Abstract
The products of the cellulose synthase A (CESA) gene family are thought to function as isoforms of the cellulose synthase catalytic subunit, but for most CESA genes, the exact role in plant growth is still unknown. Assessing the function of individual CESA genes will require the identification of the null-mutant phenotypes and of the gene expression profiles for each gene. Here, we report that only four of 10 CESA genes, CESA1, CESA2, CESA3, and CESA9 are significantly expressed in the Arabidopsis embryo. We further identified two new mutations in the RADIALLY SWOLLEN1 (RSW1/CESA1) gene of Arabidopsis that obstruct organized growth in both shoot and root and interfere with cell division and cell expansion already in embryogenesis. One mutation is expected to completely abolish the enzymatic activity of RSW1(CESA1) because it eliminated one of three conserved Asp residues, which are considered essential for β-glycosyltransferase activity. In this presumed null mutant, primary cell walls are still being formed, but are thin, highly undulated, and frequently interrupted. From the heart-stage onward, cell elongation in the embryo axis is severely impaired, and cell width is disproportionally increased. In the embryo, CESA1, CESA2, CESA3, and CESA9 are expressed in largely overlapping domains and may act cooperatively in higher order complexes. The embryonic phenotype of the presumed rsw1 null mutant indicates that the RSW1(CESA1) product has a critical, nonredundant function, but is nevertheless not strictly required for primary cell wall formation.
Plant cell shape is a key determinant in plant morphogenesis and is in turn strongly influenced by the organization of the cell wall. Within the cell wall, cellulose microfibrils constitute the major load-bearing structures, and their oriented deposition confers differential extensibility to the wall (Carpita and Gibeaut, 1993; McCann and Roberts, 1994). Presumably under the influence of the microtubular cytoskeleton, microfibrils in the primary wall of growing cells are deposited perpendicularly to the prospective elongation axis (Wyatt and Carpita, 1993; Wymer and Lloyd, 1996). Once wrapped around a cell in parallel hoops, the largely inelastic microfibrils are thought to efficiently restrict cell expansion to a single dimension. In organs along the apical-basal axis, such as stems and roots, oriented microfibrils allow cells to expand anisotropically, and in young meristematic tissues, they may even play a role in establishing new growth directions and forming new organs (Green, 1994). Because of the central role of cellulose microfibrils in shaping plant morphology at the cellular and at the organismal level, their formation is probably subject to complex developmental controls.
Cellulose, a paracrystalline form of H-bonded β-(1,4)-Glc chains (McCann and Roberts, 1991; Carpita and Gibeaut, 1993), is thought to be synthesized by membrane-bound complexes. Although these complexes can readily be visualized as rosette-terminal complexes, cellulose synthesis activity is difficult to assay in vitro. This problem has hampered biochemical approaches to identify cellulose-synthesizing enzymes (Delmer, 1999). Cellulose synthase genes have been eventually discovered genetically in bacteria, and divergent homologs in plants have been identified (for review, see Cutler and Somerville, 1997; Saxena and Brown, 2000). Members of the cellulose synthase A (CESA) gene family in higher plants are believed to encode isoforms of the cellulose synthase catalytic subunit, but for most of these genes, the precise role in developing plants remains to be characterized (Delmer, 1999).
Knowledge of the functions of individual CESA genes in specific tissues or cell types will probably provide invaluable analytical and biotechnological tools for a better understanding and control of parameters underlying plant morphogenesis and physiology. However, no loss-of-function mutants are presently available for most CESA genes, and for some of the available mutants, it is not clear to what extent they reduce the gene function. For example, two mutations have been reported for RADIALLY SWOLLEN1, RSW1(CESA1), one of two CESA genes implicated by mutation in primary cell wall formation (Arioli et al., 1998; Gillmor et al., 2002). One of these mutations is temperature sensitive and, at high temperatures, is associated with reduced cellulose synthesis, disassembly of cellulose synthase complexes, and radial expansions in young shoot and root organs (Arioli et al., 1998; Williamson et al., 2001). A presumably stronger mutation has very recently been reported to affect embryo shape at room temperature, but in this mutant as well, it is not clear whether the RSW1(CESA1) gene function is completely eliminated (Gillmor et al., 2002). A second CESA gene in primary cell wall formation, PROCUSTE1, PRC1(CESA6), is required specifically in roots and dark-grown hypocotyls (Desnos et al., 1996; Fagard et al., 2000). CESA genes in secondary cell wall formation were identified through mutations associated with xylem defects in inflorescence stems and define the IRREGULAR XYLEM (IRX) genes IRX1(CESA8) and IRX3(CESA7) (Taylor et al., 1999, 2000). Finally, mutations conferring resistance to the herbicide isoxaben (ixr) were found in the CESA3 and PRC1(CESA6) genes (hence also referred to as IXR1 and IXR2 genes, respectively; Scheible et al., 2001; Desprez et al., 2002).
Given the size of the gene family and the likely functional overlap among its members, it will take considerable effort to sort out the roles of individual genes. The complexity of this analysis can be reduced through the isolation of null mutations in individual CESA genes and through the identification of stages, in which only a subset of the gene family is expressed. Here, we report the isolation of new, strong mutations in the Arabidopsis RSW1(CESA1) gene, one of which seems to completely abolish the enzymatic activity of the gene product and is associated with extreme defects in primary cell wall and cell shape already in embryos. In this presumed null mutant, residual cellulose synthesis will probably reflect the activity of other CESA genes. In this context, we show that only three other CESA genes, CESA2, CESA3, and CESA9, are significantly expressed in the embryo. These genes are expressed in vastly overlapping domains, suggesting that none of them has a function restricted to specific embryonic tissues or stages.
RESULTS
Identification of New Mutations in RSW1(CESA1)
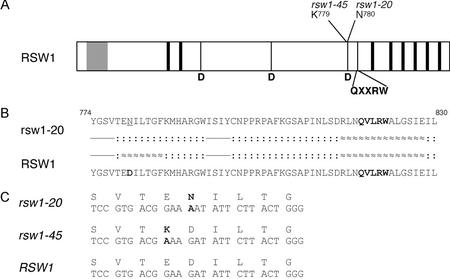
In a screen for embryo cell shape mutants, we isolated two allelic mutations that were associated with dramatic distortions in seedling morphology (Fig. 1A). Upon outcrossing to wild type, abnormalities were observed in approximately 25% of the F2 individuals (176 and 90 of 525 and 297, respectively) consistent with the recessive inheritance of the seedling morphology trait. Phenotype and map position (flanking RFLP markers g8300 and mi431) suggested that the mutations correspond to new alleles of the RSW1(CESA1) gene, which was confirmed in complementation tests with the temperature-sensitive allele rsw1-1 (Arioli et al., 1998). To identify the molecular lesions in the two mutants (rsw1-20 and rsw1-45), we determined the genomic sequence of both mutant alleles along with the Landsberg erecta (Ler) wild-type strain in four independent DNA pools and localized single-point mutations in each of the two mutant alleles. Sequence analysis of the deduced mutant gene products revealed that the mutation in the phenotypically stronger rsw1-20 mutation converted the third of three Asp residues within the conserved glycosyltransferase “D,D,D,QXXRW” motif to Asn (Fig. 2, A and C). Moreover, secondary structure analysis predicted a disruption of the local α-helix made up of residue 779 to 785 in the rsw1-20 mutant (Fig. 2B). These structural features strongly suggest complete loss of enzymatic activity of the mutant gene product (Saxena and Brown, 1997; Saxena et al., 2001; see “Discussion”). The mutation in allele rsw1-45 affected an adjacent, yet less conserved, position and presumably did not disrupt the local α-helical structure (data not shown). The fact that both mutants display similar phenotypic features at different levels of severity provides genetic evidence that all features described below are attributable to loss of RSW1(CESA1) function.
