Abstract
The rainbow trout genetic linkage groups have been assigned to specific chromosomes in the OSU (2N = 60) strain using fluorescence in situ hybridization (FISH) with BAC probes containing genes mapped to each linkage group. There was a rough correlation between chromosome size and size of the genetic linkage map in centimorgans for the genetic maps based on recombination from the female parent. Chromosome size and structure have a major impact on the female:male recombination ratio, which is much higher (up to 10:1 near the centromeres) on the larger metacentric chromosomes compared to smaller acrocentric chromosomes. Eighty percent of the BAC clones containing duplicate genes mapped to a single chromosomal location, suggesting that diploidization resulted in substantial divergence of intergenic regions. The BAC clones that hybridized to both duplicate loci were usually located in the distal portion of the chromosome. Duplicate genes were almost always found at a similar location on the chromosome arm of two different chromosome pairs, suggesting that most of the chromosome rearrangements following tetraploidization were centric fusions and did not involve homeologous chromosomes. The set of BACs compiled for this research will be especially useful in construction of genome maps and identification of QTL for important traits in other salmonid fishes.
RAINBOW trout (Oncorhynchus mykiss) is a member of the family Salmonidae, which underwent an ancestral tetraploidization event ∼90–100 million years ago (Allendorf and Thorgaard 1984; Mitchell et al. 2005). This is supported by the fact that these fishes have a genome size twice that of related species and that some homeologous chromosome arms still exchange segments as a result of quadrivalent formations in male meiosis (Allendorf and Danzmann 1997). Most teleosts have a karyotype of 24–25 acrocentric (single-armed) chromosome pairs, but rainbow trout and many other salmonids have a large number of metacentric chromosome pairs and ∼100 rather than 50 chromosome arms, suggesting that many centric fusions occurred during the radiation following tetraploidization.
Several genetic linkage maps have been constructed for rainbow trout (Sakamoto et al. 2000; Robison et al. 2001; Nichols et al. 2003; Zimmerman et al. 2004; Danzmann et al. 2005) or are in progress (C. Rexroad, personal communication). The most detailed are the OSU × Arlee male map based on doubled haploids (Nichols et al. 2003) and the sex-specific maps based on two crosses done at the University of Guelph (Sakamoto et al. 2000; Danzmann et al. 2005) in which markers are traced from both male and female parents. Synteny between the genetic maps from the OSU × Arlee cross and the crosses from the University of Guelph has been established by the mapping of shared microsatellite markers (Nichols et al. 2003). Syntenic relationships of the OSU × Clearwater doubled haploid map with the other maps has also been established using shared microsatellite or AFLP markers (K. M. Nichols and J. J. DeKoning, unpublished results). All genetic markers used in this study were mapped in one of these crosses.
The karyotype of rainbow trout has been characterized, including identification of sex chromosomes (Thorgaard 1977), localization of ribosomal RNAs (Phillips and Hartley 1988), and documentation of intraspecific variation in diploid chromosome number (2n = 58–64) (Thorgaard 1983). Banding patterns obtained with various fluorochromes (Phillips and Hartley 1988) and restriction enzymes (Lloyd and Thorgaard 1988) have been described and chromosome-specific centromeric DNAs have been identified (Reed and Phillips 1997; R. B. Phillips, M. R. Morasch and K. Keatley, unpublished results). What has been lacking is the matching of specific chromosome pairs with genetic linkage groups.
The assignment of markers on the genetic map to specific chromosomes is of special interest in rainbow trout because the male and female maps are so different (Sakamoto et al. 2000). The female:male (F:M) recombination ratio is >10:1 in regions near the centromeres, but 1:10 in regions near the telomeres. There is also considerable variation among chromosomes in these rates (Danzmann et al. 2005).
In this article we used the technique of fluorescence in situ hybridization (FISH) to assign linkage groups to specific chromosomes in the OSU strain (2n = 60) of rainbow trout and to orient the linkage maps on each chromosome pair. To do this, bacterial artificial chromosome (BAC) clones containing markers mapped to each of the linkage groups were isolated and used as probes in the FISH experiments. In some cases, BACs were isolated first and then put on the genetic map using SNPs derived from end sequences or microsatellite loci isolated from the BACs. Chromosomes were identified using a combination of relative size, chromosome arm ratios, and centromere probes.
MATERIALS AND METHODS
BAC library screening:
Almost all of the clones were obtained from two different trout BAC libraries, one made from the OSU XX clonal line (Phillips et al. 2003) and another from the Swanson YY clonal line (Palti et al. 2004). Filters from the OSU library were screened for type I clones using 32P-labeled cloned probes, and the PCR superpools from the Swanson library were screened for clones containing either type I loci or microsatellite loci using specific PCR primers for each locus. Four clones from the CHORIO Atlantic salmon BAC library (Thorsen et al. 2005) were also used. These were screened from filters and sent to us by W. S. Davidson (Simon Fraser University).
Microsatellite discovery from BACs:
DNA was isolated from BAC clones, fragmented with Sau3AI or sheared by sonication, subcloned into pUC19 or pST-Blue1 vectors, and transformed into DH5α or NovaBlue competent cells according to the Novagen Perfectly Blunt cloning kit (Novagen, Madison, WI). Transformed cells were plated onto LB agarose containing ampicillin and grown overnight. Colonies were transferred to nylon membranes for microsatellite repeat detection following the protocol of Sambrook et al. (2001). Plasmid DNA was isolated from overnight cultures of positive subclones and sequenced with Big Dye Terminator chemistry (Applied Biosystems, Foster City, CA) using standard M13F and M13R primers. Sequencing reactions were purified using ethanol precipitation and electrophoresed on an ABI 3100 Genetic Analyzer (ABI, Foster City, CA). Sequence quality was verified by PHRED analysis and vector sequence removed with CROSS_MATCH (CodonCode, Dedham, MA). Contig assembly, sequence alignment, and primer design were conducted with VectorNTI version 9.0 (InforMax, Frederick, MD). Primers for the microsatellites that are not in GenBank are given in the appendix.
APPENDIX.
