Abstract
The vertebrate RNA and ssDNA-binding protein Translin has been suggested to function in a variety of cellular processes, including DNA damage response, RNA transport, and translational control. The Translin-associated factor X (Trax) interacts with Translin, and Trax protein stability depends on the presence of Translin. To determine the function of the Drosophila Translin and Trax, we generated a translin null mutant and isolated a trax nonsense mutation. translin and trax single and double mutants are viable, fertile, and phenotypically normal. Meiotic recombination rates and chromosome segregation are also not affected in translin and trax mutants. In addition, we found no evidence for an increased sensitivity for DNA double-strand damage in embryos and developing larvae. Together with the lack of evidence for their involvement in DNA double-strand break checkpoints, this argues against a critical role for Translin and Trax in sensing or repairing such DNA damage. However, Drosophila translin is essential for stabilizing the Translin interaction partner Trax, a function that is surprisingly conserved throughout evolution. Conversely, trax is not essential for Translin stability as trax mutants exhibit normal levels of Translin protein.
THE vertebrate RNA and single-stranded DNA (ssDNA)-binding protein, Translin/testes-brain–RNA-binding protein (TB–RBP), binds to consensus sequences within recombination hotspot regions associated with chromosomal translocations in lymphoid malignancies and has been suspected of mediating chromosomal translocations in such tumors and in solid tumors (Aoki et al. 1995; Hosaka et al. 2000 and references therein).
Indications that Translin might be involved in sensing or repairing DNA damage were found while treating HeLa cells with DNA-damaging agents. After treatment with mitomycin C or etoposide, the amount of nuclear Translin greatly increased, suggesting a signaling pathway for the active nuclear transport of Translin that is initiated by exposure to DNA-damaging agents (Kasai et al. 1997). However, so far no evidence could be found for the direct involvement of Translin in DNA damage repair. Furthermore, exposure of mice embryonic fibroblasts (MEFs) from TB–RBP-deficient mice with DNA-damaging agents did not reveal differences between wild-type and TB–RBP null MEFs in terms of cell survival or number of DNA breaks and gaps (Yang et al. 2004).
Translin-associated factor X (Trax) was identified in a two-hybrid screen for Translin-interacting proteins and by immunoprecipitation experiments (Aoki et al. 1997; Wu et al. 1999). Trax shares conserved sequence similarities with Translin, and Trax orthologs have been found in virtually all species that also have Translin. The idea that Translin and Trax may play a role in cell proliferation is supported by a variety of studies that investigated the effect of Translin or Trax depletion in different cell types. MEFs cultured from TB–RBP-deficient mice grow more slowly than MEFs from heterozygous littermates (Yang et al. 2004). In addition, reduction of Translin or Trax by RNA interference slows cell growth rates of NIH3T3 cells, and reduction of Trax in HeLa cells slows growth rate and progression through G2/M (Yang et al. 2004; Yang and Hecht 2004). Consistent with this observation, overexpression of Translin leads to the opposite effect—acceleration of cell proliferation (Ishida et al. 2002).
Translin has also been identified as an RNA-binding protein that binds a variety of brain and testes RNAs. Accordingly, it is thought to play a role in the subcellular transport and/or translational control of its target RNAs in these tissues (Han et al. 1995a; Kobayashi et al. 1998; Morales et al. 1998; Muramatsu et al. 1998; Wu and Hecht 2000; Yang et al. 2003). Unlike Translin, Trax does not bind nucleic acids directly, but might be part of the RNA- or DNA-binding complex, thereby modulating the nucleic-acid-binding affinity of Translin (Chennathukuzhi et al. 2001; Finkenstadt et al. 2002; Gupta et al. 2005).
Our interest in mRNA localization, cell cycle regulation, and DNA damage response led to our analyzing the role of these evolutionarily conserved genes in Drosophila. Because the results from vertebrate Translin and Trax revealed little concrete evidence about the function of these proteins in vivo, we wanted to analyze Translin in an invertebrate model system in which molecular pathways are often less redundant and where it may be simpler to reveal Translin and Trax functions. Furthermore, the availability of mutants in the two genes would allow us to directly test whether translin (trsn) and Trax are functionally redundant for an essential process.
MATERIALS AND METHODS
Generation of fusion genes, mutants, and fly stocks:
Flies expressing C-terminal Translin and Trax GFP (or GFP derivatives) fusions were generated as described earlier (Pare and Suter 2000). A detailed description of cloning steps involved in generating constructs for transgenic flies is provided in the data supplement at http://www.genetics.org/supplemental/.
To create a translinnull mutant (Δtrsn), we mobilized an EPgy2 P element (pEY06981) that inserted in the last exon of the translin gene (Bellen et al. 2004). By bidirectional imprecise excision, a small deficiency was created that entirely removed the coding region of the gene as well as adjacent nontranscribed sequences. The neighboring genes CPTI and CG17765 are not affected by this deletion. The deficiencies Df(2R)stan2 and Df(3R)Exel6174, covering either translin or trax genomic loci were obtained from the Bloomington Stock Center (stock nos. 596 and 7653, respectively). Translin and trax mutant fly strains described in this article were of the following genotypes (unless otherwise noted) and were kept as stocks: traxW151* has been described as H813 by Schuetze et al. (2004), and hemizygous flies were analyzed as w; traxW151* ru st e ca/Df(3R)Exel6174, Δtrsn: w; Δtrsn/Df(2R)stan2 b pr, and Δtrsn;traxW151*: w; Δtrsn/Df(2R)stan2 b pr; traxW151* ru st e ca/Df(3R)Exel6174.
Generation of antibodies against Drosophila Translin and Trax:
The open reading frames of the translin and trax cDNAs were cloned into the pGEX-5X-1 (GE Healthcare) expression vector to produce GST-tagged fusion proteins, and the induced fusion proteins were purified using the GST fusion purification kit (GE Healthcare). Short C-terminal peptides of the Translin and Trax proteins were also synthesized (Sheldon Biotechnology Centre, McGill University, Montreal) and used to immunize rabbits. Anti-Translin sera were affinity purified against bacterially expressed full-length Translin–maltose binding-protein (MBP) and pMAL-vector (New England Biolabs, Beverly, MA) and conjugated to a cyanogen bromide-activated Sepharose column (GE Healthcare). Anti-Trax sera were purified against full-length Trax-MBP fusion protein immobilized on nitrocellulose membranes.
