Abstract
In all eukaryotes, cell cycle progression is controlled by cyclin-dependent kinases (CDKs) whose activity is regulated at several levels including inhibition by CDK inhibitors. Here, we report a comparative molecular and functional analysis of the tobacco (Nicotiana tomentosiformis) CDK inhibitor, NtKIS1a, and its spliced variant, NtKIS1b. The C-terminal end of NtKIS1a shares strong sequence similarity with mammalian CIP/KIP inhibitors, which is not the case for NtKIS1b. Consistent with this, NtKIS1a but not NtKIS1b inhibits in vitro the kinase activity of CDK/cyclin complexes, and tobacco (Nicotiana tabacum) D-type cyclins and an A-type CDK are NtKIS1a, but not NtKIS1b, interacting partners. Although both NtKIS1a and NtKIS1b transcripts are mainly found in flowers and more precisely in stamens, NtKIS1b transcript levels are cell cycle regulated, whereas those of NtKIS1a remain constant during the cell cycle. NtKIS1a and NtKIS1b fused to fluorescent proteins are localized in the nucleus when transiently expressed in onion epidermal cells. Furthermore, there is no competition for their nuclear localization when they are simultaneously overexpressed. In vitro competition toward CDK kinase activity suggests that NtKIS1b is a strong competitor of NtKIS1a. Arabidopsis plants overexpressing NtKIS1a-green fluorescent protein (GFP) or NtKIS1b-GFP fusion proteins were obtained. In these plants, the fusion proteins are still localized in the nucleus. Interestingly, NtKIS1a-GFP-overexpressing plants display strong morphological modifications and a reduced CDK kinase activity, whereas NtKIS1b-GFP-overexpressing plants display a wild-type phenotype including a wild-type CDK kinase activity. Our results strongly suggest that the inhibition of the kinase activity is responsible for the phenotypic modifications.
The precise control of cell cycle progression is critical for coherent development because perturbation of this control leads to phenomena such as oncogenesis in animals. However, developmental programs adopted by plants differ markedly from those of animals. Plant development is mostly postembryonic, and throughout their life, plants form new organs by virtue of meristematic cells that are permanently able to divide. However, key cell cycle regulators discovered in yeast and animals appear remarkably conserved in plants (Huntley and Murray, 1999; Mironov et al., 1999; den Boer and Murray, 2000). Thus, in all eukaryotes, the core of the cell cycle machinery consists of complexes composed of a catalytic subunit, a cyclin-dependent kinase (CDK), and a regulatory subunit, a cyclin. Two major types of plant CDKs are revealed by sequence analysis, yeast complementation assays and expression patterns. Whereas canonical PSTAIR-containing A-type CDKs resemble CDK1 and CDK2 from mammals and CDC28/cdc2+ from budding and fission yeast, respectively, B-type CDKs have plant specific features, notably a cell cycle phase-specific expression that usually peaks in the G2 phase (Segers et al., 1997; Mironov et al., 1999; Joubes et al., 2000). Three major classes of cyclins, A-, B-, and D-types, are similarly found in plants according to their sequence similarities and expression pattern (Renaudin et al., 1996; Mironov et al., 1999). Regulatory mechanisms controlling the kinase activity of plant CDK/cyclin complexes are not as well documented. The recent discovery of CDK inhibitors (CKIs) in plants (Wang et al., 1997; Lui et al., 2000; De Veylder et al., 2001; Jasinski et al., 2002) is of great importance because they may represent a direct regulatory subunit of CDKs and thus allow the integration of antimitogenic signals.
In mammals, CKIs have been classified into two distinct families based on their structure and CDK targets (for review, see Pavletich, 1999). The four members of the INK4 family (p16INK4a, p15INK4b, p18INK4c, and p19INK4d) specifically form binary complexes with and inhibit CDK4-6. Although they appear to be structurally redundant and equally potent as inhibitors, they are differentially expressed, for example during mouse development (Zindy et al., 1997). The three members of the CIP/KIP family (p21CIP1, p27KIP1, and p57KIP2) form ternary complexes with and inhibit a broad spectrum of CDK/cyclin complexes (Nakayama and Nakayama, 1998). However, CIP/KIP inhibitors play a dual role because they also participate in the assembly and stabilization of CDK4-6/cyclin D complexes (LaBaer et al., 1997; Sherr and Roberts, 1999). In addition, p21CIP1 interferes with cell cycle progression by inhibiting DNA replication through its interaction with proliferating cell nuclear antigen (PCNA; Goubin and Ducommun, 1995; Chen et al., 1996).
In plants, sequences displaying similarities with animal CIP/KIP inhibitors have been identified in Arabidopsis, Chenopodium rubrum, pea (Pisum sativum), cotton (Gossypium hirsutum), rice (Oryza sativa), and tobacco (Nicotiana tabacum). The seven CKIs from Arabidopsis (KRP1-KRP7) present similarities in their C-terminal end with the CDK-binding/inhibitory domain of p27KIP1 (Wang et al., 1997; Lui et al., 2000; De Veylder et al., 2001; Zhou et al., 2002). All the KRPs interact with D-type cyclins but not with B-type CDKs. In addition, some of the KRPs interact with A-type CDKs, whereas others do not. KRP1 (ICK1) and KRP2 (ICK2) are shown to inhibit in vitro the kinase activity of plant CDK/cyclin complexes (Wang et al., 1998; Lui et al., 2000). The overexpression of Arabidopsis KRP1 (ICK1), KRP2 (ICK2), KRP6 (ICK4), C. rubrum, and N. tabacum (NtKIS1a; accession no. AJ297906) CKIs all produce similar phenotypes in Arabidopsis (Wang et al., 2000; De Veylder et al., 2001; Jasinski et al., 2002; Zhou et al., 2002), which consists in a reduced overall growth, serrated leaves, and modified flowers.
Here, we report the isolation through a yeast two-hybrid screen of the tobacco CKI named NtKIS1a. The deduced polypeptide displays sequence similarity with mammalian CKIs. A second cDNA named NtKIS1b (accession no. AJ297907) was isolated and shown to arise by an alternative splicing of a NtKIS1 (accession no. AJ297904) pre-mRNA. Interestingly, this NtKIS1b spliced form lacks a C-terminal part of the protein compared with NtKIS1a and, thus, does not display sequence similarities with mammalian CKIs. In this paper, we report a comparative analysis of NtKIS1a and NtKIS1b. We show that both proteins have antagonistic biochemical functions and different interacting partners. Their expression patterns during cell cycle and in different organs were analyzed. Transient expression of NtKIS1a and NtKIS1b fused to fluorescent proteins in onion epidermal cells allows their subcellular localization to be determined. Finally, a comparison of NtKIS1a-green fluorescent protein (GFP) and NtKIS1b-GFP-overexpressing Arabidopsis plants demonstrates that only the overexpression of NtKIS1a-GFP reduced the CDK kinase activity and strongly impairs plant development.
