Abstract
Cellular responses to DNA damage and inhibited replication are evolutionarily conserved sets of pathways that are critical to preserving genome stability. To identify new participants in these responses, we undertook a screen for regulators that, when present on a high-copy vector, alter expression of a DNA damage-inducible RNR3-lacZ reporter construct in Saccharomyces cerevisiae. From this screen we isolated a plasmid encoding two closely related paralogs, WTM1 and WTM2, that greatly increases constitutive expression of RNR3-lacZ. Moderate overexpression of both genes together, or high-level expression of WTM2 alone from a constitutive promoter, upregulates RNR3-lacZ in the absence of DNA damage. Overexpressed, tagged Wtm2p is associated with the RNR3 promoter, indicating that this effect is likely direct. Further investigation reveals that Wtm2p and Wtm1p, previously described as regulators of meiotic gene expression and transcriptional silencing, amplify transcriptional induction of RNR3 in response to replication stress and modulate expression of genes encoding other RNR subunits.
MAINTAINING the integrity of the genome is essential to the survival and success of an organism. All prokaryotic and eukaryotic cells react to damage to their DNA or inhibited replication with coordinated actions designed to block progression through the cell cycle, repair any damage, and protect against future insults. Part of this response includes the transcriptional induction or repression of a set of genes involved in such processes as DNA metabolism, DNA repair, and cell cycle regulation (Zhou and Elledge 2000). In the budding yeast Saccharomyces cerevisiae, one of the genes most highly induced by DNA damage and replication inhibition is RNR3, encoding a large subunit of ribonucleotide reductase (RNR) (Ruby and Szostak 1985; Yagle and McEntee 1990). RNR3 is highly similar to the cell-cycle-regulated RNR1 gene (80% identity at the amino acid level) but RNR3 is expressed primarily in response to genotoxic and replication stress (Huang et al. 1998). Recent evidence suggests that Rnr3p is less susceptible to allosteric inhibition than Rnr1p, possibly allowing the rapid production of deoxynucleotides for DNA damage repair (Domkin et al. 2002). RNR2 and RNR4, encoding small catalytic subunits of the RNR enzyme complex, are also DNA damage inducible.
Mammalian cells exhibit DNA damage and replication stress responses that share many features with those of S. cerevisiae, many of whose players have been linked to cancer susceptibility (for review, see Melo and Toczyski 2002). Interestingly, one of the human genes induced by DNA damage is p53R2, a homolog of yeast RNR2 and RNR4 (Tanaka et al. 2000). Failure to induce p53R2 results in sensitivity to genotoxic agents such as ultraviolet (UV) radiation and adriamycin (Tanaka et al. 2000). The numerous RNR regulatory mechanisms activated in both yeast and humans upon DNA damage or inhibited replication, including transcriptional induction, production of alternate subunits, and changes in subcellular localization (Yao et al. 2003), attest to the importance of this enzyme in these responses.
The partially characterized pathway leading to induction of RNR3, RNR2, and RNR4 after DNA damage or replication stress relies heavily on a transcriptional repressor protein known as Crt1p or Rfx1p that binds directly to their upstream regions. A kinase cascade including the Mec1p, Rad53p, and Dun1p protein kinases culminates in the phosphorylation of Crt1p, resulting in derepression of Crt1 target genes (Huang et al. 1998; Zhou and Elledge 2000). Experimental evidence suggests, however, that there are regulators of the transcriptional response to DNA damage that remain unidentified (Huang et al. 1998; Gasch et al. 2001; Horak et al. 2002). Thus we undertook a screen to identify new regulators of the DNA damage transcriptional response, specifically the canonical DNA damage-inducible gene RNR3. This screen identified two potential novel RNR3 regulators, WD40 repeat-containing transcription modulator (Wtm)1p and Wtm2p. The Wtm proteins are a family of three proteins in S. cerevisiae that are believed to have roles in transcriptional regulation and silencing (Pemberton and Blobel 1997). The third member of this family, Ume1p (Wtm3p), has recently been found to be associated with the Sin3p/Rpd3p histone deacetylase complex (Gavin et al. 2002; Ho et al. 2002; Kurdistani et al. 2002; Mallory and Strich 2003), and direct binding of Ume1p to Rpd3p contributes to repression of meiotic genes (Mallory and Strich 2003).
We demonstrate here that Wtm1p and Wtm2p also influence RNR3 expression. Simultaneous overexpression of WTM1 and WTM2 leads to increased transcription of RNR3 independent of DNA damage or replication inhibition. When WTM2 alone is overexpressed at very high levels, even in the absence of Wtm1p, even greater expression of RNR3 occurs in conjunction with Wtm2p's association with the RNR3 promoter. Deletion of WTM2 attenuates RNR3-lacZ induction by some types of stress, implicating this gene in the DNA damage and replication stress response. wtmΔ mutations further influence transcription of both RNR2 and RNR1 reporters, implying a possible broader role in transcriptional regulation.
MATERIALS AND METHODS
Screen for regulators:
Yeast strain TSR30-23, containing two to four copies of the RNR3-lacZ fusion at the RNR3 locus, has been described (Ruby and Szostak 1985). This strain was transformed with a library of yeast genomic sequences on a 2μ plasmid vector (YEp13) (Nasmyth and Reed 1980), and transformants were screened for increased or decreased β-galactosidase activity. A total of 13,800 colonies resulting from the 20 independent transformations were replica plated onto X-gal indicator plates. Among these, a total of 15 colonies were either darker or lighter blue than the control strain containing empty vector.
To ascertain whether altered reporter activity resulted from the introduced plasmids, strains were analyzed for cosegregation of the altered phenotype with the plasmid LEU2 marker. Strains were grown on nonselective media to bring about plasmid loss and leu− subclones were rescreened for β-galactosidase activity. The plasmid present in high-activity strain T19, pSR50, was isolated by transforming bacteria with T19 DNA as previously described (Guthrie and Fink 1991) and selecting for ampicillin resistance. The plasmid was retested for activation activity in strain TSR30-15, containing a single copy of the RNR3-lacZ reporter integrated at the RNR3 locus. The region from the BglII site in YOR228C to the StuI site between WTM2 and WTM1 was subcloned and sequenced and found to be identical to the reported genomic sequence.
Oligodeoxynucleotides:
The following oligodeoxynucleotides were synthesized: oSG1 (5′-GTTTATTACGTAGTAAAAGTTGCATG-3′), oSG8 (5′-TAGGATCCAGAAGGAAACACTCAAGG-3′), oSG9 (5′-AGGCTCGAGACGCTGACACGAAAAACGAA-3′), oSG12 (5′-GCGGATCCATGGCGAAAAGCAAATCCAG-3′), oSG14 (5′-CCGCTCGAGTTAGCGGCCGCAATCGTCGTAACCTCTGCCAAT-3′), oSG15 (5′-GCCGTGGCTAGTTTCTTCTTA-3′), oSG16 (5′-CGTAGGCAGATAACTTGGCTT-3′), oSR121 (5′-AAGCAGCTTTACAGATCAATGGCGG-3′), oSR122 (5′-CGCCCTCCTTACTCATTGAGAAAAAGG-3′) oSR325 (5′-ACTACGGCGCCAAGATGAAGCGACGATGGAA-3′), oSR326 (5′-CGTCGCTTAAGATTCTTTGCTCACCGAAGGAT-3′), oSR405 (5′-AATAGGATCCATGCCAAAAAAGGTTTGGAAATCA-3′), oSR406 (5′-GTGGCTCGAGTTACTATTCGCTTTCCTCGGTATA-3′), oSR463 (5′-TTAAGTCTAGAGCTGGC-3′), oSR464 (5′-CAGCTCTAGACTTAAGGCC-3′), Act1-699 (5′-GCCTTCTACGTTTCCATCCA-3′), and Act1-r851 (5′-AAGAGTAACCACGTTCACTCAAGAT-3′).
Plasmid constructions:
A total of 5.4 kb of the pSR50 insert on a ClaI–XhoI fragment was subcloned into ClaI/SalI-cleaved YEp24 to generate plasmid pSR53 and to remove the URA3 marker. The 1.7-kb BamHI HIS3 fragment from pSZ63 (Orr-Weaver et al. 1981) was then inserted into the BamHI site of pSR53 to create pSR54. Plasmid pSR61 was made by cutting pSR54 with XhoI and BstEII, filling in the ends, and religating.
Because pSR54 lacks both a matched vector control and a convenient multicloning site, an alternate plasmid containing the same insert was generated for further manipulation and analysis. Plasmid pSG12 was generated by inserting the BamHI–NarI fragment from pSR53, containing the entire genomic insert as well as part of the tetracycline resistance gene from YEp24, into BamHI/ClaI-cleaved pRS424 (Christianson et al. 1992). Plasmid pSG11 was created by single-stranded mutagenesis (Kunkel et al. 1991) using oligonucleotide oSG1 to change the first two codons of YOR228C to stop codons and introduce a SnaBI site for screening. The 0.6-kb NcoI–NdeI fragment from pSG11, containing the mutated region, was inserted into NcoI/NdeI cleaved pSG12 to create pSG13 and sequenced in both pSG12 and pSG13. Plasmid pSG3 was generated by inserting the 5.1-kb EagI–BamHI segment from pSR54 into 2μ vector pRS425 (Christianson et al. 1992). pSG5 was then made by cutting pSG3 with BsaBI and StuI and religating; pSG6 was made by cutting pSG3 with SmaI and PmlI and religating.
pSE788 and pSE836, 2μ plasmids containing RNR2(UAS)-CYC1-lacZ and RNR1(UAS)-CYC1-lacZ reporters, respectively, were gifts from Steve Elledge (Elledge and Davis 1989; Zhou and Elledge 1992). To generate a matching RNR3(UAS)-CYC1-lacZ reporter, the upstream activating sequence (UAS) of RNR3 (from −646 to −114 relative to the major transcription start site) was amplified by PCR with oligos oSG8 and oSG9. The amplified DNA was cut with BamHI and XhoI and inserted into BglII/XhoI-cut pSE836 to replace the RNR1 UAS and generate pSG20.