Immunostainings:
Immunohistochemical stainings were done as described earlier (Suter and Steward 1991). Rabbit polyclonal antisera against full-length Translin and against a C-terminal peptide of Trax were used at a dilution of 1:400 and 1:200, respectively. The secondary OregonGreen488-conjugated anti-rabbit antibody (Molecular Probes, Eugene, OR) was used at a dilution of 1:2000. During the final washing steps, nuclei and actin filaments were stained for 20 min with 2.5 μg/ml Hoechst 33258 (Molecular Probes) and 1 unit/ml rhodamine-conjugated phalloidin (Molecular Probes), respectively. The ovaries were embedded in 60% glycerol and analyzed by confocal microscopy using a Leica TCS-SP2.
Western blots:
Ovary and testis extracts were prepared by dissecting ovaries and testes in Ringer's solution and freezing them directly in 2× SDS sample buffer. Ovaries and testes were homogenized by vortexing and boiling for 10 min. Protein samples were separated on 12% SDS–PAGE gels and transferred onto nitrocellulose membranes. To detect three different proteins simultaneously on the blots, the membranes were cut according to the molecular weights of the proteins to be detected and probed with the appropriate antibodies. Rabbit polyclonal antisera against full-length Translin protein and the C-terminal peptide of Trax were used at a dilution of 1:750 and 1:250, respectively. Horseradish-conjugated anti-rabbit secondary antibody (GE Healthcare) was used at a dilution of 1:5000. The blots were probed with ECL reagents (GE Healthcare) for 1 min and the chemiluminescence was detected with ECL films.
In situ hybridization:
For in situ hybridizations, parts of wingless and hairy cDNAs corresponding to coding regions and 5′-UTR were cloned into pBS(KS+), linearized with SacI, and transcribed with T3 polymerase (Stratagene, La Jolla, CA) in the presence of digoxygenin-rUTP (Roche) to generate digoxygenin-labeled antisense RNA probes. In situ hybridizations on embryos were in principle done according to Hughes and Krause (1999) with noted modifications. After hybridization, embryos were treated with RNAse A and T1 in 2× SSC for 15 min at 37°. The buffer was then changed in several steps from SSC to 1× maleic acid buffer (MAB) and embryos were blocked in 1× MAB containing 20% donkey serum (Chemicon) and 2% Roche blocking reagent (Roche). The digoxygenin-labeled probes were detected with a sheep-antidigoxygenin (Roche) antibody and Cy3-labeled donkey-anti-sheep F(ab′)2 fragments of IgG (Jackson ImmunoResearch Laboratories, West Grove, PA). During the final washing steps the DNA-staining dye Hoechst 33258 (Molecular Probes) was applied. The embryos were embedded in 60% glycerol and analyzed by confocal microscopy.
Tests for recombination frequency and chromosome nondisjunction:
To determine the recombination frequency of Trax-deficient flies, females of the genotypes b pr cn bw/+ + + +; traxW151*/Df(3R)Exel6174 and b pr cn bw/+ + + +; Df(3R)Exel6174/TM6B Tb ca (control) were mated to b pr cn bw males and the progeny were scored for recombination events in the b-pr, pr-cn, and cn-bw intervals independently. Flies with cn and bw markers have white eyes, which mask the pr phenotype. Therefore, the recombined chromosomes + pr cn bw and + + cn bw cannot be discriminated from each other in this assay. They were scored as b-pr and also as pr-cn crossovers, so these recombination frequencies appear slightly higher than they actually were. Similarly, one result of a double crossover, namely b + cn bw, was scored as parental. However, because the distances between b-pr and pr-cn are small (6 and 3 MU, respectively), such events are rare and barely influence the overall result.
To determine the frequency of female chromosome nondisjunction in translin and trax mutant flies, females of the genotypes w/w; Δtrsn/Df(2R)stan2 b pr, w/w; traxW151*/Df(3R)Exel6174, and w/w; Df(3R)Exel6174/TM6B Tb ca (control) were crossed to wild-type males. Female and male progeny were scored for exceptional white-eyed w/w/Y (XXY) females and red-eyed +/0 (XO) males.
To determine the frequency of male chromosome nondisjunction in translin and trax mutant flies, males of the genotypes w/BsY; Δtrsn/Df(2R)stan2 b pr, w/BsY; Δtrsn/Sp or Df(2R)stan2 b pr/CyO (control), +/BsY; traxW151*/Df(3R)Exel6174, and +/BsY; Df(3R)Exel6174/TM6B Tb ca (control) were crossed to wild-type or w; +; + females, respectively. Progeny were scored for exceptional Bar of Stone +/w/BsY females and red-eyed +/0 (for translin mutants) or white-eyed w/0 (for trax mutants) males, respectively.
Irradiation experiments:
Fly embryos were collected on apple juice plates at 25° and aged to reach the desired cell cycle stages. Then they were irradiated with a dose of 6 Gy (half-lethal dose), transferred onto fresh standard corn food, and incubated at 25°. For survival tests, pupae and surviving flies were scored. Tests for the embryonic cell cycle checkpoint were done as described in Masrouha et al. (2003).
RESULTS
Structure and expression of Drosophila Translin and Trax:
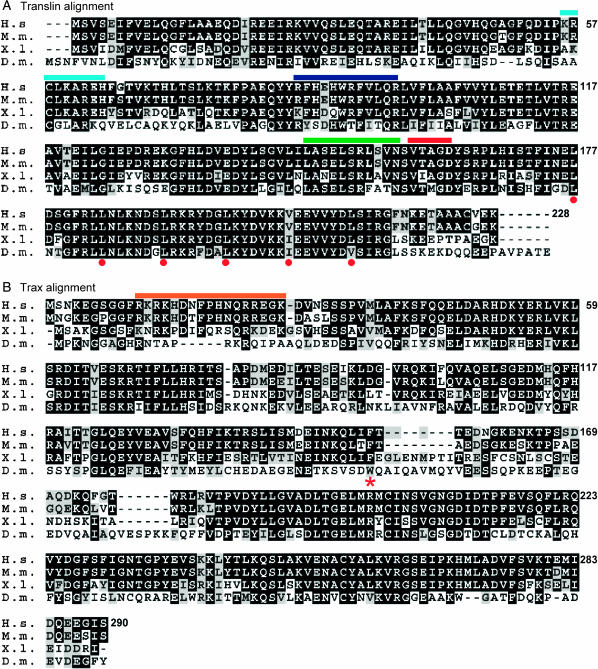
The amino acid sequences of human, mouse, and Xenopus Translin and Trax were aligned with the sequences from Drosophila (Figure 1, A and B). While the vertebrate Translin proteins share identities between 81% (human and Xenopus sequences) and 98% (human and mouse sequences), the Drosophila Translin protein is still well conserved and shares 52% identity with the vertebrate one. Note that the degree of identity between the Drosophila and vertebrate proteins is higher in the C-terminal part compared to the N-terminal half of the protein, except for the extreme C-terminal part of the Drosophila protein, which differs in sequence and length from the vertebrate proteins. The C-terminal half contains the putative leucine zipper domain (depicted in Figure 1), which is required for Translin homodimerization and probably also for nucleic acid binding (Wu et al. 1998; Aoki et al. 1999). Both the nuclear export signal (green) and the putative GTP-binding site (red) seem to be conserved in the Drosophila protein, although the GTP-binding activity of the Drosophila Translin seems to be lower than that of one of the mammalian orthologs (Sengupta et al. 2006). In contrast, the two basic regions (light and dark blue in Figure 1) that have been shown to be required for nucleic acid binding in the mouse protein (Chennathukuzhi et al. 2001) are less conserved in Drosophila Translin and a recent publication (Sengupta et al. 2006) found that, in contrast to the mammalian orthologs, Drosophila Translin does not exhibit ssDNA- or RNA-binding affinity under their experimental conditions.