RESULTS
A Tobacco cDNA Encoding a CKI-Related Protein and Its Spliced Variant
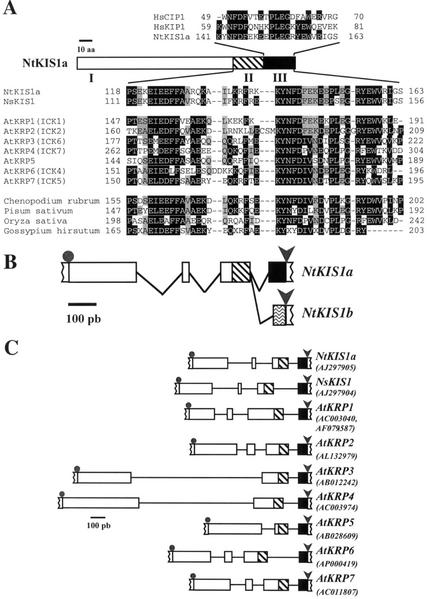
A N. tabacum A-type CDK, Nicta;CDKA;1, was used as a bait to screen a two-hybrid cDNA library of a N. tabacum cv Bright-Yellow 2 (BY-2) cell suspension. Among the positive clones, three contained different lengths of a 5′-truncated open reading frame further used as a probe to screen a N. tabacum cv Xanthi cDNA library (Galvez et al., 1996). Two cDNAs of 775 and 983 bp in length were obtained and named NtKIS1a and NtKIS1b for Nicotiana tomentosiformis (see below) kinase interacting subunit a/b. NtKIS1a is representative of three identical independent clones and contains an open reading frame predicted to encode a polypeptide of 163 amino acids with a molecular mass of 18.3 kD. The presence of an in-frame stop codon in the 5′-untranslated region, 27 nucleotides upstream of the first initiation codon, indicates that we obtained a full-length open reading frame. Comparison of the NtKIS1a deduced amino acid sequence with related proteins (see below) prompted us to define three domains: domain I (residues 1–117), domain II (residues 118–140), and domain III (residues 141–163; Fig. 1A). NtKIS1a displays similarities to Arabidopsis CKIs (KRP1-7; Wang et al., 1997; Lui et al., 2000; De Veylder et al., 2001), to a C. rubrum CDK interacting protein, and to deduced amino acid sequences of pea and rice cDNAs and a cotton expressed sequence tag (EST). The similarity between these proteins essentially resides at their C-terminal end (Fig. 1A) and consists of a 46-amino acid highly conserved region (domains II+III), the rest of the protein (domain I) being divergent. In addition, a part of this conserved C-terminal region (domain III) also displays strong similarity to the domain identified in the animal CIP/KIP inhibitors as the CDK interaction/inhibition domain (Fig. 1A; Chen et al., 1996; Russo et al., 1996). However, in animal CIP/KIP proteins, this domain is localized in the N-terminal region. Finally, the region containing the conserved LFG motif involved in the binding of CIP/KIP inhibitors with cyclins (Russo et al., 1996) is absent in NtKIS1a protein. The motifs 4, 5, and 6 present in some Arabidopsis KRP proteins (De Veylder et al., 2001) are similarly all absent in the tobacco proteins.
Figure 1.
NtKIS1 sequence analysis. A, The amino acid sequence deduced from NtKIS1a is schematically represented with its three domains: domain I (residues 1–117; white box), domain II (residues 118–140; hatched box), and domain III (residues 141–163; black box; also in B and C). Alignment of the domain III with human CIP/KIP inhibitors is shown above (HsCIP1: L25610; HsKIP1: U10906). Alignment of domains II+III with plant-related proteins is shown below. NsKIS1 corresponds to the polypeptide deduced from the GenScan-predicted open reading frame of N. sylvestris genomic sequence (http://bioweb.pasteur.fr/seqanal/interfaces/genscan.html). AtKRP1 to AtKRP7 correspond to the Arabidopsis polypeptides deduced from cDNA sequences. Correspondences with ICKs are given in brackets. C. rubrum, pea, and rice correspond to polypeptides deduced from cDNA sequences (AJ002173, AB029483, and AC069145). Cotton corresponds to the polypeptide deduced from an incomplete EST (AI728644). Identical or similar amino acids present in more than 50% and present in 30% to 50% of the proteins are highlighted, respectively, in black and gray. B, The exon-intron organization of the N. tomentosiformis NtKIS1 genomic sequence, which results from the comparison with NtKIS1a and NtKIS1b cDNAs, is schematically represented with a potential alternative splicing of the third intron (exons, boxes; introns, lines). Gray dots and arrows indicate, respectively, start and stop codons (also in C). Waved box represents the fourth exon in NtKIS1b different from NtKIS1a domain III defined above. C, The exon-intron organizations deduced from the different genomic sequences are compared. Accession numbers are indicated.
Comparison at the nucleotide level of NtKIS1b with NtKIS1a reveals a complete identity except that NtKIS1b exhibits a 218-bp extension of the 5′-untranslated region and a small 19-bp deletion at the end of the open reading frame generating a frameshift and a premature stop codon. NtKIS1b is predicted to encode a 154-amino acid polypeptide with a molecular mass of 17 kD. Because of the frameshift that modifies the C terminus of the protein, NtKIS1b has no similarity to animal CIP/KIP inhibitors. The nucleotide sequence identity of NtKIS1a and NtKIS1b, especially in their common 5′- and 3′-untranslated regions, suggests that they are derived from the same gene. Using specific primers and PCR amplification, the genomic sequence spanning the entire cDNAs was cloned from the genomic DNA of the two parents of N. tabacum, i.e. N. tomentosiformis and N. sylvestris. Alignment of genomic sequences with NtKIS1a and NtKIS1b cDNAs shows that the two cDNAs completely match the N. tomentosiformis sequence, whereas a lot of mismatches appear with the N. sylvestris sequence. These observations demonstrate that NtKIS1a and NtKIS1b both derived from the N. tomentosiformis gene and not from that of N. sylvestris. From this alignment, a gene structure with four exons-three introns is revealed. Moreover, it appears that NtKIS1a and NtKIS1b could be generated through an alternative splicing of the third intron using the same donor site and two different acceptor sites (Fig. 1B). However, when using Netplantgene, a program originally developed for Arabidopsis gene structure prediction (http://genome.cbs.dtu.dk/netpgene/cbsnetpgene.html), the alternative acceptor site for NtKIS1b is not detected, but the first intron is not predicted either. Thus, NtKIS1b could represent an alternatively spliced variant of NtKIS1a.
Finally, a comparison of gene structures deduced from alignments of genomic sequences with their corresponding cDNAs (except for NsKIS1 [accession no. AJ297905] for which we used the predicted open reading frame) was done. Although a four exons-three introns is the most commonly observed structure, AtKRP3, AtKRP4, and AtKRP5 represent three exceptions with a three exons-two introns structure (Fig. 1C). Interestingly, as already noticed by Lui et al. (2000), this also shows that the position of thelast intron is invariably conserved in Arabidopsis, N. tomentosiformis, and N. sylvestris and that the last exon always strictly encodes the same domain (domain III) except for the variant NtKIS1b (Fig. 1C).
NtKIS1a Is a CKI in Vitro, whereas NtKIS1b Prevents NtKIS1a Inhibition
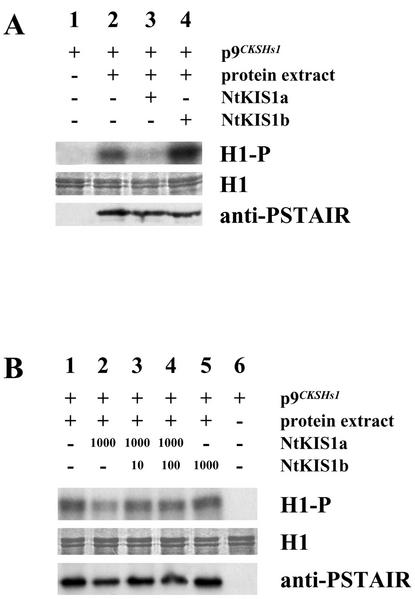
Sequence analysis indicates that NtKIS1a shares similarities with the mammalian CIP/KIP inhibitors and the Arabidopsis proteins KRPs, of which KRP1 (ICK1) and KRP2 (ICK2) were shown in vitro to have inhibitory properties on the kinase activity of plant CDK/cyclin complexes (Wang et al., 1997, 1998; Lui et al., 2000). To test whether NtKIS1a and NtKIS1b have inhibitory properties, although NtKIS1b lacks the potential CDK interacting domain, their effect on the kinase activity of CDK/cyclin complexes was assessed. Purified NtKIS1a and NtKIS1b recombinant proteins prepared as described in “Materials and Methods,” were added to affinity-purified CDK/cyclin complexes from BY-2 cells refreshed for 24 h. These complexes display a strong kinase activity (Fig. 2A, lane 2), this activity being correlated to the presence of a 34-kD protein recognized by an antibody raised against the highly conserved CDK PSTAIR motif. Addition of the purified NtKIS1a protein to the affinity-purified complexes resulted in a 60% decrease of the kinase activity (Fig. 2A, lane 3), whereas NtKIS1b addition had no effect or even a contrary effect because an increase was observed (Fig. 2A, lane 4). In both cases, a 34-kD protein recognized by an anti-PSTAIR antibody was still detected (Fig. 2A, lanes 3 and 4). These results suggest that NtKIS1a has inhibitory properties on the kinase activity of CDK/cyclin complexes from tobacco BY-2 cells, whereas NtKIS1b is an inactive variant probably as the result of its modified C-terminal end.