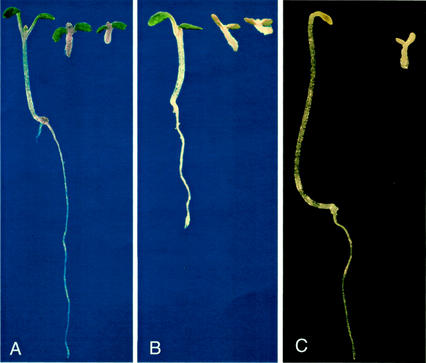
Figure 1.
Seedling phenotype of wild-type and rsw1 mutants 5 d after germination. A and B, Seedlings germinated in the light at 22°C (A) and 31°C (B). Genotypes from left to right are Ler wild type, rsw1-45, and rsw1-20. C, Ler wild-type and rsw1-20 mutant seedlings germinated at 22°C in the dark.
Figure 2.
Mutations in the RSW1(CESA1) gene. A, Schematic diagram of RSW1(CESA1) protein indicating position and identity of the predicted altered amino acid residues in the rsw1-20 and rsw1-45 mutants. Gray, Conserved zinc finger; black, transmembrane domains. Bottom, Conserved amino acids in β-glycosyltranserases. B, Predicted amino acid sequence and secondary structure of rsw1-20 mutant and wild-type gene product. Bold, Conserved residues or motifs; dashed, sheet; dotted, coil; waved, helix. C, Nucleotide and amino acid sequence in mutant and wild-type alleles. Deviations from wild-type sequence are in bold, and amino acids are in one-letter code.
Seedling Phenotypes
Seedlings of light-germinated rsw1-20 and rsw1-45 mutants appeared short and stout with an uneven surface because of irregularly shaped and swollen cells in the epidermis of both hypocotyl and cotyledons (Figs. 1A and 4F). In rsw1-20 mutants, hypocotyl length was dramatically reduced (27% of that of wild type; Table I), and the diameter was increased by approximately one-third at the basal end. In rsw1-45 mutants, the reduction in hypocotyl length was somewhat less pronounced (42% of that of wild type; Table I). Cotyledon size was also dramatically reduced, particularly in rsw1-20 mutants. Barely any root growth was observed in either mutant. All other analyzed defects, including internal cellular defects at various stages, were less severe in rsw1-45 than in rsw1-20 mutants (data not shown), suggesting a residual gene activity in rsw1-45 mutants. Unless otherwise noted, we will restrict the description of mutant phenotypes to the presumed null mutant rsw1-20.
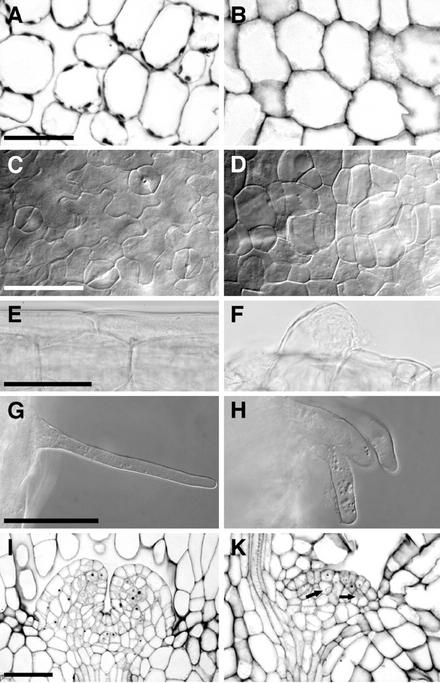
Figure 4.
Tissues in wild-type (A, C, E, G, and I) and rsw1-20 (B, D, F, H, and K) light-germinated seedlings 5 d after germination. A and B, Cross sections through cotyledon mesophyll. Intercellular spaces are strongly reduced, and no air cavities are formed in the mutant. C and D, Abaxial epidermis of cotyledons. Note the rounded cell shape and the absence of guard cells in the mutant. E and F, Hypocotyl epidermis cells, very regularly shaped in the wild type and generally irregular in the mutant. Within the irregular epidermal layer, some cells become extremely large. G and H, Root hairs are generally shorter and often thicker in the mutant. I and K, Shoot apical meristem, median section. Note fewer cells, incomplete cell divisions (arrows), and a highly irregular cell pattern in the mutant meristem. Except for a separation of the epidermal layer, no zonation is visible at this stage. Bright-field (A, B, E, F, I, and K) and whole-mount images with differential contrast optics (C, D, G, and H). Scale bars = 50 μm in A through H and 75 μm in I and K.
Table I.
Effects of temperature and light on seedling phenotype
| Conditions | Hypocotyl Length ± sd
|
|||||
|---|---|---|---|---|---|---|
| Ler | No. | rsw1-20 | No. | rsw1-45 | No. | |
| mm | ||||||
| 22°C Light | 3.12 ± 0.35 | 13 | 0.84 ± 0.17 | 12 | 1.33 ± 0.17 | 16 |
| 31°C Light | 3.03 ± 0.40 | 15 | 0.98 ± 0.40 | 7 | 1.13 ± 0.16 | 8 |
| 22°C Dark | 8.90 ± 1.79 | 16 | 1.00 ± 0.14 | 20 | n.d. | |
n.d., Not detected.
We assessed hypocotyl elongation in rsw1-20 mutants germinated in the dark. Hypocotyls of wild-type seedlings expand to a greater length when germinated in the dark than in the light. At least one cellulose synthase isoform is selectively required in this expansion process (Desnos et al., 1996; Fagard et al., 2000). Hypocotyls of both light- and dark-germinated mutant seedlings remained extremely short (Fig. 1C; Table I). Therefore, RSW1(CESA1) gene function is stringently required under both conditions, indicating that no gene activated specifically in the dark can substitute for missing RSW1(CESA1) activity (see “Discussion”).
The two new mutations in the RSW1(CESA1) gene can assist in classifying the previously identified rsw1-1 allele. This allele being temperature sensitive, the more severe defects seen at high temperature (31°C) could be attributable to temperature-sensitive properties of the mutant gene product. As an alternative, however, it is possible that the increased demand for cellulose synthesis at higher temperatures enhances abnormalities whenever the RSW1(CESA1) function is reduced. Additional alleles of the RSW1 gene allow us to distinguish between these possibilities. At 31°C, rsw1-45 mutant seedlings resemble rsw1-1 mutant seedlings (Williamson et al., 2001), suggesting approximately similar levels of residual gene activity in both mutants. However, whereas rsw1-1 mutants become largely normalized and grow long roots at room temperature (Williamson et al., 2001), the morphology of rsw1-20 and rsw1-45 was nearly identical at 22°C and 31°C (Fig. 1, A and B; Table I). This result implies that the temperature sensitivity of the rsw1-1 allele is largely, if not entirely, attributable to a temperature-dependent activity of the rsw1-1 mutant gene product.
