Abstract
Recent studies have revealed that the major genes of the mammalian sex determination pathway are also involved in sex determination of fish. Several studies have reported QTL in various species and strains of tilapia, regions contributing to sex determination have been identified on linkage groups 1, 3, and 23. Genes contributing to sex-specific mortality have been detected on linkage groups 2, 6, and 23. To test whether the same genes might control sex determination in mammals and fishes, we mapped 11 genes that are considered putative master key regulators of sex determination: Amh, Cyp19, Dax1, Dmrt2, Dmrta2, Fhl3l, Foxl2, Ixl, Lhx9, Sf1, and Sox8. We identified polymorphisms in noncoding regions of these genes and genotyped these sites for 90 individuals of an F2 mapping family. Mapping of Dax1 joined LG16 and LG21 into a single linkage group. The Amh and Dmrta2 genes were mapped to two distinct regions of LG23. The Amh gene was mapped 5 cM from UNH879 within a QTL region for sex determination and 2 cM from UNH216 within a QTL region for sex-specific mortality. Dmrta2 was mapped 4 cM from UNH848 within another QTL region for sex determination. Cyp19 was mapped to LG1 far from a previously reported QTL region for sex determination on this chromosome. Seven other candidate genes mapped to LG4, -11, -12, -14, and -17.
TELEOSTS display a variety of mechanisms for sex determination and sex differentiation (reviewed in Devlin and Nagahama 2002; Godwin et al. 2003). Primary sex determination in most species is genetic (Valenzuela et al. 2003). Nevertheless, sex differentiation of fishes is remarkably plastic and is determined by both genetic and environmental factors in many species (Baroiller et al. 1999; Baroiller and D'Cotta 2001). A few species even undergo sex change in response to behavioral cues (Devlin and Nagahama 2002). Sex differentiation can be modified by environmental factors (hormones, temperature, growth rate, and social environment) and should therefore be regarded as a complex “threshold trait” (Mittwoch 2006).
It is believed that this variety of primary signals regulates one or a few ancient molecular pathways of differentiation (Zarkower 2001). The major genes of the mammalian sex determination pathway, including Sox9, Wt1, Dax1, Sf1, otCYP19, Wnt4, Dmrt1, and Amh, have been detected in many vertebrate species (Schartl 2004; Rodriguez-Mari et al. 2005). The structure of these genes is conserved, but their regulation is variable (Yi and Zarkower 1999). Sry is considered the master key regulator (MKR) of sex determination in mammals, but this gene has not been found in most other vertebrates. Sry-related (Sox) genes have recently been studied in various fish species (e.g., Galay-Burgos et al. 2004; Koopman et al. 2004; Hett and Ludwig 2005; Hett et al. 2005). Sox genes encode a family of transcription factors that are involved in sex determination and other developmental processes in vertebrates (Galay-Burgos et al. 2004).
No common MKR has been found in nonmammalian vertebrates. Instead, it appears that different MKRs are involved among closely related species and strains (Kallman 1984; Volff and Schartl 2001; von Hofsten and Olsson 2005). Gene expression studies in nonmammalian vertebrates have shown that the expression of Dmrt1 and Sox9 throughout the sex-determining period is testis specific (Shan et al. 2000; Baron et al. 2005; Yao and Capel 2005). Environmental temperature modifies the expression level of the otCYP19 differently in males and females (Western and Sinclair 2001; Murdock and Wibbels 2003). It is therefore reasonable to study the effects of sex differentiation genes as candidates for the MKR of sex determination in fish species.
Although genetic factors probably regulate sex determination in most fishes, relatively few teleosts have karyotypically distinct sex chromosomes (Arkhipchuck 1995). Many other species have genetic sex determination, but the sex chromosomes are still in early stages of differentiation, before distinct differences in length or gene content have arisen. Both XY and WZ gonosomal systems have evolved repeatedly in various groups (Devlin and Nagahama 2002). In many species, additional autosomal loci contribute to sex determination (Kosswig 1964).
The sex chromosomes of tilapia, a group of African cichlid fishes widely used in aquaculture, are still at an early stage of differentiation. There are no gross morphological differences in any chromosome pair (Majumdar and McAndrew 1983; Kornfield 1984; Crosetti et al. 1988). A variety of evidence suggests that sex determination is principally monofactorial in tilapias (Wohlfarth and Wedekind 1991). The hypothesized sex-chromosome systems suggest that some species have the XX:XY system (Oreochromis mossambicus, O. niloticus) whereas other have the WZ:ZZ system (O. aureus, O. macrochir, O. urolepis hornorum). The primary support for these hypotheses comes from breeding animals that were sex reversed by hormone treatments, as discussed by Lee et al. (2004). Male heterogametic systems were suggested in O. mossambicus and O. niloticus because crosses of sex reversed (XX) males with normal (XX) females produce only females (Clemens and Inslee 1968; Jalabert et al. 1971). In O. aureus, mating between sex-reversed (ZZ) females and normal (ZZ) males usually results in 100% male offspring, but slight deviations have been observed (Hopkins et al. 1979; Mair et al. 1987; Lahav 1993; Rosenstein and Hulata 1994). These exceptions indicate that other genetic and environmental factors may also contribute to sex determination.