Figure 2.
In vitro effect of purified recombinant NtKIS1a and NtKIS1b proteins on the kinase activity of BY-2 CDK/cyclin complexes. A, Histone H1 phosphorylation is shown (H1-P). Protein extract of 24-h refreshed BY-2 cells was added (+, lanes 2–4) or not (−, lane 1) to p9CKSHs1 beads to purify CDK/cyclin complexes. Histone H1 (25 μg) was added in all lanes (H1). Five hundred nanograms of NtKIS1a (lane 3), 500 ng of NtKIS1b (lane 4), or buffer (lanes 1 and 2) was further added. Histone H1K activity was monitored as described in “Materials and Methods.” Proteins bound to p9CKSHs1 beads were recovered and immunoblotted with an antibody raised against the conserved CDK PSTAIR motif (anti-PSTAIR). B, Histone H1 phosphorylation is shown (H1-P). Protein extract of 24-h refreshed BY-2 cells was added (+, lanes 1–5) or not (−, lane 6) to p9CKSHs1 beads to purify CDK/cyclin complexes. Histone H1 (25 μg) was added in all lanes (H1). One thousand nanograms of NtKIS1a (lanes 2–4), 10 ng of NtKIS1b (lane 3), 100 ng of NtKIS1b (lane 4), 1,000 ng of NtKIS1b (lane 5), or buffer (lanes 1 and 6) was further added. Histone H1K activity was monitored as described in “Materials and Methods.” Proteins bound to p9CKSHs1 beads were recovered and immunoblotted with an antibody raised against the conserved CDK PSTAIR motif (anti-PSTAIR).
To test the role of NtKIS1b toward NtKIS1a, the CDK kinase activity was measured in competition experiments mixing NtKIS1b with NtKIS1a proteins. Addition of NtKIS1b (Fig. 2B, lane 3 and 4) prevents the inhibitory effect of NtKIS1a alone (Fig. 2B, lane 2), even with a NtKIS1b:NtKIS1a ratio of 1:100 (Fig. 2B, lane 3). This result suggests that NtKIS1b represents a strong competitor of NtKIS1a that “protects” the CDK kinase activity from the inhibitory action of NtKIS1a.
Interacting Partners of NtKIS1a and NtKIS1b
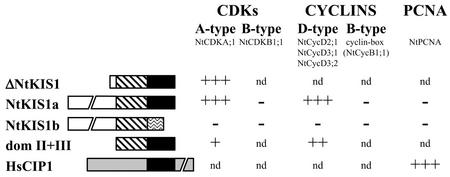
A 5′-truncated version of NtKIS1a (ΔNtKIS1) was originally obtained from the two-hybrid screen with Nicta;CDKA;1 as a bait, suggesting that residues 112 to 163 of NtKIS1a are sufficient for the interaction with Nicta;CDKA;1. NtKIS1b, lacking domain III compared with NtKIS1a, had no detectable interaction with Nicta;CDKA;1 (Fig. 3). Because the entire NtKIS1a protein was confirmed to interact with Nicta;CDKA;1, it is concluded that domain III is required for the interaction with Nicta;CDKA;1. The region, which shares a strong similarity to other plant-related proteins (domains II+III), displays a weakly detectable interaction with Nicta;CDKA;1 (Fig. 3). It is important to note that this region differs from ΔNtKIS1 in only five amino acids. NtKIS1a and NtKIS1b do not interact with the B-type CDK Nicta;CDKB1;1 (Fig. 3).
Figure 3.
Determination of NtKIS1a- and NtKIS1b-interacting partners in two-hybrid assays. NtKIS1a and NtKIS1b are schematically represented according to the three domains described in Figure 1. For each two-hybrid plasmid combination, three independent yeast transformants were analyzed as described in “Materials and Methods.” Growth on +3-AT media and 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-gal) filter assays gave the same results and are summarized in this figure. +++, ++, and + correspond respectively to a strong, moderate, and weak growth and blue color. − corresponds to no growth and white color. nd, Not determined. All constructs were tested alone as controls, and none gave rise to reporter gene expression (data not shown). HsCIP1 corresponds to the human CKI p21. Its N-terminal domain that presents similarities to NtKIS1a domain III is black boxed, the rest of the protein being different (gray box).
In animals, CIP/KIP inhibitors can form ternary complexes with CDK/cyclin by interacting with both the CDK and the cyclin subunit, whereas the members of the INK4 inhibitor family only bind to the CDK (Pavletich, 1999). Therefore, NtKIS1a and NtKIS1b were tested for their interaction with different cyclins. Three D-type cyclins and one B-type cyclin from N. tabacum were studied. A strong expression of the reporter genes was observed when NtKIS1a was cotransformed in yeast either with Nicta;CycD2;1, Nicta;CycD3;1, or Nicta;CycD3;2 cyclin (Fig. 3). The absence of reporter gene expression when NtKIS1b was cotransformed with the three D-type cyclins strongly suggests that the NtKIS1a domain III (absent in NtKIS1b) is also necessary for interaction with D-type cyclins. Domains II+III from NtKIS1a were sufficient for interactions with D-type cyclins (Fig. 3). Preliminary results indicated that expression of the full-length B-type cyclin Nicta;CycB1;1 was toxic in yeast (not shown), as was also reported for a Xenopus sp. B-type cyclin (Kong et al., 2000). Thus, a construct encoding the N-terminal part of the cyclin box (residues 180–280) was used. It contained the cyclin conserved residues that were shown to be involved in the binding of human cyclin A with p27KIP1 (Russo et al., 1996). This domain had no detectable interaction with NtKIS1a nor NtKIS1b in the two-hybrid system (Fig. 3).
Among the CIP/KIP CKIs, p21CIP1 is characterized by its ability to interact with PCNA. The N. tabacum PCNA was obtained by screening a cDNA library with an Arabidopsis EST (accession no. Z34137) and further sequenced (accession no. AJ012662). The predicted protein is 90% similar to the human PCNA and displays a strongly conserved p21CIP1-binding domain (Gulbis et al., 1996). The capacity of NtPCNA to interact with the human p21CIP1 was confirmed in the two-hybrid system (Fig. 3). No reporter gene expression was detected when either NtKIS1a or NtKIS1b was cotransformed with NtPCNA (Fig. 3).
NtKIS1a and NtKIS1b Display a Different Expression Pattern
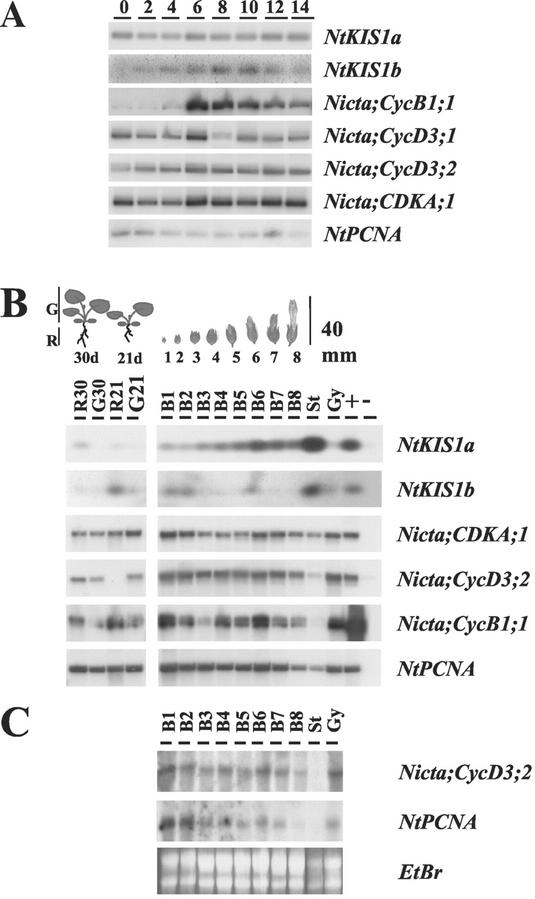
Cell cycle regulation of NtKIS1a and NtKIS1b transcripts was addressed. BY-2 cells were released from an aphidicolin treatment and a 14-h time-course experiment was performed. The expression of different cell cycle genes was followed in parallel with the expression of NtKIS1a and NtKIS1b through an reverse transcriptase (RT)-PCR analysis. RT-PCR allows separation of the contribution of NtKIS1a and NtKIS1b by the use of specific primers. NtKIS1a expression is constant during the cell cycle, whereas NtKIS1b transcript oscillates with a high level between 6 and 10 h after aphidicolin release (Fig. 4A). Because Nicta;CycB1;1, known as a G2-M transition marker (Planchais et al., 1997), appears at the same time, it strongly suggests that the NtKIS1b transcript level peaks at the G2-M transition.