Figure 1.—
Translin and Trax amino acid alignments. The amino acid sequences of human (NP_004613), mouse (NP_035780), Xenopus (AAF65620), and Drosophila (AAM50730). (A) Translin proteins were aligned using Multalin (Corpet 1988) and edited using the public-domain program BOXSHADE. The human (NP_005990), mouse (NP_058605), and Xenopus (AAH54180) (B) Trax amino acid sequences were similarly aligned with the Drosophila Trax protein sequence (DQ448818). The following protein residues have been marked in the alignment: nuclear export signal (NES) (green), GTP-binding site (red), basic regions in Translin (light and dark blue), and NLS (orange). Leucine residues of the Translin leucine zipper are marked by red dots. The tryptophane 151, which is replaced by a stop codon in the traxW151* nonsense mutation, is marked by an asterisk. Amino acid identities given in the text were revealed by Blast 2 sequences (Tatusova and Madden 1999).
Compared to Translin, the vertebrate Trax proteins are less conserved with identities ranging from 64% (between human and Xenopus sequences) to 90% (human and mouse sequences). We isolated a Trax cDNA from an ovary cDNA library (Larochelle and Suter 1995). Interestingly, this clone differs from one isolated from adult heads (GH01922; Rubin et al. 2000) by the removal of an intronic sequence and by a usage of a different polyadenylation site. Furthermore, the annotated fly genome also lists a transcript in which this intron is partially removed, but with a different splice acceptor site (CG5063-PB). Because of the splice differences, the corresponding predicted TRAX proteins also differ in size and sequence. The ovarian cDNA described here encodes the largest protein and the one with the best match to the human sequence (Figure 1B). This Drosophila Trax amino acid sequence shares 36% identity with human Trax. Vertebrate Trax contains a bipartite nuclear localization sequence (NLS), which maps to amino acid sequences 11–27 (orange in Figure 1) and was shown to be required for nuclear localization of the mouse protein (Cho et al. 2004). This feature is not conserved in Drosophila Trax, and NucPred (Heddad et al. 2004) and PredictNLS programs (Cokol et al. 2000) also did not reveal any NLS in Drosophila Trax.
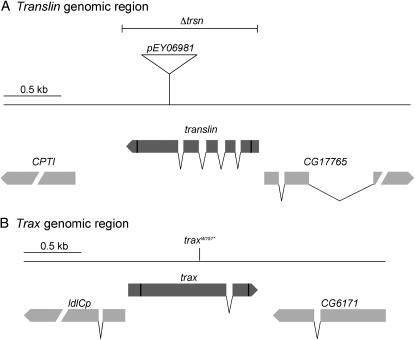
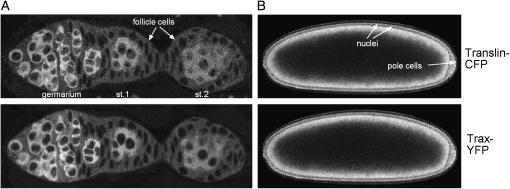
The Drosophila translin and trax genes are located on the second and third chromosome, respectively. Schematics of the genomic organization of the translin and trax regions are depicted in Figure 2, A and B. To analyze the in vivo expression and localization of Translin and Trax, we generated C-terminal Translin–cyan fluorescent protein (CFP) and Trax–yellow fluorescent protein (YFP) fusion constructs. Since vertebrate Translins have been implicated in the functioning of RNA localization processes, we focused our expression analysis on ovaries and young embryos, in which well-documented transport of RNAs and proteins takes place. Analysis of Translin and Trax protein distribution revealed that they are expressed during oogenesis with the highest rates in the germarium (Figure 3A). The primarily cytoplasmic Translin and Trax signal decreases in the egg chambers during later oogenesis stages. But in blastoderm-stage embryos relatively high levels of Translin and Trax can be detected in the cytoplasm (Figure 3B).
Figure 2.—
Genomic organization of (A) translin (chromosome 2R; 47A11) and (B) trax (chromosome 3R; 88F1) regions. The location of the pEY06981 P-element insertion and the region deleted in the translinnull excision mutant (Δtrsn) are depicted. Note that the excision of the P element was bidirectional. The position of the traxW151* nonsense point mutation is depicted in the schematic.
Figure 3.—
In vivo localization of Translin-CFP and Trax-YFP in (A) early oogenesis and in (B) syncytial blastoderm-stage embryos. Germarium, stage 1 and 2 egg chambers, and somatic follicle cells are marked. Nuclei and pole cells are indicated by arrows. (Top) Translin–CFP. (Bottom) Trax–YFP. Ovaries were dissected in halocarbon oil and immediately analyzed by confocal microscopy (left). Embryos were dechorionated before imaging (right).
Functional analysis of Translin and Trax:
Strain pEY06981 carries a P-element insertion in the 3′ exon of translin (Bellen et al. 2004). As judged from Western blotting and immunohistochemistry using polyclonal antisera against the full-length Drosophila Translin, these flies do not express detectable amounts of Translin. Nevertheless, we created a true translin-deficient mutant by mobilizing this P element and isolating an imprecise P-element excision that created a short deficiency that removed the entire translin gene (Δtrsn Figure 2). The analysis of the translin P-element insertion and the translin null allele revealed that homo- or hemizygous translin mutant flies are viable and fertile and exhibit no obvious mutant phenotype. Under standard laboratory conditions Drosophila translin is thus dispensable for viability and fertility.