Primer sequences for microsatellites isolated from BACs
| Locus | BACa | Forward primer | Reverse primer |
|---|---|---|---|
| OMM3000 | B10 | GAGGTGTGGAAGGGGAATAGG | AAAGATGTTGGGCTTGGCA |
| OMM3001 | C1 | AAATGGATGATGACTGTACTA | CACACATCTCTTTGTGACA |
| OMM 3012 | B8 | TTCTCCAGGTCCTACTCCAAGT | TTTTGGAGATGAGGTGAGGG |
| OMM 3018 | E1 | CATTGGGCCCTGAGTACAGT | CACCTCTGCCAATCTAGCAA |
| OMM 3020 | F1 | CGGACACCCTGACAAGATAAC | GACAGGGACGTGACAGTGAA |
| OMM 3032 | 1MT320A01 | TGACAGTTGGGCCCTTGTAAG | GCCGGGGATAGGAATTCAAT |
| OMM 3044 | E2 | TCTCTCCCTTGTTCCCCTGA | TCCCCACAGCATAGCATGAG |
| OMM3054 | 1MT288H17 | TGAGCAAGAGAACGAGAGCG | CCTCAGGACCATCAACGACA |
Clones numbered with letters only were random BACs isolated from the OSU BAC library. 1MT320A01 is from a clone containing Myd118-1, and 1MT288H17 is from a clone containing ID1B from the Swanson BAC library.
Genotyping:
Microsatellite loci were genotyped on the ABI 3100 using fluorescently labeled forward primers. Several microsatellite loci, obtained directly from random BACs as described above, are named with numbers in the OMM3000 series (Rexroad et al. 2005). Genotyping of offspring was done in two doubled haploid panels: OSU × Arlee (Young et al. 1998; Nichols et al. 2003) and OSU × Clearwater (Nichols et al. 2004). To correlate the OSU × Clearwater map with the OSU × Arlee map, at least one marker from the OSU × Arlee map was mapped onto the OSU × Clearwater panel for each linkage group. The doubled haploid panels were constructed by androgenesis performed on hybrids between two homozygous clonal lines, so all offspring were homozygous, which facilitated scoring of SNPs as described below.
Genotyping of SNPs in type I genes was done using several methods, including RFLP analysis of amplified products, SSCP, and the ABI PRISM SNaPshot Multiplex system. For the SNaPshot method, first we designed primers to 3′ regions from either GenBank or the The Institute for Genome Research databases for the genes of interest. These regions were amplified in OSU, Clearwater, and Arlee parental lines to look for SNPs. After sequencing the PCR product and detecting the SNP, a primer was designed immediately 5′ of the SNP, and single-base extension was performed using the SNaPshot ready mix containing fluorescently labeled ddNTPs. The extended restriction products were detected on our ABI 3100 sequencer. Information on the SNPs can be found in the NCBI SNP database (dbSNP). SNPs genotyped in the Phillips laboratory can be located using RPHILLIPS and SNPs genotyped by Krista Nichols (in Myd118-2, LDHB, TRH, THSHA) can be located using NICHOLSLAB_PURDUE.
In situ hybridization and karyotyping:
Chromosome preparations were obtained from blood of the OSU strain (2n = 60) by methods described previously (Reed and Phillips 1995). Briefly, the buffy coat was isolated from whole blood and placed in minimal essential media with pen-strep, l-glutamine, 10% fetal calf serum, and 200 μg/ml lipopolysaccharide and cultured for 6 days at 20°. Cells were collected by centrifugation and resuspended in 0.075 m KCl for 30 min and then fixed in 3:1 methanol acetic acid. Cell suspensions were dropped onto clean slides and allowed to dry on a slide warmer with humidity at 40°.
BAC clones were labeled with spectrum orange (Vysis) and digoxigenin (Roche) as recommended by the manufacturer. Hybridization with fluorochrome-labeled dUTPs was done as suggested by the manufacturer (Vysis) with minor modifications. Briefly, chromosome preparations were made the day before use and left to dry on a slide warmer at 40° overnight. Just prior to hybridization, the slides were denatured in a 70% formamide solution at 73° for 5 min. The probe was prepared by adding labeled DNA with human placental DNA and rainbow trout CoT DNA (for blocking) to the Vysis hybridization solution and denatured at 73° for 5 min. Hybridizations were allowed to proceed under a sealed coverslip in a humidified chamber at 37° overnight. The next day the slides were washed first with 0.3% NP40 in 0.4× SSC at 73° for 30 sec and then with 0.1% NP40 in 2× SSC at room temperature for 1 min. Antibodies to digoxigenin (1/100 dilution in PBS) were applied and slides incubated at 37° for 45 min, according to the manufacturer's instructions. Primary and secondary antibodies to spectrum orange (1/100 and 1/200 dilution in PBS) were used to amplify the signal in many experiments. Slides were counterstained with DAPI/antifade (Vysis).
Slides were examined using an Olympus BX60 microscope and photographed with a Sensys 1400 digital camera. Images were captured with Cytovision software (Applied Imaging, Santa Clara, CA) and selected karyotypes were prepared using Genus software (Applied Imaging). Chromosome pairs were identified using relative size, centromere staining, chromosome arm ratios, and centromere probes. Chromosomes were assorted according to size using the software described above and adjustments were made by hand to conform with the standard chromosome arm ratios and DAPI staining of centromeres. Final identification of chromosomes of similar size and morphology was done using a combination of centromere probes (Reed et al. 1998) in different colors. Dual hybridizations with these centromere probes and BAC clones containing genes from specific linkage groups (in two different colors) were done to confirm these assignments.
The orientation of the genetic map on the chromosomes was determined by comparing the location of markers on the genetic map and their location on the chromosomes. Once this orientation of the genetic map was determined (see Table 1), it was usually possible to assign homeologous regions to a specific chromosome arm (Table 3).
TABLE 1.