Anatomy of rsw1-20 Mutants
Abnormal development in rsw1-20 mutant embryos occurred often as early as at the first division of the embryo proper. However, abnormalities before the heart stage were observed only in a portion of the mutants. For example, irregular horizontal divisions of apical cells were observed in early embryos (13.3% embryos from heterozygous parent, n = 90), but overall, the sequence of early cell divisions remained unchanged (Fig. 3, F–H). From the heart stage onward, cell shape alterations and cell wall interruptions were observed in approximately 25% of the progeny of heterozygous plants (21.2% embryos from heterozygous parent, n = 151), suggesting that the RSW1(CESA1) function became crucial in each mutant individual. A slight, but significant radial expansion of cells in hypocotyls of mutant embryos was observed at heart stage (Fig. 3, H and I) and became more pronounced during torpedo and bent-cotyledon stages (Fig. 3, K and M). At bent-cotyledon stage, radial cell expansions in the stele were recognizable, but rather subtle, whereas the radial expansion of the ground tissue and epidermal layers was rather extreme (Fig. 3M). Moreover, cells in these layers were often highly vacuolated and separated by incomplete cell walls. Interestingly, the overall cell arrangement in the hypocotyl and radicle, including the cell numbers in the apical-basal dimension, were almost unchanged, resulting in the formation of very flat cells (Fig. 3M). In cotyledons, by contrast, primarily the length of the organ was affected, and cotyledon cell numbers in the mutant were adjusted to the altered organ size (Fig. 3M). Cell shapes in the cotyledons were far less abnormal than in hypocotyls and radicles.
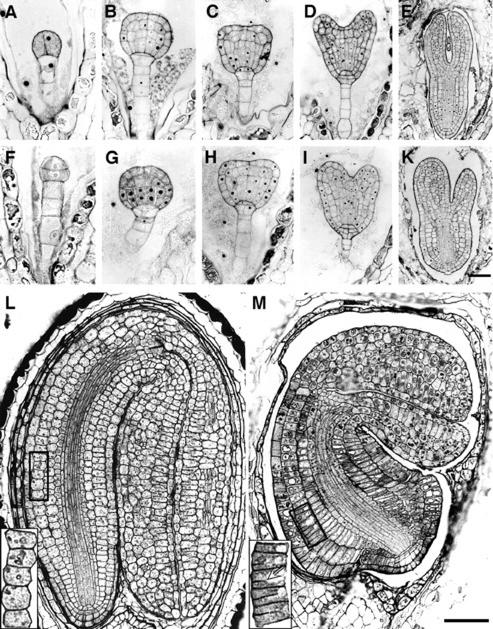
Figure 3.
Wild-type (A–E and L) and rsw1-20 mutant (F–K and M) embryogenesis. A and F, Octant stage; defects in early embryos include abnormally oriented cell divisions. B through D and G through I, Globular and heart stages; largely normal cell arrangement, but mutant cells, particularly in the ground tissue of the hypocotyl, are generally somewhat shorter and radially expanded. E and K, Torpedo stage; pronounced length-to-width changes are evident in cells along the length of the embryo, particularly in the outer layers. L and M, Bent-cotyledon stage; short cotyledons with fewer cells along the length (for example, in the palisade mesophyll layer), which are, however, relatively normal in shape. In the hypocotyl and radicle, by contrast, the number of cells in the longitudinal dimension is nearly normal, whereas the length to width ratio of cells in nonvascular tissues is extremely distorted (compare wild-type and mutant cell number and cell shape in the outer cortex layer, enlarged in inlets). Arrow in inlet points at one of many incomplete cell divisions in the outer layers of the mutant hypocotyl. Scale bars = 20 μm in A, B, F, and G, same magnification; 50 μm in C, D, H, and I, same magnification; 75 μm in E and K; and 100 μm in M and L, same magnification.
At germination, the total cell wall area increased very rapidly in wild-type seedlings and to a lesser extent also in the mutant. We did not observe a more frequent appearance of incomplete cell walls as a result of this expansion, and cell walls in mutant seedlings were apparently not more disorganized than in bent-cotyledon-stage embryos (data not shown). Most conspicuously, mutant cells in all seedling tissues remained extremely tightly packed allowing for considerably less intercellular space. This was particularly evident in cells of the mesophyll, where the abundant intercellular cavities present in wild type were absent in the mutant (Fig. 4, A and B). Mutant cells also failed to attain certain complex morphologies. Epidermal pavement cells did not display the typical interdigitated pattern, and stomatal guard cells were not produced at all (Fig. 4, C and D). On vegetative leaves, no trichomes were observed (data not shown). In contrast to trichomes, root hairs are exclusively generated by tip growth, and the mutant remarkably produced root hairs of approximately one-half the normal length (Fig. 4, G and H). Also transmission of the rsw1-20 allele through mutant pollen was not reduced. Together, these two observations may reflect a reduced requirement for RSW1(CESA1) in tip-growing cells.
In the mutant shoot apical meristem, cell divisions appeared highly irregular and were often incomplete (Fig. 4, I and K). Except for a discernable epidermal layer, no reproducible cell layer or internal zonation was apparent. Within 19 d of culture, seedlings produced an average of 7.6 extremely small vegetative leaves (sd = 1.6, n = 28). In the mutant primary root meristem, cell arrangements were normal, but cell dimensions were extremely distorted (Fig. 3M). Primary root growth was extremely limited, and no lateral roots were formed.
Cellular Defects in rsw1 Mutant Embryos
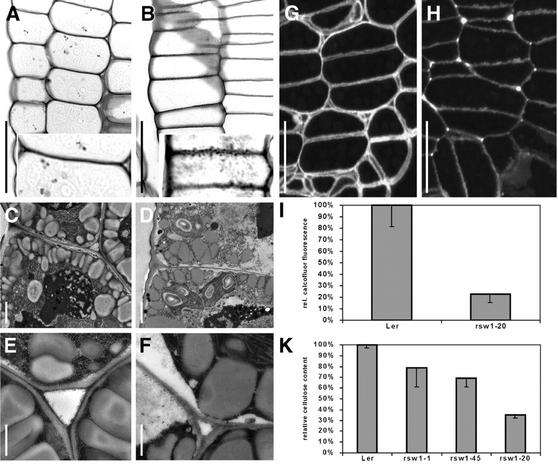
Differences in primary cell wall texture could be visualized by staining of the cell wall polysaccharides. As shown in Figure 5, A and B, mutant cell walls had a granular appearance with short projections to the inside. When viewed with the transmission electron microscope, markedly thinner, but again uneven, cell walls were observed in all analyzed tissues of the mutant, particularly in cells of the epidermis (Fig. 5, C–F). Most conspicuously, the size of intercellular spaces was strongly reduced in all tissues (Fig. 5, A and B). The intercellular spaces in between tissue layers and particularly at wall junctions were partially filled with excess wall material (Fig. 5, B and F).
Figure 5.
