Abstract
The light-harvesting proteins (Lhca) of photosystem I (PSI) from four monocot and five dicot species were extracted from plant material, separated by reversed-phase high-performance liquid chromatography (HPLC) and subsequently identified on the basis of their intact molecular masses upon on-line hyphenation with electrospray ionization mass spectrometry. Although their migration behavior in gel electrophoresis was very similar, the elution times among the four antenna types in reversed-phase-HPLC differed significantly, even more than those observed for the light-harvesting proteins of photosystem II. Identification of proteins is based on the good agreement between the measured intact molecular masses and the values calculated on the basis of their nucleotide-derived amino acid sequences, which makes the intact molecular masses applicable as intact mass tags. These values match excellently for Arabidopsis, most probably because of the availability of high-quality DNA sequence data. In all species examined, the four antennae eluted in the same order, namely Lhca1 > Lhca3 > Lhca4 > Lhca2. These characteristic patterns enabled an unequivocal assignment of the proteins in preparations from different species. Interestingly, in all species examined, Lhca1 and Lhca2 were present in two or three isoforms. A fifth antenna protein, corresponding to the Lhca6 gene, was found in tomato (Lycopersicon esculentum). However PSI showed a lower heterogeneity than photosystem II. In most plant species, Lhca2 and Lhca4 proteins are the most abundant PSI antenna proteins. The HPLC method used in this study was found to be highly reproducible, and the chromatograms may serve as a highly confident fingerprint for comparison within a single and among different species for future studies of the PSI antenna.
Photosynthetic electron transport in higher plants is driven by photosystems I and II (PSI and PSII). Each photosystem has an associated light-harvesting complex (LHCI and LHCII), which functions to deliver the excitation energy deriving from light absorption to the reaction centers (Steinback et al., 1985). These light-harvesting complexes are located within the thylakoid membrane of the chloroplast and are encoded from nuclear genes. In both photosystems, the antenna proteins contain three α-helices and are membrane-embedded proteins. The PSII major antenna proteins (LHCII) are the most abundant membrane proteins complexes on earth, accounting for up to 50% of the total chlorophyll in the thylakoid membrane, and have been well studied.
The LHCI, on the contrary, account for up to about 20% of the total chlorophyll, and this has hindered their detection and isolation (Scheller et al., 2001). Thus, because of the relatively recent identification of LHCI and isolation of the genes encoding its apoproteins, information on its structure and biogenesis is very limited. Mullet et al. (1980) have identified four thylakoid membrane proteins (molecular mass 21,500–24,500 D) from peas (Pisum sativum), which are involved in the PSI peripheral antennae. Four different families of genes have been cloned in tomato (Lycopersicon esculentum; Hoffman et al., 1987; Pichersky et al., 1987, 1988, 1989; Schwartz et al., 1991). At least two pigmented subcomplexes of LHCI have been fractionated from a PSI preparation by either non-denaturating gel electrophoresis or Suc gradient ultracentrifugation: LHCIa and LHCIb (Lam et al., 1984; Bassi and Simpson, 1987; Preiss et al., 1993). The LHCIa (also called LHCI-680, because it shows a fluorescence maximum between 680 and 690 nm) contains primarily two apoproteins of molecular mass 24,000 and 21,500 D, the products of the Lhca3 and Lhca2 genes, respectively (Lam et al., 1984; Bassi and Simpson, 1987; Ikeuchi et al., 1991; Knoetzel et al., 1992), although recent studies render this classification incorrect (Ganeteg et al., 2001). The LHCIb (also called LHCI-730, because it shows a fluorescence maximum at 730 nm) consists of a doublet of apoproteins with molecular mass of about 20,000 D in barley (Hordeum vulgare; Lam et al., 1984; Bassi and Simpson, 1987; Knoetzel et al., 1992), the products of Lhca 1 and Lhca 4 genes (Knoetzel et al., 1992; Anandan et al., 1993).
These antenna proteins seem to be organized as dimers and are associated to the backbone of PSI, a heterodimer consisting of two subunits named PSI-A and PSI-B (Jansson et al., 1996). In a recent study (Boekema et al., 2001), it was estimated that the PSI antenna contained maximally eight monomeric units of LHCI or six under particular conditions. Regarding the supramolecular organization, PSI seems to occur as monomeric pigment-proteins (Boekema et al., 2001), whereas it is generally accepted that the PSII antenna occurs as oligomers in situ. The organizational differences observed in the PSII have been related to a different efficiency of light energy transfer to the photochemical reaction center, providing the system with a way to regulate photosynthetic efficiency under the various light or stress conditions that green plants are subjected to (Dekker et al., 1999; Zolla et al., 2000). In this context, nothing equivalent has been reported for PSI, and the functional role of this heterogeneous group of proteins is unknown.
To date, with the exception of Arabidopsis (Jansson, 1999), few genes encoding the PSI proteins have been reported. As a consequence, it has not been possible to obtain, by sequence comparison, the identification of LHCI in different species. The protein components of the PSI antenna system are traditionally resolved by SDS-PAGE into four closely migrating protein bands displaying apparent molecular masses in the range of 20,000 to 24,000 D (Jansson et al., 1996). However, it is well known that most of these values diverge from the molecular masses calculated for the individual LHC proteins on the basis of their nucleotide-derived amino acid sequences (Matsuoka et al., 1987; Schwartz and Pichersky, 1990), because this mass determination by SDS-PAGE is based on the assumption that fully denatured proteins hydrophobically bind a constant amount of SDS.
The advent of electrospray ionization (ESI) for the soft ionization of biological macromolecules has greatly enhanced the role of protein mass spectrometry (MS) in structural biochemistry (Hancock et al., 1999; Griffiths, 2000; Li and Assmann, 2000), in proteomic studies (Peltier et al., 2000) and for photosynthetic proteins (Sharma et al., 1997a, 1997b, 1997c; Zheleva et al., 1998; Whitelegge et al., 1998). With accuracies of mass determination routinely achievable in the 100 to 200 ppm range (Premstaller et al., 2001), the protein molecular masses can serve as highly specific tags for protein identification, especially when supplemented by additional information such as protein hydrophobicity or pI (Wall et al., 2001).
In this communication, we report on the application of a reversed-phase chromatographic separation system for the fractionation of the different antenna protein components of PSI to establish a reference system for their identification on the basis of liquid chromatographic profiles. Identification of the proteins in the reference chromatograms is accomplished through ESI-MS, which has evolved into one of the most powerful analytical techniques for the characterization and identification of proteins, including applications for PSII antenna proteins (Corradini et al., 2000) and PSII core proteins (Sharma et al., 1997a, 1997b, 1997c; Zheleva et al., 1998). A correlation between Lhca genes and gene products is attempted.
RESULTS
Extraction of Antenna Proteins and SDS-PAGE Analysis
For each species examined, leaves were collected at different periods of the year to average out any seasonal effects. Before extraction, leaves were harvested at night in the dark to minimize any light effects. Experimental conditions, such as incubation time and detergent concentration for thylakoid extraction, were kept the same for all species whenever possible to ensure comparability of the results. PSI was isolated from thylakoid membranes of leaves by centrifugation in Suc gradient. For each set of experiments, equal sample volumes were loaded on the Suc gradient for reasons of comparability.
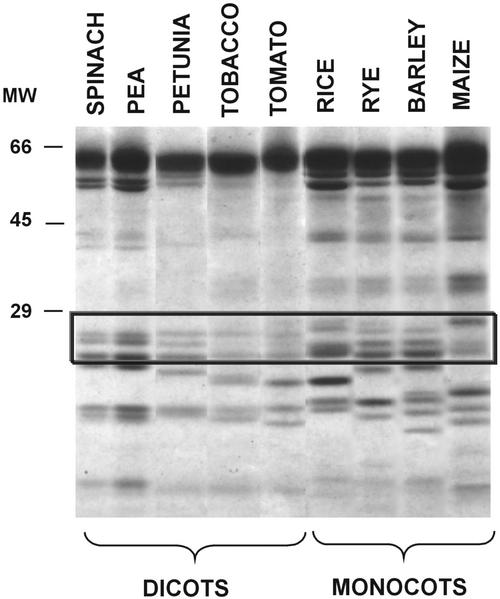
Figure 1 reports SDS-PAGE analysis of the proteins of PSI from five dicot and four monocot species. The antenna proteins of PSI were identified, as commonly performed (Croce et al., 1996; Jansson et al., 1996) by their apparent molecular masses of 20,000 to 25,000 D. It was observed that in all species examined, there were at least four bands corresponding to the four expected antenna proteins that show similar electrophoretic mobilities. Estimation of the relative stoichiometry of the antenna proteins from the intensities of the gel bands is difficult, and sometimes, low abundant proteins remain undetected because of the narrow dynamic range of detection in the stained gels. Moreover, identification of the antenna proteins is prone to error because of the known inaccuracy of apparent masses derived from gel analyses. Small differences in terms of protein abundance appear in monocots with respect to dicots.
