (Molecular Probes, Eugene, OR). A Storm PhosphorImager (Amersham-Pharmacia Biotech) was used to detect fluorescent signals on the gel.
Phylogenetic Reconstruction
Alignments of full-length CPR amino acid sequences were generated using the ClustalW 1.4 algorithm and were manually optimized using the SeqPup 0.6 multiple sequence editor. Phylogenetic analyses were performed using the Phylogenetic Analysis Using Parsimony 4.0 software. The divergent N-terminal regions (first 59 positions in the alignment) were excluded. Parsimony analyses were executed using the tree-bisection-reconnection algorithm. Characters were reweighted for optimized rescaled consistency index. For bootstrap analysis (1,000 replicates), characters were sampled equally, but weights were applied. The accession numbers of the sequences used are as follows: Arabidopsis CPR1, X66016; Arabidopsis CPR2, AF325101; guinea pig (Cavia porcellus) CPR, D10498; Madagascar periwinkle (Catharanthus roseus) CPR, CAA49446; California poppy (Eschscholzia californica) CPR, U67186; Jerusalem artichoke (Helianthus tuberosus) CPR1, CAA81209; Jerusalem artichoke CPR2, CAA81210; human (Homo sapiens) CPR, AF258341; mouse (Mus musculus) CPR, D17571; tobacco (Nicotiana tabacum) CPR, JE0230; rice (Oryza sativa) CPR, OSJNEB0022F16.04; opium poppy (Papaver somniferum) CPR, T10720; parsley (Petroselinum crispum) CPR1, AAB97737; parsley CPR2, AAB97736; pea (Pisum sativum), AAC09468; hybrid poplar CPR1, AF302496; hybrid poplar CPR2, AF302497; hybrid poplar CPR3, AF302498; Douglas fir (Pseudotsuga menziesii), Z49767; bread wheat (Triticum aestivum) CPR1, AJ303373; bread wheat CPR2, AF123610; spring vetch (Vicia sativa), S37159; and mung bean (Vigna radiata), A47298.
ACKNOWLEDGMENTS
We thank Dr. Daisaku Ohta (Research Institute for Biological Science, Okayama, Japan) for providing us Arabidopsis CPR cDNAs, and Dr. Daniela Werck-Reichhart (Centre National de la Recherche Scientifique Unité Propre de Recherche, Université Louis Pasteur, Strasbourg, France) and Dr. Beverley Green (University of British Columbia) for useful discussions. We also thank Ms. Milly Sin-Yan So (Simon Fraser University, Burnaby, BC, Canada) for help with construction of CPR-GFP fusions as part of an undergraduate-directed studies project.
Footnotes
This work was supported by the Natural Science and Engineering Research Council of Canada (grant to C.J.D.) and by the University of British Columbia (University Graduate Fellowship to D.-K.R.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.008011.