For overexpression of WTM2 under the control of the GPD promoter, the coding region was amplified by PCR from genomic DNA with oligos oSG12 and oSG13, cut with BamHI and XhoI, and inserted into the BamHI/SalI-cleaved vector pG-3 (Schena et al. 1991) to create pSGX. For overexpression of TAP-tagged WTM2 under the control of the GPD promoter, the same region was amplified from plasmid pSG3 with oligos oSG12 and oSG14, which introduced a unique NotI site just before the translational stop codon of WTM2. This product was cut with BamHI and XhoI and inserted into BamHI/SalI-cleaved pG-3 to create plasmid pSGY. DNA encoding the TAP tag flanked by NotI sites was then amplified with oSR325 and oSR326 using plasmid pFA62X (Gould et al. 2004) as template and cloned into the NotI site of pSGY to create pJW1. Plasmid pSR336 encoding TAP-tagged, truncated, mutant Wtm2p was constructed by cutting pJW1 with ApaI and SacII and ligating in hybridized oligos oSR463/464. For overexpression of WTM1 by the GPD promoter in plasmid pKL6, the same cloning strategy was used as for pSGX except that the WTM1 ORF was amplified with oligos oSR405 and oSR406 with pSG7 as template.
All plasmids created by in vitro mutagenesis or PCR were sequenced.
Yeast strains and media:
Most strains used in this work were derived from DSR741-3B (Ruby and Szostak 1985), an S288C derivative, and are listed in Table 1. Null mutations in the WTM genes were generated by gene replacement (Rothstein 1983). wtm1Δ∷HIS3 mutants contain the HIS3 gene between bases 56 and 1186 of the 1312-bp ORF. wtm2Δ∷LEU2 mutants contain the LEU2 gene between bases 107 and 613 of the 1405-bp ORF. In wtm1wtm2Δ∷HIS3 strains (wtmΔ12), the region from base 107 of WTM2 to 994 of WTM1 is replaced by the HIS3 gene. All strains with integrated mutations were confirmed by Southern analysis (data not shown). Media for yeast strain manipulations were made as described (Guthrie and Fink 1991). Hydroxyurea (HU) (Sigma, St. Louis) was made up as a 1-m stock solution in water and stored at −20°; this solution was added directly to the culture to the desired concentration.
TABLE 1.
Strains used in this study
| Strain name | Genotype | Source |
|---|---|---|
| DSR741-3B | MATaleu2-2,112 his3-11,15 trp1 ura3 | Ruby and Szostak (1985) |
| Strains derived from DSR741-3B | ||
| TSR30-15 | RNR3-lacZ (1 copy) | Ruby and Szostak (1985) |
| TSR30-23 | RNR3-lacZ (2–4 copies) | Ruby and Szostak (1985) |
| T19 | RNR3-lacZ (2–4 copies) + pSR50 | Ruby and Szostak (1985) |
| TSG1 | RNR3-lacZ (1 copy), wtm1wtm2Δ∷HIS3 | This study |
| TSG10 | RNR3-lacZ (1 copy) | This study |
| TSG12 | RNR3-lacZ (1 copy), wtm1Δ∷HIS3 | This study |
| TSG19 | RNR3-lacZ (1 copy) + pSG5 (WTM1) + pRS423 | This study |
| TSG20 | RNR3-lacZ (1 copy) + pSG5 (WTM1) + pSR61 (WTM2) | This study |
| TSG21 | RNR3-lacZ (1 copy) + pRS425 + pRS423 | This study |
| TSG23 | RNR3-lacZ (1 copy) + pRS425 + pSR61 (WTM2) | This study |
| TSG41 | pRS424 | This study |
| TSG42 | pSG12 (WTM2 + WTM1) | This study |
| TSG78 | pSG20 (RNR3-CYC1-lacZ) + pRS424 | This study |
| TSG80 | pSG20 (RNR3-CYC1-lacZ) + pSG12 (WTM2 + WTM1) | This study |
| TSG85 | RNR3-lacZ (1 copy) + pRS425 + pG-3 | This study |
| TSG86 | RNR3-lacZ (1 copy) + pRS425 + pSGX (GPD-WTM2) | This study |
| TSG88 | RNR3-lacZ (1 copy) + pSG5 (WTM1) + pG-3 | This study |
| TSG89 | RNR3-lacZ (1 copy) + pSG5 (WTM1) + pSGX (GPD-WTM2) | This study |
| TSG94 | RNR3-lacZ (1 copy) + pG-3 | This study |
| TSG95 | RNR3-lacZ (1 copy) + pSGX (GPD-WTM2) | This study |
| TSG97 | RNR3-lacZ (1 copy), wtm1wtm2Δ∷HIS3 + pG-3 | This study |
| TSG98 | RNR3-lacZ (1 copy), wtm1wtm2Δ∷HIS3 + pSGX (GPD-WTM2) | This study |
| TSG100 | RNR3-lacZ (1 copy), pSGX (GPD-WTM2) | This study |
| TSG107 | pSGX (GPD-WTM2) | This study |
| TSG125 | DIN7-lacZ (1 copy) + pRS423 | This study |
| TSG126 | DIN7-lacZ (1 copy) + pSR54 (WTM2 + WTM1) | This study |
| TSG133 | pSGX (GPD-WTM2) | This study |
| TSG134 | pG-3 | This study |
| TSG1257 | RNR3-lacZ (1 copy), wtm2Δ∷LEU2 | This study |
| TSR2048 | RNR3-lacZ (1 copy) + pSR54 (WTM2 + WTM1) | This study |
| TSR2051 | RNR3-lacZ (1 copy) + pRS423 | This study |
| TSR2147 | pSE788 (RNR2-CYC1-lacZ) + pRS424 | This study |
| TSR2148 | pSE836 (RNR1-CYC1-lacZ) + pRS424 | This study |
| TSR2150 | pSE788 (RNR2-CYC1-lacZ) + pSG12 (WTM2 + WTM1) | This study |
| TSR2151 | pSE836 (RNR1-CYC1-lacZ) + pSG12 (WTM2 + WTM1) | This study |
| TSR2167 | pSE788 (RNR2-CYC1-lacZ) | This study |
| TSR2169 | pSE836 (RNR1-CYC1-lacZ) | This study |
| TSR2170 | wtm1wtm2D∷HIS3 + pSE788 (RNR2-CYC1-lacZ) | This study |
| TSR2172 | wtm1wtm2D∷HIS3 + pSE836 (RNR1-CYC1-lacZ) | This study |
| TSR2386 | wtm2Δ∷LEU2 + pG-3 | This study |
| TSR2387 | wtm2Δ∷LEU2 + pJW1 (GPD-WTM2-TAP) | This study |
| TSR2425 | pLGSD5 (GAL10-CYC1-lacZ) + pRS424 | This study |
| TSR2427 | pLGSD5 (GAL10-CYC1-lacZ) + pSG12 (WTM2 + WTM1) | This study |
| TSR2441 | RNR3-lacZ + pKL6 (GPD-WTM1) | This study |
| TSR2451 | wtm2Δ∷LEU2 + pSR336 (GPD-wtm2-TAP) | This study |
| Y80 | MATa, can1-100, ade2-1, his3-11, leu2-3,112, trp1-1, ura3-1 | Huang et al. (1998) |
| Strains derived from Y80 | ||
| Y301 | rad53-21 | Huang et al. (1998) |
| TSG46 | RNR3-lacZ (1 copy) + pRS424 | This study |
| TSG47 | RNR3-lacZ (1 copy) + pSG13 (WTM2 + WTM1) | This study |
| TSG48 | rad53-21, RNR3-lacZ (1 copy) + pRS424 | This study |
| TSG49 | rad53-21, RNR3-lacZ (1 copy) + pSG13 (WTM2 + WTM1) | This study |
β-Galactosidase assays:
Unless otherwise indicated, cells were grown overnight in YPD or appropriate selective medium containing 2% glucose to an OD600 of 1–4, diluted to an OD600 of 0.3, and allowed to continue growing for 3 hr at 30° prior to any experimental treatment. After the time period indicated, cells were collected by centrifugation and cell pellets frozen on dry ice and stored at −80°. β-Galactosidase activity in the cell pellets was determined by the glass bead method and expressed in Miller units as previously described (Ruby et al. 1983). The general linear model (GLM) ANOVA procedure and Dunnet's comparisons to control (Figure 3A) or Tukey's multiple-comparison method (all others) contained in Minitab (Version 13) were used to analyze activity data. To equalize the variances, data were log transformed prior to analysis.