Figure 1.
SDS-PAGE analysis of the proteins extracted from the thylakoid membranes of various dicots and monocots.
Separation and Identification of the PSI Antenna Proteins by Reversed-Phase HPLC-ESI-MS
Attempts to separate the antenna proteins from the other protein components comprising PSI by using zwitterionic detergents (Croce et al., 1998) revealed that a significant amount of Lhca2 (less than 15%–20%) remained tightly bound to the core complex (L. Zolla, A.M. Timperio, and S. Rinalducci, unpublished data), which may be revealed by HPLC and not by SDS-PAGE. This results in a significant change of relative stoichiometry; therefore, we decided to analyze the whole set of PSI proteins. In a previous paper (Zolla and Timperio, 2000), it was demonstrated that most of the PSI proteins in spinach (Spinacia oleracea) could be separated by reversed-phase HPLC. The antenna proteins represented the main peaks during the first 20 to 50 min of elution, whereas the PsaA and PsaB core proteins eluted around 60 min. Nevertheless, some of the other core proteins eluted in the same elution window as the antenna proteins, but they usually remain undetected by UV absorption and do not interfere with the determination of the highly abundant antenna proteins, especially in diluted sample preparations. However, injection of isolated antenna prepared by a second Suc gradient (Croce et al., 1998) onto the column gave a similar chromatographic profile compared with that observed by injection of the whole PSI, supporting the hypothesis that antenna proteins are well resolved without interference from the other core proteins of PSI.
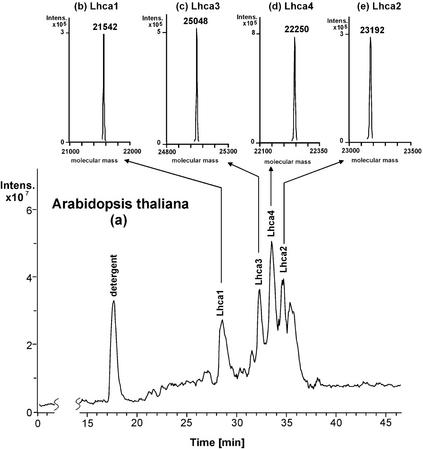
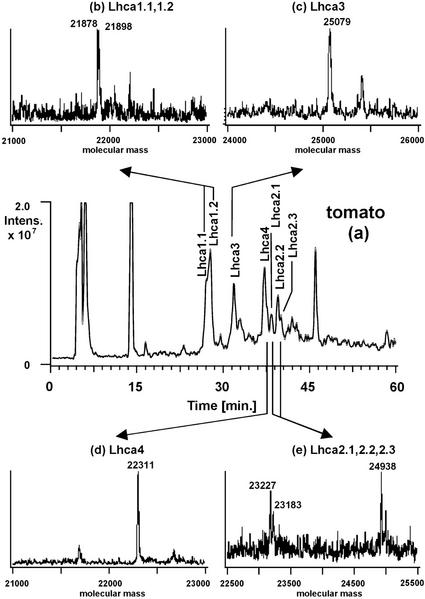
For the determination of protein intact molecular masses by ESI-MS, the column effluent was split post-column with 50 μL min−1 entering the mass spectrometer and 950 μL min−1 going to the UV or fluorescence detector. Most of the UV or fluorescence peaks showed a corresponding peak in the reconstructed ion chromatograms, which facilitated the identification of the components in the UV or fluorescence trace by means of the mass spectra extracted from the reconstructed ion chromatograms (RIC). Figures 2 and 3 show as examples the reconstructed ion chromatograms and the extracted and deconvoluted ESI mass spectra of PSI from Arabidopsis and tomato, two species where all four antenna genes have been cloned and sequenced (Hoffman et al., 1987; Pichersky et al., 1987, 1988, 1989; Schwartz et al., 1991; Jensen et al., 1992; Wang et al., 1994; Jansson, 1999). Arabidopsis was chosen for the limited numbers of Lhc genes and recent high-quality DNA sequence data, whereas tomato was chosen for the higher numbers of known genes.
Figure 2.
Reconstructed ion chromatogram of the protein components of the PSI antenna system from Arabidopsis. Column, Vydac Protein C-4 (250 × 4.6 mm i.d.); mobile phase, 45-min linear gradient from 38.75% to 68.7% (v/v) acetonitrile in water containing 0.05% (v/v) trifluoroacetic acid (TFA); flow rate, 1.0 mL min−1; flow of column effluent entering the mass spectrometer, 50 μL min−1; detection, ESI-MS; scan 500 to 2,000 atomic mass units; injection volume, 100 μL.
Figure 3.
Reconstructed ion chromatogram of the protein components of the PSI antenna system from tomato. Conditions as in Figure 2.
In the case of Arabidopsis, analysis of the reconstructed ion chromatograms (Fig. 2) revealed that the peak eluting from the column at 18 min and showing a singly charged pseudomolecular ion at m/z 512 in the mass spectrum is n-dodecyl-β-maltoside (DM) used as detergent. On the contrary, the deconvolution analysis of the other peaks (Fig. 2, b–e) indicated the presence of several proteins with molecular masses in the range of 21,000 to 25,000 D as expected for Arabidopsis PSI antenna proteins (Jansson, 1999). Each UV peak contained only one protein, and the molecular masses are collected in Table I. The identity of each protein was established by comparison of the measured intact molecular mass with the molecular mass predicted from the DNA sequence. It was observed that the intact molecular masses are easily assigned, because the native masses of the proteins match very well the values expected. In the case of Lhca3, the measured molecular mass is 112 D higher than the calculated one. However, one has to take into account that this protein type contains an unknown chemical group that blocks the amino-terminal amino acid (Jansson, 1994). Interestingly, the difference between observed and calculated masses is less than 0.03%, giving confidence in the protein identification performed by intact molecular mass measurements, as already successfully achieved for PSII components (Sharma et al., 1997b; Gómez et al., 2002 ).
Table I.
Comparison of PSI antenna protein molecular masses determined by HPLC-ESI-MS with the protein masses expected from DNA sequence in Arabidopsis and tomato
| Species | Measured Mass (±sd)a | Calculated Massb | Mass Deviationc | Protein Identificationd | Accession No.e |
|---|---|---|---|---|---|
| D | % | ||||
| Arabidopsis | 21,542 ± 0.3 | 21,543 | 0.004 | Lhca1 | M85150 |
| 25,048 ± 0.8 | 24,936 | ? | Lhca3 | U01103 | |
| 22,250 ± 0.2 | 22,255 | 0.02 | Lhca4 | M63931 | |
| 23,192 ± 0.6 | 23,200 | 0.03 | Lhca2 | AF134120 | |
| Tomato | 21,878 ± 0.9 | 21,879 | 0.004 | Lhca1.1 | 1402358A |
| 21,898 ± 1.4 | 21,851 | 0.2 | Lhca1.2 | P12360 | |
| 25,070 ± 1.5 | 25,110f or 26,128 | ? | Lhca3 | P27522 | |
| 22,310 ± 1.3 | 22,336 | 0.1 | Lhca4 | S14305 | |
| 22,286 | S14306 | ||||
| 24,938 ± 1.5 | 24,834 or 23,079f | 0.4 | Lhca2.1 | P10708 | |
| 23,184 ± 0.5 | 0.4 | Lhca2.2 | |||
| 23,226 ± 1.0 | Lhca2.3 | ||||
The mean ± sd of three experiments is presented.
Calculated average mass of the unchanged assigned gene product shown in identification column.
Percent difference between expected and observed masses. ?, The unknown group blocking the N-terminal amino acid of Lhca3 does not allow determination of the mass deviation.
Protein IDs in quotes indicate assignments made by comparison between measured and calculated molecular masses.
Accession no. in GenBank, National Center for Biotechnology Information, and Swiss-Prot databases.
Values according to alignment of Jansson (1994).