Figure 4.
Expression patterns of NtKIS1a and NtKIS1b. A, BY-2 cells were released from an aphidicolin synchronization (0), and cell samples were collected every 2 h during 14 h (2–14). Total RNA was prepared, and RT-PCR was performed using specific primers from NtKIS1a, NtKIS1b, Nicta;CycB1;1, Nicta;CycD3;1, Nicta;CycD3;2, Nicta;CDKA;1, and NtPCNA. B, N. tabacum roots (R) and green tissues (G) of 21 (21d)- and 30 (30d)-d-old seedlings and flower buds at eight different development stages (1–8) were used to prepare total RNA. Lanes R30, G30, R21, and G21 are RT-PCR products obtained from RNA samples of the roots and green tissues of 21- and 30-d-old seedlings. Lanes B1 to B8 are RT-PCR products obtained from RNA samples of the corresponding flower buds. St and Gy are RT-PCR products obtained from RNA samples of B8 stamen and gyneocium, respectively. Lane + is a positive control in which a plasmid harboring the full-length corresponding cDNA is used as a template in the PCR reaction. Lane − is a negative control in which an empty plasmid is used except for NtKIS1a and NtKIS1b where a plasmid harboring NtKIS1b and NtKIS1a full-length cDNAs, respectively, was used. C, N. tabacum flower buds at the eight development stages described in B as well as Stamen (St) and gyneocium (Gy) from B8 were used to prepare total RNA. Fifty micrograms of RNA were used to perform a northern blot, and the membrane was hybridized with Nicta;CycD3;2 and NtPCNA as probes. Equal RNA loading was controlled by ethidium bromide staining (EtBr).
The level of NtKIS1a and NtKIS1b transcripts was also monitored by RT-PCR in several different tissues of N. tabacum plants. It included roots and green tissues (leaves plus stems) of 21- and 30-d-old seedlings, flower buds at eight different developmental stages, named 1 to 8 as already described by Koltunow et al. (1990), stamens, and gyneocium from stage 8. As a control, northern analyses were conducted in parallel using the same RNA samples and were probed with Nicta;CycD3;2 and NtPCNA (Fig. 4C). Results are correlated with that of RT-PCR. Nicta;CDKA;1 was studied because its encoded protein is a potent NtKIS1a-interacting partner, and its expression did not show significant variations (Fig. 4B). NtKIS1a and NtKIS1b were detectable at a very low level in roots and green tissues from all seedlings tested (Fig. 4B). Compared with NtKIS1b, NtKIS1a was strongly expressed, and its signal clearly increased with flower bud aging (Fig. 4B). Its maximum expression was found in the stamens that probably largely contributed to the expression observed in the corresponding B8 flower buds. Like NtKIS1a, NtKIS1b expression showed its highest level in the stamen from B8 flower buds but does not seem to follow the same expression pattern as NtKIS1a.
NtKIS1a and NtKIS1b Are Localized in the Nucleus
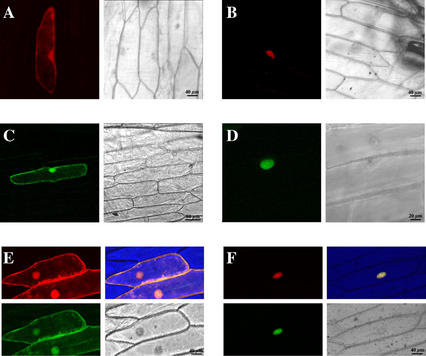
To gain insight into the subcellular localization of NtKIS1a and NtKIS1b proteins, their cDNAs were fused to a cDNA encoding respectively the DsRed or the soluble modified GFP (SmGFP) fluorescent protein. Onion epidermal cells were transiently transformed with these constructs and with control constructs corresponding to the DsRed or the SmGFP alone. The subcellular localization was determined using confocal microscopy analysis. As expected, the DsRed and the SmGFP alone are not addressed to a specific compartment and are detected in the cytosol as well as in the nucleus (Fig. 5, A and C). Interestingly, both NtKIS1a-DsRed and NtKIS1b-GFP were exclusively detected in the nucleus, indicating an efficient nuclear targeting (Fig. 5, B and D). To investigate whether NtKIS1b could misplace the NtKIS1a protein and vice versa, cotransformation was performed. As shown in Figure 5F, NtKIS1a-DsRed and NtKIS1b-GFP are still localized in the nucleus when simultaneously overexpressed in onion epidermal cells, showing that NtKIS1a and NtKIS1b do not exclude each other from the nucleus.
Figure 5.
NtKIS1a/b-GFP fusion proteins are localized in the nucleus of onion epidermal cells. A through D, Views of onion epidermal cells transformed with SmGFP (A), DsRed (C; control constructs), NtKIS1a-DsRed (B), and NtKIS1b-GFP (D). Views of onion epidermal cells transformed simultaneously with the two control constructs (E) or with the two NtKIS1a-DsRed and NtKIS1b-GFP constructs (F).
NtKIS1a-GFP Fusion Protein Is Localized in the Nucleus in Planta and Strongly Impairs Arabidopsis Development
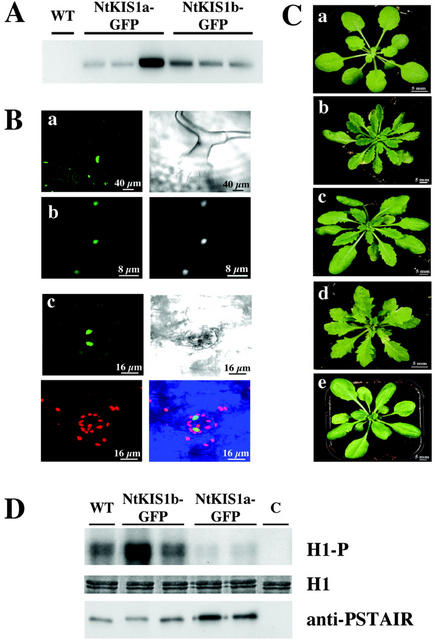
Taking advantage of the efficient NtKIS1a/b-GFP fusion constructs, the effects of their overexpression were compared in Arabidopsis. The presence of both constructs was checked on genomic DNA in the kanamycin-resistant T1 plantlets (not shown) and their effective overexpression was monitored using RT-PCR analysis (Fig. 6A). The subcellular localization of NtKIS1a/b-GFP protein fusions was assessed in these transgenic lines through confocal microscopic observations. Several tissues were inspected, such as leaf epidermis, leaf trichomes, and petal epidermis. In all cases, the two fusion proteins were localized in the nucleus (Fig. 6B). The colocalization of GFP and Hoechst fluorescences confirms the nuclear addressing of the two fusion proteins (Fig. 6B, b; data not shown). This observation confirms in planta what was previously observed in onion epidermal cells by transient expression. Furthermore, these observations demonstrate clearly that these transgenic plants effectively overproduced the fusion proteins.
Figure 6.