Schuetze et al. (2004) mapped the H813 female-sterile mutation from the Tübingen collection (Tearle and Nüsslein-Volhard 1987) to the trax region. We therefore sequenced the trax allele in this mutant and found a point mutation that generates an in-frame stop codon at amino acid position 151 (traxW151*, Figures 1 and 2). No Trax protein can be detected in homo- or hemizygous trax mutant flies by Western blot or immunostaining using polyclonal antisera against full-length Trax (data not shown). Semiquantitative RT–PCR analysis revealed very low trax mRNA levels in mutant flies, suggesting that the premature stop codon leads to the at least partial degradation of the mutant mRNA (data not shown). However, the female-sterile effect is not due to the absence of functional Trax protein, as hemizygous trax mutant flies are fertile in contrast to homozygous females. Similar to the translin mutant, the analysis of the hemizygous trax point mutation revealed that trax is also dispensable under standard laboratory conditions. Since Drosophila Trax shares homology with Translin (31% identity over large parts, 22% over the entire protein; data not shown), these two proteins may be functionally redundant and replace each other in single mutants. However, the analysis of the Δtrsn;traxW151* double mutant revealed that these are viable and fertile, indicating that the two proteins do not have any essential role for which they function redundantly.
Drosophila translin functions to stabilize Trax:
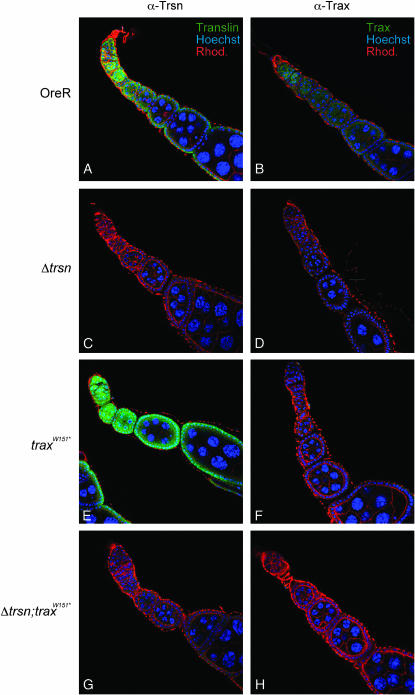
We next analyzed the distribution of Translin and Trax in ovaries from wild-type and translin and trax mutants using antibodies against these proteins (Figure 4). In wild-type ovaries, Translin can be detected throughout oogenesis with highest expression levels in the germarium and early stage egg chambers, similar to what has been observed for the Translin–CFP fusion protein (compare Figures 3A and 4A). During later stages, Translin staining can still be detected in the follicle cells that surround the ovariole, whereas the staining in the germline decreases. Similar to the Translin–CFP fusion protein, the endogenous protein seems to reside mainly in the cytoplasm and only weak nuclear staining can be detected. We note that the Translin signal is not enriched in the developing oocyte. A similar distribution is apparent for Trax (Figure 4B). In ovaries from translin-deficient flies (Δtrsn), no specific staining for Translin can be detected anymore, indicating that our antibody is very specific in immunostainings (Figure 4C). In addition to the loss of Translin staining, Trax staining also disappeared in Δtrsn ovaries (Figure 4D). It thus appears that Drosophila translin is essential in obtaining normal Trax levels and loss of Translin protein leads to loss of Trax proteins. Interestingly, this function of translin has been conserved through evolution, because similar results have also been reported for Translin knockout mice (Chennathukuzhi et al. 2003). In contrast to loss of Drosophila translin, loss of Drosophila Trax protein does not lead to a reduction of Translin protein (Figure 4, E and F). Neither Translin nor Trax can be detected by immunofluorescence in Δtrsn;traxW151* double-mutant ovaries (Figure 4, G and H).
Figure 4.—
Translin and Trax immunostainings with ovaries. Ovaries from wild-type, Δtrsn, traxW151*, and Δtrsn; traxW151* mutant females (indicated on the left) were subjected to immunostainings with either α-Translin or α-Trax antibodies (shown in green and indicated at the top). The nuclei were stained with Hoechst (blue) and actin filaments are visualized by rhodamine–phalloidin staining (red).
Other functions of Drosophila translin and trax are less evident as suggested by the viability and fertility of the mutants. Loss of Translin (and Trax) proteins does not seem to have an effect on oogenesis. Ovarioles from mutant flies look normal and the egg-laying rate of Translin-deficient females is not reduced compared to that of wild-type females. This result contrasts with results obtained in mice, where TB–RBP knockout females have been reported to be subfertile with reduced litter sizes (Chennathukuzhi et al. 2003).
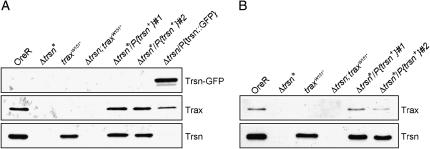
Using Western blots, we compared levels of Translin and Trax proteins in wild-type ovaries and testes with the ones in Translin and/or Trax-deficient tissues (Figure 5). Translin and Trax can be detected in ovaries (Figure 5A) and testes from wild-type flies (Figure 5B). No Translin can be detected in ovaries and testes from Translin-deficient flies and, similar to what has been observed in the immunostainings, Trax levels are lost or greatly reduced in these ovaries and testes. Translin levels are unaffected in ovaries from traxW151* mutants. Therefore, as opposed to the requirement for translin for maintaining normal levels of Trax, Translin protein levels do not depend on trax. Neither Translin nor Trax can be detected in ovaries or testes from Δtrsn;traxW151* double mutants. The low Trax protein levels found in translin mutants can be rescued by reintroducing a translin+ construct into this mutant background (Figure 5, A and B). Trax levels can also be rescued to a certain extent by reintroducing a Translin-GFP fusion construct that is expressed under the control of the translin promoter (Figure 5A). This would indicate that Translin–GFP is at least partially functional. However, a very weak Translin band can be detected, which might correspond to a degradation product of the Translin–GFP fusion. Therefore, it cannot be ruled out that part of the Trax stabilization might be due to the presence of this wild-type size degradation product.
Figure 5.—
Detection of Translin and Trax in ovaries and testes by Western blot. Ovarian (A) and testis (B) proteins from wild-type, Δtrsn*, traxW151* and Δtrsn; traxW151* mutant flies as well as Δtrsn*/P{strsn+}#1 and 2 and Δtrsn/P{trsn∷GFP} flies were separated by SDS–PAGE and transferred to membranes. These were then cut into three pieces according to the expected sizes of the proteins to be detected. Individual sections were probed with either α-Translin or α-Trax antibodies. An asterisk indicates that flies were homozygous for the Δtrsn chromosome.
