Abstract
Centromere H3 proteins (CenH3's) are variants of histone H3 specialized for packaging centromere DNA. Unlike canonical H3, which is among the most conserved of eukaryotic proteins, CenH3's are rapidly evolving, raising questions about orthology and conservation of function across species. To gain insight on CenH3 evolution and function, a phylogenetic analysis was undertaken on CenH3 proteins drawn from a single, ancient lineage, the Fungi. Using maximum-likelihood methods, a credible phylogeny was derived for the conserved histone fold domain (HFD) of 25 fungal CenH3's. The collection consisted mostly of hemiascomycetous yeasts, but also included basidiomycetes, euascomycetes, and an archaeascomycete. The HFD phylogeny closely recapitulated known evolutionary relationships between the species, supporting CenH3 orthology. The fungal CenH3's lacked significant homology in their N termini except for those of the Saccharomyces/Kluyveromyces clade that all contained a region homologous to the essential N-terminal domain found in Saccharomyces cerevisiae Cse4. The ability of several heterologous CenH3's to function in S. cerevisiae was tested and found to correlate with evolutionary distance. Domain swapping between S. cerevisiae Cse4 and the noncomplementing Pichia angusta ortholog showed that species specificity could not be explained by the presence or absence of any recognized secondary structural element of the HFD.
CENTROMERE H3 proteins (CenH3's) are variants of histone H3 specialized for packaging centromere DNA. Originally discovered in mammals and Saccharomyces cerevisiae (Sullivan et al. 1994; Stoler et al. 1995), CenH3's also have been characterized genetically in Schizosaccharomyces pombe (Takahashi et al. 2000), Caenorhabditis elegans (Buchwitz et al. 1999), Drosophila (Henikoff et al. 2000), Arabidopsis (Talbert et al. 2002), and Xenopus (Edwards and Murray 2005). Phylogenomic analysis suggests that they are universal in eukaryotes (Malik and Henikoff 2003). CenH3's are thought to play a key role in determining centromere identity, although the mechanism by which they are specifically deposited on centromere DNA and excluded from noncentromere DNA is not known (Sullivan 2001; Mellone and Allshire 2003; Henikoff and Dalal 2005). Indeed, except for the simple “point” centromeres of S. cerevisiae and its close relatives, eukaryotic centromere DNAs are ill-defined, and no unique, centromere-specific DNA sequence has been identified (Sullivan et al. 2001).
Core histones are among the most conserved eukaryotic proteins, and their evolutionary origins can be traced to the Archaea (Malik and Henikoff 2003). Histone H3 in particular is highly conserved—the H3's of human and Arabidopsis thaliana differ at only 8 of 136 amino acid positions. In contrast, CenH3's, while clearly variants of H3, are significantly diverged, both from H3 and from each other (Malik and Henikoff 2003). Homology between H3 and CenH3 is limited to the C-terminal histone fold domain (HFD), where amino acid identity is ∼50%. The HFD, a structural motif shared by all of the core histones, consists of three α-helical regions connected by loop segments (Arents et al. 1991). H3 and CenH3 have an additional helix, the N-helix, located on the N-terminal side of helix 1. The most reliable bioinformatic criterion for distinguishing CenH3's from H3 is the presence in CenH3's of a longer loop 1 in the HFD (Malik and Henikoff 2003). As loop 1 directly contacts the nucleosomal DNA, this modification may confer DNA binding specificity to CenH3 nucleosomes (Vermaak et al. 2002). H3 (and by analogy CenH3) plays a central role in organizing nucleosomal structure: homotypic contacts mediated by helices 2 and 3 of the H3 HFD are integral to (H3–H4)2 tetramer formation and establish the nucleosomal twofold axis; the first step in nucleosome assembly is the association of an (H3–H4)2 tetramer with DNA; and H3 makes two separate DNA contacts, at loop 1 and at the C-terminal side of the N-helix, where the polypeptide chain exits the nucleosome (Luger et al. 1997).
Phylogenetic analysis has shown that CenH3 proteins are rapidly evolving. In addition to a near complete lack of homology in their N termini, the HFDs of even closely related species are significantly diverged. For example, mouse and human CenH3's are 78% identical in their HFD and 70% identical overall, in contrast to mouse and human H3's, which are 96% conserved over their entire length. Amino acid conservation in the HFDs of distantly related species (e.g., human and S. cerevisiae) falls to ∼50%. The high sequence divergence is evident in phylogenetic trees characterized by multiple short branches and few strongly supported nodes (Malik and Henikoff 2003). One interpretation is that CenH3's have arisen multiple times during the course of evolution; i.e., they are not orthologous. A previous study found that yeast CenH3 phylogeny was not congruent with species phylogeny, leading the authors to conclude that CenH3's were invented at least three times during fungal evolution (Malik and Henikoff 2003). But paradoxically, CenH3's appear to be more or less interchangeable between even distantly related species. The respective CenH3's from S. cerevisiae (Cse4), C. elegans (HCP-3, DH6H3), and human (CENP-A) localize to pericentric heterochromatin when expressed in Drosophila cells, and Cse4 and HCP-3 localize to centromeric regions in HeLa cells (Henikoff et al. 2000). Wieland et al. (2004) found that S. cerevisiae Cse4 functionally complements lethal CENP-A defects induced by RNAi in human tissue culture cells. The species specificity observed among Drosophila CenH3's may be exceptional, in that it can be attributed to the rapidly evolving centromere DNAs within the Drosophila lineage (Vermaak et al. 2002). These results are explained if, despite significant divergence at the level of primary amino acid sequence, CenH3's have retained common determinants of structure that allow for their specialized function in organizing centromeric chromatin.
The CenH3 protein of S. cerevisiae is Cse4. In most respects, Cse4 is typical of the CenH3's of higher eukaryotes: it is 61% identical to S. cerevisiae H3 in the HFD, it is specifically localized to centromeric DNA throughout the cell cycle, and it is essential for accurate chromosome segregation at mitosis (Stoler et al. 1995; Meluh et al. 1998; Keith et al. 1999). Cse4 differs from other CenH3's in that it contains a protein domain in its N terminus that is essential when Cse4 is expressed at wild-type levels (Chen et al. 2000). The essential domain, known as the essential N-terminal domain (END), appears to be required at a step prior to Cse4 incorporation into centromere chromatin, because overexpression of the Cse4 HFD bypasses the requirement for the END (Morey et al. 2004). In other systems, no essential function of the N terminus has been defined. The N termini of CENP-A and Cid (the Drosophila CenH3) are not required for CenH3 centromere targeting (Sullivan et al. 1994; Vermaak et al. 2002), but both N termini contain DNA minor groove binding motifs (Malik et al. 2002), and the CENP-A N terminus directs the targeting of other kinetochore proteins (Van Hooser et al. 2001).
The Fungi are a major lineage of Eukaryota that became established ∼1 billion years ago (Feng et al. 1997). A diverse kingdom, the Fungi are divided among several classification divisions, two of which, the Basidiomycota (mushrooms, rusts, smuts) and Ascomycota (sac fungi, yeasts), are sister groups, each believed to be monophyletic (Bruns et al. 1992). Ascomycota split from the Basidiomycota ∼400 million years ago (Berbee and Taylor 1993; Sipiczki 2000; Rokas et al. 2005). The Ascomycota account for ∼75% of all described fungi, including several well-known biological model organisms: Neurospora crassa, a euascomycete; S. pombe, an archaeascomycete; and S. cerevisiae, a hemiascomycete. The Archaeascomycota diverged before the separation of Euascomycota and Hemiascomycota (Berbee and Taylor 1993; Sipiczki 2000); thus, S. pombe is more distantly related to S. cerevisiae than are N. crassa and related filamentous fungi. The biological, medical, and industrial importance of many fungi has led to an increasing database of fungal whole-genome sequences. This development prompted us to draw on the information to undertake a phylogenetic study of fungal CenH3's. Three major questions were addressed: Can a credible phylogeny of CenH3 protein sequences be derived? Is the CenH3 phylogeny congruent with fungal species phylogeny? To what extent is the rapid evolution of CenH3's manifested at the functional level? The results suggested that fungal CenH3's are orthologous. In contrast to findings in nonfungal systems, we found CenH3 function in yeasts to be species specific in that Cse4 function in S. cerevisiae is supplied only by CenH3's from closely related hemiascomycetes. We suggest that fungal CenH3 evolution is best explained by a covarion evolutionary model.