LITERATURE CITED
- Allina SM, Pri-Hadash A, Theilmann DA, Ellis BE, Douglas CJ. 4-Coumarate:coenzyme A ligase in hybrid poplar. Properties of native enzymes, cDNA cloning, and analysis of recombinant enzymes. Plant Physiol. 1998;116:743–754. doi: 10.1104/pp.116.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsuta Y, Okuda K. Isolation of rat liver mitochondrial ferredoxin and its reductase active in the 5β-cholestane-3α, 7α, 12α-triol 26-hydroxylase. J Biol Chem. 1978;253:4653–4658. [PubMed] [Google Scholar]
- Batard Y, Hehn A, Nedelkina S, Schalk M, Pallett K, Schaller H, Werck-Reichhart D. Increasing expression of P450 and P450-reductase proteins from monocots in heterologous systems. Arch Biochem Biophys. 2000;379:161–169. doi: 10.1006/abbi.2000.1867. [DOI] [PubMed] [Google Scholar]
- Benveniste I, Lesot A, Hasenfratz MP, Kochs G, Durst F. Multiple forms of NADPH-cytochrome P450 reductase in higher plants. Biochem Biophys Res Commun. 1991;177:105–112. doi: 10.1016/0006-291x(91)91954-b. [DOI] [PubMed] [Google Scholar]
- Benveniste I, Salaun JP, Durst F. Phytochrome-mediated regulation of a monooxygenase hydroxylating cinnamic acid in etiolated pea seedlings. Phytochemistry. 1978;17:359–363. [Google Scholar]
- Cabello-Hurtado F, Batard Y, Salaun JP, Durst F, Pinot F, Werck-Reichhart D. Cloning, expression in yeast, and functional characterization of CYP81B1, a plant cytochrome P450 that catalyzes in-chain hydroxylation of fatty acids. J Biol Chem. 1998;273:7260–7267. doi: 10.1074/jbc.273.13.7260. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cukovic D, Ehlting J, VanZiffle JA, Douglas CJ. Structure and evolution of 4-coumarate:coenzyme A ligase (4CL) gene families. Biol Chem. 2001;382:645–654. doi: 10.1515/BC.2001.076. [DOI] [PubMed] [Google Scholar]
- Dong MS, Yamazaki H, Guo Z, Guengerich FP. Recombinant human cytochrome P450 1A2 and an N-terminal-truncated form: construction, purification, aggregation properties, and interactions with flavodoxin, ferredoxin, and NADPH-cytochrome P450 reductase. Arch Biochem Biophys. 1996;327:11–19. doi: 10.1006/abbi.1996.0086. [DOI] [PubMed] [Google Scholar]
- Durst F, Nelson DR. Diversity and evolution of plant P450 and P450-reductases. Drug Metabol Drug Interact. 1995;12:189–206. doi: 10.1515/dmdi.1995.12.3-4.189. [DOI] [PubMed] [Google Scholar]
- English S, Greenaway W, Whatley FR. Analysis of phenolics of Populus trichocarpabud extracts by GC-MS. Phytochemistry. 1991;30:531–533. [Google Scholar]
- Feldmann KA. Cytochrome P450s as genes for crop improvement. Curr Opin Plant Biol. 2001;4:162–167. doi: 10.1016/s1369-5266(00)00154-0. [DOI] [PubMed] [Google Scholar]
- Franke R, Hemm MR, Denault JW, Ruegger MO, Humphreys JM, Chapple C. The Arabidopsis REF8gene encodes the 3-hydroxylase of phenylpropanoid metabolism. Plant J. 2002;30:47–59. doi: 10.1046/j.1365-313x.2002.01266.x. [DOI] [PubMed] [Google Scholar]
- Froehlich JE, Itoh A, Howe GA. Tomato allene oxide synthase and fatty acid hydroperoxide lyase, two cytochrome p450s involved in oxylipin metabolism, are targeted to different membranes of chloroplast envelope. Plant Physiol. 2001;125:306–317. doi: 10.1104/pp.125.1.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuchi-Mizutani M, Mizutani M, Tanaka Y, Kusumi T, Ohta D. Microsomal electron transfer in higher plants: cloning and heterologous expression of NADH-cytochrome b5 reductase from Arabidopsis. Plant Physiol. 1999;119:353–362. doi: 10.1104/pp.119.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D, St. Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray-Mitsumune M, Molitor EK, Cukovic D, Carlson JE, Douglas CJ. Developmentally regulated patterns of expression directed by poplar PAL promoters in transgenic tobacco and poplar. Plant Mol Biol. 1999;39:657–669. doi: 10.1023/a:1006148715050. [DOI] [PubMed] [Google Scholar]