Several studies have identified genetic markers linked to sex determination in tilapia. Lee et al. (2003) identified an XY system on LG1 in a strain of O. niloticus. In a strain of O. aureus, epistatic interactions were observed between a WZ system on LG3 and the XY system on LG1 (Lee et al. 2004). Two distinct QTL for sex determination in tilapias were reported on LG23 in a hybrid cross between O. aureus and O. mossambicus (Cnaani et al. 2003, 2004). A variety of sex-determining systems was demonstrated for these closely related species. The pattern of epistasis among the alleles of the different systems identified on LG1 and LG3 is complex and leads to the interesting appearance of effects on other chromosomes (e.g., LG23). QTL for sex-specific mortality were detected on LG2, -6, and -23 in an inbred line of O. aureus (Palti et al. 2002; Shirak et al. 2002). These last two studies reported an increase in the viability of a gynogenetic line through four successive generations of meiogynogenetic O. aureus. This improvement was likely the result of purging of deleterious alleles. However, significant distortions from expected Mendelian segregation were identified for three unlinked markers: UNH159 (LG2), UNH216 (LG23), and UNH231 (LG6). These authors suggested that the fixation of deleterious alleles in the inbred line occurred because they were closely linked to sex-determining loci. Alternatively, it may be that sex-specific mortality is part of the natural polygenic sex determination system in tilapia with some intra- and interloci allelic combinations that result in unviable individuals.
Dmrt1, Dmo, and Wt1 were previously considered as candidates for MKRs, but their mapping position did not overlap with QTL regions for sex determination or sex-specific mortality in tilapia (Lee et al. 2005). Thus, we adopted the following criteria to select candidate genes for mapping: (1) MKRs could be Doublesex/Mab-3-related transcription factors (DMRTs) or their close downstream/upstream genes in the sex determination pathway (Matsuda et al. 2002; Koopman and Loffler 2003; Haag and Doty 2005); (2) MKRs could be orthologs of sex-determining mammalian genes, such as Sry, which regulates transcription of Sox9 (McElreavey et al. 1993; Graves et al. 1998); and (3) MKRs could include aromatase and its upstream genes in the sex determination pathway (Desvages and Pieau 1992; Sarre et al. 2004; Gardner et al. 2005). Here we map 11 genes in tilapia that might be considered master key regulators of sex determination and compare their locations to previously identified QTL for sex determination.
MATERIALS AND METHODS
Mapping family:
Breeding was performed in the laboratory of G. Hulata at the Volcani Center, Agricultural Research Organization, Bet Dagan, Israel. A red O. niloticus male derived from the University of Stirling stock (McAndrew and Majumdar 1983) was crossed with a normally colored O. aureus female from an Israeli strain (Lee et al. 2004). Fingerlings were shipped to the University of New Hampshire, where they were raised to sexual maturity (mean weight, 149 g) and DNA was extracted. The F2 family consisted of 156 offspring of which 90 individuals were used for genotyping (Lee et al. 2005).
Orthologous sequences of human sex-determining genes:
Sequences of 11 human proteins were BLASTN searched against the NCBI (http://www.ncbi.nlm.nih.gov), The Institute for Genome Research (TIGR; http://tigrblast.tigr.org), and RBEST (http://reprobio.nibb.ac.jp) databases to detect putative orthologs in tilapia, other cichlids, or other fish species (Table 1).
TABLE 1.
Selected human proteins
| Gene symbol | Annotation | Accession no. |
|---|---|---|
| Amh | Anti-Müllerian hormone | AAC25614 |
| Dax1 | Dosage-sensitive sex reversal gene 1 | AAC13875 |
| Dmrt2 | Doublesex/Mab-3-related transcription factor 2 | AAF86293 |
| Dmrta2 | Doublesex/Mab-3-related transcription factor family A2 | CAI23011 |
| Fhl2 | Four and a half LIM domains 2 | CAG33718 |
| Foxl2 | Forkhead box L2 transcription factor | AAY21823 |
| Ixl | Intersex like | NP060062 |
| Lhx9 | LIM homeobox protein 9 | CAH71761 |
| otCYP19 | Ovarian type cytochrome P450, family 19 | NP_000094 |
| Sf1 | Steroidogenic factor 1 | AAH38446 |
| Sox8 | SRY-box-containing gene 8 | AAF37424 |
Primer design:
The exon–intron boundaries in the detected sequences were predicted by their sequence comparison to the relevant Homo sapiens, Mus musculus, Takifugu rubripes, or Danio rerio genomic sequences. Primers were designed in adjacent exons using the “Primer3” program (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). When initial primers did not provide sufficient quality of sequence for one or both grandparents, a second set of internally nested primers was designed on the basis of sequence information obtained using the initial primers (Table 2).
TABLE 2.