Cell wall structure and composition in wild-type and rsw1-20 mutant embryos. A and B, Periodic acid-Schiff's stained longitudinal sections in the hypocotyl of bent-cotyledon stage embryos. Note the presence of intercellular spaces in the wild type (A), which are absent in the mutant (B), and the uneven, granular appearance of mutant cell walls (inset). C thorough F, Epidermal cell walls from hypocotyls of bent-cotyledon stage embryos of wild type (C and E) and rsw1-20 (D and F). Note that mutant cell walls are thinner and that the middle lamella is usually not discernible. Intercellular spaces in the wild type (E) are usually filled with wall material in rsw1-20 mutants (F). G and H, Calcofluor stained cross sections in the hypocotyl of bent-cotyledon stage wild-type (G) and rsw1-20 mutant (H) embryos. Overall reduced staining in mutant walls indicates reduced β-glucan content. Note the uneven staining intensity in mutant cell walls. I, Densitometric quantification of cell wall β-glucan content measured on digital images from calcofluor-stained hypocotyl cells of bent-cotyledon stage embryos. K, Spectrophotometrical determination of β-glucan content in wall preparations of wild-type and mutant seedlings after germination and growth for 7 d at 31°C. A and B, Bright-field optics images; C through F, transmission electron micrographs; and G and H, epifluorescence optics. Scale bars = 20 μm in A, B, G, and H; insets, 2× magnification; 1 μm in C and D; and 500 nm in E and F.
To explore cell wall composition, we chose calcofluor staining to visualize biochemically wall composition across the entire embryo. Calcofluor specifically stains weakly or non-substituted β-glucans, which are probably represented by cellulose in these sections (Hughes and McCully, 1975). As shown in Figure 5, G and H, mutant walls again varied in thickness, and the staining was correspondingly nonuniform, in contrast to the even staining of wild-type cell walls. Furthermore, far less stainable material accumulated in the mutant. Quantification of the relative calcofluor fluorescence in identically treated sections from mutant and wild-type bent-cotyledon-stage hypocotyl indicated that mutant cell wall β-glucan content was reduced to 22.5% ± 4.2% of the wild-type value (Fig. 5I). We measured the corresponding β-glucan content in material extracted from mutant seedlings as 30% of that of the wild-type value (Fig. 5K). Importantly, parallel measurements of available rsw1 mutants grown at 31°C under identical conditions indicated that β-glucan content is lowest in rsw1-20 mutants (Fig. 5K).
Expression Profiles of CESA Genes
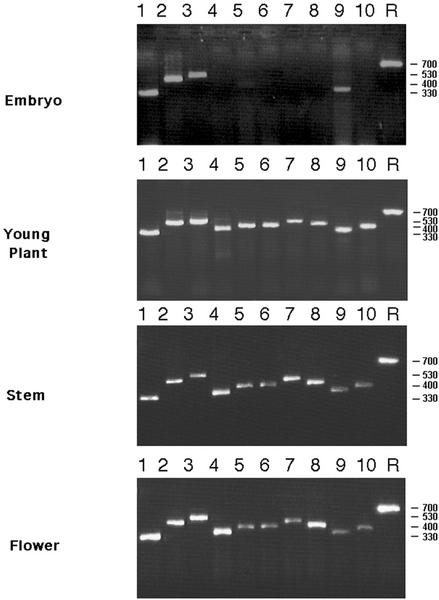
Residual cellulose synthesis in the presumed null-mutant rsw1-20 may reflect the activity of cellulose synthase isoforms, encoded by other CESA genes. To identify candidate CESA genes with overlapping functions, we assessed the expression of 10 Arabidopsis CESA genes in embryos at three postembryonic stages by semiquantitative reverse transcription (RT)-PCR. As shown in Figure 6, all 10 CESA genes were expressed at the three postembryonic stages (young plant, stem, and flower), whereas only CESA1, CESA2, CESA3, and CESA9 were significantly expressed in the embryo. In addition, very weak embryonic transcript levels were reproducibly detected for CESA5.
Figure 6.
CESA gene expression in embryos, young plants, and mature plants. Semiquantitative RT-PCR of total RNA prepared from embryos, seedlings, inflorescence stems, and flowers. Lanes 1 to 10, CESA1 to CESA10 RT-PCR products; R, approximately evenly expressed ROC1 gene (Lippuner et al., 1994); numbers on the right indicate the size of the PCR products in base pairs.
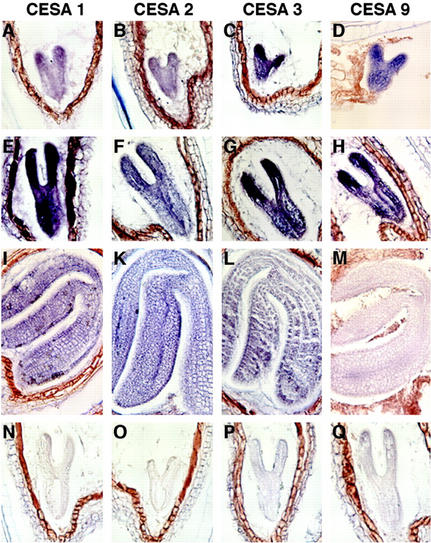
The absence of detectable embryonic expression for CESA4, CESA6, CESA7, CESA8, and CESA10 suggests that they are not required for embryonic primary wall formation (see “Discussion”). For CESA2, CESA3, CESA5, and CESA9 genes expressed along with RSW1(CESA1) in the embryo, no embryonic mutant defects have been reported. To gain insight into the roles of these four genes in embryogenesis, we determined their embryonic mRNA expression domains by in situ hybridization to sectioned embryos. No CESA5 transcripts were detected at any stage (data not shown), indicating that CESA5 expression was below the detection threshold in all analyzed stages and tissues (see “Discussion”). As shown in Figure 7, CESA1 was expressed throughout the embryo in the late-heart and torpedo stage, and was still visible at the bent-cotyledon stage. CESA2 expression had a very similar spatial and temporal pattern but at a somewhat reduced expression level, whereas expression of CESA3 and CESA9 appeared stronger in cotyledons than in hypocotyls and roots. The expression of all four genes decreased toward the bent-cotyledon stage, at which CESA9 was barely detectable. Overall, the expression of none of the four genes was restricted to specific cell types or stages, suggesting that they may collectively contribute to cell wall formation throughout the embryo.
Figure 7.
Expression pattern of CESA gene mRNA in wild-type (Col-0) embryos at heart/early torpedo (A–D), late torpedo (E–H), and bent-cotyledon (I–M) stage. Hybridization with antisense (A–M) and sense probes (N–Q). A, E, I, and N, RSW1(CESA1); B, F, K, and O, CESA2; C, G, L, and P, CESA3; and D, H, M, and Q, CESA9.
DISCUSSION
Great efforts are currently being made to identify the role of individual CESA genes (for review, see Delmer, 1999; Richmond, 2000; Richmond and Somerville, 2001). The results of these investigations could be relevant for many crop species, because the high degree of conservation among orthologs from different plant species compared with paralogs within a given species suggests conserved functional specialization within the gene family (Holland et al., 2000). The identification of null mutations in CESA genes and of developmental stages with reduced genetic complexity can greatly facilitate this analysis. Here, we have identified a highly probable null mutation in the RSW1(CESA1) gene and show that only a subset of CESA genes is expressed along with RSW1(CESA1) in the Arabidopsis embryo. Therefore, we suggest embryogenesis as a suitable stage of reduced CESA functional complexity to study the genetics of primary cell wall formation. As a first step, we have recorded CESA gene expression patterns during embryogenesis.