MATERIALS AND METHODS
Phylogenetic analysis:
DNA sequences for all of the H3's and most of the CenH3 proteins analyzed were obtained by blastp or tblastn searches of public databases: Saccharomyces Genome Database (http://www.yeastgenome.org), National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov), Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/kegg), Genolevures (http://cbi.labri.fr/Genolevures), The Genome Sequencing Center at Washington University Medical School (http://genomeold.wustl.edu/projects/yeast), Fungal Genome Initiative (http://www.broad.mit.edu/annotation/fungi), and DOE Joint Genome Institute (http://genome.jgi-psf.org). For Pichia farinosa, Kluyveromyces marxianus, and S. servazzii, only partial gene sequences were available (Genolevures), and the complete genes were amplified by inverse PCR of genomic DNA prepared from Agricultural Research Service Culture Collection reference strains Y-7553, Y-8281, and Y-12661, respectively. The P. angusta gene was amplified from DNA of ARSCC reference strain Y-2214. DNA sequencing of the protein open reading frame revealed several synonymous codon changes between it and the gene from P. angusta type strain CBS 4732 used in the Genolevures project (Blandin et al. 2000). Automated DNA sequencing was performed on primary PCR products.
Protein sequence alignments were made using the online implementation of clustalW (Chenna et al. 2003) at http://www.ebi.ac.uk/clustalw. Output for figures was generated by Jalview (Clamp et al. 2004). The HFD alignment was adjusted manually to maintain alignment of the conserved helical domains and to minimize the number of gaps in the highly variable loop 1 regions. Phylogenetic model testing and maximum-likelihood calculations were performed using the proml program of PHYLIP version 3.65, available at http://evolution.genetics.washington.edu/phylip.html (Felsenstein 2004) and the codeml program of PAML version 3.15 (Yang 1997). Heuristic tree searches were made using MrBayes version 3.1.2 (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003) and proml. MrBayes runs were terminated after 5×105 generations or when the standard deviation of split frequencies fell below 0.01. The posterior distribution was sampled every 100 generations after discarding the first 25% of samples. MrBayes implements both Jones–Taylor–Thornton (JTT) (Jones et al. 1992) and Whelan–Goldman (WAG) (Whelan and Goldman 2001) models of amino acid substitution. While higher likelihoods were obtained using WAG rates, Bayes' factor tests (Kass and Raftery 1995) indicated that the evidence in favor of the WAG model was not decisive, and likelihood-ratio tests of bootstrap samples (codeml) showed that the difference was not significant (P = 0.42). Since both models yielded maximum-likelihood trees of identical topology, and WAG is not implemented in proml, JTT rates were used for all subsequent model testing and likelihood calculations. Kishino–Hasegawa–Templeton (KHT) tests of alternate tree topology (Kishino and Hasegawa 1989) were made using codeml except for gamma + I models, in which case proml was used. Substitution rates under a gamma model of variable rates across sites were calculated by codeml using 10 rate categories. Tests for positive selection were made using codeml (NSsites = 1 2 7 8). TreeViewPPC version 1.6.6 (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html) was used to draw trees. Sequence logos were generated by WebLogo (Crooks et al. 2004). Statistical tests were made using Prism version 4.0 software (GraphPAD, San Diego).
Heterologous CenH3 expression in S. cerevisiae:
Plasmid pRB294 (CEN-ARS-TRP1) carries wild-type S. cerevisiae CSE4 modified by the incorporation of a triple-hemagglutinin (HA) epitope within the N terminus (Morey et al. 2004). Native versions of S. servazzii and K. marxianus Cse4 orthologs were expressed by cloning the respective open reading frames (ORFs) into pRB294, fusing at the initiator methionine (NsiI site). S. pombe, P. farinosa, and P. angusta orthologs were expressed as fusions of the respective HFDs to the complete N terminus of S. cerevisiae Cse4. The fusions, obtained by recombinant PCR (i.e., the two halves of the desired construct, overlapping by 18–24 bp, were obtained in separate PCRs and then joined in a third PCR using outside primers), were cloned into the pRB294 vector. P. angusta–S. cerevisiae HFD chimeras were constructed similarly; all carry the complete triple-HA-tagged S. cerevisiae N terminus. Functional complementation of the cse4Δ∷kanMX4 null allele was tested using a plasmid shuffle assay. The tester strain was R332-5D (MATa his3Δ1 leu2Δ0 ura3Δ0 trp1Δ63 cse4Δ∷kanMX4 [pRB163]), which contains wild-type CSE4 on the CEN-ARS-URA3 plasmid pRB163 (Morey et al. 2004). Plasmids to be tested were introduced by transformation, and Trp+ colonies were picked, diluted, and plated on medium containing 5-fluoroorotic acid (FOA) to score loss of the URA3 marker (pRB163). Alleles unable to complement cse4Δ∷kanMX4 do not give rise to FOAr colonies, because pRB163 loss is lethal. An analogous cse4Δ∷kanMX4 tester strain, R411-11B (MATα his3Δ1 leu2Δ0 ura3Δ0 trp1Δ63 hht1-hhf1Δ∷HIS3 HHT2-HHF2Pang cse4Δ∷kanMX4 [pRB163]), was used to test P. angusta Cse4 function in the presence of P. angusta H4. Details of the strain construction are provided in supplemental data at http://www.genetics.org/supplemental/. Expression of heterologous CenH3 proteins was verified by Western blot analysis (Morey et al. 2004) of cells grown in selective medium. Fluctuation assays for mitotic chromosome missegregation were performed as described (Hegemann et al. 1988), using tester strain KC405 (Keith et al. 1999). All media, growth conditions, and yeast genetic procedures were as described (Morey et al. 2004).
RESULTS
HFD phylogeny:
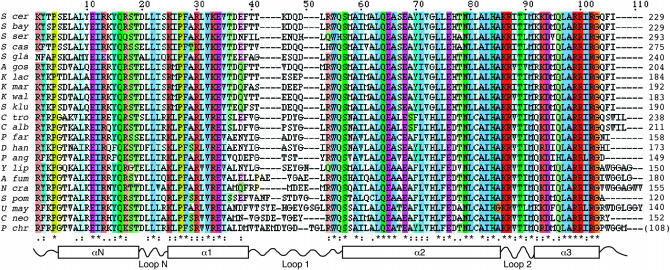
DNA sequences encoding Cse4 orthologs from 25 fungi were obtained from public databases or by cloning and sequencing. The collection consisted of three basidiomycetes (Phanerochaete chrysosporium, Cryptococcus neoformans, and Ustilago maydis) and 22 ascomycetes, including S. cerevisiae and its close sensu stricto relatives S. mikatae, S. bayanus, S. paradoxus, and S. kudriavzevii; the Saccharomyces sensu lato yeasts S. servazzii, S. kluyveri, and S. castellii; the Kluyveromyces species K. lactis, K. marxianus, K. waltii, and their relative Ashbya gossypii (Eremothecium gossypii); three Candida species, Candida glabrata, C. albicans, and C. tropicalis; and other distant Saccharomyces relatives P. farinosa, P. angusta, Debaryomyces hansenii, and Yarrowia lipolytica. Other Ascomycota were the archaeascomycete S. pombe and the euascomycetes Aspergillus fumigatus and N. crassa. Figure 1 shows an alignment of the HFD region of the Cse4 homologs (S. mikatae, S. kudriavzevii, and S. paradoxus were omitted, because they do not differ from S. bayanus in the HFD). The proteins are identical at 34 of 111 sites of the alignment (30.6%).