- Hawes C, Saint-Jore C, Martin B, Zheng HQ. ER confirmed as the location of mystery organelles in Arabidopsis plants expressing GFP! Trends Plant Sci. 2001;6:245–246. doi: 10.1016/s1360-1385(01)01980-x. [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Yamada K, Shimada T, Matsushima R, Nishizawa NK, Nishimura M, Hara-Nishimura I. A proteinase-storing body that prepares for cell death or stresses in the epidermal cells of Arabidopsis. Plant Cell Physiol. 2001;42:894–899. doi: 10.1093/pcp/pce144. [DOI] [PubMed] [Google Scholar]
- Helliwell CA, Sullivan JA, Mould RM, Gray JC, Peacock WJ, Dennis ES. A plastid envelope location of Arabidopsis ent-kaurene oxidase links the plastid and endoplasmic reticulum steps of the gibberellin biosynthesis pathway. Plant J. 2001;28:201–208. doi: 10.1046/j.1365-313x.2001.01150.x. [DOI] [PubMed] [Google Scholar]
- Hertzberg M, Aspeborg H, Schrader J, Andersson A, Erlandsson R, Blomqvist K, Bhalerao R, Uhlen M, Teeri TT, Lundeberg J et al. A transcriptional roadmap to wood formation. Proc Natl Acad Sci USA. 2001;98:14732–14737. doi: 10.1073/pnas.261293398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu WJ, Kawaoka A, Tsai CJ, Lung J, Osakabe K, Ebinuma H, Chiang VL. Compartmentalized expression of two structurally and functionally distinct 4-coumarate:CoA ligase genes in aspen (Populus tremuloides) Proc Natl Acad Sci USA. 1998;95:5407–5412. doi: 10.1073/pnas.95.9.5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull AK, Vij R, Celenza JL. Arabidopsiscytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc Natl Acad Sci USA. 2000;97:2379–2384. doi: 10.1073/pnas.040569997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilan Z, Ilan R, Cinti DL. Evidence for a new physiological role of hepatic NADPH:ferricytochrome (P-450) oxidoreductase. Direct electron input to the microsomal fatty acid chain elongation system. J Biol Chem. 1981;256:10066–10072. [PubMed] [Google Scholar]
- Iyanagi T, Mason HS. Some properties of hepatic reduced nicotinamide adenine dinucleotide phosphate-cytochrome c reductase. Biochemistry. 1973;12:2297–2308. doi: 10.1021/bi00736a018. [DOI] [PubMed] [Google Scholar]
- Joshi CP, Zhou H, Huang X, Chiang VL. Context sequences of translation initiation codon in plants. Plant Mol Biol. 1997;35:993–1001. doi: 10.1023/a:1005816823636. [DOI] [PubMed] [Google Scholar]
- Kiefer E, Heller W, Ernst D. A simple and efficient protocol for isolation of functional RNA from plant tissue rich in secondary metabolites. Plant Mol Biol Rep. 2000;18:33–39. [Google Scholar]
- Knaff DB, Hirasawa M. Ferredoxin-dependent chloroplast enzymes. Biochim Biophys Acta. 1991;1056:93–125. doi: 10.1016/s0005-2728(05)80277-4. [DOI] [PubMed] [Google Scholar]
- Koopmann E, Hahlbrock K. Differentially regulated NADPH:cytochrome P450 oxidoreductases in parsley. Proc Natl Acad Sci USA. 1997;94:14954–14959. doi: 10.1073/pnas.94.26.14954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis NG, Yamamoto E. Lignin: occurrence, biogenesis and biodegradation. Annu Rev Plant Physiol Plant Mol Biol. 1990;41:455–496. doi: 10.1146/annurev.pp.41.060190.002323. [DOI] [PubMed] [Google Scholar]
- Lu AY, Junk KW, Coon MJ. Resolution of the cytochrome P-450-containing ω-hydroxylation system of liver microsomes into three components. J Biol Chem. 1969;244:3714–3721. [PubMed] [Google Scholar]