Loss of Gene Function in rsw1-20 Mutants
The structural properties of processive β-glycosyltransferases have been studied extensively (for summary, see Saxena and Brown, 1997; Saxena et al., 2001). Three Asp residues within the “D,D,D,QXXRW” motif are conserved from bacteria to plants. Although it is not finally resolved which of the Asp residues serve as bases during the catalytic reaction, two should be required for two glycosidic linkages formed simultaneously or sequentially during the synthesis of cellulose. Moreover, only the third Asp residue, which is affected in rsw1-20 mutants, is located within “domain B,” the characteristic domain of processive β-glycosyltransferases (Saxena et al., 2001). Finally, site-directed mutagenesis in bacteria has directly demonstrated that the exchange of any of the three Asp residues results in a reduction of β-glycosyltransferase activity to less than 1% of the wild-type level (Saxena et al., 2001). We conclude that rsw1-20 mutants lack all β-glycosyltransferase activity provided by the RSW1(CESA1) locus and can therefore serve as a suitable genetic background to assess the contribution of other cellulose synthase genes. The presumed null-mutant phenotype supports a pivotal role of RSW1(CESA1) in primary wall formation but also indicates that loss of RSW1(CESA1) gene function does not obstruct embryogenesis and even allows for early stages of vegetative development.
RSW1(CESA1) Function in Cell Division and Cell Expansion
Cell wall defects in rsw1-20 embryos are extremely severe in late-stage embryos and in apical meristems. By contrast, cell elongation in germinating mutants does not seem to be associated with additional wall disruptions. Therefore, cell wall integrity seems to be stringently dependent on RSW1(CESA1) activity in rapidly dividing cells but less critical in the elongation of nondividing cells. This differential requirement could reflect the fact that rsw1 mutations do not interfere with cell division but strongly restrict cell expansion and thereby reduce the requirement for cellulose synthesis selectively in expanding cells.
Mutations in RSW1(CESA1) restrict cell expansion during germination in light and in darkness, indicating that no germination-specific cellulose synthase activity can substitute for the RSW1(CESA1) function. It has previously been noted that the activities of the RSW1(CESA1) product and the dark-growth-specific cellulose synthase isoform PRC1(CESA6) are not redundant (Desnos et al., 1996; Fagard et al., 2000). Because both rsw1 and prc1 homozygous single mutants are impaired in cell expansion and double heterozygous individuals look normal, both proteins could possibly constitute necessary components of a higher order complex (Fagard et al., 2000). Our observations are consistent with this interpretation for the action of both gene products in elongating hypocotyls. However, RSW1(CESA1) must also be able to act in the absence of PRC1(CESA6), because PRC1(CESA6) is not expressed in the embryo. Therefore, the strongest argument for a simultaneous nonredundant action of the two gene products is the inability of RSW1(CESA1) to substitute for the PRC1(CESA6) function in the elongating hypocotyl. The reciprocal inability of PRC1(CESA6) to substitute for the RSW1(CESA1) function could simply reflect the irreversibility of cell wall defects in rsw1 mutants.
Cell Type-Specific Requirement for RSW1(CESA1) Function
Plant cells in axial organs, such as stems and roots, deviate most extremely from isotropic cell shapes. Axially organized microfibrils in these cells have been genetically implicated in the acquisition of anisotropic cell shapes by mutations that affect cell wall composition and simultaneously lead to more isotropic cell shapes (Arioli et al., 1998; Nicol et al., 1998; Lane et al., 2001; Lukowitz et al., 2001; Gillmor et al., 2002). Dramatic reductions in the cellulose content of embryonic cell walls associated with mutations in the α-glucosidase I gene KNOPF (KNF) as well as in RSW1(CESA1) have recently been described to cause corresponding shifts toward isotropic embryo shape (Gillmor et al., 2002). The mutations in KNF indicate a requirement for N-glycosylation of a component or substrate in cellulose synthesis. In knf mutants, the cellulose content of embryo cell walls is reduced to approximately 14% of the wild-type values, and mutant embryos adopt a nearly spherical shape. The newly identified rsw1-2 allele also affects embryo shape but apparently less severely than rsw1-20 mutations, suggesting that there is some residual gene activity in these mutants. The availability of multiple allelic mutations with related embryonic defects increases the reliability of functional interpretations drawn from the phenotype.
In rsw1-20 mutants, the normal width to length ratio in the embryonic hypocotyl of approximately 1:4 gradually approaches 1:<2 at late embryonic stages (Fig. 3M). Isotropic organ shape seems to be the biophysically most favorable condition, adopted whenever cell walls fail to acquire differential extensibility (Gillmor et al., 2002), but local modulations of this effect deserve consideration. First, there seems to be a differential effect on cells in the main axis of the plant as opposed to cells in lateral shoot organs. Although elongation of the hypocotyl is extremely reduced and associated with considerable radial expansion in several tissue layers, no consistent radial expansion was observed in cotyledons.
Second, particularly strong epidermal defects have consistently been observed in cell wall mutants and most probably reflect the particular strain encountered by walls on the outer surface of the plant (for summary, see Nicol and Höfte, 1998). In rsw1-20 mutants, instead of a smooth layer of interdigitated pavement cells, an uneven cobblestone surface is generated, and cells of complex morphology, such as trichomes and stomatal guard cells, are not formed. The highly uneven cell shape in the epidermis suggests that reduced cellulose synthase activity is associated with stochastic variations in wall stability (Desnos et al., 1996), which, in turn, should be incompatible with the acquisition of complex cell shapes, such as trichomes and guard cells. These effects might be further enhanced in the Ler genetic background. Therefore, even the seemingly cell type-specific defects in the epidermis may result from a general weakening of the wall structure.
Dissecting Overlapping CESA Functions
Assigning functions to individual members of large gene families eventually requires the analysis of multiple loss-of-function mutants. Expression studies cannot replace, but potentially facilitate, this genetic analysis. For genes such as cellulose synthase genes that are expected to act locally, probably cell autonomously, expression studies can reduce the complexity of future mutant analysis, because only genes co-expressed at the same stage or tissue need to be examined for potentially redundant functions.
Expression profiles of CESA genes have been studied or deduced from expressed sequence tag frequencies in several species, but CESA expression in Arabidopsis embryos has not been reported yet (Fagard et al., 2000; Holland et al., 2000; Dhugga, 2001; Richmond and Somerville, 2001; Scheible et al., 2001). We noticed that transcripts of only five of 10 classified Arabidopsis CESA genes were detected by RT-PCR in RNA from embryos, of which one had only an extremely low expression level. By contrast, transcripts of all 10 genes were readily detectable at all postembryonic stages. We observed similar expression profiles in three independent assays, but rather than focusing on the relative transcript abundance of CESA genes at various stages, we aimed at detecting minute amounts of embryonic expression. No transcripts of CESA4, CESA6, CESA7, CESA8, and CESA10 were found even after extending the number of amplification cycles up to 32 in RT-PCR of embryonic RNA. The absence of CESA4, CESA6, CESA7, CESA8, and CESA10 transcripts from the embryonic RNA pool is consistent with previously assigned roles for the IRX3(CESA7) and IRX1(CESA8) genes in secondary wall formation and for PRC1(CESA6) in cell expansion at germination (Taylor et al., 1999, 2000; Fagard et al., 2000). On the basis of their expression pattern, we propose that all five genes (CESA4, CESA6, CESA7, CESA8, and CESA10) are not involved in embryonic cell wall formation, which also may be the case for CESA5. Expression levels of CESA5 are far below those of any other embryonically expressed CESA gene in all cell types and could be gratuitous.