In tomato, our analysis (Fig. 3) revealed that the first two partially resolved peaks contained two proteins having molecular masses of 21,878 and 21,898 D, respectively, the third peak a protein with a molecular mass of 25,079 D, the fourth main peak a protein of molecular mass 22,311 D and finally the last three peaks revealed the presence of three proteins of molecular mass 23,183, 23,227, and 24,938 D, respectively. By comparison of the measured molecular masses with those deduced from DNA sequence (see Table I), the identification of each antenna protein was performed for this species too. The observed mass deviations of some proteins obviously were significantly larger than 0.02%, which is characteristic for intact molecular mass measurements using quadrupole ion trap mass spectrometers. Hence, identification of the proteins is based upon the criterion that the measured molecular mass falls within the range of molecular masses calculated from known DNA sequences of the different types of antenna proteins.
Lhca1 protein was easily assigned because it corresponds exactly to the molecular masses deduced from DNA sequence of the cloned Lhca1 gene (21,879 and 21,851 D; Hoffman et al., 1987; Pichersky et al., 1987). Lhca3 protein comes close to the expected mass of the mature protein (25,110; Jansson, 1994). Lhca4 fits well with the two Lhca4 genes (22,336 and 22,286 D; Schwartz et al., 1991), whereas in the case of Lhca2, some apparent discrepancy is observed, which will be discussed later. Thus, for both species presented, the good agreement between measured and calculated molecular masses allowed an easy protein identification. On the other hand, no posttranslational modifications have ever been documented in the antenna proteins of PSI, with the exception of Lhca3 amino-terminal blocking. As a consequence, it is reasonable to suggest an assignment of an experimental molecular mass to one of the four types of antenna proteins, if it fits into the range of mass values expected from the DNA sequence.
Table II summarizes the experimental and expected molecular masses, obtained from the SWISS-PROT and NCBI databases, of the PSI antenna proteins for four monocots and four dicots, respectively. Thus, for most species, the peak identities were assigned based on the match of measured and calculated masses. On the contrary, for proteins where the DNA sequence is not reported, assignment was performed by comparison with proteins of other species having similar mass and similar hydrophobicity. In fact, the measured molecular masses are in four narrow ranges as expected from the DNA sequences: Lhca1 from 21,170 to 22,140 D, Lhca2 from 23,160 to 23,372 D, Lhca3 from 24,808 to 25,548 D, and Lhca4 from 22,124 to 22,311 D. These ranges differ significantly, thus supporting unambiguous assignment. The proteins showing a molecular mass measured over 23,000 were identified as Lhca2 and the protein with molecular masses above 25,000 D as Lhca3. In the case of Lhca1 and Lhca4, we used the elution time (see below) as an additional parameter for a better classification. In fact, in all dicots, proteins eluting at short retention times show a molecular mass below 22,000 D, and they might be easily identified as Lhca1; but in the case of monocots, the molecular mass is more than 22,000 D, which may be confused with Lhca4, but their elution at short times allowed us to recognize them as Lhca1.
Table II.
Average of experimental and expected molecular masses of PSI antenna proteins in various plant species
| SPECIES | Lhca 1
|
Lhca 2
|
Lhca 3
|
Lhca 4
|
||||
|---|---|---|---|---|---|---|---|---|
| Measured | Calculated | Measured | Calculated | Measured | Calculatedd | Measured | Calculated | |
| D | ||||||||
| Monocots | ||||||||
| Rye 1 | 22,138 | 23,194 | 25,534 | 22,264 | ||||
| Rye 2 | 22,124 | 23,238 | ||||||
| Rye 3 | 23,268 | |||||||
| Rice | 22,070 | 23,190 | 23,424 | 22,300 | ||||
| AAB65793a | ||||||||
| Barley 1 | 22,124 | 22,117 | 23,164 | 23,161 | 25,548 | 22,302 | 22,302 | |
| AAF23819a | CAA59049a | AAF90200a | ||||||
| Barley 2 | 22,140 | 23,208 | ||||||
| Maize 1 | 21,870 | 23,326 | 24,808 | 22,172 | ||||
| Maize 2 | 23,372 | |||||||
| Dicots | ||||||||
| Tomato 1 | 21,878 | 21,879 | 24,938 | 24,834 | 25,079 | 25,110c | 22,311 | 22,336 |
| 1402358Aa | P10708b | or | S14305a | |||||
| or | 26,128 | |||||||
| Tomato 2 | 21,898 | 21,851 | 23,183 | 23,079c | P27522b | 22,286 | ||
| P12360b | S14306a | |||||||
| Tomato 3 | 23,227 | |||||||
| Petunia 1 | 21,896 | 23,160 | 23,016c | 25,174 | 22,124 | |||
| or | ||||||||
| 24,772 | ||||||||
| Petunia 2 | 21,878 | P13869b | 22,260 | |||||
| Tobacco | 21,850 | 21,882 | 23,282 | 25,012 | 22,238 | |||
| CAA45523a | ||||||||
| or | ||||||||
| 21,956c | ||||||||
| Pea | 21,170 | 23,340 | 24,292 | 25,282 | 25,297c | 22,226 | 22,128 | |
| CAA57492a | AAF13731a | |||||||
sd measured masses range between ±1.5 and ±2.7.
Accession no. in NCBI database.
Accession no. in SWISS-Prot database.
Values according to the alignment of Jansson (1994).
Values reported were calculated without any chemical group blocking the N terminus.
In all species examined, Lhca1 and Lhca2 showed more than one protein with molecular masses very close to each other, whereas Lhca3 and Lhca4, with the exception of Lhca4 of petunia (Petunia hybrida), were present only in a single copy. As a consequence, the different proteins must be considered as isomeric forms of Lhca1 and Lhca2, as indicated in the chromatograms using the indexed labels Lhca1.1, Lhca1.2, etc., according to the nomenclature proposed by Jansson (1999) to identify the numerous Arabidopsis genes and isoforms found in PSII from different species (Huber et al., 2001).
Analysis of PSI Protein Preparations from Different Plants and Reproducibility of Chromatographic Profiles
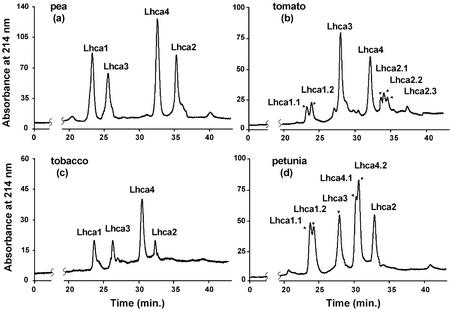
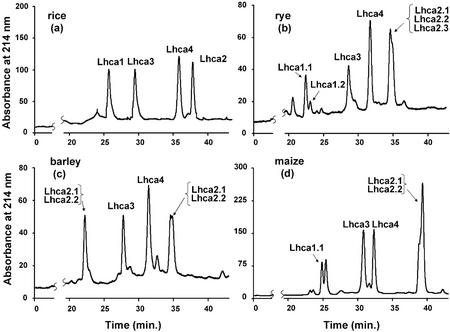
After the peaks observed in the UV or fluorescence chromatograms have been correlated with the protein components identified by ESI-MS, the ranges of retention times and the chromatographic patterns of the PSI antenna proteins are now characterized. Figure 4 reports the chromatograms and corresponding protein identification of four representative dicot species, including pea, tobacco (Nicotiana tabacum), tomato, and petunia, whereas Figure 5 shows the chromatograms for four representative monocot species, such as rice (Oryza sativa), rye (Secale cereale), barley (Hordeum vulgare), and maize (Zea mays). Because it was observed that the chromatograms detected both by UV absorption at 214 nm and by fluorescence emission at 330 nm upon excitation at 280 nm were very similar, only the UV chromatograms are presented in the figures.
Figure 4.
Chromatographic fingerprints of the PSI antenna proteins from the following dicot plant species: a, pea; b, tomato; c, tobacco; and d, petunia. Column, Vydac Protein C-4 (250 × 4.6 mm i.d.); mobile phase, 45-min linear gradient from 40% to 65% (v/v) acetonitrile in water containing 0.05% (v/v) TFA; flow rate, 1.0 mL min−1; detection UV, 214 nm; injection volume, 20 to 50 μL.
Figure 5.
Chromatographic fingerprints of the PSI antenna proteins from the following monocot plant species: a, rice; b, rye; c, barley; and d, maize. Conditions as in Figure 4.