Figure 3.—
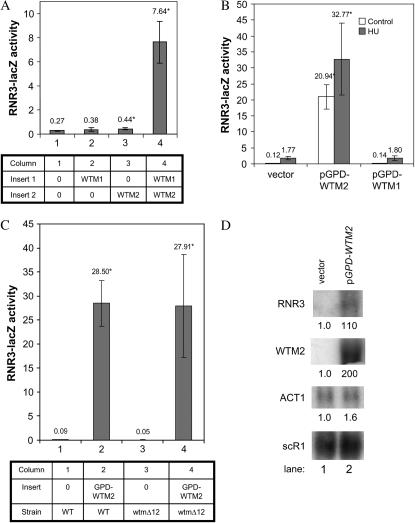
Expression levels of both WTM1 and WTM2 influence RNR3-lacZ expression. (A) Overexpression of WTM1 and WTM2 genes individually on high-copy (2μ) vectors indicates that both genes together maximally stimulate RNR3-lacZ expression. Each strain contains a single, integrated copy of RNR3-lacZ and two high-copy plasmids, either empty vector or a plasmid encoding a single WTM gene (as indicated in the table). These strains were grown to midlog phase in selective medium and assayed for β-galactosidase activity. The means and standard deviations of five independent isolates are plotted. Effect relative to empty vector: WTM1, not statistically significant; WTM2, P = 0.04; WTM1 + WTM2, P < 0.001. (B) High-level expression of WTM2 alone increases RNR3-lacZ expression. Strains containing a single, integrated copy of RNR3-lacZ and pGPD-WTM1 (a high-copy plasmid with WTM1 under control of the constitutive GPD promoter), pGPD-WTM2 (a high-copy plasmid with WTM2 under control of the constitutive GPD promoter), or a control vector were assayed for β-galactosidase activity. The means and standard deviations of three independent isolates are plotted: P < 0.001 for effect of GPD-WTM2; GPD-WTM1 effect is not statistically significant. (C) Stimulation of RNR3-lacZ by high-level WTM2 expression does not require the endogenous WTM1 gene. Wild-type or wtmΔ12 cells containing an integrated RNR3-lacZ reporter were transformed with plasmid pGPD-WTM2 and assayed for β-galactosidase activity. The means and standard deviations of four independent isolates are plotted: P < 0.001 for pGPD-WTM2; effect of wtmΔ12 mutation on pGPD-WTM2 stimulation is not statistically significant. (D) High-level expression of WTM2 alone increases RNR3 gene expression. Strains containing high-copy plasmid pGPD-WTM2 or empty vector were grown to midlog phase and their RNAs were extracted and subjected to Northern blot analysis. The same blot was probed sequentially for RNR3, WTM1, WTM2, ACT1, and scR1 (a loading control). Numbers below each band indicate the mean of three independent isolates relative to untreated vector control and normalized to scR1. Coefficients of variation were ≤23%. P-values for pGPD-WTM2 vs. vector: RNR3, P = 0.01; WTM2, P = 0.01; ACT1 is not significantly different. Strains used: (A) TSG21, TSG19, TSG23, and TSG20; (B) TSG94, TSG95, and TSR2441; (C) TSG94, TSG95, TSG97, and TSG98; (D) TSG133 and TSG134.
Northern blots:
RNA was extracted as previously described (Vijayraghavan et al. 1989) and sample integrity was assayed on a Bioanalyzer according to the manufacturer (Agilent). RNA samples were fractionated by electrophoresis in a 1.25% agarose gel with formaldehyde, transferred to Genescreen (New England Nuclear, Boston), and hybridized as previously described (Maniatis et al. 1982; Ruby 1999). Probes were labeled with 32P-dATP (ICN) by random priming (Feinberg and Vogelstein 1983). Quantitative analysis was performed with a STORM imager (Molecular Dynamics, Sunnyvale, CA) and ImageQuant software. GLM ANOVA in Minitab was used to compare RNA levels.
Chromatin immunoprecipitation assays:
Three independent yeast isolates containing pGPD-WTM-TAP and one or two negative control strains (with either untagged or no Wtm2p) were used for each of two independent chromatin preparations and chromatin immunoprecipitation (ChIP) repetitions. Crosslinking with dimethyl adipimidate (Pierce, Rockford, IL) for 1 hr followed by formaldehyde for 16–18 hr was performed as described for Ume1/Wtm3p (Kurdistani et al. 2002; Kurdistani and Grunstein 2003). All subsequent lysate preparatory steps were performed at 4°. Whole-cell lysates were prepared by glass bead breakage in ice-cold lysis buffer (200 mm KCl, 1% Triton X-100, 0.1% sodium deoxycholate, 20 mm Tris pH 7.4, and 5 mm MgCl2) supplemented with a 100-fold dilution of protease inhibitor HALT (Pierce). The lysate was drained from the glass beads by centrifugation in a clinical centrifuge for 1 min at 3000 rpm after which the crosslinked chromatin was sedimented at 40,000 × g for 3 min (Kurdistani and Grunstein 2003). The crosslinked chromatin was resuspended in lysis buffer, sonicated to an average size of 500 bp, and separated from debris by centrifugation for 1 hr at 40,000 × g. Supernatant aliquots were diluted 100-fold in water and measured spectrophotometrically at 260 nm. Immunoprecipitations were performed with 450 absorbance units of sample incubated overnight at 4° with 30 μl of IgG-Sepharose-6 beads (Amersham, Arlington Heights, IL) equilibrated in lysis buffer. After four washes in wash buffer (300 mm KCl, 1% Triton X-100, 0.1% sodium deoxycholate, 20 mm Tris–HCl, pH 7.4, and 5 mm MgCl2), the beads were incubated in elution buffer (0.1 m NaHCO3, 1% SDS, and 0.2 m NaCl) for 1 hr at 65° and then 1 hr at 75° to elute the bound material while simultaneously reversing the formaldehyde crosslinks (Solomon and Varshavsky 1985). Nucleic acids in the supernatant were precipitated, then treated sequentially with RNAseA and proteinase K as described (Kuo and Allis 1999), after which they were phenol extracted and ethanol precipitated. Inputs (110 absorbance units) were diluted in elution buffer and similarly heated and processed. Detection was by PCR with HotStarTaq polymerase (QIAGEN, Valencia, CA) and oligo pairs oSG15/16, oSR121/122, and ActI-699/851r for 1 cycle of 95° for 10 min, 50° for 45 sec, and 72° for 1 min followed by 24 cycles of 95° for 1 min, 50° for 45 sec, and 72° for 1 min. Template dilutions showed that all amplifications were in the linear range. PCR products were fractionated by agarose gel electrophoresis and visualized by ethidium bromide staining. Dilutions of PCR products were measured by Southern blot hybridization with radiolabeled probes, scanning with a STORM imager, and quantitating with ImageQuant software. Fold enrichment was calculated as the ratio of the levels for Wtm2-TAP vs. those for the negative control. The one-sample t-test and ANOVA in Minitab were used to compare fold enrichments to the null predicted ratio of one and for multiple comparisons, respectively.
For one ChIP repetition, two and three dilutions of the selected and total input samples, respectively, were also analyzed by real-time PCR with Sybr green master mix containing ROX in a 7900 HT cycler (Applied Biosystems, Foster City, CA) and quantitated via the standard curve method according to the supplier. Standard and dissociation curves showed that the primer pairs had equal efficiency and that each pair produced a single PCR product, respectively. Comparable results were obtained with real-time PCR as with Southern hybridization of PCR products.
RESULTS
Screen for RNR3 regulators:
To identify potential trans-acting regulators of the RNR3 (DIN1) gene, we initiated a screen for high-copy activators or repressors of RNR3 transcription. A library of yeast sequences on a 2μ plasmid vector (YEp13) (Nasmyth and Reed 1980) was used as this vector is maintained at 20–50 copies per cell (Old and Primrose 1994). The library was introduced into a yeast strain containing multiple copies of an RNR3-lacZ reporter integrated at the RNR3 chromosomal locus (Ruby and Szostak 1985), and transformants were screened for increased or decreased β-galactosidase activity. Among 15 strains with altered reporter activity, 4 strains were identified in which this activity segregated with the plasmid marker. One transformant in particular, T19, exhibited much higher β-galactosidase activity than the parent strain even in the absence of a DNA-damaging agent and was chosen for further analysis. The plasmid pSR50 was isolated from this strain and observed to cause a clear increase in RNR3-lacZ activity on X-gal plates upon retransformation into a yeast strain with a single copy of the integrated RNR3-lacZ reporter (data not shown). Furthermore, levels of endogenous RNR3 transcript were elevated in the presence of this plasmid in a strain lacking the reporter (data not shown, but see Figure 2).
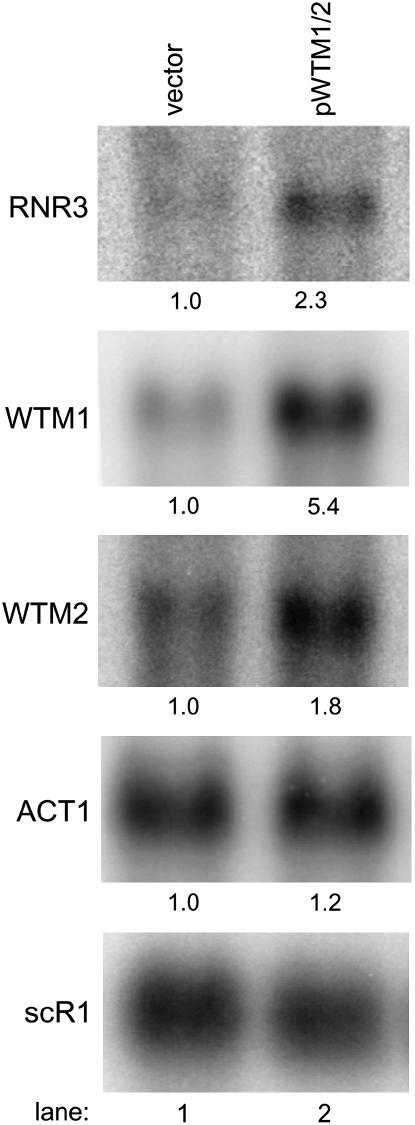
Figure 2.—
High-copy WTM1 and WTM2 increase expression of the native RNR3 gene. Cells containing empty vector (lane 1) or a plasmid encoding both WTM1 and WTM2 (lane 2) and grown in selective medium were harvested in midlog phase for RNA extraction and Northern blot analysis. The same blot was probed sequentially for RNR3, WTM1, WTM2, ACT1, and scR1 (a loading control). Numbers below each band indicate the mean of three independent isolates run on the same gel relative to untreated vector control and normalized to scR1; all coefficients of variation were <30%. P-values for pWTM1/2 vs. vector: RNR3, P = 0.003; WTM1, P < 0.001; WTM2, P = 0.001; ACT1, not statistically significant. Strains used: TSG41 and TSG42.