Analysis of NtKIS1a/b-GFP fusion protein overproduction in planta. A, RT-PCR analysis was performed as described in “Materials and Methods.” Lanes WT, NtKIS1a-GFP and NtKIS1b-GFP are RT-PCR products obtained from RNA samples of the corresponding plants. B, a, NtKIS1a-GFP localization in trichome (left, GFP fluorescence; right, transmission). b, NtKIS1a-GFP localization in petal epidermal cells (left, GFP fluorescence; right, Hoescht fluorescence). c, NtKIS1b-GFP localization in stomatal cells (top left, GFP fluorescence; top right, transmission; bottom left, background chlorophyll autofluorescence; and bottom right, superposition of GFP and chlorophyll fluorescences and transmission). C, Rosettes from three T1 35S::NtKIS1a-GFP independent lines, displaying respectively a weak (b), a medium (c), and a strong (d) serrated leaf phenotype, are compared with 35S::NtKIS1b-GFP (e) or WT (a) rosette. D, Protein extracts from WT, two 35S::NtKIS1a-GFP, two 35S::NtKIS1b-GFP Arabidopsis lines, or buffer (C) were added to p9CKSHs1 beads to purify CDK/cyclin complexes. Histone H1 phosphorylation (H1-P) was monitored, and equal loading of Histone H1 (H1) was controlled. Proteins bound to p9CKSHs1 beads were recovered and immunoblotted with an antibody raised against the conserved CDK PSTAIR motif (anti-PSTAIR).
Among the 25 T1 NtKIS1a-GFP-overexpressing plants analyzed, 18 display abnormal serrated leaves (Fig. 6B). In addition, there is a gradient in the serration, and three categories of phenotypes are revealed according to the strength of the serration: weak, medium, and strong (Fig. 6C). Not only was the leaf margin affected but the flower morphology was also modified, resulting in sterility in the strongest phenotypes (not shown). Furthermore, NtKIS1a-GFP-overexpressing plants display all the phenotypes previously described for transgenic Arabidopsis lines overexpressing NtKIS1a not fused to GFP (Jasinski et al., 2002), including an increase in leaf epidermis cell size and a block of the endoreduplication phenomenon (results not shown). Moreover, the CDK kinase activity measured in two 35S::NtKIS1a-GFP lines was significantly decreased compared with wild type (WT; Fig. 6D). These results show that NtKIS1a-GFP overexpression induces the same phenotype as NtKIS1a overexpression, demonstrating that GFP does not impair NtKIS1a function(s).
Among the 24 T1 NtKIS1b-GFP-overexpressing plants analyzed, all display a WT phenotype as already observed for the overexpression of NtKIS1b not fused to the GFP (Jasinski et al., 2002). Furthermore, the CDK kinase activity measured in two 35S::NtKIS1b-GFP lines remains as in WT (Fig. 6D). Therefore, the overexpression of the alternatively spliced form, which does not interact in the two-hybrid system with CDK and cyclins and does not modify the CDK kinase activity, is not able to induce phenotypic modifications.
DISCUSSION
Eukaryotes have developed several regulatory mechanisms to control cell cycle progression. Among them, small proteins called CKIs are able to inhibit the kinase activity of the core of the cell cycle, the CDK/cyclin complexes. In plants, CKIs have been identified in Arabidopsis, C. rubrum, pea, rice, and cotton. Here, we report the characterization of the tobacco CKI, named NtKIS1a, compared with its spliced variant NtKIS1b. As for all other plant CKI-related proteins, NtKIS1a shares a short domain of approximately 20 amino acids with the animal CIP/KIP CKIs. Consistent with this, the purified NtKIS1a protein has an inhibitory effect on the kinase activity of p9CKSHs1 affinity-bound CDK/cyclin complexes, like its Arabidopsis counterparts KRP1 (ICK1) and KRP2 (ICK2; Wang et al., 1997; Lui et al., 2000). In addition, two-hybrid assays show that NtKIS1a interacts with D-type cyclins and with an A-type CDK. Although it is not yet established that Nicta;CDKA;1 and the three tested tobacco D-type cyclins are interacting partners in planta, our two-hybrid results suggest that NtKIS1a is a potential regulator of these A-type CDK/cyclin D complexes. Consistent with this, our previous results strongly suggest that NtKIS1a is able to interact in planta with the Arath;CycD3;1 D-type cyclin (Jasinski et al., 2002).
In animals, the ability to interact with PCNA distinguishes p21CIP1 from p27KIP1 (Waga et al., 1994). Our two-hybrid assays suggest that NtPCNA and NtKIS1a are not interacting partners, which is consistent with the absence of a PCNA in NtKIS1a as found in p21CIP1 (Goubin and Ducommun, 1995; Chen et al., 1996). However, NtPCNA, which displays a strongly conserved p21CIP1-binding motif (Zhang et al., 1998), is shown to interact with the human p21CIP1, suggesting that a tobacco p21CIP1 counterpart might exist.
The remarkable conservation between animals and plants of the domain III in NtKIS1a suggests a role for this domain in the common protein function, i.e. its inhibitory function. However, as already mentioned for Arabidopsis by Lui et al. (2000), this domain has a C-terminal position versus an N-terminal position in animal CIP/KIP proteins. Comparison of NtKIS1a with other related proteins reveals the presence of a plant-specific domain, named domain II in NtKIS1a. This domain appears necessary in combination with domain III both for NtKIS1a interaction with D-type cyclins and A-type CDK. The domain I in NtKIS1a appears variable among the plant-related protein family. No consensus sequence was evident and no significant similarities were found with any sequence in the databases. However, domain I may be important in determining the specific pathway in which each plant CKI is involved. Consistent with this is the number of related proteins found in Arabidopsis, which may reflect a specialization for each member. In animals, CIP/KIP are divergent in their C-terminal halves. By using p27KIP1 C-terminal as a bait in a two-hybrid screen, Tomoda et al. (1999) isolated a mouse cDNA strongly similar to the human Jab1 and have shown that p38Jab1 functions as a negative regulator of p27KIP1 by promoting its degradation in a 26S proteasome-dependent manner. Thus, plant domain I may contain information for protein-specific degradation and/or subcellular localization. Consistent with this hypothesis, using the software psort (http://psort.nibb.ac.jp/), a nuclear targeting signal is predicted to be in the domain I for NtKIS1a and NtKIS1b.
The comparison of the intron-exon organization of the Arabidopsis, the N. tomentosiformis, and the N. sylvestris genomic sequences confirms the complete conservation of the last intron position reported by Lui et al. (2000) for AtKRP1-4. As a result, domain III is always encoded by the last exon in the plant genes. Such conserved intron positioning could suggest a regulatory role for this intron. A second cDNA, highly similar to NtKIS1a, was isolated and named NtKIS1b. Both the comparison of NtKIS1a and NtKIS1b cDNAs with each other and their comparison with genomic sequences from N. sylvestris and N. tomentosiformis show that NtKIS1a and NtKIS1b are derived from the same N. tomentosiformis gene. Therefore, it suggests that NtKIS1b is generated through an alternative splicing. An increasing number of alternative splicing cases is emerging in plants (for review, see Brown and Simpson, 1998). It represents an important posttranscriptional regulatory mechanism because it allows the modulation of the subcellular or tissue specificity of the transcript, its environment-dependent response, and also the biochemical function of the encoded polypeptide. In our case, the alternative splice site affects the invariably conserved last intron-exon junction and suppresses 19 bp from the last exon compared with NtKIS1a, inducing a frameshift. The resulting polypeptide lacks the conserved domain III. Consistent with this, NtKIS1b has no detectable interaction with Nicta;CDKA;1 and the three D-type cyclins in two-hybrid assays. The purified NtKIS1b protein similarly has no inhibitory properties in vitro on the kinase activity of CDK/cyclin complexes. Furthermore, NtKIS1a/b mixing experiments suggested that NtKIS1b is a strong competitor of NtKIS1a toward the inhibition of CDK kinase activity. Moreover, NtKIS1a and NtKIS1b display differential expression patterns during the cell cycle, with a constitutive expression for NtKIS1a, whereas the NtKIS1b transcript level peaks at the G2-M transition. It strongly suggests that the alternative splicing, from which NtKIS1a and NtKIS1b are derived, is cell cycle regulated, allowing the expression of NtKIS1b at the G2-M transition. Their antagonistic in vitro biochemical function and their differential expression pattern during the cell cycle may suggest that NtKIS1b could compete with NtKIS1a to allow a stronger CDK kinase activity at the G2-M transition. However, the mechanism by which NtKIS1b “protects” the CDK kinase activity from the inhibitory effect of NtKIS1a remains to be elucidated. Furthermore, one must consider that NtKIS1a and NtKIS1b proteins have domains I+II in common and may also compete for the same putative partners, other than CDK and cyclins.