Each sample of PSI antenna isolated from each plant was analyzed in triplicate by reversed-phase HPLC to evaluate the reproducibility of the chromatographic profile and the resolution of the antenna proteins. Moreover, triplicate analyses were performed on three different preparations of the same species. All relative standard deviations of retention times of resolved proteins were smaller than 0.4% for the main peaks observed. This high reproducibility of the chromatographic separation derived from both the same and different preparations was also corroborated by visual inspection of the chromatographic patterns and has promoted confidence in reversed-phase HPLC as a reliable method for detecting differences in the protein components of PSI antenna isolated from different plants.
A comparison of the retention times of the major antenna proteins revealed that Lhca1 always eluted first as the most hydrophilic antenna protein, in contrast to Lhca2, which is the most hydrophobic protein. The elution order of the four different types of antenna proteins was Lhca1 > Lhca3 > Lhca4 > Lhca2 in all species examined. These characteristic patterns allow an unequivocal assignment of each proteins and a highly confident and reproducible fingerprint for comparison within a single and among different species. In fact, the resolution of the antenna proteins of PSI by reversed-phase HPLC is significantly better than that possible with SDS-PAGE. By SDS-PAGE, the pattern is Lhca1 > Lhca3 > Lhca4 > Lhca2 for maize but conversely not for spinach or barley, depending also on electrophoretic system used (Ikeuchi et al., 1991; Croce et al., 1996; Jansson et al., 1996).
Finally, reversed-phase HPLC-ESI-MS revealed isoforms of the PSI antenna proteins. Some of the isoforms were separable by reversed-phase HPLC because of their differing hydrophobicity (e.g. Lhca1.1 from Lhca1.2 in tomato or petunia, or Lhca2.1 from Lhca2.2 in tomato and barley), whereas some others co-eluted in one HPLC peak but were distinguished by ESI-MS (e.g. Lhca1.1 from Lhca1.2 in barley, or Lhca2.1 from Lhca2.2 in maize).
Comparison of the Hydropathic Protein Profiles and the Theoretical Retention Coefficients
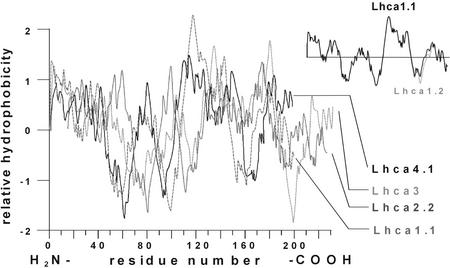
To correlate chromatographic retention with the hydrophobic properties of the antenna proteins, hydropathic profiles were calculated for some proteins of known amino acid sequence. Figure 6 compares the hydropathic profiles for the four Lhca antenna proteins from tomato using the Kyte-Doolittle method (Kyte and Doolittle, 1982). It was observed that the hydrophobicity expected for the four PSI antennae types is Lhca1 > Lhca3 > Lhca4 > Lhca2, as observed in HPLC. Moreover they differ significantly in their hydrophobic properties, with dissimilarities not confined to small regions of the proteins. The inset in Figure 6 compares the hydropathic profiles of the two isoforms of Lhca1 in tomato, for which two genes have been cloned and sequenced (Hoffman et al., 1987; Pichersky et al., 1987). Although the profiles revealed only sight differences, the two proteins are well separated by HPLC. This indicates the high selectivity of reversed-phase HPLC and its capability of separating membrane proteins that differ by only few amino acids.
Figure 6.
Hydrophatic profiles calculated from the DNA sequences of Lhca1.1, Lhca2.2, Lhca3, and Lhca4.1 in tomato. Inset, The hydrophatic profiles of two isoforms Lhca1.1 and Lhca1.2 in tomato calculated from the DNA sequences.
DISCUSSION
Suitability of Reversed-Phase HPLC as Reference Method
The complete resolution of the PSI antenna proteins from four dicots and four monocots, together with their detection and identification has been successfully obtained by the combined use of reversed-phase HPLC coupled on-line with ESI-MS. In all species examined, the antenna proteins are well separated by HPLC, especially Lhca1 and Lhca4, which have similar electrophoretic mobilities in SDS-PAGE (Jansson et al., 1996) with inevitable difficulties in their identification and quantification.
Moreover, the protein identification by HPLC is independent of experimental conditions, whereas Ikeuchi et al. (1991) reported that mobility order of Lhca apoproteins in SDS-urea-PAGE at 25°C was II, IV<I< III, quite different from that observed at 4°C (I<IV<II<III), revealing how dangerous it is to compare proteins based on their mobilities on different SDS-PAGE systems.
The reversed-phase HPLC method is also less expensive and technically demanding than two-dimensional electrophoresis, which has to be used for improved separation of the antenna proteins, and it eliminates the need for time-consuming antibody titration to determine the relative quantity of each component (Jansson et al., 1996). Using this strategy, the identification of each PSI antenna protein is unequivocal, because of their markedly different molecular masses and the high accuracy of mass determinations by ESI-MS.
Significant differences were observed in the elution times among the four different antenna types of PSI, which were more pronounced than those observed for the light-harvesting proteins of PSII. In all species examined, the elution order of the four different types of antenna proteins of Lhca1 > Lhca3 > Lhca4 > Lhca2 is indicative of highly conserved amino acid sequence among species and may serve for an unequivocal assignment of each protein and for a highly confident and reproducible fingerprint for comparison within a single and among different species. However, from the different order in which the LHCI proteins are resolved by HPLC, it can be inferred that the hydrophobic character of these membrane proteins is not directly related to their molecular mass.
Reversed-phase HPLC separates the antenna proteins on the basis of their different hydrophobicities resulting in differences in their elution times. In fact, each species exhibits a unique chromatographic pattern that reflects the varying hydrophobicity and stoichiometry of the light-harvesting proteins. To render the method suitable for comparative purposes, each protein present in the UV chromatogram has to be identified. Although several methods such as electrophoresis, immunoblotting, and amino acid sequencing may be used (Zolla et al., 1999), the great advantage of the HPLC method rests within the possibility of on-line hyphenation to ESI-MS to obtain molecular mass data suitable for identification (Corradini et al., 2000; Zolla and Timperio, 2000), which was missing in the pioneering works using HPLC (Damm and Green, 1994; Zolla et al., 1997). On the other hand the protein identification performed by HPLC-ESI-MS (Corradini et al., 2000) agreed with that performed by immunoblotting and/or amino acid microsequence (Zolla et al., 1999). Once identification has been performed by ESI-MS, the UV chromatograms reported here can be used as standards for other researchers using this method because of the high reproducibility of both the chromatographic separation and retention of proteins, regardless of sample batch or harvesting season.
From the genes reported in the literature, it was deduced that the PSI antenna proteins comprise a similar number of amino acids, ranging from 199 in Lhca4 to 233 to 234 in Lhca3. As a consequence, the expected molecular masses ranged from 21,500 D for Lhca1 to 25,035 D for Lhca3 (Jansson, 1994), which was corroborated by the experimental data summarized in Tables I and II. In Arabidopsis, barley, and tomato, there is an excellent correspondence between the measured and expected molecular masses for most of the proteins with a mass deviations of typically less than 0.02%. The use of intact molecular measurements for an unequivocal assignment of each protein to its respective gene was successfully applied for the analysis of PSII core components (Sharma et al., 1997a, 1997b, 1997c) and integral membrane proteins, including the D1 and D2 subunits of PSII (Whitelegge et al., 1998). Thus, in the species where some discrepancy is observed, we believe that this may be attributable to errors in DNA sequence determination, especially using the chemical methods of more than 10 years ago or, where the differences are relatively large, to different cleavage or to posttranslational modifications of the mature protein.
This paper also reports for the first time, to our knowledge, the real molecular mass of each PSI antenna protein in eight different species, including those proteins whose genes have not yet been cloned. A comparison of the values given in Table II reveals that Lhca1 proteins in most monocots show higher masses than in dicots. Moreover, in all species examined, the number of isomeric proteins identified is lower than three, confirming that in PSI the heterogeneity is less marked than in PSII (Huber et al., 2001), as expected from the total of number of genes cloned. Thus, a number of seven homologous genes in petunia for Lhca2 (Stayton et al., 1987) is most probably an overestimation, or maybe some of the genes are not expressed.