Assignment of rainbow trout genetic linkage groups to chromosomes
| LG | Chromosome | Orientation | Arm | Marker | Clone | Genotyped by |
|---|---|---|---|---|---|---|
| 1 | Sex | p/q | 1p | 5sRNA | ||
| 1q | B4 | B4o | Felip et al. (2004) | |||
| 1q | Scar163 | 171H7s | Felip et al. (2004) | |||
| 1q | A6 | A6o | This study (OxC) | |||
| 2 | 13 | p/q | 13q | GH2 | 25K21o | Nichols et al. (2003) |
| 13q | OMM1232 | 337O14s | This study (OxA) | |||
| 13q | OMM3006b | C11o | Nichols et al. (2003) | |||
| 3 | 14 | p/q | 14p | OMM3044b | E2o | This study (OxA) |
| 14q | MHC1B | 20C13o, 63M2o | Phillips et al. (2003) | |||
| 4 | 25a | q/c | 25cen | 10H19 | 10H19o | |
| 25q | G6 | G6o | Dual hybridization | |||
| 25q | TCRβ | 270C12o | Nichols et al. (2003) | |||
| 5 | 22 | q/p | 22cen | 10H19 | 10H19o | |
| 22q | OMM1010 | 231J20s | Nichols et al. (2003) | |||
| 22q | CTLAV | 106I2o | This study (OxC) | |||
| 6 | 1 | q/p | 1p | Myd118-2 | 142F2s | This study (OxC) |
| 1p | D2 | D2o | Dual hybridization | |||
| 1q | Fgf6 | 122J17o | Nichols et al. (2003) | |||
| 1q | G5 | G5o | Dual hybridization | |||
| 1q | MetA | 85O16a | This study (OxA) | |||
| 7 | 15 | p/q | 15p | OMM1087 | 341I3s | Dual hybridization |
| 15q | Omy7INRA | 197M11s | Nichols et al. (2003) | |||
| 8 | 5 | q/p | 5p | OMM3032(Myd118-1) | 471P3s | This study (OxC) |
| 5q | OMM1195 | 107J7s | Danzmann et al. (2005) | |||
| 9 | 12 | q/p | 12q | GH1 | 167I21o | Sakamoto et al. (2000) |
| 12q | OMM1192 | 316J1s | This study (OxA) | |||
| 10 | 6 | q/p | 6p | OMM1017 | 214D9s | Nichols et al. (2003) |
| 6p | OMM1204 | 113I17s | Danzmann et al. (2005) | |||
| 11 | 27a | q/c | 27q | Somatolactin | 193J21o | Nichols et al. (2003) |
| 12 | 7 | q/p | 7p | CD28 | 104G8o | Dual hybridization |
| 7p | OMM1236 | 165C24s | This study (OxA) | |||
| 7cen | 10H19 | 10H19o | ||||
| 7q | OMM3054(ID1B) | 288H17s | This study (OxC) | |||
| 7q | NrampB2 | 203C15o | Dual hybridization | |||
| 13 | 28a | c/q | 28q | OMM3000b | B10o | Dual hybridization |
| 28q | OMM1020 | 239K12s | Nichols et al. (2003) | |||
| 14 | 19 | p/q | 19q | BHMS281 | 198E23a | Danzmann et al. (2005) |
| 19p | OMM1134 | 271M12s | Danzmann et al. (2005) | |||
| 15 | 21 | p/q | 21p | LDH-B | 176H21s | This study (OxC) |
| 21q | B1 | B1o | This study (OxA) | |||
| 16 | 18 | q/p | 18q | MHCIa | 24K3o | Phillips et al. (2003) |
| 18q | TAPBP1 | 3O11o | Landis et al. (2006) | |||
| 17 | 20 | q/p | 20p | 18S rDNA | 54E18o | Nichols et al. (2003) |
| 20q | OMM1135 | 132B19s | Nichols et al. (2003) | |||
| 18 | 26a | q/p | 26q | ProC | 126P8o | This study (OxC) |
| 26q | B2 | B2o | Dual hybridization | |||
| 19 | 11 | p/q | 11p | OMM3020b | F1o | This study (OxC) |
| 11cen | 10H19 | 10H19o | ||||
| 11q | OMM3042b | C9o | This study (OxC) | |||
| 20 | 10 | q/p | 10p | NrampB1 | 146I11o | Dual hybridization |
| 10p | OMM1348 | 117A19s | Danzmann et al. (2005) | |||
| 10q | p53 | 26H22o | Nichols et al. (2003) | |||
| 21 | 9 | q/p | 9p | OMM1145 | 520I2s | Nichols et al. (2003) |
| 9cen | 10H19 | 10H19o | ||||
| 9q | Ssa197 | 121A9a | Danzmann et al. (2005) | |||
| 22 | 16 | p/q | 16cen | 10H19 | 10H19o | |
| 16q | TRH | 175K5s | This study (OxC) | |||
| 16q | OMM1264 | 115J2s | Danzmann et al. (2005) | |||
| 23 | 8 | q/p | 8p | TRSHA | 354I9s | This study (OxC) |
| 8cen | 66L6 | 66L6o | ||||
| 8q | OMM1295 | 142H6s | This study (OxA) | |||
| 8q | OMM1329 | 169O4s | Danzmann et al. (2005) | |||
| 24 | 4 | p/q | 4p | histone | 116H7a | Dual hybridization |
| 4cen | 66L6 | 66L6o | ||||
| 4q | OMM3012b | B8o | This study (OxC) | |||
| 4q | OMM3064(ID1C) | 327F15s | Danzmann et al. (2005) | |||
| 25 | 29a | q/c | 29cen | 10H19 | 10H19o | |
| 29q | G9 | G9o | This study (OxA) | |||
| 26 | 24a | q/c | 24cen | 55cen | 55D21o | |
| 24q | Omi66 | 366K10s | Danzmann et al. (2005) | |||
| 27 | 2 | p/q | 2p | TAPBPRa | 12I24,16E7o | Landis et al. (2006) |
| 2p | TAP1 | 34E19,68M15o | Phillips et al. (2003) | |||
| 2q | OMM3001b | C1o | Nichols et al. (2003) | |||
| 2q | MetB | 127C24a | Nichols et al. (2003) | |||
| 2q | F9 | F9o | This study (OxA) | |||
| 29 | 17 | q/p | 17p | MHC2 | 4C02o | Phillips et al. (2003) |
| 17cen | 10h19 | 10H19o | ||||
| 17q | OMM1300 | 318G19s | Danzmann et al. (2005) | |||
| 30 | 23a | q/c | 23q | CarbE19 | 86E19o | Dual hybridization |
| OMM3018b | E1o | This study (OXC) | ||||
| 31 | 3 | q/p | 3p | TAPBPRb | 12I24o, 16E7o | Landis et al. (2006) |
| 3p | Oneu102 | 18G23a | Danzmann et al. (2005) | |||
| 3q | OMM1080 | 312E5s | Danzmann et al. (2005) |
o, OSU library; s, Swanson library; a, Atlantic salmon CHORIO library.