- Meijer AH, Lopes Cardoso MI, Voskuilen JT, de Waal A, Verpoorte R, Hoge JH. Isolation and characterization of a cDNA clone from Catharanthus roseusencoding NADPH:cytochrome P-450 reductase, an enzyme essential for reactions catalysed by cytochrome P-450 monooxygenases in plants. Plant J. 1993;4:47–60. doi: 10.1046/j.1365-313x.1993.04010047.x. [DOI] [PubMed] [Google Scholar]
- Mellerowicz EJ, Baucher M, Sundberg B, Boerjan W. Unraveling cell wall formation in the woody dicot stem. Plant Mol Biol. 2001;47:239–274. [PubMed] [Google Scholar]
- Meyer K, Shirley AM, Cusumano JC, Bell-Lelong DA, Chapple C. Lignin monomer composition is determined by the expression of a cytochrome P450-dependent monooxygenase in Arabidopsis. Proc Natl Acad Sci USA. 1998;95:6619–6623. doi: 10.1073/pnas.95.12.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani M, Ohta D. Two isoforms of NADPH:cytochrome P450 reductase in Arabidopsis thaliana. Gene structure, heterologous expression in insect cells, and differential regulation. Plant Physiol. 1998;116:357–367. doi: 10.1104/pp.116.1.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani M, Ohta D, Sato R. Isolation of a cDNA and a genomic clone encoding cinnamate-4-hydroxylase from Arabidopsis and its expression manner in planta. Plant Physiol. 1997;113:755–763. doi: 10.1104/pp.113.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moniz de Sa M, Subramaniam R, Williams FE, Douglas CJ. Rapid activation of phenylpropanoid metabolism in elicitor-treated hybrid poplar (Populus trichocarpa Torr and Gray × Populus deltoidesMarsh) suspension-cultured cells. Plant Physiol. 1992;98:728–737. doi: 10.1104/pp.98.2.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus HE, Emes MJ. Nonphotosynthetic metabolism in plastids. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:111–140. doi: 10.1146/annurev.arplant.51.1.111. [DOI] [PubMed] [Google Scholar]
- Nisimoto Y. Localization of cytochrome c-binding domain on NADPH-cytochrome P-450 reductase. J Biol Chem. 1986;261:14232–14239. [PubMed] [Google Scholar]
- O'Keefe DP, Tepperman JM, Dean C, Leto KJ, Erbes DL, Odell JT. Plant expression of a bacterial cytochrome P450 that catalyzes activation of a sulfonylurea pro-herbicide. Plant Physiol. 1994;105:473–482. doi: 10.1104/pp.105.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda KI. Liver mitochondrial P450 involved in cholesterol catabolism and vitamin D activation. J Lipid Res. 1994;35:361–372. [PubMed] [Google Scholar]
- Omura T, Sander E, Estabrook RW, Cooper DY, Rosenthal O. Isolation from adrenal cortex of a nonheme iron protein and a flavoprotein functional as a reduced triphosphopyridine nucleotide-cytochrome P-450 reductase. Arch Biochem Biophys. 1966;117:660–673. [Google Scholar]
- Omura T, Sato R. The carbon monoxide-binding pigment of liver microsomes: I. Evidence for its hemoprotein nature. J Biol Chem. 1964;239:2370–2378. [PubMed] [Google Scholar]
- Pearl IA, Darling SF. Phenolic extractives of the leaves of Populus balsamifera and of P. trichocarpa. Phytochemistry. 1971;10:2844–2847. [Google Scholar]
- Pompon D, Louerat B, Bronine A, Urban P. Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol. 1996;272:51–64. doi: 10.1016/s0076-6879(96)72008-6. [DOI] [PubMed] [Google Scholar]