Figure 1.—
HFD alignment. The alignment of 22 fungal CenH3 HFDs is shown in ClustalX format and coloring, with color intensity proportional to the conservation index. Secondary structural elements inferred from the three-dimensional structure of histone H3 are diagrammed schematically below the alignment. Four conserved positions N-terminal to the N-helix are included in the alignment. (The P. chrysosporium sequence is incomplete upstream of the region encoding the HFD.)
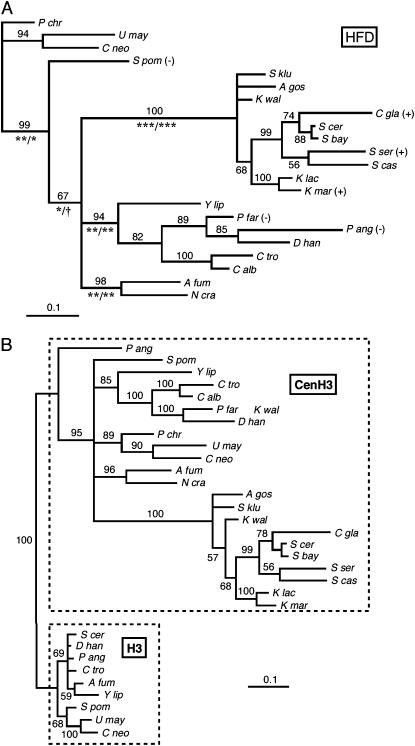
An HFD phylogeny, derived by maximum likelihood, is shown in Figure 2A. The tree is reasonably well resolved, with strong support for major branches. In particular, all of the hemiascomycetes cluster on two branches of a trifurcation that also includes the euascomycete branch (A. fumigatus, N. crassa). Nine fungal canonical H3 sequences were added to the CenH3 alignment, and the tree search was rerun under the same model and conditions. The resulting maximum-likelihood tree resolved the H3's and CenH3's into separate monophyletic groupings, consistent with the existence of a single common ancestor to all of the fungal CenH3's. The rapid evolution of CenH3's, relative to H3's, is apparent from the total lengths of the respective subtrees. Branching order of the CenH3 taxa in the combined H3/CenH3 tree differed somewhat from that of the CenH3 tree of Figure 2A—notably, P. angusta branches basally to the other CenH3's—but changing the topology of the CenH3 branch of the Figure 2B tree to match that of Figure 2A did not result in a significant likelihood change as assessed by KHT test (P = 0.30). In contrast, moving P. angusta CenH3 into the H3 branch of the tree resulted in a significant decrease in likelihood (P = 0.022).
Figure 2.—
Fungal CenH3 and H3 phylogeny. (A) CenH3 HFD phylogeny. The phylogram shows the majority rule consensus tree produced by MrBayes, running a fixed-rate model with WAG rates (see materials and methods). Numbers above the branches are the percentage of times that branch was present in the posterior distribution of trees. Confidence intervals for the lengths of major branches were estimated under both fixed-rate and gamma substitution models. Symbols below the branches indicate, for each model, respectively, the level of significance at which the confidence interval excludes zero branch length (***, P = 0.001; **, P = 0.01; *, P = 0.05; †, P = 0.10). Plus (+) and minus (−) signs indicate whether or not the Cse4 ortholog complements Cse4 function in S. cerevisiae. (B) Nine fungal H3 sequences were added to the CenH3 HFD alignment, and a phylogeny was determined exactly as in A.
The fungal CenH3 HFD tree (Figure 2A) was obtained using a fixed-rate model of amino acid substitution. Subsequent model testing showed that the likelihood of the data was significantly higher under evolutionary models where substitution rates were allowed to vary across sites (Yang 1994; Felsenstein and Churchill 1996). Maximum-likelihood estimates of α, the shape parameter of the gamma distribution used to estimate rates, were consistently <1, indicating that most sites have very low substitution rates (or are nearly invariable), while some have high rates, as is the case exactly. Including a class of invariant sites (a so-called gamma+I model), increased the likelihood further; however, gamma+I models are problematic when α < 1, because the “invariant” sites are not reliably distinguished from the “nearly invariant” sites accommodated by α < 1 (Gu 1999). (The model-testing data are provided in supplemental Table S1 at http://www.genetics.org/supplemental/.) Importantly, tree searches under either gamma or gamma+I models yielded trees having the same topology as that of Figure 2A, although branch credibilities decreased and 95% confidence intervals for branch lengths increased.
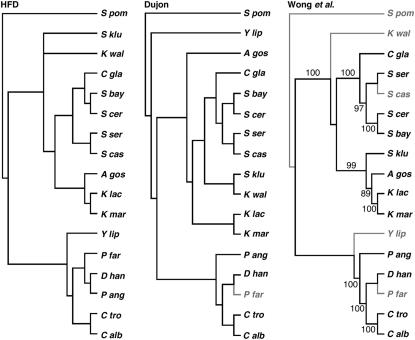
While the data set is small considering the long evolutionary history it samples, the HFD tree recapitulates the accepted phylogeny of these species (Taylor et al. 1993; Kurtzman 2003; Wong et al. 2003; Scannell et al. 2006). Focusing only on the hemiascomycetes, for which a considerable amount of molecular phylogenetic data exist, likelihood tests were carried out to compare the HFD tree with phylogenies derived from rRNA and genomic sequencing. Figure 3 shows the trees tested. The left-hand tree is that of Figure 2A, but including only the 16 hemiascomycetes plus S. pombe. On the basis of additional data presented below, A. gossypii was placed on the K. marxianus/K. lactis branch. The middle tree in Figure 3 is that proposed by Dujon (2005) based on multiple gene comparisons (Kurtzman 2003; Diezmann et al. 2004). The right-hand tree is from Wong et al. (2003) and is based on combined 5S, 18S, 5.8S, and 26S rRNAs. Pairwise KHT tests were made between the CenH3 tree and each of the others, and the data fit all trees equally well; i.e., there was no significant difference in the likelihood of the data under any of the models (supplemental Table S1 at http://www.genetics.org/supplemental/). Comparing the three trees using a Shimodaira–Hasegawa test, which corrects for multiple comparisons (Shimodaira and Hasegawa 1999), resulted in even higher P-values (data not shown). To assess the sensitivity of the calculated likelihoods to perturbations of the tree, KHT tests were made on trees in which specific branches were altered. Grouping S. servazzii with the sensu stricto yeasts resulted in likelihood differences at the bounds of statistical significance, while the more radical change of forcing P. angusta onto the Saccharomyces/Kluyveromyces branch resulted in likelihood differences of high significance (trees A and B, supplemental Table S1).
Figure 3.—
Tree topology tests. The left-hand cladogram shows the topology of the HFD tree with A. gossypii placed on the K. lactis/K. marxianus branch as described in the text. The 17-taxa tree includes only the hemiascomycetes plus S. pombe. The middle- and right-hand cladograms show phylogenies proposed by Dujon (2005) and Wong et al. (2003), respectively, based on multigene alignments. Species shown in shaded type were not included in the respective studies. Numbers on the Wong et al. tree indicate the reported percentage of bootstrap support for branches.