In situ hybridization was used to further dissect expression profiles of embryonically expressed CESA genes. These expression profiles were found to largely overlap, and all four genes were generally expressed throughout the embryo up to the bent-cotyledon stage, which could reflect their collective requirement throughout the embryo. Cellulose synthase complexes are presumably heteromeric at any stage, and a minimum of at least two types of subunits has been proposed (Scheible et al., 2001). Our results suggest that no more than four CESA gene products are required for the formation of a functional complex in the embryo and that even a complex made up of only the active products of CESA2, CESA3, and CESA9 could retain limited functionality. This conclusion is based on the observation that despite marked cell wall defects, primary cell walls are formed in rsw1-20 mutants. The recessive segregation of the rsw1-20 mutant phenotype indicates that the mutant RSW1(CESA1) product is either not incorporated or tolerated as an inactive component in the cellulose synthase complex.
Because CESA2, CESA3, and CESA9 are coexpressed with RSW1(CESA1) in virtually all embryonic tissues, their inability to substitute for RSW1(CESA1) function in these tissues, indicates functional divergence at the level of the gene products. By contrast, the degree to which CESA2, CESA3, and CESA9 can substitute for each other's function is unclear. The products of CESA2 and CESA9 share extensive sequence similarity and the absence of identified mutants as well as of dramatic defects in CESA2 antisense lines (Burn et al., 2002) could be attributable to extensive functional overlap between these two genes. Both genes are also related to PRC1(CESA6), which in turn has been shown to act nonredundantly with RSW1(CESA1) during germination (Fagard et al., 2000). The fourth coexpressed gene, CESA3(IXR1) seems to have a unique function in higher order complexes, because CESA3 antisense plants have recently been shown to be impaired in cell elongation in the postembryonic shoot and because overexpression of CESA3 did not normalize the rsw1-1 mutant phenotype (Burn et al., 2002). Taken together, at least three of the four CESA products coexpressed in most cells of the embryo seem to have unique functions in cellulose synthase complexes, and one possible scenario is therefore that ideally all four gene products are present in a complex, but that the loss of CESA2 and CESA9 would have relatively minor consequences. Given the reduced complexity of CESA function in the highly reproducible cell pattern in Arabidopsis embryogenesis, this stage seems to be best suited to genetically trace the requirement for each subunit in primary cell wall formation.
MATERIALS AND METHODS
Plant Growth Conditions
Mutants rsw1-20 and rsw1-45 were isolated from ethyl methyl sulfonate mutagenesis of Ler seeds of Arabidopsis (Jürgens et al., 1991). Seedlings were germinated and grown on one-half Murashige and Skoog (1962) medium supplemented with 1.5% (w/v) Suc at the indicated temperatures. For germination in the dark, seeds were cold-treated for 48 h and then exposed to fluorescent white light (80 μm m−2 s−1) for 2 h to synchronize germination. Subsequently, plates were wrapped in three layers of aluminum foil. For culture in the light, seeds were cold-treated for 48 h and then placed under continuous fluorescent white light (80 μm m−2s−1). Embryos were isolated from plants that were grown under long-day (16 h) light cycles in growth chambers (Conviron, Manitoba, Canada) at the indicated temperatures. Unless stated otherwise, wild type refers to the Ler line.
Measurement of Hypocotyl Length
Growth of seedlings was arrested by adding an aqueous solution of 0.4% (w/w) formaldehyde. Hypocotyls and roots were spread on agar plates, and hypocotyl lengths were measured on digitally captured images.
Light Microscopy
Samples for light microscopy were processed as described by Beeckman et al. (2000). To quantify the β-glucan content of cell walls, transverse sections (2 μm) of seeds embedded in LR White (London Resin, Basingstoke, UK) were made from wild type and mutant. The sections were stained with 0.07% (w/w) calcofluor white M2R (Sigma-Aldrich, St. Louis) in phosphate-buffered saline for 10 min, rinsed in phosphate-buffered saline, and mounted in Vectashield (Vector Laboratories, Burlingame, CA). Computer images of well-oriented cross sections through the hypocotyl were captured using an inverted (Axiovert 100M) confocal microscope (LSM510, Zeiss, Jena, Germany) equipped with an UV (364 nm) laser (Coherent Enterprise, LPS-Lasersysteme, Mössingen, Germany) and HFT UV 375/BP 385–470 as filter combination. Average fluorescence of cell walls of cortical cells from 10 independent mutant and wild-type embryos was determined by measuring the average pixel density of selected cell walls using the ImageJ analysis freeware software version 1.26 ImageJ (developed by Wayne Rasband, Research Services Branch, National Institute of Mental Health, Bethesda, MD). The same software was used to determine the cross-sectional area of selected cell walls. Relative fluorescence was reported as integrated density, which is the product of the cross-sectional area and the average fluorescence of cell walls. Polysaccharides in cell walls were visualized through periodic acid-thiocarbohydrazide silver proteinate staining (PATAg; Roland and Vian, 1991).
Measurement of Cellulose Content
Wild-type and rsw1-20 seeds were germinated and grown in continuous light at 31°C. Seedlings were harvested and frozen in liquid nitrogen 7 d after germination. Crude cell walls were extracted as described by Reiter et al. (1993). Plant samples of 20 mg of wall material (dry weight) were processed together with 20 mg of filter paper (cellulose content: 80% as a standard). Cellulose was isolated and solubilized by the method of Updegraff (1969). Cellulose content was deduced from Glc concentration measured by the anthrone method as described by Scott and Melvin (1953). Three independent measurements were performed.
Transmission Electron Microscopy
Specimens for electron microscopy were fixed in 2.5% (w/w) glutaraldehyde and 4% (w/v) paraformaldehyde in cacodylate buffer for 4 d at 4°C, post-fixed in 1% (v/v) OsO4 for 2.5 h at 4°C, and contrasted with 1% (v/v) uranyl acetate at 4°C; ultra-thin sections were analyzed with a transmission electron microscope (Siemens, Karlsruhe, Germany).
Sequence Analysis
Approximately 4 kb of genomic DNA encompassing the entire RSW1(CESA1) transcription unit defined by the external primer sequences (5′-gagatgccgtattgaatcgg and 3′-catggaatcaccttaactgcc) were amplified in multiple PCR reactions on DNA from the Ler background line and from homozygous rsw1-20 and rsw1-45 mutants. PCR fragments were directly sequenced by the dideoxy chain termination method (Sanger et al., 1977) on an ABI377 automated sequencer (PerkinElmer Instruments, Norwalk, CT). Single-base pair deviations from Ler in the two mutant lines were confirmed in four independent DNA samples of each genotype. No other sequence deviations in mutant lines were reproduced from independent DNA samples. Secondary structure prediction in protein sequences was performed with the PREDATOR program (http://mips.gsf.de/mips/staff/frishman; Frishman and Argos, 1997) using default parameters.