- Ro DK, Mah N, Ellis BE, Douglas CJ. Functional characterization and subcellular localization of poplar (Populus trichocarpa × Populus deltoides) cinnamate 4-hydroxylase. Plant Physiol. 2001;126:317–329. doi: 10.1104/pp.126.1.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosco A, Pauli HH, Priesner W, Kutchan TM. Cloning and heterologous expression of NADPH-cytochrome P450 reductases from the Papaveraceae. Arch Biochem Biophys. 1997;348:369–377. doi: 10.1006/abbi.1997.0374. 0. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schoch G, Goepfert S, Morant M, Hehn A, Meyer D, Ullmann P, Werck-Reichhart D. CYP98A3 from Arabidopsis thalianais a 3′ hydroxylase of phenolic esters, a missing link in the phenylpropanoid pathway. J Biol Chem. 2001;276:36566–36574. doi: 10.1074/jbc.M104047200. [DOI] [PubMed] [Google Scholar]
- Shen AL, Kasper CB. Role of acidic residues in the interaction of NADPH-cytochrome P450 oxidoreductase with cytochrome P450 and cytochrome c. J Biol Chem. 1995;270:27475–27480. doi: 10.1074/jbc.270.46.27475. [DOI] [PubMed] [Google Scholar]
- Shet MS, Sathasivan K, Arlotto MA, Mehdy MC, Estabrook RW. Purification, characterization, and cDNA cloning of an NADPH-cytochrome P450 reductase from mung bean. Proc Natl Acad Sci USA. 1993;90:2890–2894. doi: 10.1073/pnas.90.7.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons DL, Lalley PA, Kasper CB. Chromosomal assignments of genes coding for components of the mixed-function oxidase system in mice. Genetic localization of the cytochrome P-450PCN and P-450PB gene families and the NADPH-cytochrome P-450 oxidoreductase and epoxide hydratase genes. J Biol Chem. 1985;260:515–521. [PubMed] [Google Scholar]
- Subramaniam R, Reinold S, Molitor EK, Douglas CJ. Structure, inheritance, and expression of hybrid poplar (Populus trichocarpa × Populus deltoides) phenylalanine ammonia-lyase genes. Plant Physiol. 1993;102:71–83. doi: 10.1104/pp.102.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban P, Mignotte C, Kazmaier M, Delorme F, Pompon D. Cloning, yeast expression, and characterization of the coupling of two distantly related Arabidopsis thalianaNADPH-cytochrome P450 reductases with P450 CYP73A5. J Biol Chem. 1997;272:19176–19186. doi: 10.1074/jbc.272.31.19176. [DOI] [PubMed] [Google Scholar]
- Urban P, Werck-Reichhart D, Teutsch HG, Durst F, Regnier S, Kazmaier M, Pompon D. Characterization of recombinant plant cinnamate 4-hydroxylase produced in yeast. Kinetic and spectral properties of the major plant P450 of the phenylpropanoid pathway. Eur J Biochem. 1994;222:843–850. doi: 10.1111/j.1432-1033.1994.tb18931.x. [DOI] [PubMed] [Google Scholar]
- Vermilion JL, Ballou DP, Massey V, Coon MJ. Separate roles for FMN and FAD in catalysis by liver microsomal NADPH-cytochrome P-450 reductase. J Biol Chem. 1981;256:266–277. [PubMed] [Google Scholar]
- von Heijne G, Steppuhn J, Herrmann RG. Domain structure of mitochondrial and chloroplast targeting peptides. Eur J Biochem. 1989;180:535–545. doi: 10.1111/j.1432-1033.1989.tb14679.x. [DOI] [PubMed] [Google Scholar]
- Watson CJ, Froehlich JE, Josefsson CA, Chapple C, Durst F, Benveniste I, Coolbaugh RC. Localization of CYP86B1 in the outer envelope of chloroplasts. Plant Cell Physiol. 2001;42:873–878. doi: 10.1093/pcp/pce110. [DOI] [PubMed] [Google Scholar]
- Werck-Reichhart D, Hehn A, Didierjean L. Cytochromes P450 for engineering herbicide tolerance. Trends Plant Sci. 2000;5:116–123. doi: 10.1016/s1360-1385(00)01567-3. [DOI] [PubMed] [Google Scholar]
- Yamazaki H, Ueng YF, Shimada T, Guengerich FP. Roles of divalent metal ions in oxidations catalyzed by recombinant cytochrome P450 3A4 and replacement of NADPH-cytochrome P450 reductase with other flavoproteins, ferredoxin, and oxygen surrogates. Biochemistry. 1995;34:8380–8389. doi: 10.1021/bi00026a020. [DOI] [PubMed] [Google Scholar]