END homology:
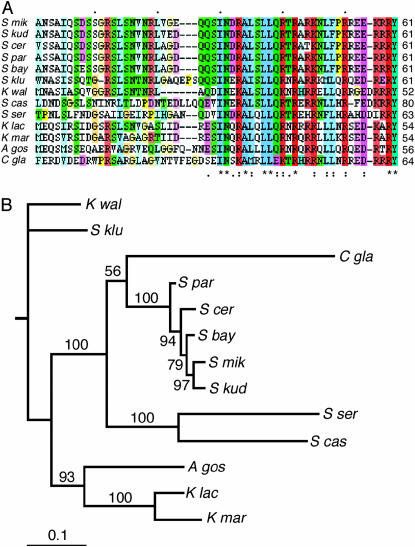
A domain homologous to the S. cerevisiae END was found in the N termini of all yeasts of the Saccharomyces/Kluyveromyces clade (Figure 4A). The strongest homology was to positions 32–61 of S. cerevisiae Cse4, corresponding almost exactly to the endpoints of the END identified by functional assay (residues 28–60) (Chen et al. 2000). Weaker homology extended an additional 30 residues on the N-terminal side of the END. The location of the END homology was variable with respect to its spacing from the HFD, ranging from 31 residues in K. lactis to 131 residues in S. servazzii. Mutational studies in S. cerevisiae have shown no strict functional constraint on this spacing; proteins having the END adjacent to or separated by >300 amino acid residues from the HFD retain wild-type Cse4 function (Chen et al. 2000). No homologies were observed within the N termini of any of the other fungi. Overall, the lengths of the N termini are quite variable, ranging from 20 amino acids (S. pombe) to 194 amino acids (S. servazzii).
Figure 4.—
END homology. (A) An alignment of the END homology regions present in the N termini of S. cerevisiae and close relatives is shown in ClustalX format and coloring, with color intensity proportional to the conservation index. (B) A phylogeny of the Saccharomyces/Kluyveromyces clade obtained by Bayesian inference using an alignment containing both HFD and END sequences (13 taxa, 157 sites). The phylogram shows the majority rule consensus tree produced by MrBayes, running a fixed-rate model with WAG rates (see materials and methods). Numbers above the branches are the percentage of times that branch was present in the posterior distribution of trees.
Adding the END region alignment to the HFD alignment enabled us to derive an improved phylogeny for the Saccharomyces/Kluyveromyces branch of the tree (Figure 4B). The HFDs of this 13-taxa group are identical in length, allowing alignment without gaps. In addition, inclusion of the END allowed resolution of the five sensu stricto species that have essentially identical HFDs. The resulting tree topology was the same as the original HFD tree except A. gossypii resolved onto the K. lactis/K. marxianus branch. Model testing showed that the data were more likely under gamma or gamma+I substitution models, not surprising since the HFD comprises 111 of the 157 total sites of the alignment, but again tree searches under variable-sites models yielded trees identical to or nearly identical to the fixed-rate tree. In contrast to the case for the entire HFD tree, the use of gamma models for this 13-taxa group resulted in improved branch credibilities (data not shown).
Variable-sites analysis:
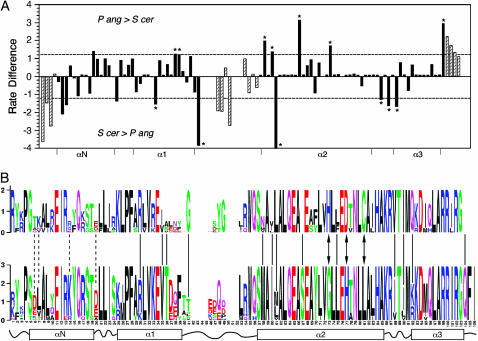
In analyzing the Saccharomyces/Kluyveromyces clade separately from the rest of the HFD tree, we noted that estimates of α under gamma models were consistently lower for this major branch than values of α estimated for the entire 22-taxa tree. Gu has shown that this situation occurs when the locations of variable positions differ for different branches of the tree (Gu 1999). To confirm this, substitution rates were estimated separately for the two major homophyletic groupings of hemiascomycetes, the S. cerevisiae clade (10 taxa) and the P. angusta clade (6 taxa). The difference in estimated substitution rate was calculated for each position of the alignment (Figure 5A). The rate differences were not normally distributed (Kolmogorov–Smirnov test, P < 10−4); rather, there was an excess of values in the tails of the distribution (kurtosis = 2.6). Ignoring the four N-terminal residues lying outside of the N-helix and positions in loop 1 and the extreme C terminus where alignment gaps introduce uncertainty in the likelihood calculations (hatched bars in Figure 5A), there were 17 positions where the rate difference between branches was greater than one standard deviation from the mean. They were equally divided between positions more variable in the S. cerevisiae branch and those more variable in the P. angusta branch. Thirteen of the 17 sites (asterisks in Figure 5A) were invariant in one lineage and variable in the other. The differentially variable sites, identified in Figure 5B with respect to the consensus sequence of each branch, were located throughout the HFD and not restricted to any specific secondary structural element. In addition, three invariant positions were identified that differed in amino acid between branches (arrows in Figure 5B). Thus, in addition to the significant number of positions that are 100% conserved over the entire HFD tree, additional residues are coconserved in different branches of the tree.
Figure 5.—
Variable-sites analysis. (A) The substitution rate at each site was estimated separately for the P. angusta and S. cerevisiae branches of the HFD tree, and the rate difference plotted vs. position. Dashed lines indicate the standard deviation of the difference calculated over all sites. Asterisks denote positions that are invariable in one branch but variable in the other. Positions N-terminal to the HFD or where gaps are present in the alignment are designated with hatched bars. The boundaries of the HFD helical domains are indicated on the x-axis. (B) The consensus sequences of proteins of the P. angusta (top) and S. cerevisiae (bottom) clades are shown in logo format (Schneider and Stephens 1990). Solid and dashed lines indicate positions where the substitution rate difference varies by more than one standard deviation from the mean (loop 1 and C-terminal residues excluded), with the solid lines corresponding to positions that are invariant in one branch or the other. Arrows denote positions that are invariant in both branches but different in amino acid.
Complementation of S. cerevisiae Cse4 function by heterologous CenH3 proteins:
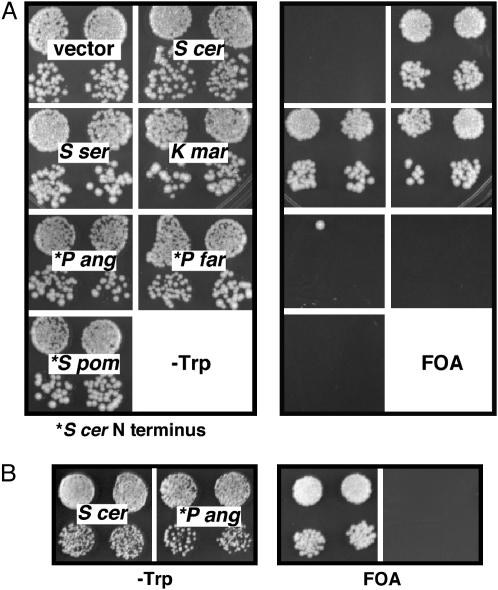
Several of the CenH3 orthologs were tested for their ability to complement Cse4 function in S. cerevisiae. All were expressed using the promoter and 5′ upstream region of S. cerevisiae CSE4. In cases where the proteins lacked the END homology, the HFDs were fused to the complete S. cerevisiae N terminus, which also carried an HA epitope tag. K. marxianus and S. servazzii orthologs complemented S. cerevisiae cse4Δ; S. pombe, P. farinosa, and P. angusta did not (Figure 6). Western blots showed that all of the noncomplementing proteins were expressed at levels comparable to that of S. cerevisiae Cse4 (data not shown). Although not confirmed here, Stoyan et al. (2004) showed that C. glabrata Cse4 also complements S. cerevisiae cse4Δ. Complementing ability correlated with evolutionary distance; all of the complementing and none of the noncomplementing proteins lie on the Saccharomyces/Kluyveromyces branch of the HFD tree. Mitotic chromosome segregation was analyzed in strains complemented by the K. marxianus and S. servazzii orthologs. The assay, which measures mitotic loss (or gain) of one copy of chromosome III from a diploid test strain, is sensitive to relatively small perturbations to centromere function (Hegemann et al. 1988). Mutations affecting the HFD of S. cerevisiae Cse4 display chromosome III nondisjunction rates 5- to 25-fold higher than the wild-type rate of 2×10−4 events/division (Keith et al. 1999). Nonlethal END mutations cause 10-fold elevations in loss rate (Chen et al. 2000). Here, loss rates measured for the S. servazzii and K. marxianus strains were 3.5 × 10−4 and 6.8 × 10−4 events/division, respectively, only 2- to 4-fold higher than the measured wild-type rate of 1.8 × 10−4. In all cases, the loss events were associated with chromosome gain, i.e., 2:0 segregation (nondisjunction). Thus, both orthologs provide near wild-type Cse4 function in S. cerevisiae.