Lhca1 molecular masses range from 21,170 to 22,140 D. This antenna protein is the smallest protein in all species examined, in agreement with reports for Arabidopsis (Jansson, 1999). In the case of barley, two isoforms are revealed by ESI-MS. The molecular mass of Lhca1.1 corresponds exactly to the mass derived from the gene reported in the literature (Klimmek, 1999). In the case of tomato, two genes have been cloned, and two proteins, separable by reversed-phase HPLC, were found: one showing the molecular mass expected and another 47 D greater. In tobacco, the measured mass is only 32 D less than that expected from the DNA sequence (Palomares et al., 1991), whereas the value expected by alignment with tomato (Jansson, 1994) is 150 D greater, suggesting that in tobacco the precursor cleavage is different from tomato. Interestingly, in all dicots examined the mass ranges from 21,170 to 21,898 D, whereas in most monocots, with the exception of maize, the Lhca1 masses are 300 to 400 D greater than in dicots. This justifies the previous evidence that in barley, the Lhca1 run lower than spinach Lhca4 (Knoetzel et al., 1992). Isoforms of Lhca1 were detected both in dicots and monocots.
Lhca2 molecular masses range from 23,160 to 23,372 D in all species examined. In barley, the value measured by ESI-MS (23,164 D) corresponds within 0.02% to that expected 23,161 D (Knoetzel, 1995). In the case of rice, only one PSI gene is reported in the literature (Lee et al., 1997) without any identification. Because it shows a high similarity with the Lhca2 gene of barley, their alignment assuming the same starting point of the mature protein yields an expected molecular mass of 23,424 D, which reasonably corresponds to the measured value of 23,190 D. In tomato, three isoforms eluting in three separate HPLC peaks were found. ESI-MS analysis revealed that one of them shows a molecular mass of 24,938 D, which is close to the cloned cab 7 gene (24,834 D; Pichersky et al., 1988), whereas the other two show molecular masses of 23,183 and 23,227 D, which are close to the value proposed by Jansson (1994; 23,079 D), assuming that the mature proteins starts at amino acid 60. Interestingly, the gene cloned (Pichersky et al., 1988) gives a protein of molecular mass 24,834 D, whereas the mature protein should have a molecular mass of 23,079 D (Jansson, 1994), which is close to the mass of the second isoform. The only possible explanation of this apparent discrepancy is the presence of more than one Lhca2 gene having different lengths. This hypothesis is supported by the recent evidence that in the case of Arabidopsis and canola (Brassica napus; Jansson, 1999), two possible genes of Lhca2 have been suggested: one called “normal Lhca2” (expected molecular mass, 23,200 D), which shares 92% and 85% of the amino acids with its homologs from tomato and Scots pine (Pinus sylvestris) and an “unusual Lhca2 gene” (called also Lhca6; Jansson, 1999) having a molecular mass of 24,549 D, which encodes a protein with only 68% identity. Thus, it may be postulated that the isoform of the Lhca2 protein with molecular mass of more than 24,000 D represents the product of the unusual Lhca2 gene (the Lhca6 gene). If this is true, the question raised from the genomic data “is the unusual Lhca2 gene a nonexpressed pseudogene, or do both genes have a function?” seems to have the answer that both genes are expressed. Tandem mass spectrometric sequencing of trypsin-digested antenna proteins from Arabidopsis is in progress in our lab, with the final aim to get partial sequences of the amino terminus and to confirm this hypothesis.
Lhca3 molecular masses range from 24,808 to 25,548 D. It is the largest protein showing the largest variability in molecular mass. In the case of pea, the expected molecular mass is 25,297 D (Jansson, 1994; using AAT as start amino acid sequence and without the mass of the unknown chemical group blocking the amino terminus), which is close to the measured value of 25,282 D. In tomato, the DNA sequence reported in literature aligned at a starting sequence of AST (amino acid 42) similarly gives an expected mature protein of 25,110 D (Jansson, 1994), which is very close to the molecular mass of the protein found (25,079 D).
Lhca4 molecular masses range from 22,124 to 22,311 D. In barley, the molecular mass measured corresponds within 0.02% with the molecular mass expected (22,302 D). In the case of tomato, only one protein was found (22,311 D) that ranges between the values expected from the two genes cloned: cab11 and cab12 (Schwartz et al., 1991) of 22,336 and 22,286 D, respectively. Because one of them is expressed at very low level, 100 times less the other (Schwartz et al., 1991), the less abundant protein may be not revealed by our ESI-MS measurement. Finally, in the case of pea, only one PSI gene is reported in the literature (Brosche et al., 1999) without any identification. It may be considered as Lhca4 gene, assuming a mature protein that starts at amino acid 52 with KK as in tomato and barley. With this assumption, the protein expected has a molecular mass of 22,128 D, comparing favorably with a measured mass of 22,226 D.
Total Number of Lhca Proteins and Protein Isoforms
Our analysis revealed five different Lhca gene products. Genomic analysis of Arabidopsis predicts one more gene product, Lhca5 (Jansson, 1999). However, the corresponding protein has not been experimentally detected so far, and its expression may be too low to be revealed by reversed-phase HPLC-ESI-MS. Isomeric forms of Lhca1 and Lhca2 were found in PSI of most species examined. In addition, two isoforms of Lhca4 were present in petunia. The existence of isoforms was expected in tomato, where two genes for Lhca1 have been cloned (Hoffman et al., 1987; Pichersky et al., 1987). Moreover, evidence on the possible existence of isoforms came out from SDS-PAGE analyses of Lhca2 in barley (Knoetzel et al., 1992) and spinach (Ikeuchi et al., 1991). However, Ikeuchi et al. (1991) explained the heterogeneity of the Lhca2 proteins observed in the electropherograms and in the fluorescence spectra as attributable to denaturation (Ikeuchi et al., 1991). Whereas the existence of multimeric forms in PSII was expected from molecular genetic data (Morishige and Thornber, 1991; Sigrist and Staehelin, 1994), the existence of isomeric forms in PSI antenna proteins is now clearly confirmed. The presence of Lhca protein isoforms has to be taken into account for physiological interpretation.
General Considerations
In contrast to observations with the antenna proteins of PSII (Huber et al., 2001), the dissimilarities of the hydropathic profiles of PSI antenna proteins are not localized in the amino-terminal region but are extended to the transmembrane- and carboxy-terminal region. This explains the markedly diverse retention behavior observed among the four antennae types within the same species. Lhca1 generally shows lower hydrophobicity than Lhca2. Thus, it is reasonable to hypothesize that Lhca1 is located in the outer region of the supramolecular organization, whereas Lhca2 is tightly bound to core proteins under all conditions.
On the other hand, the same types of the four antenna proteins show very similar hydrophobic properties in terms of chromatographic retention among species, which is indicative of a strongly conserved amino acid sequence. It is tempting to speculate that the observed difference in hydrophobicity among the different types might be the basis for the distinctive dimeric organization supposed for PSI (Jansson et al., 1996), in which Lhca1 interacts with Lhca4, and Lhca2 with Lhca3 or form homodimers (Boekema et al., 2001). Interestingly, Lhca1 and Lhca4 show significant different hydrophobic profiles in the amino-terminal region, whereas it is similar in Lhca2 and Lhac3. Moreover, isoforms exist only for Lhca1 and Lhca2.
The stoichiometry and hydrophobic properties of antenna proteins do not show general differences between dicots and monocots, suggesting that the supramolecular organization of PSI should be similar along the evolution of two phyla, which is in contrast to findings for PSII (Huber et al., 2001). A comparison between monocots and dicots shows a significant difference only for the molecular mass of the Lhca1 protein, which is higher in monocots. This small difference clearly cannot account for the slower protein assembling of PSI observed in dicots under light, which was interpreted by a different final organization of PSI in monocots and dicots (Dreyfuss and Thornber, 1994). The aggregation state is obviously strongly influenced by the relative stoichiometry of all components in different species.
The relative stoichiometry of the antenna system can be readily inferred from the chromatographic profiles based on the fact that the antenna proteins are strongly conserved, and it may be assumed that they have about similar extinction coefficients. Hence, the areas underlying the HPLC peaks represent approximately the real relative amounts of protein. The relative amounts of the individual antenna proteins appear to be markedly different between species and within the same species (Figs. 4 and 5), whereas the electrophoretic patterns suggest the presence of all Lhca proteins in approximately equal amounts (Fig. 1).