Translin is not required for the apical localization of hairy and wingless transcripts during embryogenesis:
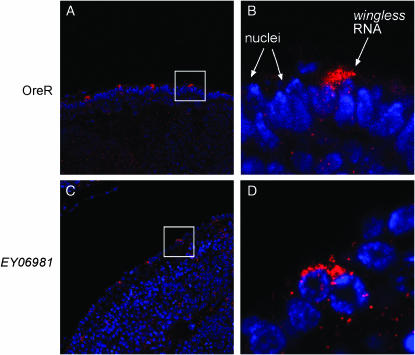
Human and mouse Translin proteins were found to associate with a variety of different so-called Y- and H-element-containing RNAs from brain and testes, including α-CAMKII, MBP, Tau, and Protamine mRNAs as well as noncoding RNAs such as BC1 (Han et al. 1995a,b; Kobayashi et al. 1998; Muramatsu et al. 1998; Severt et al. 1999). Translin has been suggested as being involved in the subcellular localization and translational regulation of its target RNAs (Morales et al. 1998; Severt et al. 1999; Yang et al. 2003). We set out to analyze this process in embryos, which contain relatively large amounts of Translin and Trax. During Drosophila embryogenesis, pair-rule and wingless transcripts become specifically localized to the apical cytoplasm above the nuclei (Figure 6, A and B). Apical wingless localization is not affected in trsnpEY06981 embryos derived from mutant mothers (Figure 6, C and D). Similarly, the proper apical localization of hairy mRNA also is not affected in Translin-deficient embryos (data not shown). translin is thus not essential for the proper apical localization of hairy and wingless transcripts during embryogenesis.
Figure 6.—
Apical localization of wingless transcripts in wild-type and translin mutant embryos. Localization of endogenous wingless transcripts of wild-type (A and B) and pEY06981(C and D) embryos was detected by in situ hybridization and is shown in red. The nuclei are labeled by Hoechst staining and are shown in blue. B and D are magnifications of the depicted areas in A and C, respectively.
Meiotic recombination and DNA damage response:
Translin and Trax have been implicated in playing a role during DNA double-strand break repair (DSBR) either by nonhomologous end joining (NHEJ) or homologous recombination (HR). In addition to the more general repair of double-strand breaks (DSBs), homologous recombination is also utilized during meiotic recombination. To test whether Drosophila Translin and Trax are involved in homologous recombination during meiosis, we analyzed whether the recombination frequency is reduced in translin or trax mutant flies. For these experiments, multiply marked chromosomes were used and the meiotic recombination rate of these markers was determined by visual inspection of the phenotype of the offspring. The calculated recombination rates for mutant flies were compared to rates of control flies and to published map distances (Table 1). While the recombination frequency between b and pr is almost identical, the number of crossovers between pr and cn is slightly increased in trax mutant females compared to wild type controls. The observed recombination frequency between cn and bw does not take into consideration double recombination events and possible effects of the third chromosomal balancer that could explain the observed differences at least in part. Because the recombination frequencies in the b-pr and pr-cn intervals are similar in trax mutants and control flies and in agreement with the published map distances, we conclude that trax does not have a critical function in meiotic recombination. Similar results were obtained for translin mutants (data not shown). Therefore translin and trax are not essential for meiotic recombination in Drosophila.
TABLE 1.
Recombination frequencies in trax mutant females
| Recombinantsb
|
Frequencyc
|
||||||
|---|---|---|---|---|---|---|---|
| Maternal genotype | Na total | b-pr | pr-cn | cn-bw | b-pr | pr-cn | cn-bw |
| b pr cn bw/+ + + +; traxW151*/Df(3R)Exel6174 | 457 | 29 | 17 | 161 | 6.35 | 3.72 | 35.23 |
| b pr cn bw/+ + + +; Df(3R)Exel6174/TM6B (control) | 550 | 36 | 12 | 246 | 6.55 | 2.18 | 44.73 |
| Expected recombination frequencies | 6 | 3 | 47 | ||||
TM6B contains Tb, ca.
N, the total number of progeny scored from a cross of females of the genotype listed and b pr cn bw males.
Recombinants of the three intervals were scored independently and scored recombination events or frequencies include all recombinant progeny in a certain interval whether or not a second recombination event was scored in a different interval.
The recombination frequency for each interval was calculated as follows: (recombinants) × 100/N. The expected recombination frequencies as deduced from the published genetic positions (Lindsley and Zimm 1992) are shown below.
As many genes involved in meiotic recombination and double-strand break repair in Drosophila are also required for proper chromosome segregation, mutations in these genes cause increased meiotic chromosome nondisjunctions (McKim et al. 2002). We therefore also tested whether the rate of chromosome nondisjunction is increased in translin and trax mutants. We analyzed the segregation of the sex chromosomes in a standard assay, which allowed us to score the exceptional females and males that were produced by such chromosome nondisjunction events. No differences in the rate of X chromosome nondisjunction were observed between control flies and translin or trax mutant females or males (Table 2). Furthermore, the overall frequency of nondisjunction events is comparable to the published wild-type nondisjunction frequency (Ghabrial et al. 1998; McKim et al. 2002 and references therein).
TABLE 2.
Female and male nondisjunction
| Maternal genotype | Normal progeny (XX and XY) | Nondisjunction progeny (XXY and X0) | % nondisjunctionc |
|---|---|---|---|
| Female nondisjunctiona | |||
| w/w; Δtrsn/Df(2R)stan2 b pr | 3591 (1819 + 1772) | 3 (0 + 3) | 0.08 |
| w/w; traxW151*/Df(3R)Exel6174 | 3037 (1709 + 1328) | 3 (1 + 2) | 0.1 |
| w/w; Df(3R)Exel6174/TM6B Tb ca (control) | 2753 (1474 + 1279) | 3 (2 + 1) | 0.11 |
| Paternal genotype | Normal progeny (XX and XBSY) | Nondisjunction progeny (XXBSY and X0) | % nondisjunctionc |
| Male nondisjunctionb | |||
| w/BsY; Δtrsn/Df(2R)stan2 b pr | 1778 (841 + 937) | 3 (0 + 3) | 0.17 |
| w/BsY; Δtrsn/Sp or Df(2R)stan2 b pr/CyO (control) | 2057 (1043 + 1014) | 4 (1 + 3) | 0.19 |
| +/BsY; traxW151*/Df(3R)Exel6174 | 3658 (1960 + 1698) | 0 (0 + 0) | — |
| +/BsY; Df(3R)Exel6174/TM6B Tb ca (control) | 2302 (1387 + 915) | 1 (0 + 1) | 0.04 |
For female nondisjunction, progeny were scored from crosses of wild-type males with females of the genotype listed. Normal chromosome segregation in females gives rise to w/+ females and w/Y males. Chromosome nondisjunction gives rise to exceptional white-eyed w/w/Y (XXY) females and red-eyed +/0 (X0) males that can be distinguished from their wild-type siblings.