Analysis at the plant organ level revealed an organ-specific expression pattern. NtKIS1a and NtKIS1b transcripts were mainly found in flower buds and especially in stamens. Organ-specific patterns of CKIs have been already reported for Arabidopsis (Wang et al., 1998; Lui et al., 2000; De Veylder et al., 2001). All members of the animal CIP/KIP family similarly display tissue-specific expression (for review, see Nakayama and Nakayama, 1998). Furthermore, NtKIS1a expression increases with flower bud aging. This increase, which is correlated with the disappearance of cell division markers like NtPCNA and Nicta;CycD3;2, is consistent with an arrest of cell cycle activities expected during cellular differentiation and suggests a role of NtKIS1a in stamen development. Because the NtKIS1 gene displays both a cell cycle- and a tissue-specific regulation, its promoter might contain cis-elements driving such a regulation.
The subcellular localization of a protein could bring an interesting clue concerning its function. Our study reveals that both NtKIS1a and NtKIS1b proteins are localized in the nucleus. To our knowledge, it is the first time that the subcellular localization of a plant CKI was addressed. In animals, the CKI p27Kip1 was shown to be addressed to the nucleus (Reynisdottir and Massague, 1997) and mislocalization (i.e. cytoplasmic) of CIP/KIP CKIs is often associated with oncogenesis (Singh et al., 1998; Winters et al., 2001). Regarding the role of NtKIS1b toward NtKIS1a, already assessed in vitro, an attractive competition hypothesis for the localization could be proposed. Thus, to test this hypothesis NtKIS1a and NtKIS1b fused to fluorescent proteins were co-expressed in onion epidermal cells. Because both fusion proteins are still localized in the nucleus when simultaneously overexpressed, it shows that NtKIS1b does not interfere or disturb NtKIS1a localization. However, in accordance with our in vitro competition experiment, transient expressions in Arabidopsis protoplasts suggest that the simultaneous localization of NtKIS1a and NtKIS1b fluorescent proteins in the nucleus promote division, whereas Arabidopsis protoplasts transformed with NtKIS1a-DsRed alone do not divide and rapidly die (data not shown).
The GFP tagging allowed us to follow in planta the overproduction of the NtKIS1a and NtKIS1b proteins. Therefore, to assess the function of the domain III, the phenotypic effects of NtKIS1a/b-GFP overexpression were compared in Arabidopsis. Similar to transient expression, NtKIS1a and NtKIS1b are observed in the nucleus in all tissues inspected. It suggests that their role is in this organelle. Moreover, NtKIS1a-GFP-overexpressing plants display strong morphological modifications and a reduced CDK kinase activity as already observed in NtKIS1a-overexpressing plants (Jasinski et al., 2002). Therefore, the fusion protein displays the same function in planta as its nonfluorescent counterpart. In contrast, NtKIS1b-GFP-overexpressing plants like NtKIS1b-overexpressing ones display a WT phenotype. Furthermore, the 35S::NtKIS1b-GFP plants display a CDK kinase activity similar to that of the WT, a result consistent with our in vitro kinase activity experiments. It strongly suggests that the phenotypic modifications observed in NtKIS1a-GFP-overexpressing plants are related to an inhibition of the CDK kinase activity. Consistent with this, all the plant CKIs that were overexpressed in Arabidopsis until now cause the same developmental abnormalities, such as an overall reduced growth, serrated leaves, abnormal flowers, larger cells, and a block of the endoreduplication phenomenon (Wang et al., 2000; De Veylder et al., 2001; Jasinski et al., 2002; Zhou et al., 2002). Thus, because all these CKIs have only domains II+III in common, it reinforces our conclusion that the phenotypic modifications are only attributable to an overall reduction of the CDK kinase activity in planta. The challenge is now to understand how a reduced CDK kinase activity impairs the plant developmental program.
MATERIALS AND METHODS
Plant Material
Tobacco plants (Nicotiana tabacum, Nicotiana tomentosiformis, and Nicotiana sylvestris) were grown in a greenhouse under long-day conditions (16 h light). Tobacco BY-2 cell suspension was maintained as described by Nagata et al. (1992). For histone H1 kinase (H1K) assays, BY-2 cells were refreshed for 24 h. For cell cycle regulation analysis, BY-2 cells were synchronized. Fifteen milliliters of a stationary tobacco BY-2 cell culture (8 d-old) was grown in 50 mL of fresh medium supplemented with 2 μg mL−1 aphidicolin (Sigma-Aldrich, St. Louis) for 20 h. The cells were subsequently washed three times and cultivated in 150 mL of fresh medium. Time-course experiments were performed on these cells for 14 h. Arabidopsis plants were grown in a greenhouse under short-day conditions (10 h light) and further transferred to long-day conditions (16 h light).
Transgene Constructs and Arabidopsis Transformation
The SmGFP (Davis and Vierstra, 1998) and the DsRed (BD Biosciences Clontech, Palo Alto, CA) fluorescent proteins were amplified by PCR, sequenced, and then cloned into the pBI221 vector (BD Biosciences Clontech), previously deleted for the GUS cDNA. NtKIS1a or NtKIS1b cDNAs were further cloned upstream of the SmGFP and DsRed cDNAs into the resulting pBI/SmGFP or pBI/DsRed vectors. The two plasmids encoding the NtKIS1a-DsRed and NtKIS1b-GFP fusion proteins were used to transiently transform onion epidermal cells. The EcoRI-HindIII cassettes from pBI/NtKIS1a-SmGFP and pBI/NtKIS1b-SmGFP were transferred into the binary plasmid pPZP111. The two pPZP111-derived plasmids encoding NtKIS1a-GFP and NtKIS1b-GFP were introduced into Agrobacterium tumefaciens (HBA105). Arabidopsis plants, ecotype Columbia (referred as WT in the text), were transformed and analyzed as described by Jasinski et al. (2002).
Yeast Two-Hybrid Screen and Assays
The vectors pGAD424 and pACT2 harboring the GAL4 activation domain (AD), the vector pGBT9 harboring the GAL4 binding domain (BD), and the yeast strain HF7c were obtained from BD Biosciences Clontech. To construct the BD-Nicta;CDKA;1 hybrid used as a bait in the screen, a PCR fragment obtained with PfuI polymerase (Stratagene, La Jolla, CA) was first cloned into pUC18, sequenced, and further introduced into pGBT9. The GAL4 AD cDNA library of tobacco BY-2 cell suspension was constructed in pACT2 by BD Biosciences Clontech laboratories from total RNA provided by us. Yeast transformation was performed according to the protocol described in the Matchmaker two-hybrid system (BD Biosciences Clontech). Independent cotransformants (2 × 106) were screened for the HIS3 reporter gene expression on synthetic dextrose (SD) medium lacking Leu (L), Trp (T), and His (H). After 6 d of incubation, growing colonies were transferred to the same medium supplemented with 10 mm 3-aminotriazole (3-AT). 3-AT resistant colonies were tested for the LacZ reporter gene expression through X-gal filter assays. AD-plasmid was recovered from His prototrophic and blue colonies and sequenced using AD-vector specific primers. For two-hybrid assays, most of the fusions used were constructed following the same procedure as described for the bait. NtKIS1a, NtKIS1b, and NtPCNA cDNA were obtained in this work, Nicta;CycB1;1 was previously cloned in our laboratory, and D-type cyclin and Nicta;CDKB1;1 cDNAs were provided by J. Murray (Cambridge, UK). The BD-HsCIP1 construct (human cDNA encoding p21CIP1 cloned into pGBT9) was provided by B. Ducommun (Toulouse, France). For all interactions tested, yeast double transformants selected on SD medium lacking LT were cultured, adjusted to OD600 = 1, deposited in parallel as 10-μL drops on (a) SD medium lacking LT, (b) SD medium lacking LTH, (c) SD medium lacking LTH and supplemented with 10 mm 3-AT (+3-AT), and (d) SD medium lacking LT on which a filter was previously deposited. Growth was recorded and X-gal filter assay was performed after 3 d of incubation at 30°C.