Acrocentric chromosome.
Microsatellites isolated from random BAC clones described in the appendix.
TABLE 3.
Chromosomal location of homeologous regions
| Homeologous pair
|
||
|---|---|---|
| LGs | Chromosome | Duplicated markers |
| 1 and 8 | sexq and 8q | GHR1, GHR2 |
| 2 and 9 | 13q and 12q | GH1, GH2; HoxB4a/i, HoxB4a/ii; OmyIgM/iDIAS, OmyIgM/iiDIAS; OmyFGT18/iTUF, OmyFGT18/iiTUF; OmyFGT32/iTUF, OmyFGT32/iiTUF; OmyRGT40/iTUF, Omy RGT40/iiTUF; OmyRGT42/iTUF, Omy RGT42/iiTUF OMM1218/i, OMM1218/ii; OMM1258/i, OMM1258/ii; OMM1262/i, OMM1262/ii OMM1274/i, OMM1274/ii; |
| 2 and 29 | 13p and 17p | OmyFGT25TUF/i, OmyFGT25TUF/ii; Omy11/iINRA, Omy11iiINTRA; SmaBFRO1/i, SmaBFRO1/ii; SsaLEE184/i, SsaLEE184/ii; OMM1064/i, OMM1064/ii; OMM1217/i, OMM1217/ii; OMM1269/i, OMM1269/ii; OMM1330/i, OMM1330/ii |
| 3 and 16 | 14q and 16q | Hox4ai, Hox4aii; MHCIA, MHCIB; |
| 3 and 25 | 14p and 29qa | Ogo2/iUW, Ogo2/iiUW |
| 6 and 8 | 1p and 5p | Myd118-1, Myd118-2 |
| 6 and 30 | 1q and 23qa | GnRH3A, GnRH3B |
| 6 and 27 | 1q and 2q | MetA, MetB; WT1-1, WT1-2 |
| 7 and 15 | 15q and 21q | OmyRGT15/iTUF, OmyRGT15/iiTUF; Sal8/iUoG, Sal8/iiUoG; SalF41/i, SalF41/ii BHMS124/i, BHMS124/ii; OMM1164/i, OMM1164/ii |
| 9 and 13 | 13p and 28qa | OmyFGT28/iTUF, OmyFGT28/iiTUF |
| 10 and 11 | 6p and 27qa | Omy7/iDIAS, Omy7/iiDIAS |
| 10 and 18 | 6p and 26qa | OMM1197/i, OMM1197/ii; OmyCOSB/iTUF, OmyCOSB/iiTUF |
| 12 and 16 | 7p and 18p | ATP1B1B/i, ATP1B1B/ii; OmyRGT10/iTUF, OmyRGT10/iiTUF; OmyOGT5/i, OmyOGT5/ii; Ssa119/iNVH, Ssa119/iiNVH; Omy3/iINRA, Omy3/iiINRA; BHMS219/i, BHMS219/ii; OMM1167/i, OMM1167/ii; OMM1345/i, OMM1345/ii |
| 14 and 20 | 19p and 10q | Omy296/i, Omy296/ii; BHMS205/i, BHMS205ii OMM1134/i, OMM1134/ii |
| 17 and 22 | 20p and 16q | HoxB5bi, HoxB5bii; OmyRGT6/iTUF, OmyRGT6/iiTUF |
| 23 and 24 | 8p and 4p | Omy27/iINRA, Omy27/iiINRA |
| 27 and 31 | 2p and 3p | TAPBPR1, TAPBPR2; HoxA2bi, HoxA2bii; ATP1A3/i, ATP1A3/ii, OmyFGT8/iTUF, OmyFGT8/iiTUF; Omy272/iUoG, Omy272/iiUoG; BHMS254i, BHMS254ii; OMM1122i, OMM1122ii; Oneu18/iASC, Oneu18/iiASC, Oneu102/iADFG, Oneu102/iiADFG, Ssa125/iNVH, Ssa125/iiNVH, OMM1122/i, OMM1122/ii |
Markers were assigned to chromosome arms either directly from in situ hybridization results or by close genetic linkage to markers used that were localized using in situ hybridization. Genetic linkage data for markers not included in Table 1 were obtained from Danzmann et al. (2005), Gharbi et al. (2004), Leder et al. (2006), Moghadam et al. (2005), and Nichols et al. (2003).
Acrocentric chromosome.