Semiquantitative RT-PCR
Embryos of heart to bent-cotyledon stages were excised from siliques. Young plants were harvested after growth on Murashige and Skoog medium containing 1.5% (w/v) Suc at 25°C for 7 d. Stem tissue and flowers were obtained from the upper infloresence of plants grown for 3 weeks at 22°C. Total RNA was isolated with RNeasy (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Semiquantitative RT-PCR of total RNA was performed with One-Step-RT-PCR (Qiagen) according to the manufacturer's instructions. In each reaction, 100 ng of total RNA was used. The length of an intron-spanning RT-PCR product of the ACT7 transcript (McDowell et al., 1996) confirmed the absence of DNA contamination in all RNA preparations. Specificity of CESA gene PCR products was controlled by analytical restriction digests. Approximate linearity of the PCR reaction was monitored by comparing relative amounts of PCR products after 24, 26, and 28 cycles. An amplification fragment from the ROC1 transcript (Lippuner et al., 1994) served as expression standard of an evenly expressed gene. Cycling conditions were: 50°C for 30 min, followed by incubation at 95°C for 15 min, 26 cycles of 94°C for 30 s, 55°C for 45 s, and 72°C for 1 min, and one cycle at 72°C for 10 min. Primer sequences were ACT7-F, ggtgaggatattcagccacttgtctg; ACT7-R, tgtgagatcccgacccgcaagatc; ROC1F, caaacctcttcttcagtctgatagaga; ROC1R, gagtgctcattccttatttctggtag; Ath-B(CESA3) F, gttccgcagacttgccagat; Ath-B(CESA3) R, caagctacattcccgagtcca; Ath-A F, cactctcttcacgccttaacacc; Ath-A R, gaggcgacacagaattaacatcc; IRX3 F, tctgctgaggctatgctgtatgg; IRX3 R, agagacagcgaaccagatttcac; IRX1 F, taggtctcccatctgcaacact; IRX1 R, tgagcgtcttgttgttcagc; CESA4 F, gcgatgaggtcaaagacgat; CESA4 R, tctttccccaacacacttcc; CESA5 F, ggactgcctttgattgaactc; CESA5 R, tcagacaagaagaatccctcaca; CESA6 F, ttcccacggatcaaagagag; CESA6 R, gtctccaagcatgcggatt; CESA9 F, tggtttggtcgattcttcttg; CESA9 R, cagcatccattcagggtctt; CESA10 F, attcgccatcgattcaaatg; and CESA10 R, taccacaatcgttcggtgaa.
In Situ Hybridization
Blunt-end PCR products for CESA1, CESA3, CESA5, and CESA9 (primer sequences, see above) were inserted in both orientations into pBluescript (Promega, Madison, WI); for CESA2, a fragment bordered by the sequences gctaagggggaccagtgtt and caaaagaattaatttaggggtaacaaa was chosen. Orientation of the inserts was determined by analytical PCR. Sense and antisense transcripts were generated by transcription from the T7 promoter. Antisense probes were linearized in the plasmid polylinker or at gtactgttgagttcaactaccctcagaa (CESA3) and agaatctgtggtcttgactgtttaaa (CESA9) to ensure specificity. Maximum similarity to any database sequence for any antisense probe was below 10%. All probes were spotted on nylon membranes (Amersham Biosciences UK, Ltd., Little Chalfont, Buckinghamshire, UK) along with labeled RNA controls (Roche Diagnostics, Brussels) to monitor the yield of digoxigenin-labeled RNA. Sensitivity of hybridization to sections of bent-cotyledon-stage embryos was tested by hybridization of control probes. Preparation of tissue sections, hybridization, and exposure were performed as described (Jackson, 1992).
ACKNOWLEDGMENTS
We would like to thank Richard E. Williamson (Australian National University, Canberra, Australia) for providing the rsw1-1 mutant and for helpful suggestions, Gerd Jürgens and Ulrike Mayer (Universität Tübingen, Germany) and Ben Scheres (University of Utrecht, The Netherlands) for providing mutant seeds, Heiko Schoof and Gerhard Wanner (Technical University Munich, Germany) for help with protein secondary structure analysis and transmission electron microscopy and for various suggestions on the manuscript, respectively, and Rebecca Verbanck and Martine De Cock (Ghent University, Belgium), for help in preparing figures and manuscript.
Footnotes
This work was supported by the Multidisciplinary Network program of the Natural Science and Engineering Research Council of Canada, by the Interuniversity Poles of Attraction Program (Belgian State, Prime Minister's Office, Federal Office for Scientific, Technical and Cultural Affairs; grant nos. P4/15 and P5/13), and by the European Community BIOTECH Research Program (grant no. ERB–BIO4–CT96–0217).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.010603.
LITERATURE CITED
- Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Höfte H, Plazinski J, Birch R et al. Molecular analysis of cellulose biosynthesis in Arabidopsis. Science. 1998;279:717–720. doi: 10.1126/science.279.5351.717. [DOI] [PubMed] [Google Scholar]
- Beeckman T, De Rycke R, Viane R, Inzé D. Histological study of seed coat development in Arabidopsis thaliana. J Plant Res. 2000;113:139–148. [Google Scholar]
- Burn JE, Hocart CH, Birch RJ, Cork AC, Williamson RE. Functional analysis of the cellulose synthase genes CesA1, CesA2, and CesA3 in Arabidopsis. Plant Physiol. 2002;129:797–807. doi: 10.1104/pp.010931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993;3:1–30. doi: 10.1111/j.1365-313x.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- Cutler S, Somerville C. Cellulose synthesis: cloning in silico. Curr Biol. 1997;7:R108–R111. doi: 10.1016/s0960-9822(06)00050-9. [DOI] [PubMed] [Google Scholar]
- Delmer DP. Cellulose biosynthesis: exciting times for a difficult field of study. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:245–276. doi: 10.1146/annurev.arplant.50.1.245. [DOI] [PubMed] [Google Scholar]
- Desnos T, Orbovic' V, Bellini C, Kronenberger J, Caboche M, Traas J, Höfte H. Procuste1 mutants identify two distinct genetic pathways controlling hypocotyl cell elongation, respectively in dark- and light-grown Arabidopsis seedlings. Development. 1996;122:683–693. doi: 10.1242/dev.122.2.683. [DOI] [PubMed] [Google Scholar]
- Desprez T, Vernhettes S, Fagard M, Refrégier G, Desnos T, Aletti E, Py N, Pelletier S, Höfte H. Resistance against herbicide isoxaben and cellulose deficiency caused by distinct mutations in same cellulose synthase isoform CESA6. Plant Physiol. 2002;128:482–490. doi: 10.1104/pp.010822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhugga KS. Building the wall: genes and enzyme complexes for polysaccharide synthases. Curr Opin Plant Biol. 2001;4:488–493. doi: 10.1016/s1369-5266(00)00205-3. [DOI] [PubMed] [Google Scholar]