Figure 6.—
Function of orthologous Cse4 proteins in S. cerevisiae. Cse4 orthologs were expressed in S. cerevisiae and tested for their ability to complement a cse4 null allele, as observed by the ability to grow on FOA medium. Proteins indicated by an asterisk were tested as HFD fusions to the S. cerevisiae N terminus. (A) The S. cerevisiae host strain carries wild-type copies of histone H3 and H4 genes at both genomic loci. (The single FOAr colony arising among the progeny of one of the P. angusta HFD transformants was likely the result of a rare interplasmid recombination event; no FOAr colonies were observed in other replicates of this experiment.) (B) The S. cerevisiae host strain carries a single S. cerevisiae H3 gene (HHT2) and a single P. angusta H4 gene (HHF2Pang) (see materials and methods).
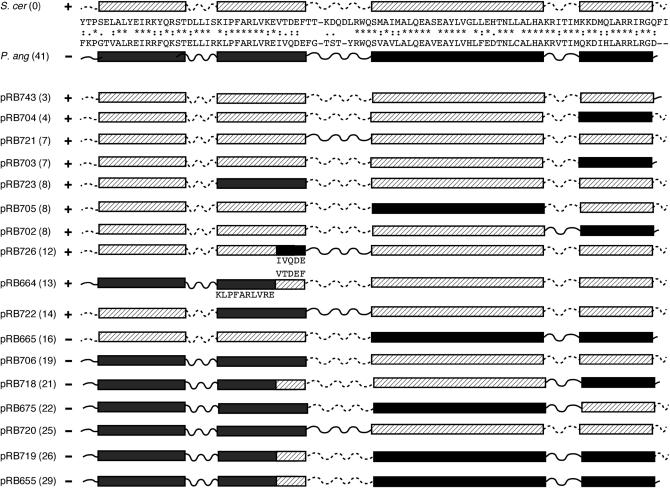
What accounts for the species specificity of some yeast CenH3's? P. angusta Cse4, the most diverged of the orthologs tested, is about as diverged from S. cerevisiae Cse4 in the HFD (59% amino acid identity) as S. cerevisiae Cse4 is from S. cerevisiae H3 (60% identity). Keith et al. (1999) tested Cse4–H3 chimeras (“domain swaps”) and found that no localized domain of Cse4 confers centromere-specific function to H3; i.e., residues specifying Cse4 function were located throughout the entire Cse4 HFD. But as Vermaak et al. (2002) pointed out, CenH3 and H3 proteins have probably evolved under quite different selective constraints, and the more appropriate test for analyzing CenH3 species specificity would be to swap comparable regions of different CenH3's. Taking this approach, these authors showed that the loop 1 region is necessary and sufficient for species-specific centromere targeting of Cid in Drosophila (Vermaak et al. 2002). To determine if the same were true for the yeasts, chimeras of P. angusta and S. cerevisiae Cse4 were constructed and analyzed for function in S. cerevisiae. P. angusta and S. cerevisiae Cse4 HFDs are equally as diverged (59% amino acid identity) as the two Cid proteins analyzed in the Drosophila study (D. melanogaster and D. bipectinata).
The chimeric proteins tested are shown in Figure 7. All contain the full-length S. cerevisiae N terminus, and all were found to be expressed at levels comparable to that of the wild-type protein (data not shown). The pRB655 and pRB726 swaps are analogous to the Cid chimeras analyzed by Vermaak et al. (2002). In contrast to the Drosophila example, substituting the S. cerevisiae loop 1 region (including five C-terminal residues of helix 1) into the P. angusta protein (pRB655) was not sufficient to confer Cse4 function in S. cerevisiae, nor did the reciprocal swap abolish function of the S. cerevisiae protein (pRB726). In fact, S. cerevisiae Cse4 tolerated exchanges of its N-helix-N loop (pRB664), helix 1 (pRB723), loop 1 (pRB721), helix 2 (pRB705), and helix 3 with or without loop 2 and the C terminus (pRB702, pRB703, pRB704), while still maintaining the ability to provide Cse4 function in the absence of wild-type protein. Conversely, no functional chimera was produced that contained P. angusta substitutions at ≥16 of the 41 amino acids that differ between the two HFD domains regardless of where the substitutions were located (Figure 7). The chimera collection is not a random sampling of all amino acid replacements possible; however, the results suggest that Cse4 function in S. cerevisiae is inversely correlated with the extent of P. angusta amino acid replacements rather than with the presence or absence of any given element of S. cerevisiae Cse4 secondary structure.
Figure 7.—
P. angusta–S. cerevisiae chimeras. Chimeric genes were constructed in which one or more regions of the S. cerevisiae Cse4 HFD (hatched boxes, dotted lines) were exchanged with the corresponding regions of the P. angusta HFD (solid boxes, solid lines). The chimeric HFDs, fused to the full-length S. cerevisiae N terminus, were expressed and tested for function in S. cerevisiae. Plus (+) and minus (−) signs indicate the ability and inability, respectively, of the chimeric genes to complement a cse4 null mutation. Numbers in parentheses are the total number of amino acid substitutions present with respect to the S. cerevisiae sequence.
In contrast to the species specificity of P. angusta CenH3, core histones H3 and H4 were found to be interchangeable between P. angusta and S. cerevisiae. P. angusta H4 differs from S. cerevisiae H4 at three positions (alanine vs. glycine at position 49, asparagine vs. serine at position 65, and alanine vs. serine at position 70). P. angusta H3 also differs at three positions from S. cerevisiae H3 (alanine vs. serine at position 32, cysteine vs. alanine at position 111, and glutamine vs. lysine at position 126). The S. cerevisiae HHT2-HHF2 locus encoding H3 and H4 was mutagenized to encode P. angusta replacement amino acids in both proteins, and viable S. cerevisiae strains were generated that expressed only P. angusta H3, only P. angusta H4, or both (supplemental Figure S1 at http://www.genetics.org/supplemental/). To test the hypothesis that P. angusta CenH3 fails to function in S. cerevisiae due to incompatibility with S. cerevisiae H4, a tester strain was constructed that expressed only P. angusta H4. The P. angusta Cse4 ortholog (with S. cerevisiae N terminus) also failed to complement Δcse4 in this strain background (Figure 6B).
DISCUSSION
A previous phylogenetic analysis of CenH3's, sampled widely from plant and animal kingdoms, found “a remarkably poor lack of resolution,” leading the authors to question the assumption of CenH3 orthology and suggest instead that CenH3's might have arisen multiple times during the course of evolution (Malik and Henikoff 2003). The results here offer an alternative view. First, fungal CenH3 HFD phylogeny gives strong support to the existence of an ancestral CenH3 protein common to all of the Ascomycota, three main branches of which—Archaeascomycetes (S. pombe), Hemiascomycetes (S. cerevisiae), and Euascomycetes (N. crassa)—were already established 300 million years ago (Berbee and Taylor 1993). Second, when canonical H3's are included in the phylogeny, the H3's and CenH3's form separate monophyletic groupings. Third, fungal CenH3 HFD phylogeny is congruent with the accepted evolutionary relationships between these species. In particular, the hemiascomycete HFD tree does not differ significantly from trees derived from multigene alignments of rRNA and other conserved proteins. While the data are not sufficient to exclude definitively other interpretations, they fully support the conclusion that the CenH3 proteins of all the Fungi, an early radiation of Eukaryota, are orthologous.