RESULTS
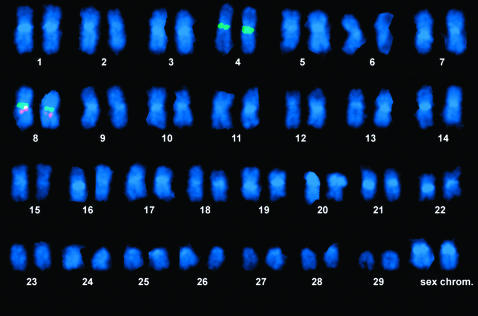
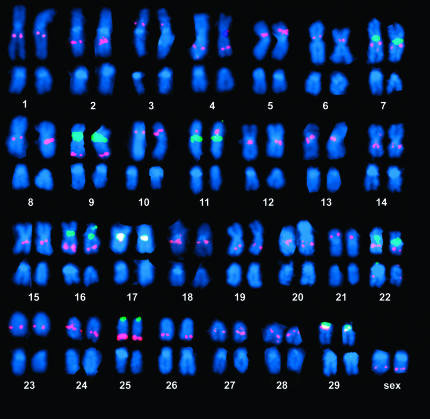
Hybridization experiments were carried out with BAC clones containing genetic markers mapped to each linkage group. Dual hybridizations with centromere probes allowed us to accurately identify each chromosome. Two different centromere probes were used extensively as an aid to chromosome identification: 66L6, which hybridizes to centromeres of chromosomes 4 and 8, and 10h19, which hybridizes to centromeres of chromosomes 7, 9, 11, 16, 17, 22, 25, and 29. Figure 1 shows a dual hybridization with the 66L6 centromere probe (labeled in green) and a BAC clone containing the microsatellite locus Omm1295 (labeled in red). Omm1295 has been mapped to LG23, so this experiment showed that LG23 corresponds to Omy8, the smaller of the two chromosome pairs that are positive with the 66L6 centromere probe. A composite of images showing the results of hybridization of one probe for each linkage group to specific chromosomes is shown in Figure 2. In these hybridizations the BAC probes are in red and the 10h19 centromere probe is in green. In every case, the sex chromosome pair from the same metaphase is shown below the chromosome containing the probe signal, to indicate relative size. The sex chromosome pair can always be identified in every metaphase because it is the only subtelocentric pair in the karyotype and the X chromosome has a bright band with the DAPI stain (used as a counterstain in the FISH experiments) on the short arm next to the centromere.
Figure 1.—
Dual hybridization with the 66L6 centromere probe (labeled in green), which hybridizes to centromeres of chromosomes Omy4 and Omy8, and a BAC clone (labeled in red) containing the microsatellite locus, Omm1295, mapped to linkage group 23, which hybridizes to Omy8.
Figure 2.—
Composite of 30 partial karyotypes showing results of dual hybridizations with the 10h19 centromere probe (labeled in green) and a BAC clone (labeled in red) containing a marker mapped to each specific linkage group. In each case, the sex chromosome pair from the same metaphase cell is shown below the chromosome pair containing the probe signal to indicate relative size, except for the sex chromosome pair itself, which is shown at the bottom right. Probes shown are Omy1:Fgf6, Omy2:Met B, Omy3:Oneu102, Omy4:IDIC, Omy5:Myd118, Omy6:Omm1204, Omy7:ID1B, Omy8:Omm1295, Omy9:Ssa197, Omy10:Omm1348, Omy11:F1, Omy12:GH1, Omy13:GH2, Omy14:MHC1B, Omy15:Omy7INRA, Omy16:Omm1264, Omy17:MHCII, Omy18:MHC1A, Omy19:BHMS281, Omy20:Omy1135, Omy21:B1, Omy22:Omm1010, Omy23:E1, Omy24:Omi66, Omy25:TCRβ, Omy26:ProC, Omy27:Somat, Omy28:Omm1020, Omy 29:G9, and Omy30 (sex chromosome pair):A6. MHCII on Omy17 and G9 on Omy29 are located very close to their respective centromeres. As a result of this, the green signal from 10h19 on these centromeres and the red signals from the MHCII probe on Omy17 and the G9 probe on Omy29 have merged to give yellow signals.
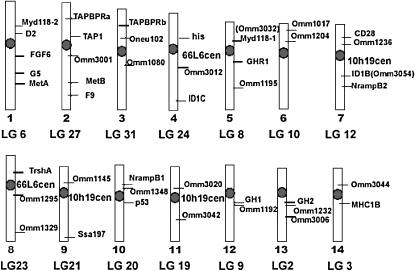
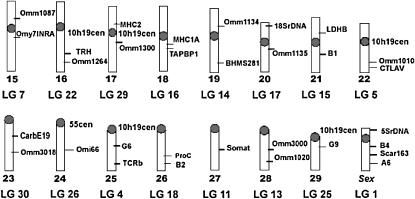
The clones that were used to assign genetic linkage groups to chromosomes, and the orientation of the chromosome map on each chromosome, are shown in Table 1. An ideogram of the rainbow trout karyotype showing location of probes used to assign linkage groups to specific chromosomes is shown in Figure 3. The relative sizes of the linkage maps (Ox A male map and female maps from lots 25 and 44 from the University of Guelph) in centimorgans and the female-to-male recombination ratios (based on lots 25 and 44) of the different linkage groups are shown in Table 2 for metacentric and acrocentric chromosomes (Sakamoto et al. 2000; Danzmann et al. 2005). For the female maps, the average genetic map distance in centimorgans of the 12 largest chromosomes was 104 cM, while the average of the 10 smallest chromosomes it was 41 cM, so there is a rough correlation between chromosome size and linkage map distance in centimorgans. Recombination ratios of female:male maps for metacentric chromosomes vary between 1 and 26.55, while ratios for acrocentric chromosomes vary between 1 and 4.18 in the sex-specific maps from the University of Guelph.
Figure 3.—
An ideogram of the rainbow trout karyotype of the OSU strain showing the location of probes mapped by in situ hybridization.
TABLE 2.