This is in agreement with the experimental evidence for a different antenna composition of PSI in different domains of the thylakoid membrane (Jansson et al., 1997), and the recent evidence that the Lhca protein composition of LHCI is flexible and varies with the light intensity during growth (Bailey et al., 2001). Our quantitative results confirm that the final protein amounts are not identical, although the mRNA levels of Arabidopsis Lhca antenna proteins are about the same under laboratory standard conditions (Jansson, 1999). It may be assessed that in most plant species, Lhca2 and Lhca4 proteins are the most abundant PSI antenna proteins, although the mRNA level in Arabidopsis is higher for Lhca3 gene (Jansson, 1999). Moreover, our data agree with the evidence that in tomato, the Lhca2 gene has the highest level of expression of all Lhca genes (Piechulla et al., 1991). On the contrary, Lhca1 represents the lowest abundant protein. This may have implications on the supramolecular organization of the chlorophyll proteins in the antenna complex, which could be different among species.
CONCLUSIONS AND OUTLOOK
The high resolving power of reversed-phase HPLC separation, together with the possibility to accurately determine the molecular mass by ESI-MS make the hyphenated technique of a very powerful method for reinterpreting many ambiguous results about the PSI antenna system deduced from SDS-PAGE and/or immunoblotting experiments. The high reproducibility of the chromatographic profiles enables the identification of the antenna proteins in different plant species on the basis of chromatographic retention times without the need for ESI-MS analysis, which may be not available. The use of HPLC with spectrophotometric detection for separation and identification of the antenna proteins also facilitates the determination of the quantitative distribution of chlorophyll a/b binding present in PSI.
In the near future, we want to investigate the environmental effects on both photosystems separating the thylakoid membrane into grana and stroma, avoiding the tedious separation by Suc-gradient ultracentrifugation. In this way, it will be possible to determine the quantitative relationship between chlorophyll a/b binding proteins present in both photosystems. This knowledge is expected to shed light on the understanding of the molecular mechanisms underlying the physiological migration of PSII antenna proteins of higher plants under light stress. It is also possible to use the reversed-phase HPLC-ESI-MS method to collect information on the presence or absence of phosphorylation of the migrating proteins, as recently performed for PSII components (Vener et al., 2001; Gómez et al., 2002 ). The last aspect touches an important area, which is to understand the molecular mechanism by which the chloroplasts introduce posttranslational protein modifications to modulate the adaptation of the photosynthetic apparatus to environmental changes.
MATERIALS AND METHODS
Chemicals
Reagent-grade phosphoric acid, magnesium chloride, sodium chloride, silver nitrate, sodium carbonate, TFA, methanol, ethanol, and formamide as well as HPLC-grade water and acetonitrile were obtained from Carlo Erba (Milan). Acrylamide, N,N′-methylene-bis-acrylamide, and all other reagents for SDS-PAGE were purchased from Bio-Rad (Segrate, Italy). Suc, Tricine, Tris, n-octyl β-d glucopyranoside, DM, and MES were obtained from Sigma (Milan). Triton X-100 was purchased from Calbiochem (San Diego).
Isolation of Chloroplast Thylakoid and PSI Membranes by Suc Gradient Ultracentrifugation
Spinach (Spinacia oleracea) leaves from a local market, were homogenized in 0.4 m sorbitol, 0.1 m Tricine, pH 7.8, 10 mm NaCl, and 5 mm MgCl2. Isolation of the thylakoid was performed as described previously (Bassi et al., 1987). Freshly prepared thylakoid was resuspended at 1 mg mL−1 chlorophyll in distilled water and solubilized by DM. Purification of PSI was performed according to the method of Croce et al. (1998). In brief, thylakoids were solubilized by DM at a final concentration of 1%. After stirring for 20 min at 4°C, the sample was centrifuged for 10 min at 20,000g, and 6-mL aliquots of the supernatant were loaded onto 0.1 to 1 m Suc gradients, containing 5 mm Tricine, pH 7.8, and 0.03% (w/v) DM. After centrifugation for 42 h at 28,000 rpm in a SW28 rotor (Beckman Coulter, Fullerton, CA) at 4°C, three green bands were distinguishable. The lower most band, containing PSI, was diluted in 5 mm Tricine, pH 7.8, and centrifuged for 3 h at 70,000 rpm in an 80 Ti rotor (Beckman Coulter). The pellet was resuspended in 5 mm Tricine, pH 7.8, and 50 mm sorbitol, frozen in liquid nitrogen, and stored at −80°C.
PAGE
Denaturating SDS-PAGE was carried out in 8% to 25% (w/v) acrylamide gradient gels prepared according to Fling and Gregerson (1986). Resolving gels contain 0.75 m Tris-HCl, pH 8.85, and 0.1% (w/v) SDS, and the stacking gels contain 0.125 m Tris-HCl, pH 6.8, and 0.1% (w/v) SDS. Gels were run at ambient temperature (20°C) for 16 h at constant power of 2 W using 0.05 m Tris/0.19 m Gly buffer, containing 0.1% (w/v) SDS. Gels were fixed and stained for 2 h in a 5:1:4 (v/v) methanol-glacial acetic acid-water mixture, containing 0.1% (w/v) Coomassie Blue. For silver-staining, gels were fixed in 50% (v/v) methanol-water and 10% (v/v) ethanol-water solutions, stained with 0.1% (w/v) silver nitrate-water solution, and developed in 3.5% (w/v) aqueous sodium carbonate containing 0.05% (v/v) formamide.
HPLC and ESI-MS
Chromatographic separations were performed using a System Gold HPLC system, consisting of two solvent delivery pumps (model 126, Beckman Coulter), a UV detector (model 168, Beckman Coulter), and a fluorescence detector (model LC 240, PerkinElmer Life Sciences, Boston). The UV absorbance was monitored at 214 nm, whereas fluorescence emission was monitored at 330 nm after excitation at 280 nm. Samples were introduced onto the column by a sample injection valve (model 210A, Beckman Coulter) with either a 20- or a 50-μL sample loop. The proteins were separated in a reversed-phase column packed with 5-μm porous butyl silica particles (250- × 4.6-mm i.d., Vydac Protein C-4, The Separation Group, Hesperia, CA). The column was operated at a flow rate of 1 mL min−1 and room temperature. All solutions were filtered through a membrane filter (type FH 0.5-μm, Millipore, Milan) and degassed by sparging with helium before use.
The system used for HPLC-ESI-MS experiments consisted of a low-pressure gradient micropump (model Rheos 2000, Flux Instruments, Basel) controlled by a personal computer, a vacuum degasser (Knauer, Berlin), and an injector (model 7125, Rhodyne, Cotati, CA) with a 100-μL sample loop. ESI-MS was performed on a triple quadrupole mass spectrometer (TSQ 7000, Thermo Finnigan, San Jose, CA) or by ion trap Esquire 3000 plus (Bruker Daltonik, Germany). The 1 mL min−1 flow through the analytical column was split post-column, with 50 μL min−1 entering the mass spectrometer and 950 μL min−1 going to the UV and fluorescence detector. For analysis with pneumatically assisted ESI, an electrospray voltage of 3 to 4 kV and a nitrogen sheath gas flow were employed. The temperature of the heated capillary was set to 200°C. Protein mass spectra were recorded by scanning the first quadrupole, the scan range was 500 to 2,000 atomic mass units.
Computation of the Aliphatic Antenna Protein Parameters
The hydropathic profiles were calculated using the Kyte-Doolittle method on DNA sequences reported in the literature (Kyte and Doolittle, 1982).
ACKNOWLEDGMENTS
We thank Dr. Sonia Troiani for technical assistance and Dr. Andrea Kiehne (Bruker Daltonik, Germany) for the help in Arabidopsis MS measurements. We also acknowledge Dr. Jaqueline Scarpa for manuscript revision and Dr. Thomma Bart (Centre of Microbial and Plant Genetics, Katholieke University, Belgium) for his generous gift of Arabidopsis plants.
Footnotes
This work was supported by Ministero dell'Università e della Ricerca Scientifica Co-Finanziamento 2001, by the Austrian Science Fund (grant no. P–13442–PHY), and by the European Community INCO-COPERNICUS Project (no. IC15 CT98–0126).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.009803.