The plasmid pSR50 contains an insert of ∼5.6 kb of yeast genomic DNA. Sequencing revealed that this plasmid contains a segment of DNA from S. cerevisiae chromosome XV that includes the complete coding sequences of the paralogous WTM2 and WTM1 genes, which are 61% identical to each other at the amino acid level and reside on the chromosome as a tandem repeat separated by 990 bp of noncoding DNA. Approximately 5.4 kb of the pSR50 insert was subcloned into another 2μ vector and transformation of this construct (pSR54) into yeast containing an RNR3-lacZ reporter reconfirmed that the presence of this sequence on a high-copy vector leads to activation of the reporter. Quantitative assays demonstrated the increase to be 36-fold in untreated cells, greater than the 16-fold increase resulting from treatment with 100 mm HU, a replication inhibitor known to activate the DNA damage response (Huang et al. 1998) (P < 0.001) (Figure 1A). Treatment of pSR54-containing cells with HU resulted in minimal added increases in reporter activity (Figure 1A), although much higher levels of activity can be elicited from this reporter with DNA-damaging agents such as methyl methanesulfonate (MMS) (data not shown). This suggests that there may be significant overlap in the mechanisms by which HU and pSR54 activate RNR3 expression.
Figure 1.—
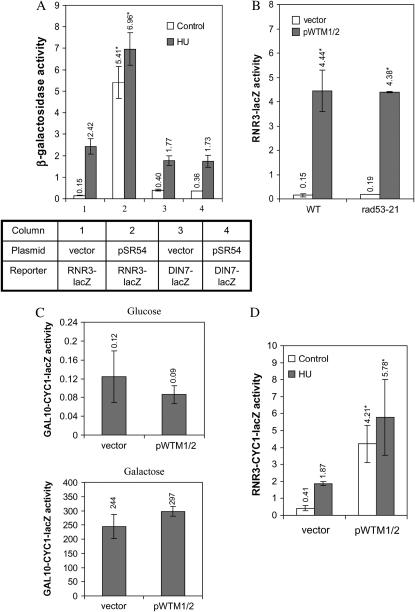
High-copy WTM1 and WTM2 increase expression of both integrated and extrachromosomal RNR3-lacZ reporters, independent of Rad53p activity. (A) Strains containing either an RNR3-lacZ or a DIN7-lacZ integrated reporter and transformed with pSR54 (a high-copy plasmid containing a genomic DNA insert encoding WTM2 and WTM1) or empty vector were grown to midlog phase in synthetic medium and then incubated for 3 hr with or without 100 mm HU. β-Galactosidase activities are the means of four independent isolates with error bars and asterisks indicating standard deviations and statistically significant differences, respectively. Effect of pSR54 on activity: P < 0.001 for RNR3-lacZ, not significant for DIN7-lacZ. (B) Isogenic rad53-21 and wild-type (wt) strains containing an integrated RNR3-lacZ reporter and either a plasmid encoding both WTM1 and WTM2 (pWTM1/2) or empty vector were assayed for β-galactosidase activity in midlog phase. Values represent means of two independent isolates; P < 0.001 for effect of pWTM1/2 on both strains. (C) Cells containing a plasmid-based GAL10-CYC1-lacZ reporter and either a plasmid expressing WTM1 and WTM2 or empty vector were grown in media containing glucose (top) or galactose (bottom) and assayed for β-galactosidase activity in midlog phase. Values represent means of three and six independent isolates for strains with empty vector and a plasmid expressing WTM1 and WTM2, respectively. Effects of pWTM1/2 and empty vector on reporter activity were not significant. (D) Cells containing a plasmid-based RNR3-CYC1-lacZ reporter and either a plasmid expressing WTM1 and WTM2 or empty vector were assayed for β-galactosidase activity in midlog phase. Values represent means of four independent isolates; P < 0.001 for pWTM1/2 vs. vector. Strains used: (A) TSR2051, TSR2048, TSG125, and TSG126; (B) TSG46, TSG47, TSG48, and TSG49; (C) TSR2425 and TSR4247; (D) TSG78 and TSG80.
One possible explanation for these observations is that the overexpression of WTM2 and/or WTM1 somehow produces DNA damage or interferes with DNA replication and thereby indirectly induces RNR3. To test this possibility, we examined the effects of pSR54 on the expression of another transcript, DIN7 (DIN3), which is inducible by a variety of DNA-damaging agents as well as replication inhibition (Ruby and Szostak 1985; Mieczkowski et al. 1997). Strains containing a DIN7-lacZ reporter exhibit no increase in β-galactosidase activity when transformed with plasmid pSR54, suggesting that the presence of this plasmid does not cause DNA damage or otherwise initiate a general DNA damage response (Figure 1A). Induction of the DIN7-lacZ reporter in the presence of HU is also unaffected by WTM1 and WTM2 overexpression (Figure 1A). Finally, induction of RNR3-lacZ by WTM1 and WTM2 overexpression is unaffected in cells containing an inactivating mutation in RAD53 (rad53-21, which eliminates kinase activity), a major upstream player in the DNA damage and replication inhibition responses (Figure 1B) (Allen et al. 1994; Huang et al. 1998). These data, along with the observation that WTM1 and WTM2 overexpression has no discernible effect on cell growth, viability, or sensitivity to agents including HU, MMS, and UV radiation (data not shown), indicate that this overexpression does otherwise induce the DNA damage response.
Another possible explanation for these observations is that overexpression of Wtm1p and Wtm2p, both WD40 repeat proteins, nonspecifically interferes with the function of Tup1p, another WD40 repeat protein. To test this hypothesis, we examined the effect of WTM1 and WTM2 overexpression on a GAL1/GAL10 reporter that is also repressed by the Ssn/Tup1 complex (reviewed in Smith and Johnson 2000). We found that expression of the GAL10-CYC1-lacZ reporter, on the 2μ plasmid pLGSD5 (Guarente et al. 1982), was unaffected by WTM gene overexpression (Figure 1C).
To further characterize the mechanism of action for WTM1 and WTM2, we constructed a 2μ plasmid with the UASs of RNR3 driving expression of the CYC1-lacZ reporter, such that no RNR3 coding sequence is present in the expressed message. This UAS region from −646 to −114 relative to the major transcription start site spans from the 3′ end of the neighboring gene FIS1 to the TATA box of RNR3, including all three Crt1p recognition sites (X-boxes). As expected, introduction of this plasmid [RNR3(UAS)-CYC1-lacZ] into yeast results in β-galactosidase activity that is inducible by HU (Figure 1D; P < 0.03) as well as the alkylating agent MMS (data not shown). Cotransformation of cells containing the RNR3(UAS)-CYC1-lacZ reporter with a plasmid encoding WTM1 and WTM2 (pWTM1/2) results in 10-fold higher β-galactosidase activity than cotransformation with empty vector (Figure 1D; P < 0.001). A matched control reporter plasmid lacking the RNR3 upstream sequences yields very low activity regardless of DNA damage, replication stress, or WTM1 and WTM2 overexpression (data not shown). This demonstrates that the RNR3 upstream sequences are both necessary and sufficient for activation by pWTM1/2.
A number of factors other than transcription can potentially influence the activity of a lacZ reporter construct (Ruby et al. 1983), so we wished to determine whether a high-copy vector containing WTM1 and WTM2 has an effect on the endogenous RNR3 gene. We measured expression of RNR3 in yeast transformed with either a plasmid harboring WTM1 and WTM2 or a matched empty vector. RNAs were extracted from cells grown to midlog phase in appropriate selective synthetic medium and analyzed by Northern blotting. Little RNR3 transcript is present in untreated cells containing empty vector, while a clear band is present in cells overexpressing WTM1 and WTM2 (Figure 2). The increase is estimated to be 2.3-fold (P = 0.003) but, given the low message level in cells containing empty vector, the effect is difficult to quantify accurately. This increase is smaller than the 30-fold activation of the lacZ reporter, but expression effects are often amplified in β-galactosidase reporter assays (Ruby and Szostak 1985; S. W. Ruby, unpublished observations), possibly due to the tetrameric structure of the active enzyme (Jacobson et al. 1994). These results demonstrate that the increased expression of RNR3-lacZ by pWTM1/2 reflects a genuine, although smaller, increase in RNR3 mRNA levels, but do not entirely rule out the possibility that there are also post-transcriptional effects.
The same blot probed with WTM1 and WTM2 gene sequences demonstrates that their transcripts are elevated fivefold (P < 0.001) and twofold (P = 0.001), respectively, in strains harboring pWTM1/2, whereas RNA levels of control genes ACT1 and scR1 are unchanged (Figure 2). ACT1 encodes actin protein and scR1 (small cytoplasmic RNA 1) (Felici et al. 1989) is a polIII transcript gene used as a loading and normalization control (Sharma et al. 2003) (Figure 2).
Dosage levels of both WTM1 and WTM2 influence RNR3-lacZ expression:
A series of experiments was undertaken to establish whether both WTM1 and WTM2 contribute to the observed RNR3-lacZ activation. Assays of several derived constructs suggested that disruption of either the WTM1 or the WTM2 regions of pSR54 interfered with increased expression of the RNR3-lacZ reporter (data not shown). When WTM1 and WTM2 were each introduced on separate plasmids with different selectable markers, RNR3 activation was comparable to the original construct (30-fold increase, P < 0.001) (Figure 3A). Cells overexpressing WTM1 or WTM2 alone exhibit relatively little change, although the WTM2 plasmid does cause a small but reproducible and statistically significant increase in RNR3-lacZ expression (P = 0.03) (Figure 3A). This implies that both WTM1 and WTM2 must be present at high copy to increase the RNR3-lacZ reporter transcription.