Chromosomes and linkage groups according to size
| Size and rank of LG
|
F:M recombination ratio
|
|||||
|---|---|---|---|---|---|---|
| Chromosome | LG | OxA | 25f | 44f | Lot 25 | Lot 44 |
| 1 | 6 | 1 | 7 | 6 | 20.05 | 7.76 |
| 2 | 27 | 21 | 2 | 3 | 6.78 | 11.67 |
| 3 | 31 | 13 | 14 | 19 | 19.39 | 3.7 |
| 4 | 24 | 27 | 4 | 1 | 1.82 | 7.21 |
| 5 | 8 | 16 | 26 | 21 | 1.0 | NS |
| 6 | 10 | 23 | 15 | 8 | 17.54 | 3.65 |
| 7 | 12 | 4 | 3 | 4 | — | — |
| 8 | 23 | 22 | 17 | 13 | 1.57 | 1.40 |
| 9 | 21 | 6 | 6 | 10 | 2.86 | 2.6 |
| 10 | 20 | 7 | 20 | 7 | 3.62 | 2.19 |
| 11 | 19 | 3 | 13 | 2 | 4.52 | 14.02 |
| 12 | 9 | 9 | 12 | 5 | 26.55 | 16.62 |
| 13 | 2 | 2 | 5 | 12 | — | — |
| 14 | 3 | 28 | 14 | 15 | 6.60 | 9.75 |
| 15 | 7 | 18 | 22 | 9 | 12.63 | 13.59 |
| 16 | 22 | 11 | 18 | 11 | 2.0 | 10.01 |
| 17 | 29 | 14 | 1 | 20 | 5.97 | 5.09 |
| 18 | 16 | 10 | 8 | 14 | — | — |
| 19 | 14 | 26 | 10 | 15 | 1.0 | 23.15 |
| 20 | 17 | 17 | 16 | 18 | 4.43 | 3.16 |
| 21 | 15 | 5 | 9 | 16 | 8.68 | 3.17 |
| 22 | 5 | 15 | 28 | 22 | 1.0 | — |
| 23 | 30 | 22 | 23 | 23 | 2.68 | 3.36 |
| 24 | 26 | 28 | 27 | — | — | 1.09 |
| 25 | 4 | 12 | — | — | — | — |
| 26 | 18 | 24 | 24 | — | — | 5.62 |
| 27 | 11 | 19 | 19 | 25 | 3.96 | .55 |
| 28 | 13 | 20 | 21 | 24 | 4.18 | 1.25 |
| 29 | 25 | 8 | 25 | 17 | 1.0 | 2.62 |
| Sex | 1 | 25 | 29 | 26 | — | — |
Data are from Nichols et al. (2003) and Danzmann et al. (2005). OxA is a male map based on doubled haploid offspring, but lot 25 and lot 44 map distances are from female maps. The female:male ratios are from Danzmann et al. (2005) and are based on males and females from lots 25 and lots 44. When the regression of the size of the chromosome with the LG map size is calculated, it is significant for both lot 25 and lot 44 (data not shown). Numbers are given only for cases in which a minimum of six markers could be evaluated.
Table 3 groups the chromosome arms and homeologous linkage groups that have been identified in several studies (Nichols et al. 2003; Danzmann et al. 2005) and this article, including information on the duplicate genes that have been mapped to these chromosome arms. Whenever the same linkage group is involved in two different homeologies, they almost always involve different chromosome arms. The only chromosome pair involved in three homeologies was the largest chromosome pair, which corresponds to linkage group 6.
DISCUSSION
The largest linkage groups usually corresponded to the largest chromosomes. In females, the linkage groups corresponding to the 12 largest chromosomes had an average size of 104 cM, while the linkage groups corresponding to the 10 smallest chromosomes had an average size of 41 cM. In males, the comparable figures were 68.4 and 36.8 cM. It is known that male recombination is greatly suppressed near the centromeres, but inflated near the telomeres. For example, TCRβ, which hybridizes to the end of chromosome 25, maps at position 97.4 cM of a total of 162.4 cM on the linkage map published by Nichols et al. (2003). Chromosome size and structure has a major effect on recombination rate differences between the sexes (Table 2). Linkage groups of bi-armed (metacentric) chromosomes had higher female:male recombination rates than linkage groups of acrocentric (single-armed) chromosomes (pairs 23–30). This is most pronounced in the largest metacentrics, so it appears to be mainly a consequence of the fact that almost all of the metacentric pairs are larger than the acrocentrics. There are only a couple of metacentric chromosomes (pairs 20–22) that are in the same size range as the acrocentric chromosomes (pairs 23–30), and these have F:M recombination ratios similar to the acrocentrics. The large difference in male and female maps for the larger chromosomes suggests that suppression of crossing over may extend out from the centromeres over a considerable portion of the chromosome in males. The observed increase in recombination in the smaller chromosomes in males relative to the larger chromosomes reduces the difference in recombination rate between the sexes for these chromosomes. It is well known that recombination in most organisms is increased in smaller chromosomes (Kong et al. 2002), probably because there is a minimum number of chiasmata required per chromosome for proper segregation.
There is one major exception to the correlation between large metacentrics and high F:M recombination ratios. This is chromosome 5, the fifth largest pair, which corresponds to LG8. In addition to having an equal F:M recombination, this LG ranks in the last quartile for size in the Guelph crosses and 16th for the O × A cross. This is the LG to which a major QTL for early maturity has been mapped in O × C (Robison et al. 2001), and a major QTL for spawning date has been mapped in the Guelph crosses (O'Malley et al. 2003). This linkage group is remarkably condensed (18 markers mapped on top of each other near the centromere of the female map) in both males and females (Danzmann et al. 2005). There is evidence based on the mapping of a duplicated pair of known genes that this linkage group may be the homeologous linkage group to the sex chromosome pair (K. M. Nichols and R. B. Phillips, unpublished results).
Salmonid fishes underwent a tetraploidization event 50–100 years ago (reviewed in Allendorf and Thorgaard 1984), so that many genes are present in duplicates. The most common diploid karyotype in teleosts is 48–50 single-armed (acrocentric) chromosomes, and salmonid fishes have diploid chromosome numbers between 54 and 92 with ∼100 chromosome arms in most species (reviewed in Phillips and Rab 2001). This suggests that most of the chromosome rearrangements following the tetraploid event were centric fusions. Rainbow trout karyotypes vary from 58 to 64, with primarily metacentric (bi-armed) chromosomes, which is consistent with this hypothesis.
Genetic maps of allozyme loci in salmonid fishes showed that up to 20% of loci are isoloci, which are still recombining in males (Wright 1983; May and Johnson 1993). These loci are usually located at the ends of linkage groups, which would correspond to telomeres (Allendorf and Danzmann 1997), and frequency of tetrasomic inheritance is increased in crosses between strains from different geographic regions. The BAC probes that hybridize to more than one chromosome pair all are found at locations from the middle of the arm to the telomere, consistent with this hypothesis. Some of these contain type I genes and others are random BACs. The precise location on the chromosome arm appears to be conserved for the duplicate loci that we mapped and the size of the homeologous chromosome arms appears to be similar (Table 3).