LITERATURE CITED
- Anandan S, Morishige DT, Thornber JP. Light-induced biogenesis of light-harvesting complex I (LHC I) during chloroplast development in barley (Hordeum vulgare): studies using cDNA clones of the 21-kilodalton and 20-kilodalton LHC I apoproteins. Plant Physiol. 1993;101:227–236. doi: 10.1104/pp.101.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S, Walters RG, Jansson S, Horton P. Acclimation of Arabidopsis thalianato the light environment: the existence of separate low light and high light responses. Planta. 2001;213:794–801. doi: 10.1007/s004250100556. [DOI] [PubMed] [Google Scholar]
- Bassi R, Hoyer-Hansen G, Barbato R, Giacometti GM, Simpson DJ. Chlorophyll-proteins of the photosystem II antenna system. J Biol Chem. 1987;262:13333–13341. [PubMed] [Google Scholar]
- Bassi R, Simpson D. Chlorophyll-protein complexes of barley photosystem I. Eur J Biochem. 1987;163:221–230. doi: 10.1111/j.1432-1033.1987.tb10791.x. [DOI] [PubMed] [Google Scholar]
- Boekema EJ, Jensen PE, Schlodder E, van Breemen JFL, van Roon H, Scheller HV, Dekker JP. Green plant photosystem I binds light-harvesting complex I on one side of the complex. Biochemistry. 2001;40:1029–1036. doi: 10.1021/bi0015358. [DOI] [PubMed] [Google Scholar]
- Brosche M, Fant C, Bergkvist SW, Strid H, Svensk A, Olsson O, Strid A. Molecular markers for UV-B stress in plants: alteration of the expression of four classes of genes in Pisum sativumand the formation of high molecular mass RNA adducts. Biochim Biophys Acta. 1999;1447:185–198. doi: 10.1016/s0167-4781(99)00154-2. [DOI] [PubMed] [Google Scholar]
- Corradini D, Huber CG, Timperio AM, Zolla L. Use of reversed-phase liquid chromatography with electrospray-mass spectrometric detection for the resolution and identification of the protein components of the photosystem II antenna system of higher plants. J Chromatogr A. 2000;886:111–121. doi: 10.1016/s0021-9673(00)00449-0. [DOI] [PubMed] [Google Scholar]
- Croce R, Zucchelli G, Garlaschi FM, Bassi R, Jennings RC. Excited state equilibration in the photosystem I-light-harvesting I complex: P700 is almost isoenergetic with its antenna. Biochemistry. 1996;35:8572–8579. doi: 10.1021/bi960214m. [DOI] [PubMed] [Google Scholar]
- Croce R, Zucchelli G, Garlaschi FM, Jennings RC. A thermal broadening study of the antenna chlorophylls in PSI-200, LHCI, and PSI core. Biochemistry. 1998;37:17355–17360. doi: 10.1021/bi9813227. [DOI] [PubMed] [Google Scholar]
- Damm I, Green BR. Separation of closely related intrinsic membrane polypeptides of the photosystem II light-harvesting complex (LHC II) by reversed-phase high-performance liquid chromatography on a poly(styrene-divinylbenzene) column. J Chromatogr A. 1994;664:33–38. doi: 10.1016/0021-9673(94)80625-X. [DOI] [PubMed] [Google Scholar]
- Dekker JP, van Roon H, Boekema EJ. Heptameric association of light-harvesting complex II trimers in partially solubilized photosystem II membranes. FEBS Lett. 1999;449:211–214. doi: 10.1016/s0014-5793(99)00442-1. [DOI] [PubMed] [Google Scholar]
- Dreyfuss BW, Thornber JP. Organization of the light-harvesting complex of photosystem I and its assembly during plastid development. Plant Physiol. 1994;106:841–848. doi: 10.1104/pp.106.3.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fling SP, Gregerson DS. Peptide and protein molecular weight determination by electrophoresis using a high-molarity Tris buffer system without urea. Anal Biochem. 1986;155:83–88. doi: 10.1016/0003-2697(86)90228-9. [DOI] [PubMed] [Google Scholar]
- Ganeteg U, Strand A, Gustafsson P, Jansson S. The properties of the chlorophyll a/b-binding proteins Lhca2 and Lhca3 studied in vivo using antisense inhibition. Plant Physiol. 2001;127:150–158. doi: 10.1104/pp.127.1.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez SM, Nishio JN, Faull KF, Whitelegge JP. The chloroplast grana proteome defined by intact mass measurements from liquid chromatography mass spectrometry. Mol Cell Proteomics. 2002;1:46–59. doi: 10.1074/mcp.m100007-mcp200. [DOI] [PubMed] [Google Scholar]
- Griffiths WJ. Nanospray mass spectrometry in protein and peptide chemistry. Exper Suppl (Basel) 2000;88:69–79. doi: 10.1007/978-3-0348-8458-7_5. [DOI] [PubMed] [Google Scholar]
- Hancock W, Apffel A, Chakel J, Hahnenberger K, Choudhary G, Traina JA, Pungor E. Integrated genomic/proteomic analysis. Anal Chem. 1999;71:742A–748A. doi: 10.1021/ac9907641. [DOI] [PubMed] [Google Scholar]
- Hoffman NE, Pichersky E, Malik VS, Castresana C, Ko K, Darr SC, Cashmore AR. A cDNA clone encoding a photosystem I protein with homology to photosystem II chlorophyll a/b-binding polypeptides. Proc Natl Acad Sci USA. 1987;84:8844–8848. doi: 10.1073/pnas.84.24.8844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber CG, Timperio AM, Zolla L. Isoforms of photosystem II antenna proteins in different plant species revealed by liquid chromatography-electrospray ionization mass spectrometry. J Biol Chem. 2001;276:45755–45761. doi: 10.1074/jbc.M106700200. [DOI] [PubMed] [Google Scholar]
- Ikeuchi M, Hirano A, Inoue Y. Correspondence of apoproteins of light-harvesting chlorophyll ab complexes associated with photosystem I to cabgenes: evidence for novel type IV apoprotein. Plant Cell Physiol. 1991;32:103–112. [Google Scholar]
- Jansson S. The light-harvesting chlorophyll a/b-binding proteins. Biochim Biophys Acta. 1994;1184:1–19. doi: 10.1016/0005-2728(94)90148-1. [DOI] [PubMed] [Google Scholar]
- Jansson S. A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci. 1999;4:236–240. doi: 10.1016/s1360-1385(99)01419-3. [DOI] [PubMed] [Google Scholar]
- Jansson S, Andersen B, Scheller HV. Nearest-neighbor analysis of higher-plant photosystem I holocomplex. Plant Physiol. 1996;112:409–420. doi: 10.1104/pp.112.1.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson S, Stefansson H, Nystrom U, Gustafsson P, Albertsson PA. Antenna protein composition of PSI and PSII in thylakoid sub-domains. Biochim Biophys Acta. 1997;1320:207–309. [Google Scholar]
- Jensen PE, Kristensen M, Hoff T, Lehmbeck J, Stummann BM, Henningsen KW. Identification of a single-copy gene encoding a type I chlorophyll a/b-binding polypeptide of photosystem I in Arabidopsis thaliana. Physiol Plant. 1992;84:561–567. [Google Scholar]
- Klimmek F (1999) Directed Submission to NCBI Data Bank, accession AAF23819
- Knoetzel J (1995) Directed Submission to NCBI Data Bank, accession CAA59049
- Knoetzel J, Svendsen I, Simpson DJ. Identification of the photosystem I antenna polypeptides in barley: isolation of three pigment-binding antenna complexes. Eur J Biochem. 1992;206:209–215. doi: 10.1111/j.1432-1033.1992.tb16918.x. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RFJ. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lam E, Ortiz W, Malkin R. Chlorophyll a/bproteins of photosystem I. FEBS Lett. 1984;168:10–14. [Google Scholar]
- Lee MC, Kim CS, Eun MY (1997) Directed Submission to NCBI Data Bank, accession AAB65793
- Li J, Assmann SM. Mass spectrometry: an essential tool in proteome analysis. Plant Physiol. 2000;123:807–809. doi: 10.1104/pp.123.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka M, Kano-Murakami Y, Yamamoto N. Nucleotide sequence of cDNA encoding the light-harvesting chlorophyll a/bbinding protein from maize. Nucleic Acids Res. 1987;15:6302. doi: 10.1093/nar/15.15.6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishige DT, Thornber JP. Correlation of apoproteins with the genes of the major chlorophyll a/b binding protein of photosystem II in Arabidopsis thaliana: confirmation for the presence of a third member of the LHC IIb gene family. FEBS Lett. 1991;293:183–187. doi: 10.1016/0014-5793(91)81182-8. [DOI] [PubMed] [Google Scholar]