Sequence divergence among the fungal CenH3 HFDs is not uniform. Loop 1 regions and C termini are extremely variable, while, of helical domains, the N-helix and helix 1 have more replacements than helices 2 and 3. Thirty-one percent of HFD residues are invariant across the entire fungal HFD phylogeny. The variability in replacement rates presumably reflects differing selective pressures acting at different sites of the protein. In the maximum-likelihood framework, the variability in amino acid replacement rates is reflected by significantly increased likelihoods for the data under evolutionary models allowing for variable rates across sites, i.e., gamma or gamma+I models. However, variable-sites models assume that the substitution rate at each site, while variable, does not change over the evolutionary history of the protein; i.e., it is the same in all branches of the phylogeny. In their covarion model of evolution, Fitch and Markowitz (1970) proposed that sites critical to a protein's function may change over time and in different lineages, causing substitution rates to vary in different branches of the phylogeny. Further, the covarion model posits that amino acid substitution at one site in the protein can affect the substitution rate at other sites with which it “covaries.” The fungal CenH3 data, albeit a limited sampling, are consistent with a covarion mechanism. Rate variability (characterized by the α-parameter of the gamma distribution) is not homogeneous over all branches of the tree. The inhomogeneity is explained by the fact that the location of variable positions differs between branches. Positions 35/36 (helix 1), 58/60/67/75 (helix 2), and 104 (C terminus) are invariant in the Saccharomyces/Kluyveromyces clade, but not in the P. angusta clade, while the opposite is true for positions 30 (helix 1), 41 (loop 1), 61 (helix 2), 88/90 (loop 2), and 92 (helix 3). Positions 73, 77, and 81 in helix 2 are a special case of covarying sites. Although invariant in both major branches of the tree, they differ in amino acid between branches. We interpret the lineage-specific coconservation of amino acid sequence to mean that when a substitution first arose at one of those positions, it created strong selective pressure for the respective amino acids at the other positions. It is not unexpected that the covarying amino acids are scattered across the HFD sequence. By analogy to the known structure of H3 within the nucleosome, CenH3 is expected to make multiple tertiary and quaternary interactions; therefore, a structural alteration in one part of the molecule could readily affect the structure at a distant site. An example of long-range interaction between Cse4 amino acids is the temperature-sensitive cse4-102 allele, which carries two replacements, leucine to serine at position 175 in loop 1 and methionine to threonine at position 217 in helix 3. Neither mutation by itself causes a centromere defect, but together they lead to mitotic arrest at the restrictive temperature (Glowczewski et al. 2000).
Plant and Drosophila CenH3 proteins evolve adaptively; i.e., they are subject to positive selection (Malik and Henikoff 2001; Cooper and Henikoff 2004). It has been suggested that the positive selection is driven by altered DNA binding specificity in response to rapidly evolving centromeric satellite DNAs in the respective organisms (Henikoff et al. 2001). Positive selection is recognized in molecular phylogenetic analyses by an increased rate of nonsynonymous codon change (Ka) relative to the rate of synonymous change (Ks). In Brassicaceae and Drosophila lineages, loop 1 codons of CenH3 genes display Ka/Ks ratios significantly >1, and it is this region of Drosophila Cid that is both necessary and sufficient for species-specific centromere localization (Vermaak et al. 2002). Previous tests on a limited number of yeast CenH3 sequences found no evidence for positive selection (Talbert et al. 2004). Positive selection tests were performed on the fungal CenH3 genes studied here, and again results were negative (R. Baker, unpublished results), although analyzing loop 1, the region where positive selection might be expected, is problematic due to uncertainty in the alignment over this highly variable region. Separate analysis of the two major branches of the yeast CenH3's, which could be aligned with no (Saccharomyces/Kluyveromyces clade) or at most two gaps (P. angusta group), also yielded no evidence for positive selection. In fact, consistent with the previous report, strong purifying selection was observed (Ka/Ks = 0.01–0.02) at all sites.
One aspect of the yeast CenH3 phylogeny that may argue for coevolution of CenH3's with centromere DNA sequence is conservation of the END homology in yeasts of the Kluyveromyces/Saccharomyces clade. These yeasts all have simple, point centromeres closely resembling those of S. cerevisiae, while centromeres of the other yeasts bear more similarity to the complex “regional” centromeres of S. pombe and higher eukaryotes (Cleveland et al. 2003). Centromeres of S. cerevisiae were the first to be cloned and sequenced (Fitzgerald-Hayes et al. 1982). They are 111–120 bp in length and consist of three conserved DNA elements (CDEs): CDEI, the degenerate octanucleotide RTCACRTG; CDEII, 79–88 bp of highly AT-rich (86–98%) DNA; and CDEIII, a conserved 24-bp sequence that binds the essential kinetochore protein complex CBF3 (Baker and Rogers 2005 and primary references therein). Centromeres of the other Kluyveromyces/Saccharomyces yeasts are similar, differing only in the orientation of CDEI, the length of CDEII, and the specific sequence of CDEIII (Heus et al. 1993; Iborra and Ball 1994; Kitada et al. 1997). In contrast, S. pombe centromeres are 40–100 kbp in length and consist of 4–7 kbp AT-rich, nonhomologous central cores flanked by repetitive sequences (Steiner et al. 1993). C. albicans centromeres resemble the S. pombe central cores, but lack the flanking repeats (Sanyal et al. 2004), while Y. lipolytica centromeres are similar in size to those of S. cerevisiae but, aside from AT-richness, lack uniquely recognizable sequence motifs (Vernis et al. 2001). All yeasts with S. cerevisiae-like CDEI–CDEII–CDEIII centromeres also have CenH3's containing the END homology in the N terminus, while the N termini of other yeast CenH3's lack the END homology or any other conserved motif. The observed conservation of the END is not due simply to lack of evolutionary distance between the sampled taxa, because no sequence conservation is observed elsewhere in the N termini, and HFD divergence (i.e., total branch length) is roughly equivalent between species of the Kluyveromyces/Saccharomyces clade and the other hemiascomycetes considered separately.
In contrast to results in other systems where CenH3 function appears to be conserved even between CenH3's separated by long evolutionary distances, fungal CenH3 HFDs are not universally interchangeable despite less divergence. In the examples tested here, the noncomplementing P. farinosa, P. angusta, and S. pombe HFDs are less diverged from S. cerevisiae (35, 41, and 40% replacements, respectively) than is human from either S. cerevisiae or D. melanogaster HFDs (49 and 62% replacements, respectively), both pairings having been found to be cross-functional (Henikoff et al. 2000; Wieland et al. 2004). One explanation for the disparity in results may be that the idiosyncratic S. cerevisiae point centromere places severe structural constraints on its CenH3. More likely, the difference can be attributed to the functional assays used to assess CenH3 function in the various experimental systems. Our test for function in S. cerevisiae is stringent: it requires the heterologous CenH3 to provide Cse4 function in the complete absence of endogenous protein. In the human and fly systems, the assay was centromere localization of the heterologous CenH3 in tissue culture cells or the ability to rescue cells from mitotic arrest upon RNAi inhibition of endogenous CENP-A. In neither case was endogenous CenH3 completely absent, nor was the accuracy of chromosome segregation measured.