For male nondisjunction, progeny were scored from crosses of wild-type (for translin) or w;+;+ (for trax) females with males of the genotype listed. Normal chromosome segregation in males containing the BSY-chromosome gives rise to w/+ females and +/BSY or w/BSY males, respectively. Male chromosome nondisjunction gives rise to exceptional Bar of Stone w/+/BSY females and red-eyed (+/0) or white-eyed (w/0) males which can be distinguished from their Bar of Stone siblings.
Female and male nondisjunction was counted as follows: (nondisjunction progeny) × 100/total progeny.
To test whether Translin and/or Trax function in a double-strand DNA damage pathway in Drosophila, we determined the survival rates of translin and trax double mutants with and without γ-irradiation during embryogenesis. Translin, trax, and translin;trax double mutants do not seem to be more sensitive to γ-irradiation as similar survival rates were observed in treated and untreated animals (data not shown). Similarly, embryos lacking Trsn and Trax (Δtrsn;traxW151* double mutants derived from mutant parents) display a normal cell cycle arrest during embryonic nuclear cycle 14 when double-strand breaks are induced by γ-irradiation, suggesting that Translin and Trax are dispensable for this DNA damage checkpoint. In addition, no nuclear transfer or enrichment of Translin–CFP and Trax–YFP fusion proteins was observed after embryo irradiation (data not shown).
DISCUSSION
Differences between Drosophila and mammalian Translin/Trax protein sequences:
Sequence alignments show that Drosophila Translin shares 52% identity with human and mouse Translins (Figure 1). The degree of identity between Drosophila and vertebrate proteins is lower in the N-terminal part of the proteins. This includes the two basic regions required for nucleic acid binding (Chennathukuzhi et al. 2001). Thus, the weak conservation of the two basic regions in Drosophila might be the reason why no RNA or DNA-binding activity has been revealed for the Drosophila Translin in gel-shift assays (Sengupta et al. 2006). However, even though the Schizosaccaromyces pombe Translin is less conserved than the Drosophila ortholog (33% identity with human Translin), it still binds single-stranded oligodeoxynucleotide and oligoribonucleotide probes (Laufman et al. 2005). In addition, in contrast to human Translin, the fission yeast protein has much higher affinities for RNA sequences than for homologous DNA sequences. Since the two groups used different RNA and ssDNA probes in gel-shift assays, it cannot be ruled out that lack of nucleic acid binding of Drosophila Translin simply reflects that Drosophila Translin recognizes RNA and DNA sequences different from the ones used.
The Translin nuclear export signal (NES), which resides in the more C-terminal part of the mammalian protein (Chennathukuzhi et al. 2001), is fairly well conserved in the Drosophila protein, suggesting that Translin might shuttle between the nucleus and the cytoplasm. In mammalian tissue culture cells, the subcellular localization of Translin and Trax seems to be interdependent and determined by their relative ratio (Cho et al. 2004). However, this shuttling interdependency is probably not conserved as Drosophila Trax does not seem to exhibit a functional NLS.
Post-transcriptional downregulation of Trax in the absence of Translin:
In this study we showed that Translin is required to maintain normal Trax levels in Drosophila. Similar results were obtained previously in TB–RBP-deficient mice and embryonic fibroblasts from such mice (Chennathukuzhi et al. 2003; Yang et al. 2004). Since truncated Translin proteins, which do not homo- or heterodimerize, do not stabilize Trax, this function is probably directly dependent on protein interaction (Yang et al. 2004). There is evidence that excess Trax is ubiquitinated and degraded in the proteasome (Yang et al. 2004). In contrast, as we showed here, Translin stability is independent of trax and this feature may also be conserved over evolution as it is also the case in S. pombe (Laufman et al. 2005).
Role of translin and trax in RNA transport and localization:
A role for Translin in RNA transport was suggested from in situ hybridization and EM–immunolocalization studies, which showed the colocalization of Translin with certain RNAs in the nuclei as well as in the cytoplasm and in intercellular bridges that interconnect developing male germ cells (Morales et al. 2002). Similarly, during Drosophila oogenesis, large amounts of RNAs, proteins, and other materials are synthesized in the nurse cells and transported through intercellular bridges, called ring canals, into the oocyte, which is (almost) transcriptionally quiescent and depends on the RNAs supplied by the nurse cells. These similarities in intercellular transport events in mouse testes and Drosophila egg chambers suggest that Translin might also be involved in RNA transport processes in Drosophila. However, the highest expression rates of Translin (and Trax) have been observed during the earliest stages of oogenesis, in the germarium, and the amount of Translin decreases continuously during the development of the egg chamber and is comparably low in stages in which the nurse-cell-to-oocyte transport is at its peak. Because RNA transport and localization processes also take place in Drosophila embryos and as these contain relatively large amounts of Translin and Trax, we focused our analysis on this developmental phase. However, the translin mutation EY06981 did not affect apical localization of wingless and hairy transcripts and we therefore concluded that translin is not essential for the proper localization of these transcripts.
Role of Translin/Trax in DSBR, meiotic HR, and double-strand DNA damage response:
Translin was initially shown to specifically bind to consensus sequences residing at the breakpoint regions of chromosomal translocations that are associated with lymphoid malignancies and solid tumors. Thus it was hypothesized that Translin might be involved in mediating chromosomal translocations. As translocations involve the union of different chromosomes, it is thought that they arose from DNA DSBs that had been substrates for the cellular DNA repair machinery (Ferguson and Alt 2001). Translin binds preferentially to single-stranded or tailed duplex DNA structures, which also occur during double-strand DNA breakage events (Sengupta and Rao 2002). Therefore, Translin might function in NHEJ, a double-strand break repair pathway which is also crucial to V(D)J recombination in developing lymphocytes. However, mice deficient for the murine Translin homolog TB–RBP show a normal development of B- and T-cells, suggesting that Translin has no essential function in NHEJ processes required for immunoglobulin or TcR rearrangements (Chennathukuzhi et al. 2003). In addition, TB–RBP-deficient MEFs do not exhibit an increased sensitivity for DNA-damaging agents or irradiation (Yang et al. 2004). Similarly, Drosophila translin and trax single and double mutants also did not show an increased sensitivity for DNA double-strand breaks. In addition, S. pombe translin and trax single or double mutants also did not exhibit an increased sensitivity for different DNA-damaging drugs (M. Claußen and B. Suter, unpublished observations).