RNA Isolation, RT-PCR, and Northern-Blot Analyses
Total RNA was isolated by using TRIzol reagent (Invitrogen, Carlsbad, CA) from various N. tabacum cv Xanthi tissues or BY-2 cells, previously frozen, and ground in a mortar. For RT-PCR analysis, first-strand cDNA was synthesized from 5 μg of total RNA using SuperscriptII RNase H-Reverse Transcriptase (Invitrogen) following the manufacturer's instruction. For PCR, 2.5 μL was used in a final volume of 25 μL. PCR products (10 μL) were run on a 1% (w/v) agarose gel and transferred to a Hybond N+ membrane (Amersham Biosciences AB, Uppsala). Northern-blot analysis was performed as described by Planchais et al. (1997). Hybridizations were performed at 62°C according to Church and Gilbert (1984). Probes for NtKIS1a and NtKIS1b consist of the coding sequence of NtKIS1a. Probes for Nicta;CDKA;1, Nicta;CycD3;1, Nicta;CycD3;2, Nicta;CycB1;1 and NtPCNA are the corresponding coding sequences.
Purification of Recombinant NtKIS1a and NtKIS1b Proteins
The full-length NtKIS1a and NtKIS1b open reading frames were cloned into pET-14b vector (Novagen), and recombinant proteins were purified from overproducing E. coli strains. The cell pellets from 500-mL cultures were resuspended in a native buffer (50 mm Tris-HCl, pH 7.2, 500 mm NaCl, 5 mm dithiothreitol [DTT], and 3.5 mg mL−1 lysozyme) and then incubated for 30 min at 20°C. Triton X-100 was added to a concentration of 1% (v/v); this was followed by ultrasound sonication by bursts of 30 s followed by cooling. The extracts were treated by 20 mg L−1 DNase I for 20 min at 37°C. The inclusion bodies overwhelmingly composed of the recombinant proteins were sedimented by centrifugation at 30,000g for 30 min at 4°C. The pellets were washed twice with Tris-buffered saline (20 mm Tris, pH 7.5, and 140 mm NaCl) containing 1% (v/v) Triton X-100 followed by centrifugation at 30,000g for 30 min at 4°C. The inclusion body pellets were solubilized in 4 mL of 50 mm Tris-HCl, pH 7.2, 6 m guanidine HCl, and 25 mm DTT by vigorous shaking. Insoluble material was removed by centrifugation at 21,000g for 20 min. The protein concentration was adjusted to 1 mg mL−1 using 50 mm Tris-HCl, pH 7.2, 6 m guanidine HCl, and 25 mm DTT. The solubilized proteins were diluted as quickly as possible one in 10 into cold folding buffer (10 mm Tris-HCl, pH 7.2, 200 mm NaCl, 400 mm Arg HCl, 10% [v/v] glycerol, 1 mm DTT, 0.1% [v/v] Tween 20, 20 μm MgCl2, and 20 μm ZnAc) and dialyzed against the same buffer (500 mL per 2-mL sample) at 4°C overnight. Samples were further dialyzed against the histone H1K assay buffer (Azzi et al., 1992) supplemented with 10% (v/v) glycerol. Samples were stored at −80°C.
Histone H1K Assay
p9CKSHs1 beads were prepared according to Azzi et al. (1992). Fifty milliliters of 24-h refreshed BY-2 cells was pelleted and ground in liquid nitrogen in 2 mL of extraction buffer (25 mm MOPS, pH 7.2, 60 mm β-glycerophosphate, 15 mm p-nitrophenylphosphate, 15 mm EGTA, 15 mm MgCl2, 1 mm DTT, 1 mm NaF, 1 mm NH4VO3, 1 mm phenylphosphate, 0.2 μg mL−1 leupeptin, and 0.3 μg mL−1 pepstatin). The resulting powder was slowly thawed on ice and centrifuged for 30 min at 18,000g at 4°C to eliminate cell debris. Protein extract (600 μg) was added to 20 μL of packed p9CKSHs1 beads previously washed with bead buffer (Azzi et al., 1992) and kept under rotation at 4°C for 1 h. After a centrifugation pulse and removal of the supernatant, beads were washed three times with bead buffer. An input of purified recombinant proteins (see figure legends) was added to the bead pellet. Tubes were kept under rotation at 4°C for 1h. After removal of the supernatant, beads were washed three times with bead buffer and used for H1K assay. Samples containing the initial 20 μL of packed beads were incubated 30 min at 30°C with 1 μCi of [γ-32P]ATP and 25 μg of histone H1 (Sigma-Aldrich) in a final volume of 30 μL of H1K assay buffer (Azzi et al., 1992). After a centrifugation pulse, Laemmli buffer was added to 30 μL of supernatant. Samples were analyzed by 12% (w/v) SDS-PAGE followed by Coomassie Blue staining to visualize histone H1 and an autoradiography to detect histone H1 phosphorylation, which was further quantified with the NIH Image 1.62 software.
Western Blotting
After H1K assays, proteins bound to p9CKSHs1 beads were recovered by boiling in the presence of Laemmli buffer, run in 12% (w/v) SDS-PAGE, and transferred to a 0.1-μm nitrocellulose membrane for 1 h in a semidry system (Millipore, Bedford, MA) at 2.5 V cm−2. The membrane was further treated as described in the ECL+Plus System protocol (Amersham Biosciences AB). The primary antibody was a monoclonal anti-PSTAIR antibody (Sigma-Aldrich), and the secondary one was a goat anti-mouse peroxidase conjugated antibody (Bio-Rad, Hercules, CA). Detection was performed by chemiluminescence using the ECL+Plus System (Amersham Biosciences AB).
Microprojectile Bombardment and Confocal Microscopy
Onion bulb scale pieces were bombarded at 650 psi using a helium biolistic device (Bio-Rad PDS-1000) with 1.5 μg of DNA coated onto 0.5 mg of 1-μm gold particles, according to the manufacturer's instructions (Bio-Rad). They were kept in a humid chamber for 2 d, and the epidermis was peeled and mounted in water for viewing in the confocal microscope (LSM510, Zeiss, Welwyn Garden City, UK).
ACKNOWLEDGMENTS
We thank J.A.H. Murray for providing the tobacco D-type cyclin and Nicta;CDKB1;1 cDNAs. The BD-HsCIP1 construct was a gift from B. Ducommun. We thank A. Perret for his helpful advice on protein purification and R. Stevens for her critical review of the manuscript. We thank J.P. Barès and G. Santé for taking care of the plants and R. Boyer for photography.
Footnotes
This work was supported by the French Ministère de l'Education Nationale, de la Recherche, et de la Technologie (grant to S.J.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.008573.