Initial, second-round, and marker primers
| Gene | Initial and second-round primers | Marker primers |
|---|---|---|
| Amh | F: AACTGAGTGCGTTCCAGGAG | MF: AGGTGACATGTGCAAGAACAG |
| R: AGTTTTCTTTGCGCGTCGTA | MR: AAGCTGCAGCGGGATACTT | |
| SRF: AGGTGACATGTGCAAGAACAG | ||
| SRR: TCTGCCGACTTCAGAACTTTT | ||
| Dax1 | F: CCGAAACAATAACAGCAGCA | MF: ACGTTGGATGGAAGTAGTTAGGTAAAGTGGC |
| R: GCGACTGGATGTAGTGCAGA | MR: ACGTTGGATGATCCTAAACTTCCACCCCTG | |
| EP: GTAAAGTGGCATGTCTGTAC | ||
| Dmrta2 | F: GAGGTGTTTGGTTCCGTCAG | MF: ACGTTGGATGAAGGCCATCAAAAGCGCATC |
| R: CTCCGTCTTTCAAAGGCTTG | MR: ACGTTGGATGGTCTGTCGCTTTACTGTCTC | |
| EP: GCATCAGCACAGAATCATAAT | ||
| Dmrt2 | F: GCCAAAAGCATCCTCGAA | MF: TGCATCAGAAGATAGACCAGTTT |
| R: GTTTCCAGCTCCTTGTCTGC | MR: CACAAATAAAGTTATTCAAACATGC | |
| SRF: AATTCCAAACACACTCGATGC | ||
| SRR: GAGGGAAACATCGACATCGT | ||
| Fhl3l | F: CGATTGGCTCAAAGTCCTTC | MF: TTGCACACGCTGTAAAAAGG |
| R: TTCTTGGCGTACAGGTTTCC | MR: TCAAAGGTCATGTGGAAATCTG | |
| Foxl2 | F: CGCAATTTGGAGGACAGTTT | MF: ACGTTGGATGGCGCACACACCAAAACAAAC |
| R: CCTTTTTCGGTGCAGCTTAC | MR: ACGTTGGATGTTCACCGATCGAGACAGAAG | |
| EP: CACCAAAACAAACAAGTCC | ||
| Ixl | F: AGAATCGCCTCCCTGAATTT | MF: AGAATCGCCTCCCTGAATTT |
| R: TGTCGATGCTCTGTGAGAGG | MR: CTGAAGTGTCGCTGCTTTTG | |
| Lhx9 | F: GCGGTGGACAAGCAGTGG | MF: ACGTTGGATGTAGCGTTTCAGTTTCACGGG |
| R: TAACGGGCTGACGCTGACTA | MR: ACGTTGGATGACTGGGTCAGTGGATTTAGG | |
| EP: ACGGGCTTGCTCCAAA | ||
| otCYP19 | F: TTTCAGCCGTTCGGTTCAG | MF: TCTCCACATGAGGTTCTTACCC |
| R: TTTCAGTGTTAGCAGGTTTAAATG | MR: TTCACATCATGCATACAGTTTGAG | |
| Sf1 | F: GCGGAAAACCAGGAGTGTAA | MF: GCGGAAAACCAGGAGTGTAA |
| R: AACTTGAAACCGTTGGATCG | MR: CAGGCAAAATGTCCCTGTTT | |
| Sox8 | F: GACTACAAGTACCAGCCTCGG | MF: GGACTGGGAGGAATGAGTGA |
| R: CTGAGCTCGGAGATGTCCAC | MR: CCTGCTGCTGTCAACAAGTC | |
| SRF: CTGGAGCAGAACTGGCTCAT | ||
| SRR: CCTGCTGCTGTCAACAAGTC |
F and R, initial forward and reverse primers; SRF and SRR, second-round forward and reverse primers; MF and MR, marker forward and reverse primers; EP, extension primers for SNP markers.
Amplification of genomic DNA in target genes:
Using primers in adjacent exons, fragments of the target genes were amplified by high-fidelity BIO-X-ACT Long DNA polymerase (Bioline, London) separated on agarose gels and stained with ethidium bromide. The desired DNA fragment was visualized with UV light and excised from the gel. DNA fragments were purified with the AccuPrep gel purification kit DNA (Bioneer, Life Science, Rockville, MD) and then sequenced on an ABI 377. The sequences were aligned and polymorphisms between the two parental species (red O. niloticus and O. aureus) were characterized.
Genotyping of microsatellite markers:
For microsatellite DNA markers, forward or reverse primers were dye labeled, and genotypes were obtained by automated sizing of fluorescently tagged polymerase chain reaction amplification products. Markers that produced alleles distinguished in their length by two or more bases were amplified using Super-Therm Taq DNA polymerase (JMR Holding, London). Markers that produced alleles distinguished in their length by one base were amplified using High Fidelity Accuzyme DNA polymerase (Bioline). Electrophoretic analysis was conducted on a 4% acrylamide gel in an ABI-377 DNA sequencer as previously described (Palti et al. 2002). The DNA fragments were automatically sized by comparison with an internal standard using Genescan (version 2.1). Genotypes of individuals were determined by Genotyper (version 2.0) and automatically exported to a database.
Genotyping of SNP markers:
Genotyping of SNP markers was performed using DNA MassArray technology (Jurinke et al. 2002). External and extension primers were designed using Sequanom's assay-design software (Table 2). This software also selected the appropriate termination mix for each of the SNPs. MALDI–TOF mass spectrometry analysis was performed using the MassArray genotype analyzer.
Gene mapping:
Genotype data for the 11 genes in the F2 mapping population were added to the published data set of 545 markers, and mapping was performed using JoinMap software (3.0) as previously described (Lee et al. 2005).
RESULTS
Identification of orthologous sequences:
Complete O. niloticus mRNA sequences were obtained from the NCBI database for most of the analyzed genes (Table 3). The sequences identified by BLAST had E-values <e−39. The E-value was relatively high only for Amh. When we repeated the search using the Amh of zebrafish (AAT77729), we identified two tilapia sequences (ONI06JC.39_H08 and ONI05ID.39_D09) with much lower E-values (2e−15 and 9e−12, respectively). The two sequences were positioned in two adjacent exons of the tilapia Amh, and one of them was similar to the human Amh. In the RBEST database, an Fhl2-like cDNA was found with an E-value of e−104. This cDNA showed a higher similarity to Fhl3 (E = e−109) in the NCBI database, which is another human gene of the FHL family. Hence, we defined the detected tilapia cDNA as part of the Fhl3l (Fhl3-like) gene.
TABLE 3.