The fact that 80% of the BACs containing duplicate genes hybridize to only one chromosomal location (Table 1 and our unpublished information) suggests that intergenic regions have diverged substantially between the two subgenomes. This has been shown directly by sequence analysis of two BAC clones containing duplicate regions in Atlantic salmon that were localized by in situ hybridization to two different chromosomes in this species (Mitchell et al. 2005). Although the same 10 protein-coding genes were found in both BACs, intergenic regions were highly diverged and contained different repetitive elements. Comparisons of the sequences of the genes in these two BACs led to an estimate of 90–110 million years for the time of the original duplication. A rainbow trout clone containing one of these same regions was also sequenced and compared to the orthologous Atlantic salmon clone. It had the same genes in the same order but the intergenic regions were also highly conserved (L. Mitchell, personal communication), which explains why many rainbow trout clones will hybridize to the orthologous loci in other salmonid species). These results suggest that diploidization occurred rapidly after the tetraploidization event and speciation occurred considerably later. It is not known why some regions are still not diploidized after ∼100 million years and why these are shared in different species.
The cytogenetic map will have a number of applications. First, it will be useful in helping investigators determine how many loci there are for duplicate genes and assist in assembling contigs of BACs for the different loci. Second, it will allow quick assignment of the linkage group without having to search for SNPs and map them in crosses. This will allow investigators to determine if their BAC clone maps to a region containing a QTL, or if multiple clones are located in homeologous regions. There is evidence that QTL are found in homeologous regions (O'Malley et al. 2003), so it will be important to continue to integrate the genetic and cytogenetic maps so that all of these regions can be identified. Third, the BACs isolated in this study will be especially useful for characterizing the chromosome rearrangements that are present in different strains of rainbow trout and related salmonids.
We have already used BAC probes isolated in this study to determine that the Clearwater and Swanson strains (2n = 58) have the same chromosome fusion involving chromosome pairs 25 and 29 (LG4 and LG 25) (Phillips et al. 2005). Hybrids between OSU and Clearwater and OSU and Swanson had 59 chromosomes and the single metacentric pair had TCRβ (LG4) on one end of one chromosome arm and G9 (LG25) near the centromere on the other arm. These strains originated from Idaho and Alaska and previous work suggested that most of the interior rainbow strains are 2n = 58 (Thorgaard et al. 1983) and may represent the ancestral rainbow trout karyotype. The crosses made at the University of Guelph (Sakamoto et al. 2000; Danzmann et al. 2005) had only 29 linkage groups and linkage group 4 was missing. Parents and offspring for these crosses are deceased and were not karyotyped. However, we believe it is likely that they were 2n = 58 fish. One marker found on LG4 in the linkage map based on OSU × Arlee (Nichols et al. 2003) was found on LG25 in these crosses (Danzmann et al. 2005). In future work we plan to isolate additional BAC clones containing mapped genes until we have one for each chromosome arm in rainbow trout. This set of reagents will be especially useful for producing a “quick map” of other salmonid species and for identifying QTL in different salmonid species.
Acknowledgments
The authors thank Barbara Wimpee, John Hansen, Yniv Palti, and Marc Noakes for screening the OSU and Swanson libraries for several clones containing type I genes and microsatellites. Heather Ligman assisted with growing BACs and mapping some of the markers in the O × A cross. John Hansen provided sequence information and primers used in mapping several type I genes. This research was supported by grant NRI 2002-2046 from the United State Department of Agriculture.
References
- Allendorf, F. W., and R. G. Danzmann, 1997. Secondary tetrasomic segregation of MDH-B and preferential pairing of homeologues in rainbow trout. Genetics 145: 1083–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allendorf, F. W., and G. H. Thorgaard, 1984. Tetraploidy and the Evolution of Salmonid Fishes. Plenum Publishing, New York.
- Danzmann, R. G., M. Cairney, W. S. Davidson, M. M. Ferguson, K. Gharbi et al., 2005. A comparative analysis of the rainbow trout genome with two other species of fish (Arctic charr and Atlantic salmon) within the tetraploid derivative Salmonidae family (subfamily: Salmoninae). Genome 48: 1037–1041. [DOI] [PubMed] [Google Scholar]
- Felip, A., A. Fujiwara, W. P. Young, P. A. Wheeler, M. Noakes et al., 2004. Polymorphism and differentiation of rainbow trout Y chromosomes. Genome 47: 1105–1113. [DOI] [PubMed] [Google Scholar]
- Gharbi, K., J. W. Semple, M. M. Ferguson and R. G. Danzmann, 2004. Linkage arrangement of K, Na ATPase genes in the tetraploid derived genome of the rainbow trout (Oncorhynchus mykiss). Anim. Genet. 35: 321–325. [DOI] [PubMed] [Google Scholar]
- Kong, A., D. F. Gudbjartsson, J. Sainz, G. M. Jonsdottir, S. A. Gudjonsson et al., 2002. A high-resolution recombination map of the human genome. Nat. Genet. 31: 241–247. [DOI] [PubMed] [Google Scholar]
- Landis, E. D., Y. Palti, J. J. DeKoning, R. B. Phillips and J. D. Hansen, 2006. Mapping and functional genomics of the TABP and TAPBPR genes. Immunogenetics 58: 56–59. [DOI] [PubMed] [Google Scholar]
- Leder, E. H., R. G. Danzmann and M. M. Ferguson, 2006. The candidate gene, Clock, localizes to a strong spawning time quantitative trait locus region in rainbow trout. J. Hered. 97: 74–80. [DOI] [PubMed] [Google Scholar]
- Lloyd, M. A., and G. H. Thorgaard, 1988. Restriction endonuclease banding of rainbow trout chromosomes. Chromosoma 96: 171–177. [Google Scholar]
- May, B., and K. R. Johnson, 1993. Composite linkage map of salmonid fishes (Salvelinus, Salmo, Oncorhynchus), pp. 309–317 in Genetic Maps: Locus Maps of Complex Genomes, edited by S. J. O'Brien. Cold Spring Harbor Laboratory Press, Woodbury, NY.