In addition to the complete coding sequences of WTM2 and WTM1, the pSR54 insert also contains much (696 of 909 bp) of the coding sequence of a divergently transcribed uncharacterized ORF, YOR228C, whose removal from the plasmid results in less robust increases in RNR3-lacZ expression (data not shown). However, when the start codon of YOR228C was mutated to a stop codon in a plasmid otherwise containing the full insert, the effect on RNR3-lacZ activation was minimal (data not shown). Thus it appears that the protein product of YOR228C is not necessary to bring about RNR3-lacZ activation, but that its coding region contains sequences important for WTM2 expression. This conclusion is bolstered by Northern analysis revealing that deletion of this region from the plasmid results in a 50% decrease in WTM2 message (data not shown).
The small increase in RNR3-lacZ when WTM2 is overexpressed alone (Figure 2A) suggests that WTM2 might be capable of acting independently. While Wtm2p is constitutively expressed at much lower levels than Wtm1p, they are thought to interact with a 1:1 stoichiometry in a large nuclear complex (Pemberton and Blobel 1997), so Wtm2p dosage could be limiting for the phenotypic effect seen when WTM1 and WTM2 are simultaneously overexpressed from their native promoters. Therefore, we constructed high-copy plasmids pGPD-WTM1 and pGPD-WTM2 in which expression of WTM1 and WTM2, respectively, is individually driven by the constitutive GPD promoter (Schena et al. 1991) and tested the ability of these plasmids to increase expression of either the integrated, single-copy RNR3-lacZ reporter or the intact RNR3 gene. While high-level expression of WTM2 dramatically increases RNR3 reporter activity, comparable high-level expression of WTM1 has no significant effect on the reporter (Figure 3B). Simultaneous overexpression of WTM1 does not further increase the RNR3-lacZ reporter expression stimulated by pGPD-WTM2 (data not shown) nor does the deletion of the chromosomal WTM1 and WTM2 genes impair it (Figure 3C). Plasmid pGPD-WTM2 also increases RNA levels from the normal chromosomal copy of RNR3 at least 100-fold (Figure 3D). Thus, when expressed at these very high levels, Wtm2p can act alone to stimulate RNR3 expression.
Normally, Rnr3p is present at less than one-tenth the levels of Rnr1p and contributes little to the synthesis and maintenance of deoxynucleotide (dNTP) pools as it has very low specific activity (Domkin et al. 2002), but the increased RNR3 expression that we observe with highly expressed WTM2 might alter dNTP levels, which would manifest as increased sensitivity or resistance to replication inhibitors or DNA-damaging treatments. Others have shown that increased dNTP pools raise resistance to some DNA-damaging treatments such as UV radiation and MMS (Chabes et al. 2003). However, cells overexpressing WTM2 on pGPD-WTM2 grow as well as wild-type cells and are comparably sensitive to HU, MMS, and UV radiation as the empty vector control (data not shown). Thus, some other factor must be limiting resistance to these treatments.
Wtm2p acts directly at the RNR3 promoter:
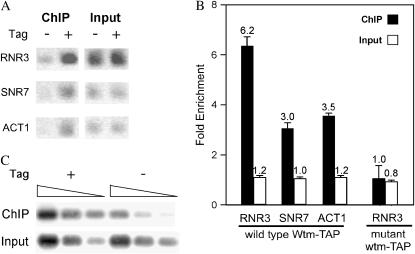
We hypothesized that increased RNR3 expression by highly expressed WTM2 could be due to a direct association of Wtm2p with the RNR3 promoter. To test this possibility, we used ChIP to investigate whether highly expressed Wtm2p localizes to the promoter of the intact, normal chromosomal copy of RNR3. We added a sequence for an epitope (TAP) tag onto the 3′ end of WTM2 expressed under the GPD promoter in the high-copy plasmid pGPD-WTM2-TAP and confirmed that the tagged protein could activate expression of the integrated RNR3-lacZ reporter to the same extent as the untagged form (data not shown). Cells containing pGPD-WTM2-TAP and a deletion of the chromosomal copy of WTM2 were then subjected to ChIP, along with two negative controls: cells with pGPD-WTM2 and therefore untagged Wtm2p or vector without WTM2. We found that the upstream region of RNR3 is enriched 6.2-fold in Wtm2-TAP-selected samples relative to the negative controls (P < 0.01) and 2-fold relative to the control genes ACT1 (encoding actin protein) and SNR7 (encoding U5 RNA) (P = 0.002) (Figure 4, A and B). These two control genes were used because they are actively transcribed in mitotically growing cells. As another control, we expressed a mutant form of the Wtm2-TAP protein in which only the first one-third of Wtm2p is present. This mutant protein, when expressed via the GPD promoter, neither increases RNR3-lacZ expression (data not shown) nor immunoprecipitates the RNR3 promoter above the levels of the negative controls (Figure 4). Thus, association of Wtm2p with the RNR3 promoter correlates with its ability to increase constitutive RNR3 expression.
Figure 4.—
Highly expressed Wtm2p associates with the upstream region of RNR3. Chromatin immunoprecipitations (ChIP's) with IgG sepharose were performed on extracts of cells with a chromosomal wtm2 deletion and plasmid pGPD-WTM2-TAP expressing epitope-tagged Wtm2p. Control extracts were from cells with untagged Wtm2p, no Wtm2p, and a truncated, mutant, epitope-tagged wtm2. (A) Quantitative PCR products from one representative ChIP experiment. Input and ChIP samples were assayed using primers for the upstream region of RNR3 and for the coding regions of SNR7 and ACT1. Products fractionated by agarose gel electrophoresis and visualized by ethidium bromide staining are shown as the inverse image. (B) Bar graph of quantitative PCR results. Dilutions of PCR products were fractionated by agarose gel electrophoresis and measured by Southern analyses. The data are represented as signals from tagged full-length, wild-type Wtm2p or mutant wtm2p samples normalized to those of untagged or no Wtm2p samples. Solid and open bars represent ChIP and input samples, respectively. The mean and standard deviations of three independent isolates, each measured in two independent chromatin preparations and experiments, are plotted for wild-type Wtm2p-TAP: P < 0.01 for RNR3, SNR7, and ACT1 enrichments, each being different from the background ratio of 1.0; P = 0.002 for RNR3 enrichment being different from those of SNR7 and ACT1; there were no statistically significant differences for input samples. Coefficients of variation were 16% for RNR3 ChIP and ≤10% for all other samples. Strains used: TSR2386, TSR2387, TSG107, and TSR2451. (C) Southern blot analysis of PCR products from amplification reactions using three different amounts of starting template. Template dilutions (from left to right: 1×, 0.25×, and 0.1×) from ChIP and total input samples from strains with full-length, wild-type Wtm2p-TAP or the untagged control were amplified by PCR and then analyzed by Southern blotting with the upstream region of RNR3 as probe. Strains used: TSR2386 and TSR2387.
Finally, we looked at the effects of HU treatment on chromatin association of Wtm2-TAP. In cells treated with HU, the association of overexpressed Wtm2-TAP with the RNR3 promoter did not change significantly (data not shown), consistent with the observation that HU exposure only moderately increases RNR3 expression when WTM2 is highly expressed (data not shown). When Wtm2-TAP is expressed at normal levels from the WTM2 chromosomal locus, we found no statistically significant association with the RNR3 promoter in either untreated or HU-treated cells (data not shown).
We conclude that when Wtm2p is highly expressed, it associates with the RNR3 upstream regulatory region and increases its transcription, rather than indirectly influencing RNR3 expression. However, other factors in addition to high levels of Wtm2p may also be involved in increasing RNR3 expression as Wtm2p associates with at least two other genes, ACT1 and SNR7, although to a lesser extent than with RNR3. WTM2 overexpression does not, however, significantly affect ACT1 or U5 RNA levels in untreated cells (Figures 2 and 3D and data not shown).
Role of the Wtm proteins in the response to replication stress:
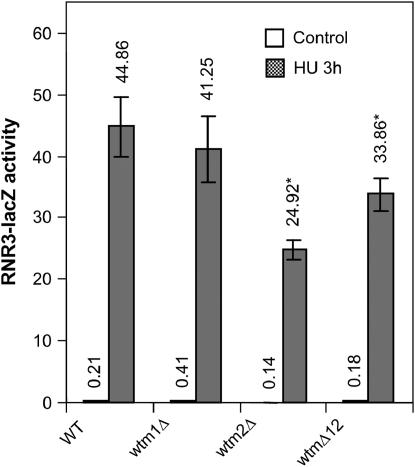
Since RNR3 is expressed at appreciable levels in mitotically growing cells only subsequent to DNA damage or replication inhibition (Ruby and Szostak 1985; Jia et al. 2002), we hypothesized that the Wtm proteins are involved in the DNA damage or replication stress responses. If the Wtm proteins are genuine regulators of RNR3 induction by stress, mutants lacking the WTM genes should exhibit defects in this response. We therefore generated wtm1Δ, wtm2Δ, and wtm1Δwtm2Δ (hereafter referred to as wtmΔ12) mutants by gene replacement (see materials and methods) and tested their ability to survive and to increase RNR3-lacZ reporter expression in response to replication stress and DNA-damaging treatments.
None of the wtmΔ strains exhibited growth defects (Pemberton and Blobel 1997) or altered sensitivity to HU, UV radiation, MMS, or ionizing radiation (data not shown). While the induction of RNR3-lacZ by HU is relatively unaffected in a wtm1Δ strain, activity is reproducibly attenuated by ∼45% in a wtm2Δ single mutant (P < 0.001) and by ∼25% in the double mutant (P < 0.002; Figure 5). This is somewhat surprising in light of data suggesting that the two proteins may act synergistically (Figure3A), but confirms our observations that Wtm2p appears to be more important in RNR3 expression than Wtm1p. Similar observations were made in cells treated with a 5-krad dose of ionizing radiation (IR), which induces double-strand breaks as well as other lesions (Birrell et al. 2002), but no difference in induction of RNR3-lacZ by the alkylating agent MMS was observed (data not shown). This is consistent with previous studies that have found that different agents can activate different pathways (Huang et al. 1998). Our data suggest that endogenous Wtm2p accounts for up to half the RNR3 induction in response to some types of stress, such as HU and IR. No change in either WTM2 or WTM1 expression was observed by Northern blot in cells treated with HU (data not shown).