Identifying sequences of tilapia, cichlids, and other fish species in public databases (human proteins vs. translated teleost sequences)
| Gene | Accession nos. | Species | Database | E-value | Identity (%) | Similarity (%) |
|---|---|---|---|---|---|---|
| Amh | ONI06JC.39_H08 | O. niloticus | RBEST | 3.0e-06 | 29 (35/117) | 41 (48/117) |
| Dax1 | AY135397 | O. niloticus | NCBI | 4.0e-54 | 46 (126/269) | 63 (170/269) |
| Dmrt2 | AY149606 | O. niloticus | NCBI | 9.0e-46 | 78 (85/108) | 86 (93/108) |
| Dmrta2 | AY149605 | O. niloticus | NCBI | 2.0e-39 | 72 (78/108) | 77 (84/108) |
| Fhl2(Fhl3l) | ONI05DB.39_H04 | O. niloticus | RBEST | 1.0e-104 | 54 (164/299) | 68 (204/299) |
| Foxl2 | AY554172 | O. niloticus | NCBI | 9.0e-70 | 62 (178/286) | 67 (192/286) |
| Ixl | TC1294 | Haplochromis | TIGR | 1.1e-50 | 73 (99/135) | 84 (114/135) |
| Lhx9 | AY534647 | O. latipes | NCBI | 1.0e-110 | 90 (208/229) | 95 (219/229) |
| BC093258 | D. rerio | 4.0e-161 | 90 (287/317) | 95 (302/317) | ||
| otCYP19 | AF472620 | O. niloticus | NCBI | 2.0e-147 | 52 (243/466) | 73 (341/466) |
| Sf1 | AB060814 | O. niloticus | NCBI | 9.0e-133 | 58 (277/474) | 70 (335/474) |
| Sox8 | AY935980 | T. rubripes | NCBI | 2.0e-76 | 61 (217/352) | 71 (252/352) |
| DQ294028 | S. salar | 3.0e-66 | 59 (203/339) | 67 (229/339) |
Intron–exon boundaries and polymorphism:
Reports for tilapia Dmrta2 (CAI23011) and otCYP19 (NP_000094) already included information on exon–intron borders. We predicted the exon–intron boundaries for the other nine genes (Table 4). Using D. rerio genomic structure, we predicted that the two nonoverlapping O. niloticus sequences for Amh are parts of the sixth and seventh exons of the same gene. To test this prediction, we designed primers in these exons and successfully amplified intron 6 and bridged these two cDNAs. Polymorphism was detected in introns in eight genes. In Dmrt2, otCYP19, and Foxl2, polymorphism was detected in translated regions (Table 4).
TABLE 4.
Accession numbers of sequenced regions, marker positions, and polymorphism
| Gene | Genomic fragment used for predictiona | No. of exons predicted | Position of marker | Accession nos.
|
Polymorphism
|
||
|---|---|---|---|---|---|---|---|
| O. niloticus | O. aureus | SNP | Microsatellites (bp) | ||||
| Amh | BX005098 (DR) | 7 | Sixth intron | AM232733 | AM232734 | 107/108 | |
| Dax1 | CAAB01000386 (TR) | 2 | Intron | AM232753 | AM232754 | C/G | |
| Dmrt2 | AJ295039 (TR) | 3 | 3′-UTR of third exon | AM232741 | AM232742 | 104/105 | |
| Dmrta2 | — | 2 | Intron | AM232736 | AM232735 | A/G | |
| Fhl3l | BX530064 (DR) | 6 | Fourth intron | AM232752 | AM232751 | 162/163 | |
| Foxl2 | AC120148 (MM) | 1 | Exon | AM232737 | AM232738 | C/A | |
| Ixl | NC000019 (HS) NC007124 (DR) | 4 | Third intron | AM232748 | AM232747 | 165/167 | |
| Lhx9 | AJ277917 AJ277918 (HS) | 4 | Third intron | AM232745 | AM232746 | A/G | |
| OtCYP19 | — | 9 | 3′UTR of ninth exon | AM232740 | AM232739 | 185/187 | |
| Sf1 | CAAB01010380 (TR) | 4 | Third intron | AM232744 | AM232743 | 169/183 | |
| Sox8 | AY688943(TR) | 3 | Second intron | AM232749 | AM232750 | 254/256 | |
TR, T. rubripes; HS, H. sapiens; MM, M. musculus; and DR, D. rerio.
Grandparental genotypes:
The genotypes of the red O. niloticus grandparent of the mapping family showed complete homozygosity for the 11 analyzed genes. O. aureus showed complete homozygosity for a different allele for each one of seven markers. The O. aureus grandparent was heterozygous for the remaining four markers (Dmrta2, Lhx9, Dax1, and Fhl3l). For each of these markers one allele was found to be common to both grandparents, possibly reflecting an impurity of the O. aureus stock.
Mapping position of the candidate genes:
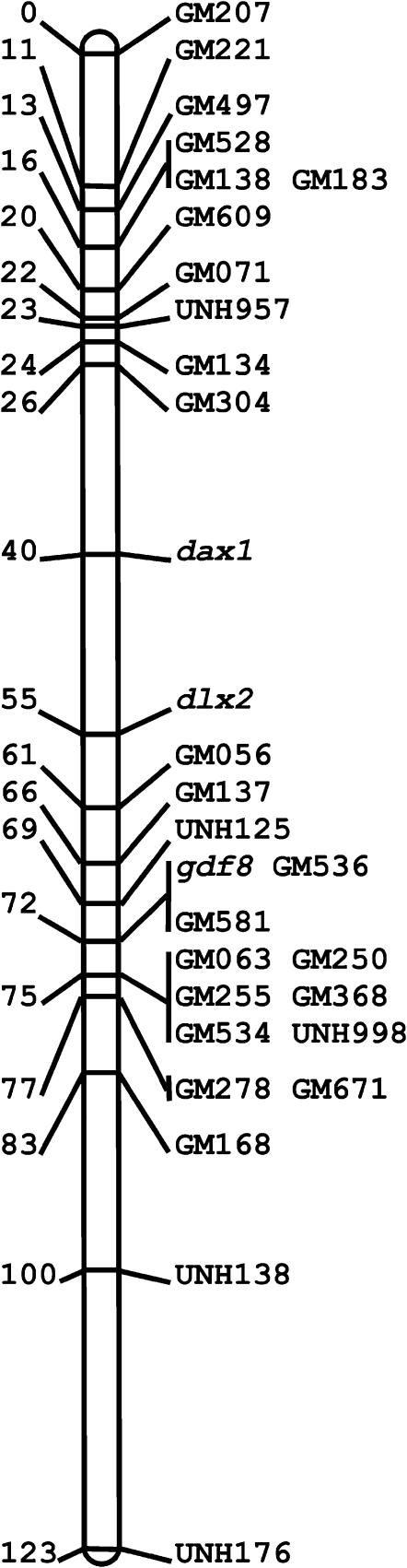
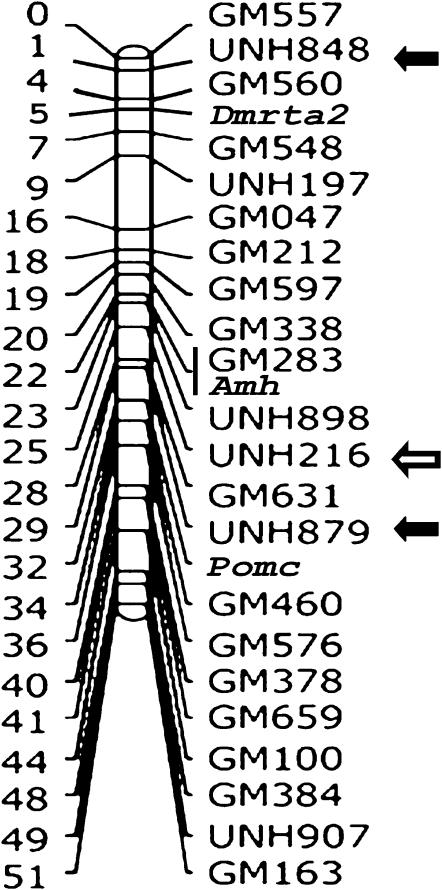
Genotypes were obtained for 76–89 individuals of the F2 mapping population. Each gene was localized to a specific location on the tilapia linkage map (Table 5). The mapping of Dax1 merged LG16 and LG21 into a single linkage group (Figure 1). The Amh and Dmrta2 genes were mapped to two distinct regions in LG23. The Amh gene mapped 5 cM from UNH879 within a QTL region for sex determination, and 2 cM from UNH216 within a QTL region for sex-specific mortality. Dmrta2 was mapped 4 cM from UNH848 within a QTL region for sex determination. Cyp19 was mapped to the start of LG1 but this region is not contained within the previously reported QTL region for sex determination. Seven other candidate genes mapped to LG4, -11, -12, -14, and -17.
TABLE 5.