A second cellular DNA double-strand break repair pathway involves the HR of free DNA ends with intact homologous sequences. As this repair process is also initiated upon the occurrence of double-strand breaks or two nearby single-strand breaks, which often contain single-stranded DNA overhangs, Translin might be involved in homologous recombination. In addition to DSBR, HR is also utilized during meiotic recombination. The fact that Drosophila translin and trax are not essential for meiotic recombination and chromosomal segregation during meiosis argues against these hypotheses.
Very recently, Stein et al. (2006) reported alterations in learning and memory, locomotor activity, and anxiety-related behavior in translin knockout mice. Similar behavioral phenotypes have been reported for mice lacking the fragile X mental retardation protein (FMRP), suggesting that Translin and FMRP may have similar functions in neurons (Stein et al. 2006 and references therein). Similarly, mutations in the Drosophila homolog dfmr1 have been implicated in affecting locomotor as well as courtship behavior (Dockendorff et al. 2002). Behavioral analysis of translin and trax mutant flies thus might show why translin and trax remained conserved during evolution even though they have no essential function under laboratory conditions. On the other hand, Translin and Trax might also be redundant with a third gene in an essential pathway, and genetic screens in a translin and trax mutant background might then reveal the factors that functionally replace Translin and Trax.
Acknowledgments
This work was supported by funds from the Swiss National Science Foundation and the Kanton Bern.
Sequence data from this article have been deposited with the EMBL/GenBank Data Libraries under accession no. DQ448818.
References
- Aoki, K., K. Suzuki, T. Sugano, T. Tasaka, K. Nakahara et al., 1995. A novel gene, Translin, encodes a recombination hotspot binding protein associated with chromosomal translocations. Nat. Genet. 10: 167–174. [DOI] [PubMed] [Google Scholar]
- Aoki, K., R. Ishida and M. Kasai, 1997. Isolation and characterization of a cDNA encoding a Translin-like protein, TRAX. FEBS Lett. 401: 109–112. [DOI] [PubMed] [Google Scholar]
- Aoki, K., K. Suzuki, R. Ishida and M. Kasai, 1999. The DNA binding activity of Translin is mediated by a basic region in the ring-shaped structure conserved in evolution. FEBS Lett. 443: 363–366. [DOI] [PubMed] [Google Scholar]
- Bellen, H. J., R. W. Levis, G. Liao, Y. He, J. W. Carlson et al., 2004. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167: 761–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chennathukuzhi, V. M., Y. Kurihara, J. D. Bray and N. B. Hecht, 2001. Trax (translin-associated factor X), a primarily cytoplasmic protein, inhibits the binding of TB-RBP (translin) to RNA. J. Biol. Chem. 276: 13256–13263. [DOI] [PubMed] [Google Scholar]
- Chennathukuzhi, V., J. M. Stein, T. Abel, S. Donlon, S. Yang et al., 2003. Mice deficient for testis-brain RNA-binding protein exhibit a coordinate loss of TRAX, reduced fertility, altered gene expression in the brain, and behavioral changes. Mol. Cell. Biol. 23: 6419–6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, Y. S., V. M. Chennathukuzhi, M. A. Handel, J. Eppig and N. B. Hecht, 2004. The relative levels of translin-associated factor X (TRAX) and testis brain RNA-binding protein determine their nucleocytoplasmic distribution in male germ cells. J. Biol. Chem. 279: 31514–31523. [DOI] [PubMed] [Google Scholar]
- Cokol, M., R. Nair and B. Rost, 2000. Finding nuclear localization signals. EMBO Rep. 1: 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet, F., 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16: 10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dockendorff, T. C., H. S. Su, S. M. McBride, Z. Yang, C. H. Choi et al., 2002. Drosophila lacking dfmr1 activity show defects in circadian output and fail to maintain courtship interest. Neuron 34: 973–984. [DOI] [PubMed] [Google Scholar]
- Ferguson, D. O., and F. W. Alt, 2001. DNA double strand break repair and chromosomal translocation: lessons from animal models. Oncogene 20: 5572–5579. [DOI] [PubMed] [Google Scholar]
- Finkenstadt, P. M., M. Jeon and J. M. Baraban, 2002. Trax is a component of the Translin-containing RNA binding complex. J. Neurochem. 83: 202–210. [DOI] [PubMed] [Google Scholar]
- Ghabrial, A., R. P. Ray and T. Schüpbach, 1998. okra and spindle-B encode components of the RAD52 DNA repair pathway and affect meiosis and patterning in Drosophila oogenesis. Genes Dev. 12: 2711–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, G. D., R. D. Makde, R. P. Kamdar, J. S. D'Souza, M. G. Kulkarni et al., 2005. Co-expressed recombinant human Translin-Trax complex binds DNA. FEBS Lett. 579: 3141–3146. [DOI] [PubMed] [Google Scholar]
- Han, J. R., W. Gu and N. B. Hecht, 1995. a Testis-brain RNA-binding protein, a testicular translational regulatory RNA-binding protein, is present in the brain and binds to the 3′ untranslated regions of transported brain mRNAs. Biol. Reprod. 53: 707–717. [DOI] [PubMed] [Google Scholar]
- Han, J. R., G. K. Yiu and N. B. Hecht, 1995. b Testis/brain RNA-binding protein attaches translationally repressed and transported mRNAs to microtubules. Proc. Natl. Acad. Sci. USA 92: 9550–9554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heddad, A., M. Brameier and R. M. MacCallum, 2004. Evolving regular expression-based sequence classifiers for protein nuclear localisation. Lecture Notes Comput. Sci. 3005: 31–40. [Google Scholar]
- Hosaka, T., H. Kanoe, T. Nakayama, H. Murakami, H. Yamamoto et al., 2000. Translin binds to the sequences adjacent to the breakpoints of the TLS and CHOP genes in liposarcomas with translocation t(12;6). Oncogene 19: 5821–5825. [DOI] [PubMed] [Google Scholar]
- Hughes, S. C., and H. M. Krause, 1999. Single and double FISH protocols for Drosophila. Methods Mol. Biol. 122: 93–101. [DOI] [PubMed] [Google Scholar]
- Ishida, R., H. Okado, H. Sato, C. Shionoiri, K. Aoki et al., 2002. A role for the octameric ring protein, Translin, in mitotic cell division. FEBS Lett. 525: 105–110. [DOI] [PubMed] [Google Scholar]
- Kasai, M., T. Matsuzaki, K. Katayanagi, A. Omori, R. T. Maziarz et al., 1997. The translin ring specifically recognizes DNA ends at recombination hot spots in the human genome. J. Biol. Chem. 272: 11402–11407. [DOI] [PubMed] [Google Scholar]
- Kobayashi, S., A. Takashima and K. Anzai, 1998. The dendritic translocation of translin protein in the form of BC1 RNA protein particles in developing rat hippocampal neurons in primary culture. Biochem. Biophys. Res. Commun. 253: 448–453. [DOI] [PubMed] [Google Scholar]
- Larochelle, S., and B. Suter, 1995. Molecular cloning of the Drosophila homologue of the rat ribosomal protein L11 gene. Biochim. Biophys. Acta 1261: 147–150. [DOI] [PubMed] [Google Scholar]
- Laufman, O., R. Ben Yosef, N. Adir and H. Manor, 2005. Cloning and characterization of the Schizosaccharomyces pombe homologs of the human protein Translin and the Translin-associated protein TRAX. Nucleic Acids Res. 33: 4128–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley, D. L., and G. G. Zimm, 1992. The Genome of Drosophila melanogaster. Academic Press, San Diego.