Figure 5.—
RNR3-lacZ induction is attenuated in wtm2 deletion mutants. Cells lacking WTM1, WTM2, or both genes were grown to midlog phase in YPD medium, incubated with or without 100 mm HU for 3 hr, and then assayed for β-galactosidase activity. Values indicate mean and standard deviation of four independent isolates: P = 0.001 and P = 0.002 for effects of wtm2Δ and wtmΔ12 vs. control on HU induction, respectively. Strains used: TSR30-15, TSG10, TSG12, TSR1257, and TSG1.
Effect on other RNR genes:
All four genes encoding ribonucleotide reductase subunits in yeast are DNA damage-inducible, and RNR2, RNR3, and RNR4 share a common regulatory mechanism by which this induction occurs. These three genes are repressed by the Crt1p (Rfx1p) protein, which binds to X-boxes in their promoters (Huang et al. 1998). Another regulatory element shared by RNR2 and RNR3 is a binding site for the repressive Rpa protein complex (Singh and Samson 1995). RNR1, on the other hand, shows significant coding sequence similarity to RNR3 but the two genes are not generally observed to be transcriptionally coregulated (Huang et al. 1998; Gasch et al. 2001).
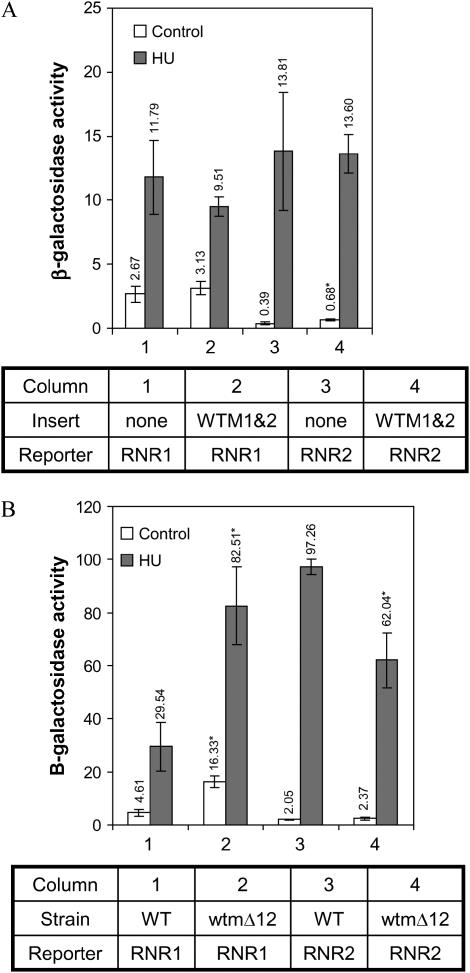
To see if overexpression of WTM1 and WTM2 has a general effect on ribonucleotide reductase production, we utilized reporter constructs to examine RNR gene expression. The plasmids pSE836 and pSE788 contain the upstream activating sequences of RNR1 or RNR2, respectively, driving expression of the CYC1-lacZ fusion gene on a 2μ plasmid (Zhou and Elledge 1992). Yeast cells containing each of these reporters in addition to plasmid pWTM1/2 overexpressing both WTM1 and WTM2 were grown to midlog phase and were either left untreated or exposed to HU for 4 hr. Overexpression of WTM1 and WTM2 results in a small (75%) but significant increase in RNR2 reporter activity in untreated cells (P = 0.03) and no change in RNR1 reporter expression (Figure 6A). For comparison, a matched reporter construct containing RNR3 upstream sequence exhibits a 10-fold increase in β-galactosidase under identical conditions (Figure 1D).
Figure 6.—
Effects of WTM1 and WTM2 dosage on RNR2 and RNR1 reporter expression. Cells containing high-copy RNR2 or RNR1 reporter plasmids were grown in selective medium to midlog phase, incubated with or without 100 mm HU, and then assayed for β-galactosidase activity. (A) Effect of high-copy WTM1/2 plasmid compared to empty vector at 4 hr incubation. Values indicate mean and standard deviation of four independent isolates; effect of pWTM1/2 is statistically significant only for a twofold increase in the RNR2 reporter in untreated cells, P = 0.03. (B) Wild-type or wtmΔ12 deletion strains were assayed for β-galactosidase activity after 3 hr incubation. Values indicate mean and standard deviation of four independent isolates: for the RNR1 reporter, P < 0.001 for effect of wtmΔ12 in both treated and untreated cells; for the RNR2 reporter, P = 0.02 for effect of wtmΔ12 on HU response. Strains used: (A) TSR2147, TSR2148, TSR2150, and TSR2151; (B) TSR2167, TSR2170, TSR2169, and TSR2172.
When WTM1 and WTM2 are deleted rather than overexpressed, the response of the RNR2 reporter is qualitatively similar to that of the RNR3 reporter: its induction by HU is attenuated 30–40% in the double mutant (P = 0.02; Figure 6B). In contrast, the double mutant exhibits a marked (2.5- to 3.5-fold) increase in RNR1 reporter expression in both untreated and HU-treated cells (P < 0.001; Figure 6B). The similarities in RNR2 and RNR3 reporter response when the WTM genes are either overexpressed or deleted suggest a common mechanism. But the fact that their deletion also affects RNR1 (Figure 6B) and IME2 (Pemberton and Blobel 1997) expression suggests they may have a more global role in gene regulation.
DISCUSSION
We demonstrate here that two of the WTM genes, previously known to influence transcriptional silencing and meiotic gene regulation (Pemberton and Blobel 1997), also modulate expression of the DNA damage and replication stress-inducible gene RNR3. We find that simultaneous overexpression of WTM1 and WTM2 produces a >30-fold increase in β-galactosidase activity in strains bearing an RNR3-lacZ reporter gene and a 2.3-fold increase in endogenous RNR3 mRNA levels, as well as a 2-fold increase in RNR2 reporter activity (Figures 1 and 6). Increased constitutive expression of the RNR3-lacZ reporter can also be achieved by high-level expression of WTM2, but not WTM1, alone. This stimulation by WTM2 occurs even in the absence of WTM1 and is associated with the presence of Wtm2p at the RNR3 promoter (Figures 3C and 4). Reciprocally, deletion of WTM2 attenuates the stress inducibility of an integrated RNR3-lacZ reporter by ∼45% (Figure 5).
The most striking effect of the two WTM genes is that they increase RNR3 expression when overexpressed together, or when WTM2 is highly expressed alone, even in the absence of DNA-damaging treatments or replication stress. For this stimulation, they may partially overcome RNR3's transcriptional repression, established and maintained mainly by a protein complex comprising Crt1p, Tup1p, and Ssn6p (Keleher et al. 1992). This complex positions nucleosomes in the upstream region of RNR3 (Li and Reese 2001), possibly via interactions with general transcription factors (Zhang and Reese 2004) and deacetylated N-terminal regions of histones H3 and H4 (Davie et al. 2002). When overexpressed, the Wtm proteins could alter any one or more of these interactions to increase RNR3 transcription. It is worth noting that Tup1p is also a WD40 repeat protein and could potentially compete with Wtm1p and Wtm2p for binding partners such as Ssn6p or Crt1p. However, Wtm1p and Wtm2p overexpression does not lead to a generalized derepression of Ssn6p/Tup1p-regulated genes, as expression of a GAL10 reporter is not affected (Figure 1C). Furthermore, the effect of highly expressed Wtm2p is unique in that individual overexpression of WTM1, or the closely related paralog UME1 (WTM3), under control of the GPD promoter does not increase RNR3 reporter expression despite the considerable similarity between their WD40 repeat regions (Figure 3B and data not shown). The promoter specificity of the effect when WTM1/2 are overexpressed together and the unique effect of highly expressed WTM2, as well as the decrease in RNR3-lacZ expression in the wtm2 deletion mutant (Figure 5), suggest that Wtm2p normally plays a role in RNR3 regulation rather than nonspecifically or artifactually interfering with Tup1p function when overexpressed. Additional experiments are required to determine the mechanism (be it derepression or activation) by which Wtm2p functions alone and in combination with Wtm1p.
There are some hints that the two WTM genes may increase RNR3 transcription by participating in chromatin remodeling. As we have shown here, Wtm2p associates with the upstream region of RNR3 when highly expressed. Others have shown that disruption of the interaction between Tup1/Ssn6 and the N termini of histones H3 and H4 partially derepresses RNR3 and RNR2 expression in the absence of stress (Edmondson et al. 1996). Although Wtm1p and Wtm2p have no clear orthologs in mammals, they are members of a subfamily of WD-repeat proteins that includes some chromatin-remodeling factors (Pemberton and Blobel 1997). For example, two members of this family, human proteins RpAp48 and RpAp46, associate with several different remodeling complexes (Loyola and Almouzni 2004). Wtm2p itself associates with the chromatin-remodeling factor Rvb1p (Jonsson et al. 2001; Ho et al. 2002), and both Wtm1p and Wtm2p are involved in regulating transcriptional silencing at some loci (Pemberton and Blobel 1997). Interestingly, the third, more distantly related yeast family member, Ume1p/Wtm3p, associates with the Rpd3p/Sin3p histone deacetylase complex (Gavin et al. 2002; Ho et al. 2002; Kurdistani et al. 2002) but does not copurify with Wtm1p and Wtm2p (Pemberton and Blobel 1997) or increase RNR3-lacZ activity when overexpressed (data not shown).