Positions of the genes on the second-generation linkage map of tilapia (Lee et al. 2005)
| Gene | No. of F2 genotypes | Linkage group | Mapping position (cM) | Overlap with a QTL regiona | Comments and references |
|---|---|---|---|---|---|
| Amh | 82 | 23 | 22 | SD, SSM | Shirak et al. (2002); Cnaani et al. (2003) |
| Dax1 | 86 | 16 and 21 | — | — | Connecting LG16 and LG21 |
| Dmrt2 | 85 | 12 | 43 | — | Located near Dmrt1 |
| Dmrta2 | 84 | 23 | 5 | SD | Cnaani et al. (2004) |
| Fhl3l | 89 | 11 | 69 | — | |
| Foxl2 | 86 | 14 | 17 | — | |
| Ixl | 83 | 14 | 41 | — | |
| Lhx9 | 89 | 17 | 13 | — | |
| OtCYP19 | 76 | 1 | 5 | — | |
| Sf1 | 80 | 12 | 6 | — | |
| Sox8 | 83 | 4 | 43 | — |
SD, sex determination; SSM, sex-specific mortality.
Figure 1.—
The joint linkage group of LG16 and LG21.
DISCUSSION
In this study we focused on analysis and mapping of 11 genes for which there is a broad consensus in the literature about their role in mammalian sex determination. Due to their high level of conservation, we were able to use the sequences of human proteins in a BLAST analysis to recover the sequences of cichlid homologs for 9 of the 11 targeted genes. Primers based on sequences of more distantly related fish species such as O. latipes, D. rerio, T. rubripes, and S. salar were also successfully used to amplify specific regions in tilapia. We used comparative sequence information to predict exon–intron boundaries and design primers and to amplify parts of the tilapia Lhx9 and Sox8 genes, even without any preliminary sequence information from tilapia.
Map positions of 2 of the 11 genes overlapped with two of the four previously reported QTL for sex determination in tilapia species and their hybrids (Cnaani et al. 2003, 2004; Lee et al. 2003, 2004). Amh and Dmrta2 mapped 5 and 4 cM from UNH879 and UNH848, respectively (Figure 2). These markers were highly associated with two different QTL for sex determination on LG23 (Cnaani et al. 2003, 2004). Moreover, Amh is located 2 cM from UNH216, which marks a QTL region for sex-specific mortality (Shirak et al. 2002). The overlap in position on LG23 of two QTL for sex determination and sex-specific mortality may be a result of accumulation of deleterious alleles near sex-determining genes as predicted by Palti et al. (2002). Thus, we propose Amh and Dmrta2 as candidate genes for master key regulators for sex determination in tilapia. Amh was differentially expressed in adult human testis germ cells among 79 tissues tested in GeneAtlas (http://symatlas.gnf.org/SymAtlas/). Likewise, Dmrta2 had the highest expression in adult ovary but expression was also high in most other tissues. Nevertheless, it is worthwhile to explore expression for both Amh and Dmrta2 during the critical stages of sex determination in tilapia at the embryo level. In mammals, Amh expression is induced by Sox9 (Vidal et al. 2001). Knockout of Amh in mice suggests that normal primary male sex determination still occurs in spite of the absence of Amh. In birds and fish, unlike in mammals, expression of Amh in the undifferentiated gonads of both sexes occurs in the absence of Sox9 (Oreal et al. 2002; Rodriguez-Mari et al. 2005). Furthermore, in birds, Amh is able to alter primary sex determination (Elbrecht and Smith 1992). In zebrafish, Amh expression peaked at 20 days postfertilization at a period when the oocytes undergo apoptosis in presumptive males, leading to the development of testes (Uchida et al. 2002). Furthermore, Amh is a strong candidate for a direct regulator of aromatase, which has been shown to inhibit aromatase biosynthesis in the rat (di Clemente et al. 1992). This hormone may play a central role in sex determination and the response to temperature changes (D'Cotta et al. 2001). In addition to Sox9, other Sox family genes, e.g., Sox8, were shown to regulate Amh for testis differentiation in mice by interaction with the Sox-binding element of Amh promoter (Schepers et al. 2003). The transcriptional regulation of teleost Amh has so far not been elucidated, but the Amh gene promoter sequence contains putative binding sites for the same transcription factors that regulate mammalian Amh (von Hofsten and Olsson 2005). Comparison of the O. aureus and O. niloticus sequences predicts a single amino acid difference in the coding region of Amh between these two tilapia species. Recently, two alternatively spliced mRNA variants were reported in O. aureus for Amh (ABB69056/7), but no gender differences were noted. Our linkage results provide additional incentive for examining the functional significance of these various forms.