One possible explanation for why CenH3's from distantly related yeasts (e.g., P. angusta) fail to function in S. cerevisiae is that they are unable to interact effectively with the other S. cerevisiae core histones, in particular H4. P. angusta H3 and H4 each differ from their S. cerevisiae counterparts at three amino acid positions; however, they are able to substitute for their S. cerevisiae counterparts either singly or together. In contrast, the P. angusta Cse4 ortholog (with S. cerevisiae N terminus) fails to complement Δcse4 in a strain background expressing only P. angusta H4. Together these results indicate that S. cerevisiae H2A–H2B dimers are compatible with P. angusta (H3–H4)2 tetramers and that the cross-species incompatibility of P. angusta Cse4 in S. cerevisiae is not due to the absence of its cognate H4.
Biochemical and genetic experiments have identified key regions of the CenH3 protein that distinguish it functionally from H3. Black et al. (2004) found that tetramers of CENP-A and histone H4 are more compact than H3–H4 tetramers and that the structural alteration is due to more rigid conformation in the loop 1–helix 2 region of CENP-A. Substitution of the corresponding loop 1–helix 2 residues into H3 was sufficient to target the chimera to centromeric chromatin in vivo, consistent with earlier findings by Shelby et al. (1997) identifying both loop 1 and helix 2 as critical for CENP-A centromere targeting. Again, the situation appears to differ in S. cerevisiae. Keith et al. (1999) found that residues critical for S. cerevisiae Cse4p function are distributed across the entire Cse4 HFD. Similarly, here we find that the ability of P. angusta–S. cerevisiae chimeric Cse4 HFDs to function in S. cerevisiae correlates with the overall divergence of the chimeric HFD from S. cerevisiae Cse4, not the presence or absence of any specific secondary structural determinant(s). Cse4 function is lost as the number of amino acid replacements increases, regardless if the replacement amino acid is from H3 or from the heterologous CenH3. This is not consistent with the notion that CenH3 evolution has converged to a universal base structure fine tuned by adaptive variations in some lineages; rather, it might appear that the P. angusta Cse4 ortholog is structurally as different from S. cerevisiae Cse4 as is H3. In general, we suggest that CenH3 evolution may be more constrained than the existing sequence divergence and apparent functional interchangeability would imply: differing lineage-specific selective constraints have produced a diversity of lineage-specific (and noninterchangeable) solutions to the problem of packaging centromeric chromatin.
Acknowledgments
The authors thank Cletus Kurtzman for supplying reference strains and 23S rRNA alignments. This work was supported by a grant from the National Institutes of Health to R.E.B. (GM61120).
References
- Arents, G., R. W. Burlingame, B. C. Wang, W. E. Love and E. N. Moudrianakis, 1991. The nucleosomal core histone octamer at 3.1 A resolution: a tripartite protein assembly and a left-handed superhelix. Proc. Natl. Acad. Sci. USA 88: 10148–10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, R. E., and K. Rogers, 2005. Genetic and genomic analysis of the AT-rich centromere DNA element II of Saccharomyces cerevisiae. Genetics 171: 1463–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbee, M. L., and J. W. Taylor, 1993. Dating the evolutionary radiations of the true fungi. Can. J. Bot. 71: 1114–1127. [Google Scholar]
- Black, B. E., D. R. Foltz, S. Chakravarthy, K. Luger, V. L. Woods, Jr. et al., 2004. Structural determinants for generating centromeric chromatin. Nature 430: 578–582. [DOI] [PubMed] [Google Scholar]
- Blandin, G., B. Llorente, A. Malpertuy, P. Wincker, F. Artiguenave et al., 2000. Genomic exploration of the hemiascomycetous yeasts: 13. Pichia angusta. FEBS Lett. 487: 76–81. [DOI] [PubMed] [Google Scholar]
- Bruns, T. D., R. Vilgalys, S. M. Barns, D. Gonzalez, D. S. Hibbett et al., 1992. Evolutionary relationships within the fungi: analyses of nuclear small subunit rRNA sequences. Mol. Phylogenet. Evol. 1: 231–241. [DOI] [PubMed] [Google Scholar]
- Buchwitz, B. J., K. Ahmad, L. L. Moore, M. B. Roth and S. Henikoff, 1999. A histone-H3-like protein in C. elegans. Nature 401: 547–548. [DOI] [PubMed] [Google Scholar]
- Chen, Y., R. E. Baker, K. C. Keith, K. Harris, S. Stoler et al., 2000. The N terminus of the centromere H3-like protein Cse4p performs an essential function distinct from that of the histone fold domain. Mol. Cell. Biol. 20: 7037–7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson et al., 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31: 3497–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clamp, M., J. Cuff, S. M. Searle and G. J. Barton, 2004. The Jalview Java alignment editor. Bioinformatics 20: 426–427. [DOI] [PubMed] [Google Scholar]
- Cleveland, D. W., Y. Mao and K. F. Sullivan, 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112: 407–421. [DOI] [PubMed] [Google Scholar]
- Cooper, J. L., and S. Henikoff, 2004. Adaptive evolution of the histone fold domain in centromeric histones. Mol. Biol. Evol. 21: 1712–1718. [DOI] [PubMed] [Google Scholar]
- Crooks, G. E., G. Hon, J. M. Chandonia and S. E. Brenner, 2004. WebLogo: a sequence logo generator. Genome Res. 14: 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diezmann, S., C. J. Cox, G. Schonian, R. J. Vilgalys and T. G. Mitchell, 2004. Phylogeny and evolution of medical species of Candida and related taxa: a multigenic analysis. J. Clin. Microbiol. 42: 5624–5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon, B., 2005. Hemiascomycetous yeasts at the forefront of comparative genomics. Curr. Opin. Genet. Dev. 15: 614–620. [DOI] [PubMed] [Google Scholar]
- Edwards, N. S., and A. W. Murray, 2005. Identification of xenopus CENP-A and an associated centromeric DNA repeat. Mol. Biol. Cell 16: 1800–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J., 2004. PHYLIP (Phylogeny Inference Package) Version 3.6. Department of Genome Sciences, University of Washington, Seattle.