Another, not mutually exclusive, possibility is that the two overexpressed WTM genes alter an as yet unknown pathway or mechanism modulating RNR gene expression. The Wtm proteins could interact with Rpa1, another repressor of RNR3 (Singh and Samson 1995), or other transcription factors implicated in RNR3 regulation such as Msn4p (Martinez-Pastor et al. 1996; Moskvina et al. 1998; Tadi et al. 1999) and Yox1p (Horak et al. 2002). Interestingly, overexpression of Msn4p, a factor involved in the general stress response, results in a sixfold increase in constitutive RNR3 RNA levels (Gasch et al. 2000). The roles of these factors in the DNA damage and replication stress responses, as well as their relationships to the Crt1/Tup1/Ssn6 complex, are not known, but it is apparent that multiple factors including Wtm1p and Wtm2p are involved in regulating RNR3 expression. While it is possible that WTM1 and WTM2 function in different cellular processes when overexpressed vs. normally expressed, the simplest explanation is that they function in the same process in either condition.
One other possibility that we considered is that perturbation of WTM levels could, by interfering with chromatin structure or some aspect of DNA metabolism, induce a DNA damage response and indirectly lead to increased RNR3 transcription. However, we think that this possibility is unlikely for several reasons. Most importantly, highly expressed Wtm2p localizes to the RNR3 gene (Figure 4). Furthermore, a DIN7-lacZ reporter that is normally induced by DNA damage (Ruby and Szostak 1985) is unaffected by WTM1 and WTM2 overexpression (Figures 1). Finally, the transcriptional upregulation of RNR3-lacZ by WTM1 and WTM2 overexpression is unaffected by the rad53-21 mutation that effectively disables the known pathway of the DNA damage and replication stress response, including RNR3 induction, by blocking kinase activity (Allen et al. 1994; Craven and Petes 2000) (Figure 1B).
The combined results from experiments in which the two WTM genes are either overexpressed or deleted suggest that these genes are normally involved in modulating expression of the RNR genes. WTM overexpression triggers increases in RNR3 and RNR2 expression in unstressed cells (Figures 1, 3, and 6), while deletion reduces RNR2 and RNR3 induction by hydroxyurea (Figure 5) and RNR3 induction by ionizing radiation (data not shown). As RNR2 and RNR3 share some regulatory factors such as Crt1p, a common regulatory role by the Wtm proteins is plausible. RNR1, previously thought to be regulated differently than the other RNR genes (Huang et al. 1998), does not respond to WTM1 and WTM2 overexpression, but it is responsive to the WTM genes as their deletion increases RNR1 expression (Figure 6). Deletion and overexpression of other regulatory factors in yeast often, but not always, result in reciprocal effects on target gene expression (Herrgard et al. 2003). Therefore, the simplest explanation is that WTM1 and WTM2 normally modulate expression of the RNR genes. Furthermore, their regulatory role is more global than that of the RNR genes, as they also affect IME2 expression and transcriptional silencing of some genes (Pemberton and Blobel 1997). Finally, at least in the case of the RNR2 and RNR3 genes, WTM1 and WTM2 also normally amplify expression in response to inhibited replication as their deletion reduces induced levels. Additional work is necessary to determine the extent of the roles of the two WTM genes in the transcriptional responses to replication stress and DNA damage.
Our evidence indicates that Wtm2p localizes to the upstream region of RNR3 and increases RNR3 transcription when highly overexpressed, even in the absence of Wtm1p. Lower-level overexpression of WTM2 has only a small effect on RNR3 reporter activity; however, this activity is dramatically enhanced by overexpression of WTM1. These data suggest that the two proteins normally cooperate, yet each has a distinct function. Indeed, a previous study has shown that these two proteins copurify in a large, nuclear complex (Pemberton and Blobel 1997; Ho et al. 2002). Furthermore, two independent recent reports showed that Wtm1p associates with and retains a complex of Rnr2/Rrn4 subunits in the nucleus in normal yeast cells and shuttles this complex between the nucleus and cytoplasm in response to DNA damage or replication inhibition (Lee and Elledge 2006; Zhang et al. 2006). Wtm2p also associates with this complex, but a wtm2 deletion has no effect on localization or shuttling of the complex. Furthermore, Lee and Elledge (2006) observed that when Wtm2p is overexpressed under control of a GAL promoter, it increases the levels of Rrn2 and Rnr4 protein, consistent with our observations here that overexpression of WTM2 increases RNR2 and RNR3 expression. The combined results suggest that Wtm2p has a function different from that of Wtm1p. Intriguingly the only remarkable difference in sequence between the two proteins is a stretch of 14 charged amino acids, 13 of which are either aspartate or glutamate. This stretch is located between the first and second putative WD repeats of Wtm2p and may have a functional role as it has been conserved among some other ascomycetes retaining the two paralogs (Gish and States 1993; Balakrishnan et al. 2005).
Although our results indicate that Wtm2p can increase transcription, previous work with fusion proteins suggested that Wtm1p and Wtm2p repress transcription (Pemberton and Blobel 1997). It could be that nonspecific steric hindrance is responsible for the repressive activity of Wtm-LexA fusion proteins when targeted to a reporter gene by LexA binding. However, chromatin remodeling complexes such as Swi/Snf and Rvb1/Rvb2 have also been observed to act as both positive and negative regulators, depending on context (Sudarsanam et al. 2000; Jonsson et al. 2001). The Wtm proteins may also have this type of dual activity: they behave like positive regulators of silenced genes at HM loci and telomeres, but like negative regulators of the meiotic gene IME2 (Pemberton and Blobel 1997) and RNR1. While Wtm2p acts as a positive regulator at the RNR3 locus, Wtm1p and Wtm2p could potentially interact with chromatin in a manner that represses transcription at other loci.
Acknowledgments
We thank J. Stam, K. Gruchalla, M. Morgan, and B. Willems for technical assistance; J. Miller for a wtm1Δ construction; A. Lichens for assistance with the initial screen; G. Kawasaki for the library DNA; S. J. Elledge for yeast strains Y80 and Y301 and plasmids pSE836 and pSE788; L. Guarente for pLGSD5; K. Yamamoto for pG-3; K. Madura for pDGL5; K. Gould for pFAX6; and J. Szostak for pSZ63 and for supporting S.R. during the early phases of this work. We also thank the Keck University of New Mexico Genomics Resource facility and the Center for Genetics and Medicine at the University of New Mexico (UNM) School of Medicine for RNA and real-time PCR analyses and for sequencing, respectively. We are grateful to J. Nickoloff, M. A. Osley, and J. Summers for comments on the manuscript. This work was funded by the HHW Trust and the W. M. Keck and UNM Foundations as well as by Research Allocation Committee grant C2138 from the Dedicated Health Research Funds and Dean Roth, both of the UNM School of Medicine.
References
- Allen, J. B., Z. Zhou, W. Siede, E. C. Friedberg and S. J. Elledge, 1994. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 8: 2401–2415. [DOI] [PubMed] [Google Scholar]
- Balakrishnan, R., K. R. Christie, M. C. Costanzo, K. Dolinski, S. S. Dwight et al., 2005. Fungal BLAST and model organism BLASTP best hits: new comparison resources at the Saccharomyces Genome Database (SGD). Nucleic Acids Res. 33: D374–D377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birrell, G. W., J. A. Brown, H. I. Wu, G. Giaever, A. M. Chu et al., 2002. Transcriptional response of Saccharomyces cerevisiae to DNA-damaging agents does not identify the genes that protect against these agents. Proc. Natl. Acad. Sci. USA 99: 8778–8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabes, A., B. Georgieva, V. Domkin, X. Zhao, R. Rothstein et al., 2003. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell 112: 391–401. [DOI] [PubMed] [Google Scholar]
- Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero and P. Hieter, 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110: 119–122. [DOI] [PubMed] [Google Scholar]
- Craven, R. J., and T. D. Petes, 2000. Involvement of the checkpoint protein Mec1p in silencing of gene expression at telomeres in Saccharomyces cerevisiae. Mol. Cell. Biol. 20: 2378–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davie, J. K., R. J. Trumbly and S. Y. Dent, 2002. Histone-dependent association of Tup1-Ssn6 with repressed genes in vivo. Mol. Cell. Biol. 22: 693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domkin, V., L. Thelander and A. Chabes, 2002. Yeast DNA damage-inducible Rnr3 has a very low catalytic activity strongly stimulated after the formation of a cross-talking Rnr1/Rnr3 complex. J. Biol. Chem. 277: 18574–18578. [DOI] [PubMed] [Google Scholar]
- Edmondson, D. G., M. M. Smith and S. Y. Roth, 1996. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev. 10: 1247–1259. [DOI] [PubMed] [Google Scholar]
- Elledge, S. J., and R. W. Davis, 1989. Identification of the DNA damage-responsive element of RNR2 and evidence that four distinct cellular factors bind it. Mol. Cell. Biol. 9: 5373–5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg, A. P., and B. Vogelstein, 1983. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132: 6–13. [DOI] [PubMed] [Google Scholar]
- Felici, F., G. Cesareni and J. M. Hughes, 1989. The most abundant small cytoplasmic RNA of Saccharomyces cerevisiae has an important function required for normal cell growth. Mol. Cell. Biol. 9: 3260–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen et al., 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11: 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch, A. P., M. Huang, S. Metzner, D. Botstein, S. J. Elledge et al., 2001. Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast atr homolog mec1p. Mol. Biol. Cell 12: 2987–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch et al., 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147. [DOI] [PubMed] [Google Scholar]
- Gish, W., and D. J. States, 1993. Identification of protein coding regions by database similarity search. Nat. Genet. 3: 266–272. [DOI] [PubMed] [Google Scholar]
- Gould, K. L., L. Ren, A. S. Feoktistova, J. L. Jennings and A. J. Link, 2004. Tandem affinity purification and identification of protein complex components. Methods 33: 239–244. [DOI] [PubMed] [Google Scholar]
- Guarente, L., R. R. Yocum and P. Gifford, 1982. A GAL10–CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc. Natl. Acad. Sci. USA 79: 7410–7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie, C., and G. R. Fink (Editors), 1991. Guide to Yeast Genetics and Molecular Biology. Academic Press, San Diego.