Figure 2.—
The tilapia linkage group 23. Mapping positions of QTL for sex determination and sex-specific mortality are denoted by solid and open arrows, respectively.
The doublesex/mab3 (DM) gene family was originally described on the basis of similarities of the Drosophila (dsx) and Caenorhabditis elegans (mab3) homologs. Apart from the analysis of DM genes in these two species, the functional role of DM-domain-containing genes has not been extensively studied (Yi and Zarkower 1999; Lei and Heckert 2002). It was therefore a surprise to find that a DM gene was responsible for sex determination in medaka (O. latipes) (Matsuda et al. 2002; Nanda et al. 2002). Dmy is a recent duplication of the Dmrt1 gene, which created a new sex chromosome in medaka ∼10 MYA (Zhang 2004). It has been suggested that other DM genes may regulate Dmrt1 in a dosage-dependent fashion in the sex-determining pathway (Nanda et al. 1999).
Guan et al. (2000) isolated two DM-containing sequences from tilapia gonads. These cDNAs are encoded by two different genes, Dmrt1 and Dmo, which are predominantly expressed in the testis and the ovary, respectively. Moreover, they showed that Dmrt1, but not Dmo, has a Sry consensus site. The map positions of these genes do not overlap with any of the defined QTL regions for sex determination (Lee et al. 2005). Dmrta2 and Dmo (Dmrta1) belong to the same DMRTA subfamily of Dmrt genes. It will be interesting to analyze Dmrta2-binding sites for Sry-related transcription factors and also to learn if Dmrta2 has regulatory effects on the expression of other DM-domain genes in tilapia. Sex-specific alternative splicing of Dmrt1 was reported in zebrafish and rice field eel (Guo et al. 2005; Huang et al. 2005). Alternatively spliced products of DM genes were also observed in human Dmrt2 (Ottolenghi et al. 2000). However, we have no such information for tilapia Dmrt genes.
Lee et al. (2005) constructed a linkage map of tilapia containing 525 microsatellite and 20 type I (gene) markers. The markers were positioned into 22 large and two small linkage groups. They predicted that the two small linkage groups (8 and 24) would eventually merge with other linkage groups to correspond with the 22 chromosomal pairs of tilapia (Lee et al. 2005). Unexpectedly, Dax1 merged two relatively large linkage groups, LG16 and LG21, and simultaneously extended the tilapia linkage map by 29 cM. It remains unlikely that LG1, LG3, and LG23, which contain the QTL for sex determination, will eventually coalesce into a single linkage group corresponding to a single chromosome.
A major problem in the maintenance of laboratory and commercial purebred stocks of tilapia is the high risk of hybridization and dilution of purebred stocks. The morphology of such hybrids is frequently indistinguishable from that of the parental species (Taniguchi et al. 1985; Macaranas et al. 1986; Mair and Little 1991). The high number of common alleles (36%) between the two lines studied here suggests the possibility that O. niloticus alleles may have introgressed into the O. aureus stock. Alternatively, this finding may also reflect common ancestral polymorphism for these closely related species (Agnese et al. 1997; Rao and Majumdar 1998).
New QTL for sex determination may emerge in hybrids due to the interactions of alleles from different species, such as those predicted by the autosomal theory (Hammerman and Avtalion 1979). The multiple QTL phenomenon may also be due to different MKRs controlling sex determination among closely related tilapia species. Although investigation of sex determination in tilapia hybrids, rather than in purebred lines, introduces significant complications (Mair et al. 1991), it also provides a unique model for the study of interactions between multiple QTL in the sex determination pathway.
Acknowledgments
We acknowledge the assistance of Y. Nagahama for providing DNA sequences for AMH in tilapia. Genotyping of SNP was performed at the Genome Knowledge Center, the Weizmann Institute of Science, and the Crown Human Genome Center, Israel. We thank Tami Koch and Edna Ben-Asher from the Weizmann Institute of Science for operating the DNA MassArray technology. This research was supported by research grant no. IS-3561-04 from the United States–Israel Binational Agricultural Research and Development Fund and by National Research Initiative grant no. 2004-36205-14205 from the U. S. Department of Agriculture Cooperative State Research, Education, and Extension Service.