LITERATURE CITED
- Azzi L, Meijer L, Reed SI, Pidikiti R, Tung HY. Interaction between the cell-cycle-control proteins p34cdc2 and p9CKShs2: evidence for two cooperative binding domains in p9CKShs2. Eur J Biochem. 1992;203:353–360. doi: 10.1111/j.1432-1033.1992.tb16557.x. [DOI] [PubMed] [Google Scholar]
- Brown JWS, Simpson CG. Splice site selection in plant pre-mRNA splicing. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:77–95. doi: 10.1146/annurev.arplant.49.1.77. [DOI] [PubMed] [Google Scholar]
- Chen IT, Akamatsu M, Smith ML, Lung FD, Duba D, Roller PP, Fornace AJ, O'Connor PM. Characterization of p21Cip1/Waf1 peptide domains required for cyclin E/Cdk2 and PCNA interaction. Oncogene. 1996;12:595–607. [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis SJ, Vierstra RD. Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol Biol. 1998;36:521–528. doi: 10.1023/a:1005991617182. [DOI] [PubMed] [Google Scholar]
- den Boer BG, Murray JA. Triggering the cell cycle in plants. Trends Cell Biol. 2000;10:245–250. doi: 10.1016/s0962-8924(00)01765-7. [DOI] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GT, Krols L, Terras F, Landrieu I, van der Schueren E, Maes S, Naudts M, Inzé D. Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell. 2001;13:1653–1668. doi: 10.1105/TPC.010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvez S, Hodges M, Decottignies P, Bismuth E, Lancien M, Sangwan RS, Dubois F, LeMarechal P, Cretin C, Gadal P. Identification of a tobacco cDNA encoding a cytosolic NADP-isocitrate dehydrogenase. Plant Mol Biol. 1996;30:307–320. doi: 10.1007/BF00020116. [DOI] [PubMed] [Google Scholar]
- Goubin F, Ducommun B. Identification of binding domains on the p21Cip1 cyclin-dependent kinase inhibitor. Oncogene. 1995;10:2281–2287. [PubMed] [Google Scholar]
- Gulbis JM, Kelman Z, Hurwitz J, O'Donnell M, Kuriyan J. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- Huntley RP, Murray JA. The plant cell cycle. Curr Opin Plant Biol. 1999;2:440–446. doi: 10.1016/s1369-5266(99)00027-8. [DOI] [PubMed] [Google Scholar]
- Jasinski S, Riou-Khamlichi C, Roche O, Perennes C, Bergounioux C, Glab N. The CDK inhibitor NtKIS1a is involved in plant development, endoreduplication and restores normal development of cyclin D3;1-overexpressing plants. J Cell Sci. 2002;115:973–982. doi: 10.1242/jcs.115.5.973. [DOI] [PubMed] [Google Scholar]
- Joubes J, Chevalier C, Dudits D, Heberle-Bors E, Inzé D, Umeda M, Renaudi JP. CDK-related protein kinases in plants. Plant Mol Biol. 2000;43:607–620. doi: 10.1023/a:1006470301554. [DOI] [PubMed] [Google Scholar]
- Koltunow AM, Truettner J, Cox KH, Wallroth M, Goldberg RB. Different temporal and spatial gene expression patterns occur during anther development. Plant Cell. 1990;2:1201–1224. doi: 10.1105/tpc.2.12.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong M, Barnes EA, Ollendorff V, Donoghue DJ. Cyclin F regulates the nuclear localization of cyclin B1 through a cyclin-cyclin interaction. EMBO J. 2000;19:1378–1388. doi: 10.1093/emboj/19.6.1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBaer J, Garrett MD, Stevenson LF, Slingerland JM, Sandhu C, Chou HS, Fattaey A, Harlow E. New functional activities for the p21 family of CDK inhibitors. Genes Dev. 1997;11:847–862. doi: 10.1101/gad.11.7.847. [DOI] [PubMed] [Google Scholar]
- Lui H, Wang H, Delong C, Fowke LC, Crosby WL, Fobert PR. The Arabidopsis Cdc2a-interacting protein ICK2 is structurally related to ICK1 and is a potent inhibitor of cyclin-dependent kinase activity in vitro. Plant J. 2000;21:379–385. doi: 10.1046/j.1365-313x.2000.00688.x. [DOI] [PubMed] [Google Scholar]
- Mironov V, De Veylder L, Van Montagu M, Inzé D. Cyclin-dependent kinases and cell division in plants: the nexus. Plant Cell. 1999;11:509–522. doi: 10.1105/tpc.11.4.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata T, Nemoto Y, Hasezawa S. Tobacco BY-2 cell line as the “HeLa” cell in the cell biology of higher plants. Int Rev Cytol. 1992;132:1–30. [Google Scholar]
- Nakayama K, Nakayama K. Cip/Kip cyclin-dependent kinase inhibitors: brakes of the cell cycle engine during development. Bioessays. 1998;20:1020–1029. doi: 10.1002/(SICI)1521-1878(199812)20:12<1020::AID-BIES8>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Pavletich NP. Mechanisms of cyclin-dependent kinase regulation: structures of Cdks, their cyclin activators, and Cip and INK4 inhibitors. J Mol Biol. 1999;287:821–828. doi: 10.1006/jmbi.1999.2640. [DOI] [PubMed] [Google Scholar]
- Planchais S, Glab N, Trehin C, Perennes C, Bureau JM, Meijer L, Bergounioux C. Roscovitine, a novel cyclin-dependent kinase inhibitor, characterizes restriction point and G2/M transition in tobacco BY-2 cell suspension. Plant J. 1997;12:191–202. doi: 10.1046/j.1365-313x.1997.12010191.x. [DOI] [PubMed] [Google Scholar]
- Renaudin JP, Doonan JH, Freeman D, Hashimoto J, Hirt H, Inzé D, Jacobs T, Kouchi H, Rouze P, Sauter M et al. Plant cyclins: a unified nomenclature for plant A-, B- and D-type cyclins based on sequence organization. Plant Mol Biol. 1996;32:1003–1018. doi: 10.1007/BF00041384. [DOI] [PubMed] [Google Scholar]
- Reynisdottir I, Massague J. The subcellular locations of p15(Ink4b) and p27(Kip1) coordinate their inhibitory interactions with cdk4 and cdk2. Genes Dev. 1997;11:492–503. doi: 10.1101/gad.11.4.492. [DOI] [PubMed] [Google Scholar]
- Russo AA, Jeffrey PD, Patten AK, Massague J, Pavletich NP. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature. 1996;382:325–331. doi: 10.1038/382325a0. [DOI] [PubMed] [Google Scholar]
- Segers G, Rouzé P, Van Montagu M, Inzé D. J.A.B.a.D. Chiatante, ed, Plant Cell Proliferation and Its Regulation in Growth and Development. Sussex, UK: John Wiley & Sons Ltd.; 1997. Cyclin-dependent kinases in plants; pp. 1–19. [Google Scholar]
- Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- Singh SP, Lipman J, Goldman H, Ellis FH, Jr, Aizenman L, Cangi MG, Signoretti S, Chiaur DS, Pagano M, Loda M. Loss or altered subcellular localization of p27 in Barrett's associated adenocarcinoma. Cancer Res. 1998;58:1730–1735. [PubMed] [Google Scholar]
- Tomoda K, Kubota Y, Kato J. Degradation of the cyclin-dependent-kinase inhibitor p27Kip1 is instigated by Jab1. Nature. 1999;398:160–165. doi: 10.1038/18230. [DOI] [PubMed] [Google Scholar]
- Waga S, Hannon GJ, Beach D, Stillman B. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature. 1994;369:574–578. doi: 10.1038/369574a0. [DOI] [PubMed] [Google Scholar]
- Wang H, Fowke LC, Crosby WL. A plant cyclin-dependent kinase inhibitor gene. Nature. 1997;386:451–452. doi: 10.1038/386451a0. [DOI] [PubMed] [Google Scholar]
- Wang H, Qi Q, Schorr P, Cutler AJ, Crosby WL, Fowke LC. ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J. 1998;15:501–510. doi: 10.1046/j.1365-313x.1998.00231.x. [DOI] [PubMed] [Google Scholar]
- Wang H, Zhou Y, Gilmer S, Whitwill S, Fowke LC. Expression of the plant cyclin-dependent kinase inhibitor ICK1 affects cell division, plant growth and morphology. Plant J. 2000;24:613–623. doi: 10.1046/j.1365-313x.2000.00899.x. [DOI] [PubMed] [Google Scholar]
- Winters ZE, Hunt NC, Bradburn MJ, Royds JA, Turley H, Harris AL, Norbury CJ. Subcellular localisation of cyclin B, Cdc2 and p21(WAF1/CIP1) in breast cancer: association with prognosis. Eur J Cancer. 2001;37:2405–2412. doi: 10.1016/s0959-8049(01)00327-6. [DOI] [PubMed] [Google Scholar]
- Zhang P, Sun Y, Hsu H, Zhang L, Zhang Y, Lee MY. The interdomain connector loop of human PCNA is involved in a direct interaction with human polymerase delta. J Biol Chem. 1998;273:713–719. doi: 10.1074/jbc.273.2.713. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Fowke LC, Wang H. Plant CDK inhibitors: studies of interactions with cell cycle regulators in the yeast two-hybrid system and functional comparisons in transgenic Arabidopsis plants. Plant Cell Rep. 2002;20:967–975. [Google Scholar]
- Zindy F, Quelle DE, Roussel MF, Sherr CJ. Expression of the p16INK4a tumor suppressor versus other INK4 family members during mouse development and aging. Oncogene. 1997;15:203–211. doi: 10.1038/sj.onc.1201178. [DOI] [PubMed] [Google Scholar]