- Herrgard, M. J., M. W. Covert and B. O. Palsson, 2003. Reconciling gene expression data with known genome-scale regulatory network structures. Genome Res. 13: 2423–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore et al., 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180–183. [DOI] [PubMed] [Google Scholar]
- Horak, C. E., N. M. Luscombe, J. Qian, P. Bertone, S. Piccirrillo et al., 2002. Complex transcriptional circuitry at the G1/S transition in Saccharomyces cerevisiae. Genes Dev. 16: 3017–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, M., Z. Zhou and S. J. Elledge, 1998. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94: 595–605. [DOI] [PubMed] [Google Scholar]
- Jacobson, R. H., X. J. Zhang, R. F. DuBose and B. W. Matthews, 1994. Three-dimensional structure of beta-galactosidase from E. coli. Nature 369: 761–766. [DOI] [PubMed] [Google Scholar]
- Jia, X., Y. Zhu and W. Xiao, 2002. A stable and sensitive genotoxic testing system based on DNA damage induced gene expression in Saccharomyces cerevisiae. Mutat. Res. 519: 83–92. [DOI] [PubMed] [Google Scholar]
- Jonsson, Z. O., S. K. Dhar, G. J. Narlikar, R. Auty, N. Wagle et al., 2001. Rvb1p and Rvb2p are essential components of a chromatin remodeling complex that regulates transcription of over 5% of yeast genes. J. Biol. Chem. 276: 16279–16288. [DOI] [PubMed] [Google Scholar]
- Keleher, C. A., M. J. Redd, J. Schultz, M. Carlson and A. D. Johnson, 1992. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell 68: 709–719. [DOI] [PubMed] [Google Scholar]
- Kunkel, T. A., K. Bebenek and J. McClary, 1991. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 204: 125–139. [DOI] [PubMed] [Google Scholar]
- Kuo, M. H., and C. D. Allis, 1999. In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods 19: 425–433. [DOI] [PubMed] [Google Scholar]
- Kurdistani, S. K., and M. Grunstein, 2003. In vivo protein-protein and protein-DNA crosslinking for genomewide binding microarray. Methods 31: 90–95. [DOI] [PubMed] [Google Scholar]
- Kurdistani, S. K., D. Robyr, S. Tavazoie and M. Grunstein, 2002. Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat. Genet. 31: 248–254. [DOI] [PubMed] [Google Scholar]
- Lee, Y. D., and S. J. Elledge, 2006. Control of ribonucleotide reductase localization through an anchoring mechanism involving Wtm1. Genes Dev. 20: 334–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B., and J. C. Reese, 2001. Ssn6-Tup1 regulates RNR3 by positioning nucleosomes and affecting the chromatin structure at the upstream repression sequence. J. Biol. Chem. 276: 33788–33797. [DOI] [PubMed] [Google Scholar]
- Loyola, A., and G. Almouzni, 2004. Histone chaperones, a supporting role in the limelight. Biochim. Biophys. Acta 1677: 3–11. [DOI] [PubMed] [Google Scholar]
- Mallory, M. J., and R. Strich, 2003. Ume1p represses meiotic gene transcription in Saccharomyces cerevisiae through interaction with the histone deacetylase Rpd3p. J. Biol. Chem. 278: 44727–44734. [DOI] [PubMed] [Google Scholar]
- Maniatis, T., E. F. Fritsch and J. Sambrook, 1982. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Martinez-Pastor, M. T., G. Marchler, C. Schuller, A. Marchler-Bauer, H. Ruis et al., 1996. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 15: 2227–2235. [PMC free article] [PubMed] [Google Scholar]
- Melo, J., and D. Toczyski, 2002. A unified view of the DNA-damage checkpoint. Curr. Opin. Cell Biol. 14: 237–245. [DOI] [PubMed] [Google Scholar]
- Mieczkowski, P. A., M. U. Fikus and Z. Ciesla, 1997. Characterization of a novel DNA damage-inducible gene of Saccharomyces cerevisiae, DIN7, which is a structural homolog of the RAD2 and RAD27 DNA repair genes. Mol. Gen. Genet. 253: 655–665. [DOI] [PubMed] [Google Scholar]
- Moskvina, E., C. Schuller, C. T. Maurer, W. H. Mager and H. Ruis, 1998. A search in the genome of Saccharomyces cerevisiae for genes regulated via stress response elements. Yeast 14: 1041–1050. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K. A., and S. I. Reed, 1980. Isolation of genes by complementation in yeast: molecular cloning of a cell-cycle gene. Proc. Natl. Acad. Sci. USA 77: 2119–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Old, R. W., and S. B. Primrose, 1994. Principles of Genetic Engineering. Blackwell Science, Cambridge, MA.
- Orr-Weaver, T. L., J. W. Szostak and R. J. Rothstein, 1981. Yeast transformation: a model system for the study of recombination. Proc. Natl. Acad. Sci. USA 78: 6354–6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton, L. F., and G. Blobel, 1997. Characterization of the Wtm proteins, a novel family of Saccharomyces cerevisiae transcriptional modulators with roles in meiotic regulation and silencing. Mol. Cell. Biol. 17: 4830–4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein, R. J., 1983. One-step gene disruption in yeast. Methods Enzymol. 101: 202–211. [DOI] [PubMed] [Google Scholar]
- Ruby, S. W., 1999. A yeast spliceosome assay. Methods Mol. Biol. 118: 365–390. [DOI] [PubMed] [Google Scholar]
- Ruby, S. W., and J. W. Szostak, 1985. Specific Saccharomyces cerevisiae genes are expressed in response to DNA-damaging agents. Mol. Cell. Biol. 5: 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby, S. W., J. W. Szostak and A. W. Murray, 1983. Cloning regulated yeast genes from a pool of lacZ fusions. Methods Enzymol. 101: 253–269. [DOI] [PubMed] [Google Scholar]
- Schena, M., D. Picard and K. R. Yamamoto, 1991. Vectors for constitutive and inducible gene expression in yeast. Methods Enzymol. 194: 389–398. [DOI] [PubMed] [Google Scholar]
- Sharma, V. M., B. Li and J. C. Reese, 2003. SWI/SNF-dependent chromatin remodeling of RNR3 requires TAF(II)s and the general transcription machinery. Genes Dev. 17: 502–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, K. K., and L. Samson, 1995. Replication protein A binds to regulatory elements in yeast DNA repair and DNA metabolism genes. Proc. Natl. Acad. Sci. USA 92: 4907–4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, R. L., and A. D. Johnson, 2000. Turning genes off by Ssn6-Tup1: a conserved system of transcriptional repression in eukaryotes. Trends Biochem. Sci. 25: 325–330. [DOI] [PubMed] [Google Scholar]
- Solomon, M. J., and A. Varshavsky, 1985. Formaldehyde-mediated DNA-protein crosslinking: a probe for in vivo chromatin structures. Proc. Natl. Acad. Sci. USA 82: 6470–6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsanam, P., V. R. Iyer, P. O. Brown and F. Winston, 2000. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97: 3364–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadi, D., R. N. Hasan, F. Bussereau, E. Boy-Marcotte and M. Jacquet, 1999. Selection of genes repressed by cAMP that are induced by nutritional limitation in Saccharomyces cerevisiae. Yeast 15: 1733–1745. [DOI] [PubMed] [Google Scholar]
- Tanaka, H., H. Arakawa, T. Yamaguchi, K. Shiraishi, S. Fukuda et al., 2000. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature 404: 42–49. [DOI] [PubMed] [Google Scholar]
- Vijayraghavan, U., M. Company and J. Abelson, 1989. Isolation and characterization of pre-mRNA splicing mutants of Saccharomyces cerevisiae. Genes Dev. 3: 1206–1216. [DOI] [PubMed] [Google Scholar]
- Yagle, K., and K. McEntee, 1990. The DNA damage-inducible gene DIN1 of Saccharomyces cerevisiae encodes a regulatory subunit of ribonucleotide reductase and is identical to RNR3. Mol. Cell. Biol. 10: 5553–5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, R., Z. Zhang, A. Xiuxiang, B. Bucci, D. L. Peristein et al., 2003. Subcellular localization of yeast ribonucleotide reductase regulated by the DNA replication and damage checkpoint pathways. Proc. Natl. Acad. Sci. USA 100: 6628–6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., and J. C. Reese, 2004. Redundant mechanisms are used by Ssn6-Tup1 in repressing chromosomal gene transcription in Saccharomyces cerevisiae. J. Biol. Chem. 279: 39240–39250. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., X. An, K. Yang, D. L. Perlstein, L. Hicks et al., 2006. Nuclear localization of the Saccharomyces cerevisiae ribonucleotide reductase small subunit requires a karyopherin and a WD40 repeat protein. Proc. Natl. Acad. Sci. USA 103: 1422–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, B. B., and S. J. Elledge, 2000. The DNA damage response: putting checkpoints in perspective. Nature 408: 433–439. [DOI] [PubMed] [Google Scholar]
- Zhou, Z., and S. J. Elledge, 1992. Isolation of crt mutants constitutive for transcription of the DNA damage inducible gene RNR3 in Saccharomyces cerevisiae. Genetics 131: 851–866. [DOI] [PMC free article] [PubMed] [Google Scholar]