References
- Agnese, J. F., B. Adepo-Gourene, E. K. Abban and Y. Fermon, 1997. Genetic differentiation among natural populations of the Nile tilapia Oreochromis niloticus (Teleostei, cichlidae). Heredity 79: 88–96. [DOI] [PubMed] [Google Scholar]
- Arkhipchuk, V. V., 1995. Role of chromosomal and genome mutations in the evolution of bony fishes. Hydrobiol. J. 31: 55–65. [Google Scholar]
- Baroiller, J.-F., and H. D'Cotta, 2001. Environment and sex determination in farmed fish. Comp. Biochem. Physiol. C Comp. Pharmacol. 130: 399–409. [DOI] [PubMed] [Google Scholar]
- Baroiller, J.-F., Y. Guiguen and A. Fostier, 1999. Endocrine and environmental aspects of sex differentation in fish. Cell. Mol. Life Sci. 55: 910–931. [Google Scholar]
- Baron, D., R. Houlgatte, A. Fostier and Y. Guiguen, 2005. Large-scale temporal gene expression profiling during gonadal differentiation and early gametogenesis in rainbow trout. Biol. Reprod. 73: 959–966. [DOI] [PubMed] [Google Scholar]
- Clemens, H. P., and T. Inslee, 1968. The production of unisexual broods of Tilapia mossambica sex reversed with methyltestosterone. Trans. Am. Fish. Soc. 97: 18–21. [Google Scholar]
- Cnaani, A., E. Hallerman, M. Ron, J. I. Weller, M. Indelman et al., 2003. Detection of a chromosomal region with two quantitative trait loci, affecting cold tolerance and fish size, in an F2 tilapia hybrid. Aquaculture 223: 117–128. [Google Scholar]
- Cnaani, A., N. Zilberman, S. Tinman, G. Hulata and M. Ron, 2004. Genome-scan analysis for quantitative trait loci in an F2 tilapia hybrid. Mol. Genet. Genomics 272: 162–172. [DOI] [PubMed] [Google Scholar]
- Crosetti, D., L. Sola, P. Brunner and S. Cataudella, 1988. Cytogenetical characterization of Oreochromis niloticus, O. mossambicus and their hybrid, pp. 143–151 in The Second International Symposium on Tilapia in Aquaculture, edited by R. S. V. Pullin, T. Bhukaswan, K. Tonguthai and J. L. Maclean. ICLARM, Manila, Philippines.
- D'Cotta, H., A. Fostier, Y. Guiguen, M. S. Govoroun and J.-F. Baroiller, 2001. Aromatase plays a key role during normal and temperature-induced sex differentiation of tilapia Oreochromis niloticus. Mol. Reprod. Dev. 59: 265–276. [DOI] [PubMed] [Google Scholar]
- Desvages, G., and C. Pieau, 1992. Time required for temperature-induced changes in gonadal aromatase activity and related gonadal structure in turtle embryos. Differentiation 52: 13–18. [Google Scholar]
- Devlin, R. H., and Y. Nagahama, 2002. Sex determination and sex differentiation in fish: an overview of genetic, physiological, and environmental influences. Aquaculture 208: 191–365. [Google Scholar]
- di Clemente, N., S. Ghaffari, R. B. Pepinsky, C. Pieau, N. Josso et al., 1992. Quantitative and interspecific test for biological activity of anti-mullerian hormone: the fetal ovary aromatase assay. Development 114: 721–727. [DOI] [PubMed] [Google Scholar]
- Elbrecht, A., and R. G. Smith, 1992. Aromatase enzyme activity and sex determination in chickens. Science 255: 467–469. [DOI] [PubMed] [Google Scholar]
- Galay-Burgos, M., L. Llewellyn, C. C. Mylonas, A. V. M. Canario, S. Zanuy et al., 2004. Analysis of the Sox gene family in the European sea bass (Dicentrarchus labrax). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 137: 279–284. [DOI] [PubMed] [Google Scholar]
- Gardner, L., T. Anderson, A. R. Place, B. Dixon and A. Elizur, 2005. Sex change strategy and the aromatase genes. J. Steroid Biochem. Mol. Biol. 94: 395–404. [DOI] [PubMed] [Google Scholar]
- Godwin, J., J. A. Luckenbach and R. J. Borski, 2003. Ecology meets endocrinology: environmental sex determination in fishes. Evol. Dev. 5: 40–49. [DOI] [PubMed] [Google Scholar]
- Graves, J. A. M., C. M. Disteche and R. Toder, 1998. Gene dosage in the evolution and function of mammalian sex chromosomes. Cytogenet. Cell Genet. 80: 94–103. [DOI] [PubMed] [Google Scholar]
- Guan, G., T. Kobayashi and Y. Nagahama, 2000. Sexually dimorphic expression of two types of DM (Doublesex/Mab-3)-domain genes in a teleost fish, the tilapia (Oreochromis niloticus). Biochem. Biophys. Res. Commun. 272: 662–666. [DOI] [PubMed] [Google Scholar]
- Guo, Y., H. Cheng, X. Huang, S. Gao, H. Yu et al., 2005. Gene structure, multiple alternative splicing, and expression in gonads of zebrafish Dmrt1. Biochem. Biophys. Res. Commun. 330: 950–957. [DOI] [PubMed] [Google Scholar]
- Haag, E. S., and A. V. Doty, 2005. Sex determination across evolution: connecting the dots. PLoS Biol. 3: e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerman, I. S., and R. R. Avtalion, 1979. Sex determination in Sarotherodon (Tilapia). II. The sex ratio as a tool for the determination of genotype. A mathematical model of autosomal and gonosomal influence. Theor. Appl. Genet. 55: 177–187. [DOI] [PubMed] [Google Scholar]
- Hett, A. K., and A. Ludwig, 2005. SRY-related (Sox) genes in the genome of European Atlantic sturgeon (Acipenser sturio). Genome 48: 181–186. [DOI] [PubMed] [Google Scholar]
- Hett, A. K., C. Pitra, I. Jenneckens and A. Ludwig, 2005. Characterization of Sox9 in European Atlantic sturgeon (Acipenser sturio). J. Hered. 96: 150–154. [DOI] [PubMed] [Google Scholar]
- Hopkins, K. D., W. L. Shelton and C. R. Engle, 1979. Estrogen sex-reversal of Tilapia aurea. Aquaculture 18: 263–268. [Google Scholar]
- Huang, X., Y. Guo, Y. Shui, S. Gao, H. Yu et al., 2005. Multiple alternative splicing and differential expression of Dmrt1 during gonad transformation of the rice field eel. Biol. Reprod. 73: 1017–1024. [DOI] [PubMed] [Google Scholar]
- Jalabert, B., P. Kammacher and P. Lessent, 1971. Determinisme du sexe chez les hybrides entre Tilapia macrochir et Tilapia nilotica. Etude de la sex-ratio dans les recroisements des hybrides de premiere generation par les especes parentes. Ann. Biol. Anim. Biochim. Biophys. 11: 155–165. [Google Scholar]
- Jurinke, C., D. van den Boom, C. R. Cantor and H. Koster, 2002. Automated genotyping using the DNA MassArray technology. Methods Mol. Biol. 187: 179–192. [DOI] [PubMed] [Google Scholar]
- Kallman, K. D., 1984. A new look at sex determination in poeciliid fishes, pp. 95–171 in Evolutionary Genetics of Fishes, edited by B. J. Turner. Plenum, New York.