- Mitchell, L., K. Lubieniecki, S. H. S. Ng, G. Cooper, R. B. Phillips et al., 2005. Characterization of paralogous regions of the Atlantic Salmon (Salmo salar) genome. Plant and Animal Genome XIII Proceedings. Applied Biosystems, Foster City, CA.
- Moghadam, H. K., M. M. Ferguson and R. G. Danzmann, 2005. Evidence for HOX gene duplication in rainbow trout, a tetraploid model species. J. Mol. Evol. 61: 804–818. [DOI] [PubMed] [Google Scholar]
- Nichols, K. M., W. P. Young, R. G. Danzmann, B. D. Robison, C. Rexroad et al., 2003. An updated genetic linkage map for rainbow trout (Oncorhynchus mykiss). Anim. Genet. 34: 102–115. [DOI] [PubMed] [Google Scholar]
- Nichols, K. M., P. A. Wheeler and G. H. Thorgaard, 2004. Quantitative trait loci analyses for meristic traits in Oncorhynchus mykiss. Environ. Biol. Fishes 69(1–4): 317–331. [Google Scholar]
- O'Malley, K. G., T. Sakamoto, R. G. Danzmann and M. M. Ferguson, 2003. Quantitative trait loci for spawning date and body weight in rainbow trout: testing for conserved effects across ancestrally duplicated chromosomes. J. Hered. 94: 273–284. [DOI] [PubMed] [Google Scholar]
- Palti, Y., S. A. Gahr, J. D. Hansen and C. E. Rexroad, 2004. Characterization of a new BAC library for rainbow trout: evidence for multi-locus duplication. Anim. Genet. 35: 130–133. [DOI] [PubMed] [Google Scholar]
- Phillips, R. B., and S. E. Hartley, 1988. Fluorescent banding patterns of the chromosomes of the genus Salmo. Genome 30: 193–197. [Google Scholar]
- Phillips, R. B., and P. Rab, 2001. Chromosome evolution in the Salmonidae (Pisces): an update. Biol. Rev. 76: 1–25. [DOI] [PubMed] [Google Scholar]
- Phillips, R. B., A. Zimmerman, M. Noakes, Y. Palti, M. Morasch et al., 2003. Physical and genetic mapping of the rainbow trout major histocompatibility regions: evidence for duplication of the class I region. Immunogenetics 91: 561–569. [DOI] [PubMed] [Google Scholar]
- Phillips, R. B., M. R. Morasch, P. A. Wheeler and G. H. Thorgaard, 2005. Rainbow trout (Oncorhynchus mykiss) of Idaho and Alaskan origin (2n=58) share a chromosome fusion relative to trout of California origin (2n=60). Copeia 2005(3): 660–663. [Google Scholar]
- Reed, K. M., and R. B. Phillips, 1995. Molecular cytogenetic analysis of the double CMA3 chromosome in lake trout, Salvelinus namaycush. Cytogenet. Cell Genet. 70: 104–107. [DOI] [PubMed] [Google Scholar]
- Reed, K. M., and R. B. Phillips, 1997. Polymorphism of the nucleolar organizer region (NOR) on the putative sex chromosomes of Arctic char (Salvelinus alpinus) is not sex related. Chromosome Res. 5: 221–227. [DOI] [PubMed] [Google Scholar]
- Reed, K. M., M. O. Dorschner and R. B. Phillips, 1998. Characterization of two salmonid repetitive DNA families in rainbow trout (Oncorhynchus mykiss). Cytogenet. Cell Genet. 79: 184–187. [DOI] [PubMed] [Google Scholar]
- Rexroad, C. E., M. F. Rodriguez, I. Coulibaly, K. Gharbi, R. G. Danzmann et al., 2005. Comparative mapping of expressed sequence tags containing microsatellites in rainbow trout (Oncorhynchus mykiss). BMC Genomics 6: 54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison, B. D., P. A. Wheeler, K. Sundin, P. Sikka and G. H. Thorgaard, 2001. Composite interval mapping reveals a major locus influencing embryonic development rate in rainbow trout (Oncorhynchus mykiss). J. Hered. 92: 16–22. [DOI] [PubMed] [Google Scholar]
- Sakamoto, T., R. G. Danzmann, K. Ghabari, P. Howard, A. Ozaki et al., 2000. A microsatellite linkage map of rainbow trout (Oncorhynchus mykiss) characterized by large sex-specific differences in recombination rates. Genetics 155: 1331–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., and D. W. Russell, 2001. Molecular Cloning: A Laboratory Manual, Ed. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Thorgaard, G. H., 1977. Heteromorphic sex chromosomes in male rainbow trout. Science 196: 900–902. [DOI] [PubMed] [Google Scholar]
- Thorgaard, G. H., 1983. Chromosomal differences among rainbow trout populations. Copeia 1983: 650–662. [Google Scholar]
- Thorgaard, G. H., F. W. Allendorf and K. L. Knudsen, 1983. Gene centromere mapping in rainbow trout: high interference over long map distances. Genetics 103: 771–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsen, J., B. Zhu, E. Frengen, K. Osegawa, P. J. deJong et al., 2005. A highly redundant BAC library of Atlantic salmon (Salmo salar): an important tool for salmon projects. BMC Genomics 6: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, J. E. J. (Editor), 1983. Meiotic Models to Explain Classical Linkage, Pseudolinkage, and Chromosome Pairing in Tetraploid Derivative Salmonid Genomes. Alan R. Liss, New York. [PubMed]
- Young, W. P., P. A. Wheeler, V. H. Coryell, P. Keim and G. H. Thorgaard, 1998. A detailed linkage map of rainbow trout produced using doubled haploids. Genetics 148: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman, A. M., J. P. Evenhuis, G. H. Thorgaard and S. S. Ristow, 2004. A single major chromosomal region controls natural killer cell-like activity in rainbow trout. Immunogenetics 55: 825–835. [DOI] [PubMed] [Google Scholar]