- Masrouha, N., L. Yang, S. Hijal, S. Larochelle and B. Suter, 2003. The Drosophila chk2 gene loki is essential for embryonic DNA double-strand-break checkpoints induced in S phase or G2. Genetics 163: 973–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim, K. S., J. K. Jang and E. A. Manheim, 2002. Meiotic recombination and chromosome segregation in Drosophila females. Annu. Rev. Genet. 36: 205–232. [DOI] [PubMed] [Google Scholar]
- Morales, C. R., X. Q. Wu and N. B. Hecht, 1998. The DNA/RNA-binding protein, TB-RBP, moves from the nucleus to the cytoplasm and through intercellular bridges in male germ cells. Dev. Biol. 201: 113–123. [DOI] [PubMed] [Google Scholar]
- Morales, C. R., S. Lefrancois, V. Chennathukuzhi, M. El-Alfy, X. Wu et al., 2002. A TB-RBP and Ter ATPase complex accompanies specific mRNAs from nuclei through the nuclear pores and into intercellular bridges in mouse male germ cells. Dev. Biol. 246: 480–494. [DOI] [PubMed] [Google Scholar]
- Muramatsu, T., A. Ohmae and K. Anzai, 1998. BC1 RNA protein particles in mouse brain contain two y-,h-element-binding proteins, translin and a 37 kDa protein. Biochem. Biophys. Res. Commun. 247: 7–11. [DOI] [PubMed] [Google Scholar]
- Pare, C., and B. Suter, 2000. Subcellular localization of Bic-D∷GFP is linked to an asymmetric oocyte nucleus. J. Cell Sci. 113(Pt. 12): 2119–2127. [DOI] [PubMed] [Google Scholar]
- Rubin, G., L. Hong, P. Brokstein, M. Evans-Holm, E. Frise et al., 2000. A Drosophila complementary DNA resource. Science 287: 2222–2224. [DOI] [PubMed] [Google Scholar]
- Schuetze, C., M. Peters, J. J. Duong, M. Cavey, R. Dorig et al., 2004. Map positions of third chromosomal female sterile and lethal mutations of Drosophila melanogaster. Genome 47: 832–838. [DOI] [PubMed] [Google Scholar]
- Sengupta, K., and B. J. Rao, 2002. Translin binding to DNA: recruitment through DNA ends and consequent conformational transitions. Biochemistry 41: 15315–15326. [DOI] [PubMed] [Google Scholar]
- Sengupta, K., R. P. Kamdar, J. S. D'Souza, S. M. Mustafi and B. J. Rao, 2006. GTP-induced conformational changes in Translin: a comparison between human and Drosophila proteins. Biochemistry 45: 861–870. [DOI] [PubMed] [Google Scholar]
- Severt, W. L., T. U. Biber, X. Wu, N. B. Hecht, R. J. DeLorenzo et al., 1999. The suppression of testis-brain RNA binding protein and kinesin heavy chain disrupts mRNA sorting in dendrites. J. Cell Sci. 112(Pt. 21): 3691–3702. [DOI] [PubMed] [Google Scholar]
- Stein, J. M., W. Bergman, Y. Fang, L. Davison, C. Brensinger et al., 2006. Behavioral and neurochemical alterations in mice lacking the RNA-binding protein translin. J. Neurosci. 26: 2184–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter, B., and R. Steward, 1991. Requirement for phosphorylation and localization of the bicaudal-D protein in Drosophila oocyte differentiation. Cell 67: 917–926. [DOI] [PubMed] [Google Scholar]
- Tatusova, T. A., and T. L. Madden, 1999. BLAST 2 sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174: 247–250. [DOI] [PubMed] [Google Scholar]
- Tearle, R. G., and C. Nüsslein-Volhard, 1987. Tübingen mutants and stock list. Dros. Inf. Serv. 66: 209–269. [Google Scholar]
- Wu, X. Q., and N. B. Hecht, 2000. Mouse testis brain ribonucleic acid-binding protein/translin colocalizes with microtubules and is immunoprecipitated with messenger ribonucleic acids encoding myelin basic protein, alpha calmodulin kinase II, and protamines 1 and 2. Biol. Reprod. 62: 720–725. [DOI] [PubMed] [Google Scholar]
- Wu, X. Q., S. Lefrancois, C. R. Morales and N. B. Hecht, 1999. Protein-protein interactions between the testis brain RNA-binding protein and the transitional endoplasmic reticulum ATPase, a cytoskeletal gamma actin and Trax in male germ cells and the brain. Biochemistry 38: 11261–11270. [DOI] [PubMed] [Google Scholar]
- Yang, J., V. Chennathukuzhi, K. Miki, D. A. O'Brien and N. B. Hecht, 2003. Mouse testis brain RNA-binding protein/translin selectively binds to the messenger RNA of the fibrous sheath protein glyceraldehyde 3-phosphate dehydrogenase-S and suppresses its translation in vitro. Biol. Reprod. 68: 853–859. [DOI] [PubMed] [Google Scholar]
- Yang, S., Y. S. Cho, V. M. Chennathukuzhi, L. A. Underkoffler, K. Loomes et al., 2004. Translin-associated factor X is post-transcriptionally regulated by its partner protein TB-RBP, and both are essential for normal cell proliferation. J. Biol. Chem. 279: 12605–12614. [DOI] [PubMed] [Google Scholar]
- Yang, S., and N. B. Hecht, 2004. Translin associated protein X is essential for cellular proliferation. FEBS Lett. 576: 221–225. [DOI] [PubMed] [Google Scholar]