- Felsenstein, J., and G. A. Churchill, 1996. A hidden Markov model approach to variation among sites in rate of evolution. Mol. Biol. Evol. 13: 93–104. [DOI] [PubMed] [Google Scholar]
- Feng, D. F., G. Cho and R. F. Doolittle, 1997. Determining divergence times with a protein clock: update and reevaluation. Proc. Natl. Acad. Sci. USA 94: 13028–13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch, W. M., and E. Markowitz, 1970. An improved method for determining codon variability in a gene and its application to the rate of fixation of mutations in evolution. Biochem. Genet. 4: 579–593. [DOI] [PubMed] [Google Scholar]
- Fitzgerald-Hayes, M., J. M. Buhler, T. G. Cooper and J. Carbon, 1982. Isolation and subcloning analysis of functional centromere DNA (CEN11) from Saccharomyces cerevisiae chromosome XI. Mol. Cell. Biol. 2: 82–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowczewski, L., P. Yang, T. Kalashnikova, M. S. Santisteban and M. M. Smith, 2000. Histone-histone interactions and centromere function. Mol. Cell. Biol. 20: 5700–5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, X., 1999. Statistical methods for testing functional divergence after gene duplication. Mol. Biol. Evol. 16: 1664–1674. [DOI] [PubMed] [Google Scholar]
- Hegemann, J. H., J. H. Shero, G. Cottarel, P. Philippsen and P. Hieter, 1988. Mutational analysis of centromere DNA from chromosome VI of Saccharomyces cerevisiae. Mol. Cell. Biol. 8: 2523–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S., and Y. Dalal, 2005. Centromeric chromatin: What makes it unique? Curr. Opin. Genet. Dev. 15: 177–184. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., K. Ahmad, J. S. Platero and B. van Steensel, 2000. Heterochromatic deposition of centromeric histone H3-like proteins. Proc. Natl. Acad. Sci. USA 97: 716–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S., K. Ahmad and H. S. Malik, 2001. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293: 1098–1102. [DOI] [PubMed] [Google Scholar]
- Heus, J. J., B. J. Zonneveld, H. Y. de Steensma and J. A. van den Berg, 1993. The consensus sequence of Kluyveromyces lactis centromeres shows homology to functional centromeric DNA from Saccharomyces cerevisiae. Mol. Gen. Genet. 236: 355–362. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck, J. P., and F. Ronquist, 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- Iborra, F., and M. M. Ball, 1994. Kluyveromyces marxianus small DNA fragments contain both autonomous replicative and centromeric elements that also function in Kluyveromyces lactis. Yeast 10: 1621–1629. [DOI] [PubMed] [Google Scholar]
- Jones, D. T., W. R. Taylor and J. M. Thornton, 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8: 275–282. [DOI] [PubMed] [Google Scholar]
- Kass, R. E., and A. E. Raftery, 1995. Bayes factors. J. Am. Stat. Assoc. 90: 773–795. [Google Scholar]
- Keith, K. C., R. E. Baker, Y. Chen, K. Harris, S. Stoler et al., 1999. Analysis of primary structural determinants that distinguish the centromere-specific function of histone variant Cse4p from histone H3. Mol. Cell. Biol. 19: 6130–6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishino, H., and M. Hasegawa, 1989. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in hominoidea. J. Mol. Evol. 29: 170–179. [DOI] [PubMed] [Google Scholar]
- Kitada, K., E. Yamaguchi, K. Hamada and M. Arisawa, 1997. Structural analysis of a Candida glabrata centromere and its functional homology to the Saccharomyces cerevisiae centromere. Curr. Genet. 31: 122–127. [DOI] [PubMed] [Google Scholar]
- Kurtzman, C. P., 2003. Phylogenetic circumscription of Saccharomyces, Kluyveromyces and other members of the Saccharomycetaceae, and the proposal of the new genera Lachancea, Nakaseomyces, Naumovia, Vanderwaltozyma and Zygotorulaspora. FEMS Yeast Res. 4: 233–245. [DOI] [PubMed] [Google Scholar]
- Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent and T. J. Richmond, 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389: 251–260. [DOI] [PubMed] [Google Scholar]
- Malik, H. S., and S. Henikoff, 2001. Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics 157: 1293–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik, H. S., and S. Henikoff, 2003. Phylogenomics of the nucleosome. Nat. Struct. Biol. 10: 882–891. [DOI] [PubMed] [Google Scholar]
- Malik, H. S., D. Vermaak and S. Henikoff, 2002. Recurrent evolution of DNA-binding motifs in the Drosophila centromeric histone. Proc. Natl. Acad. Sci. USA 99: 1449–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellone, B. G., and R. C. Allshire, 2003. Stretching it: putting the CEN(P-A) in centromere. Curr. Opin. Genet. Dev. 13: 191–198. [DOI] [PubMed] [Google Scholar]
- Meluh, P. B., P. Yang, L. Glowczewski, D. Koshland and M. M. Smith, 1998. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell 94: 607–613. [DOI] [PubMed] [Google Scholar]
- Morey, L., K. Barnes, Y. Chen, M. Fitzgerald-Hayes and R. E. Baker, 2004. The histone fold domain of Cse4 is sufficient for CEN targeting and propagation of active centromeres in budding yeast. Eukaryot. Cell 3: 1533–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas, A., D. Kruger and S. B. Carroll, 2005. Animal evolution and the molecular signature of radiations compressed in time. Science 310: 1933–1938. [DOI] [PubMed] [Google Scholar]
- Ronquist, F., and J. P. Huelsenbeck, 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Sanyal, K., M. Baum and J. Carbon, 2004. Centromeric DNA sequences in the pathogenic yeast Candida albicans are all different and unique. Proc. Natl. Acad. Sci. USA 101: 11374–11379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell, D. R., K. P. Byrne, J. L. Gordon, S. Wong and K. H. Wolfe, 2006. Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature 440: 341–345. [DOI] [PubMed] [Google Scholar]
- Schneider, T. D., and R. M. Stephens, 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18: 6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby, R. D., O. Vafa and K. F. Sullivan, 1997. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J. Cell Biol. 136: 501–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira, H., and M. Hasegawa, 1999. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 16: 1114–1116. [Google Scholar]
- Sipiczki, M., 2000. Where does fission yeast sit on the tree of life? Genome Biol. 1: REVIEWS1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner, N. C., K. M. Hahnenberger and L. Clarke, 1993. Centromeres of the fission yeast Schizosaccharomyces pombe are highly variable genetic loci. Mol. Cell. Biol. 13: 4578–4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler, S., K. C. Keith, K. E. Curnick and M. Fitzgerald-Hayes, 1995. A mutation in CSE4, and essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 9: 573–586. [DOI] [PubMed] [Google Scholar]
- Stoyan, T., and J. Carbon, 2004. Inner kinetochore of the pathogenic yeast Candida glabrata. Eukaryot. Cell 3: 1154–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, B. A., M. D. Blower and G. H. Karpen, 2001. Determining centromere identity: cyclical stories and forking paths. Nat. Rev. Genet. 2: 584–596. [DOI] [PubMed] [Google Scholar]
- Sullivan, K. F., 2001. A solid foundation: functional specialization of centromeric chromatin. Curr. Opin. Genet. Dev. 11: 182–188. [DOI] [PubMed] [Google Scholar]
- Sullivan, K. F., M. Hechenberger and K. Masri, 1994. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J. Cell Biol. 127: 581–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, K., E. S. Chen and M. Yanagida, 2000. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science 288: 2215–2219. [DOI] [PubMed] [Google Scholar]
- Talbert, P. B., R. Masuelli, A. P. Tyagi, L. Comai and S. Henikoff, 2002. Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell 14: 1053–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert, P. B., T. D. Bryson and S. Henikoff, 2004. Adaptive evolution of centromere proteins in plants and animals. J. Biol. 3: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, J. W., B. Bowman, M. L. Berbee and T. J. White, 1993. Fungal model organisms: phylogenetics of Saccharomyces, Aspergillus and Neurospora. Syst. Biol. 42: 440–457. [Google Scholar]
- Van Hooser, A. A., I. I. Ouspenski, H. C. Gregson, D. A. Starr, T. J. Yen et al., 2001. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J. Cell Sci. 114: 3529–3542. [DOI] [PubMed] [Google Scholar]
- Vermaak, D., H. S. Hayden and S. Henikoff, 2002. Centromere targeting element within the histone fold domain of Cid. Mol. Cell. Biol. 22: 7553–7561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernis, L., L. Poljak, M. Chasles, K. Uchida, S. Casaregola et al., 2001. Only centromeres can supply the partition system required for ARS function in the yeast Yarrowia lipolytica. J. Mol. Biol. 305: 203–217. [DOI] [PubMed] [Google Scholar]
- Whelan, S., and N. Goldman, 2001. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18: 691–699. [DOI] [PubMed] [Google Scholar]
- Wieland, G., S. Orthaus, S. Ohndorf, S. Diekmann and P. Hemmerich, 2004. Functional complementation of human centromere protein A (CENP-A) by Cse4p from Saccharomyces cerevisiae. Mol. Cell. Biol. 24: 6620–6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, S., M. A. Fares, W. Zimmermann, G. Butler and K. H. Wolfe, 2003. Evidence from comparative genomics for a complete sexual cycle in the ‘asexual’ pathogenic yeast Candida glabrata. Genome Biol. 4: R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z., 1994. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J. Mol. Evol. 39: 306–314. [DOI] [PubMed] [Google Scholar]
- Yang, Z., 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13: 555–556. [DOI] [PubMed] [Google Scholar]