- Fagard M, Desnos T, Desprez T, Goubet F, Refregier G, Mouille G, McCann M, Rayon C, Vernhettes S, Höfte H. PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell. 2000;12:2409–2423. doi: 10.1105/tpc.12.12.2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frishman D, Argos P. Seventy-five percent accuracy in protein secondary structure prediction. Proteins. 1997;27:329–335. doi: 10.1002/(sici)1097-0134(199703)27:3<329::aid-prot1>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Gillmor CS, Poindexter P, Lorieau J, Palcic MM, Somerville C. α-Glucosidase I is required for cellulose biosynthesis and morphogenesis in Arabidopsis. J Cell Biol. 2002;156:1003–1013. doi: 10.1083/jcb.200111093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green PB. Connecting gene and hormone action to from pattern and organogenesis: biophysical transductions. J Exp Bot Suppl. 1994;45:1775–1788. [Google Scholar]
- Holland N, Holland D, Helentjaris T, Dhugga KS, Xoconostle-Cazares B, Delmer DP. A comparative analysis of the plant cellulose synthase (CesA) gene family. Plant Physiol. 2000;123:1313–1323. doi: 10.1104/pp.123.4.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A, McCully M. The use of an optical brightener in the study of plant structure. Stain Technol. 1975;50:319–329. doi: 10.3109/10520297509117082. [DOI] [PubMed] [Google Scholar]
- Jackson D. In situ hybridization in plants. In: Gurr SJ, McPherson MJ, Bowles DJ, editors. Molecular Plant Pathology: A Practical Approach. I. Practical Approach Series. Oxford: IRL Press; 1992. pp. 163–174. [Google Scholar]
- Jürgens G, Mayer U, Torres Ruiz RA, Berleth T, Miséra S. Genetic analysis of pattern formation in the Arabidopsis embryo. Development. 1991;Suppl 1:27–38. [Google Scholar]
- Lane DR, Wiedemeier A, Peng L, Höfte H, Vernhettes S, Desprez T, Hocart CH, Birch RJ, Baskin TI, Burn JE et al. Temperature-sensitive alleles of RSW2 link the KORRIGAN endo-1,4-β-glucanase to cellulose synthesis and cytokinesis in Arabidopsis. Plant Physiol. 2001;126:278–288. doi: 10.1104/pp.126.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippuner V, Chou IT, Scott SV, Ettinger WF, Theg SM, Gasser CS. Cloning and characterization of chloroplast and cytosolic forms of cyclophilin from Arabidopsis thaliana. J Biol Chem. 1994;269:7863–7868. [PubMed] [Google Scholar]
- Lukowitz W, Nickle TC, Meinke DW, Last RL, Conklin PL, Somerville CR. Arabidopsis cyt1 mutants are deficient in a mannose-1-phosphate guanylyltransferase and point to a requirement of N-linked glycosylation for cellulose biosynthesis. Proc Natl Acad Sci USA. 2001;98:2262–2267. doi: 10.1073/pnas.051625798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann MC, Roberts K. Architecture of the primary cell wall. In: Lloyd CW, editor. The Cytoskeletal Basis of Plant Growth and Form. London: Academic Press; 1991. pp. 109–129. [Google Scholar]
- McCann MC, Roberts K. Changes in cell wall architecture during cell elongation. J Exp Bot. 1994;45:1683–1691. [Google Scholar]
- McDowell JM, An Y-Q, Huang S, McKinney EC, Meagher RB. The Arabidopsis ACT7 actin gene is expressed in rapidly developing tissues and responds to several external stimuli. Plant Physiol. 1996;111:699–711. doi: 10.1104/pp.111.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Nicol F, His I, Jauneau A, Vernhettes S, Canut H, Höfte H. A plasma membrane-bound putative endo-1,4-β-d-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J. 1998;17:5563–5576. doi: 10.1093/emboj/17.19.5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol F, Höfte H. Plant cell expansion: scaling the wall. Curr Opin Plant Biol. 1998;1:12–17. doi: 10.1016/s1369-5266(98)80121-0. [DOI] [PubMed] [Google Scholar]
- Reiter W-D, Chapple CCS, Somerville CR. Altered growth and cell walls in a fucose-deficient mutant of Arabidopsis. Science. 1993;261:1032–1035. doi: 10.1126/science.261.5124.1032. [DOI] [PubMed] [Google Scholar]
- Richmond T. Higher plant cellulose synthases. Genome Biol. 2000;1:3001–3005. doi: 10.1186/gb-2000-1-4-reviews3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond TA, Somerville CR. Integrative approaches to determining Csl function. Plant Mol Biol. 2001;47:131–143. [PubMed] [Google Scholar]
- Roland JC, Vian B. General preparation and staining of thin sections. In: Hall JL, Hawes C, editors. Electron Microscopy of Plant Cells. London: Academic Press; 1991. pp. 1–66. [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena IM, Brown RM. Identification of cellulose synthase(s) in higher plants: sequence analysis of processive β-glycosyltransferases with the common motif 'D, D, D35Q(R,Q) XRW'. Cellulose. 1997;4:33–49. [Google Scholar]
- Saxena IM, Brown RM., Jr Cellulose synthases and related enzymes. Curr Opin Plant Biol. 2000;3:523–531. doi: 10.1016/s1369-5266(00)00125-4. [DOI] [PubMed] [Google Scholar]
- Saxena IM, Brown RM, Dandekar T. Structure-function characterization of cellulose synthase: relationship to other glycosyltransferases. Phytochemistry. 2001;57:1135–1148. doi: 10.1016/s0031-9422(01)00048-6. [DOI] [PubMed] [Google Scholar]
- Scheible W-R, Eshed R, Richmond T, Delmer D, Somerville C. Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis ixr1 mutants. Proc Natl Acad Sci USA. 2001;98:10079–10084. doi: 10.1073/pnas.191361598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott AS, Melvin EH. Determination of dextran with anthrone. Anal Chem. 1953;25:1656–1661. [Google Scholar]
- Taylor NG, Laurie S, Turner SR. Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell. 2000;12:2529–2539. doi: 10.1105/tpc.12.12.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Scheible W-R, Cutler S, Somerville CR, Turner SR. The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell. 1999;11:769–779. doi: 10.1105/tpc.11.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updegraff DM. Semimicro determination of cellulose in biological materials. Anal Biochem. 1969;32:420–424. doi: 10.1016/s0003-2697(69)80009-6. [DOI] [PubMed] [Google Scholar]
- Williamson RE, Burn JE, Birch R, Baskin T, Arioli T, Betzner AS, Cork A. Morphology of rsw1, a cellulose-deficient mutant of Arabidopsis thaliana. Protoplasma. 2001;215:116–127. doi: 10.1007/BF01280308. [DOI] [PubMed] [Google Scholar]
- Wyatt SE, Carpita NC. The plant cytoskeleton: cell-wall continuum. Trends Cell Biol. 1993;3:413–417. doi: 10.1016/0962-8924(93)90022-s. [DOI] [PubMed] [Google Scholar]
- Wymer C, Lloyd C. Dynamic microtubules: implications for cell wall patterns. Trends Plant Sci. 1996;1:222–228. [Google Scholar]