- Mullet JE, Burke JJ, Arntzen CJ. Chlorophyll proteins of photosystem I. Plant Physiol. 1980;65:814–822. doi: 10.1104/pp.65.5.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomares R, Herrmann RG, Oelmuller R. Different blue-light requirement for the accumulation of transcripts from nuclear genes for thylakoid proteins in Nicotiana tabacum and Lycopersicon esculentum. J Photochem Photobiol B. 1991;11:151–162. doi: 10.1016/1011-1344(91)80257-i. [DOI] [PubMed] [Google Scholar]
- Peltier J, Friso G, Kalume DE, Roepstorff P, Nilsson F, Adamska I, van Wijk KJ. Proteomics of the chloroplast: systematic identification and targeting analysis of lumenal and peripheral thylakoid proteins. Plant Cell. 2000;12:319–341. doi: 10.1105/tpc.12.3.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichersky E, Brock TG, Nguyen D, Hoffman NE, Piechulla B, Tanksley SD, Green BR. A new member of the CAB gene family: structure, expression and chromosomal location of Cab-8, the tomato gene encoding the type III chlorophyll a/b-binding polypeptide of photosystem I. Plant Mol Biol. 1989;12:257–270. doi: 10.1007/BF00043203. [DOI] [PubMed] [Google Scholar]
- Pichersky E, Hoffman NE, Bernatzky R, Piechulla B, Tanksley SD, Cashmore AR. Molecular characterization and genetic mapping of DNA sequences encoding the type I chlorophyll a/b-binding polypeptide of photosystem I in Lycopersicon esculentum(tomato) Plant Mol Biol. 1987;9:205–216. doi: 10.1007/BF00166457. [DOI] [PubMed] [Google Scholar]
- Pichersky E, Tanksley SD, Piechulla B, Stayton MM, Dunsmuir P. Nucleotide sequence and chromosomal location of Cab-7, the tomato gene encoding the type II chlorophyll a/b-binding polypeptide of photosystem I. Plant Mol Biol. 1988;11:69–71. doi: 10.1007/BF00016015. [DOI] [PubMed] [Google Scholar]
- Piechulla B, Kellmann JW, Pichersky E, Schwartz E, Forster HH. Determination of steady-state mRNA levels of individual chlorophyll a/b binding protein genes of the tomato cabgene family. Mol Gen Genet. 1991;230:413–422. doi: 10.1007/BF00280298. [DOI] [PubMed] [Google Scholar]
- Preiss S, Peter GF, Anandan S, Thornber JP. The multiple pigment-proteins of the photosystem I antenna. Photochem Photobiol. 1993;57:152–157. [Google Scholar]
- Premstaller A, Oberacher H, Walcher W, Timperio A-M, Zolla L, Chervet J-P, Cavusoglu N, Van Dorsellaer A, Huber CG. High-performance liquid chromatography-electrospray ionization mass spectrometry using monolithic capillary columns for proteomic studies. Anal Chem. 2001;73:2390–2396. doi: 10.1021/ac010046q. [DOI] [PubMed] [Google Scholar]
- Scheller HV, Jensen PE, Haldrup A, Lunde C, Knoetzel J. Role of subunits in eukaryotic photosystem I. Biochim Biophys Acta. 2001;1507:41–60. doi: 10.1016/s0005-2728(01)00196-7. [DOI] [PubMed] [Google Scholar]
- Schwartz E, Pichersky E. Sequence of two tomato nuclear genes encoding chlorophyll a/b-binding proteins of CP24, a PSII antenna component. Plant Mol Biol. 1990;15:157–160. doi: 10.1007/BF00017734. [DOI] [PubMed] [Google Scholar]
- Schwartz E, Shen D, Aebersold R, McGrath JM, Pichersky E, Green BR. Nucleotide sequence and chromosomal location of Cab11 and Cab12, the genes for the fourth polypeptide of the photosystem I light-harvesting antenna (LHCI) FEBS Lett. 1991;280:229–234. doi: 10.1016/0014-5793(91)80299-i. [DOI] [PubMed] [Google Scholar]
- Sharma J, Panico M, Barber J, Morris HR. Characterization of the low molecular weight photosystem II reaction center subunits and their light-induced modifications by mass spectrometry. J Biol Chem. 1997a;272:3935–3943. doi: 10.1074/jbc.272.7.3935. [DOI] [PubMed] [Google Scholar]
- Sharma J, Panico M, Barber J, Morris HR. Purification and determination of intact molecular mass by electrospray ionization mass spectrometry of the photosystem II reaction center subunits. J Biol Chem. 1997b;272:33153–33157. doi: 10.1074/jbc.272.52.33153. [DOI] [PubMed] [Google Scholar]
- Sharma J, Panico M, Shipton CA, Nilsson F, Morris HR, Barber J. Primary structure characterization of the photosystem II D1 and D2 subunits. J Biol Chem. 1997c;272:33158–33166. doi: 10.1074/jbc.272.52.33158. [DOI] [PubMed] [Google Scholar]
- Sigrist M, Staehelin LA. Appearance of type 1, 2, and 3 light-harvesting complex II and light-harvesting complex I proteins during light-induced greening of barley (Hordeum vulgare) etioplasts. Plant Physiol. 1994;104:135–145. doi: 10.1104/pp.104.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stayton MM, Brosio P, Dunsmuir P. Characterization of a full-length petunia cDNA encoding a polypeptide of the light-harvesting complex associated with photosystem I. Plant Mol Biol. 1987;10:127–137. doi: 10.1007/BF00016150. [DOI] [PubMed] [Google Scholar]
- Steinback KE, Arntzen CJ, Bogorad L. The physical organization and genetic determinants of the photosynthetic apparatus of chloroplasts. In: Steinback KE, Bonitz S, Arntzen CJ, Bogorad L, editors. Molecular Biology of the Photosynthetic Apparatus. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1985. pp. 1–19. [Google Scholar]
- Vener AV, Harms A, Sussman MR, Vierstra RD. Mass spectrometric resolution of reversible protein phosphorylation in photosynthetic membranes of Arabidopsis thaliana. J Biol Chem. 2001;276:6959–6966. doi: 10.1074/jbc.M009394200. [DOI] [PubMed] [Google Scholar]
- Wall DB, Parus SJ, Lubman DM. Comparison of the capabilities of liquid isoelectric focusing-one-dimensional nonporous silica reversed-phase liquid chromatography-electrospray ionization time-of-flight mass spectrometry and liquid isoelectric focusing-one-dimensional sodium dodecyl sulfate polyacrylamide gel electrophoresis mass mapping for the analysis of intact protein molecular mass. J Chromatogr B. 2001;763:139–148. doi: 10.1016/s0378-4347(01)00382-6. [DOI] [PubMed] [Google Scholar]
- Wang J, Zhang H, Goodman HM. An Arabidopsiscab gene homologous to Cab-8 of tomato. Plant Physiol. 1994;104:297. doi: 10.1104/pp.104.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelegge JP, Gundersen CB, Faull KF. Electrospray-ionization mass spectrometry of intact intrinsic membrane proteins. Protein Sci. 1998;7:1423–1430. doi: 10.1002/pro.5560070619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheleva D, Sharma J, Panico M, Morris HR, Barber J. Isolation and characterization of monomeric and dimeric CP47-reaction center photosystem II complexes. J Biol Chem. 1998;273:16122–16127. doi: 10.1074/jbc.273.26.16122. [DOI] [PubMed] [Google Scholar]
- Zolla L, Bianchetti M, Timperio AM, Corradini D. Rapid resolution by reversed-phase high-performance liquid chromatography of the thylakoid membrane proteins of the photosystem II light-harvesting complex. J Chromatogr A. 1997;779:131–138. [Google Scholar]
- Zolla L, Rinalducci S, Timperio AM. Dynamics of light-harvesting protein stoichiometry adjustment by light quality in chloroplasts. In: Pandalay SG, editor. Recent Research Developments in Bioenergetics. Vol. 1. Trivandrum, India: Transworld Research Network; 2000. pp. 9–32. [Google Scholar]
- Zolla L, Timperio AM. High performance liquid chromatography-electrospray mass spectrometry for the simultaneous resolution and identification of intrinsic thylakoid membrane. Proteins. 2000;41:398–406. doi: 10.1002/1097-0134(20001115)41:3<398::aid-prot110>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- Zolla L, Timperio AM, Testi MG, Bianchetti M, Bassi R, Manera F, Corradini D. Isolation and characterization of chloroplast photosystem II antenna of spinach by reversed-phase liquid chromatography. Photosynth Res. 1999;61:281–290. [Google Scholar]