- Koopman, P., and K. Loffler, 2003. Sex determination: the fishy tale of Dmrt1. Curr. Biol. 13: R177–R179. [DOI] [PubMed] [Google Scholar]
- Koopman, P., G. Schepers, S. Brenner and B. Venkatesh, 2004. Origin and diversity of the Sox transcription factor gene family: genome-wide analysis in Fugu rubripes. Gene 328: 177–186. [DOI] [PubMed] [Google Scholar]
- Kornfield, I. L., 1984. Descriptive genetics of cichlid fishes, pp. 519–616 in Evolutionary Genetics of Fishes, edited by B. J. Turner. Plenum, New York.
- Kosswig, C., 1964. Polygenic sex determination. Experientia 20: 190–199. [DOI] [PubMed] [Google Scholar]
- Lahav, E., 1993. Use of sex-reversed females to produce all-male tilapia (Oreochromis aureus) fry. Isr. J. Aquac. 45: 131–136. [Google Scholar]
- Lee, B.-Y., D. J. Penman and T. D. Kocher, 2003. Identification of a sex-determining region in Nile tilapia (Oreochromis niloticus) using bulked segregant analysis. Anim. Genet. 34: 379–383. [DOI] [PubMed] [Google Scholar]
- Lee, B.-Y., G. Hulata and T. D. Kocher, 2004. Two unlinked loci controlling the sex of blue tilapia (Oreochromis aureus). Heredity 92: 543–549. [DOI] [PubMed] [Google Scholar]
- Lee, B.-Y., W.-J. Lee, J. T. Streelman, K. L. Carleton, A. E. Howe et al., 2005. A second-generation genetic linkage map of tilapia (Oreochromis spp.). Genetics 170: 237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, N., and L. L. Heckert, 2002. Sp1 and Egr1 regulate transcription of the Dmrt1 gene in Sertoli cells. Biol. Reprod. 66: 675–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaranas, J. M., N. Taniguchi, M. J. R. Pante, J. B. Capili and R. S. V. Pullin, 1986. Electrophoretic evidence for extensive hybrid gene introgression into commercial Oreochromis niloticus in the Philippines. Aquac. Fish Manage. 17: 249–258. [Google Scholar]
- Mair, G. C., and D. C. Little, 1991. Population control in farmed tilapias. Naga 14: 8–13. [Google Scholar]
- Mair, G. C., D. J. Penman, A. Scott, D. O. F. Skibinski and J. A. Beardmore, 1987. Hormonal sex reversal and the mechanisms of sex determination in Oreochromis, pp. 289–300 in Proceedings of the World Symposium on Selection, Hybridization and Genetic Engineering in Aquaculture, Vol. II, edited by K. Tiews. Heenemann Verlagsgesellschaft, Berlin.
- Mair, G. C., A. G. Scott, D. J. Penman, D. O. F. Skibinski and J. A. Beardmore, 1991. Sex determination in the genus Oreochromis. 2. Sex reversal, hybridisation, gynogenesis and triploidy in O. aureus Steindachner. Theor. Appl. Genet. 82: 153–160. [DOI] [PubMed] [Google Scholar]
- Majumdar, K. C., and B. J. McAndrew, 1983. Relative DNA content of somatic nuclei and chromosomal studies in three genera, Tilapia, Sarotherodon, and Oreochromis of the tribe Tilapiini (Pisces, Cichlidae). Genetica 68: 175–188. [Google Scholar]
- Matsuda, M., Y. Nagahama, A. Shinomiya, T. Sato, C. Matsuda et al., 2002. Dmy is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417: 559–563. [DOI] [PubMed] [Google Scholar]
- McAndrew, B. J., and K. C. Majumdar, 1983. Tilapia stock identification using electrophoretic markers. Aquaculture 30: 249–261. [Google Scholar]
- McElreavey, K., E. Vilain, N. Abbas, I. Herskowitz and M. Fellous, 1993. A regulatory cascade hypothesis for mammalian sex determination: SRY represses a negative regulator of male development. Proc. Natl. Acad. Sci. USA 90: 3368–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittwoch, U., 2006. Sex is a threshold dichotomy mimicking a single gene effect. Trends Genet. 22: 96–100. [DOI] [PubMed] [Google Scholar]
- Murdock, C., and T. Wibbels, 2003. Expression of Dmrt1 in a turtle with temperature-dependent sex determination. Cytogenet. Genome Res. 101: 302–308. [DOI] [PubMed] [Google Scholar]
- Nanda, I., Z. Shan, M. Schartl, D. W. Burt, M. Koehler et al., 1999. 300 million years of conserved synteny between chicken Z and human chromosome 9. Nat. Genet. 21: 258–259. [DOI] [PubMed] [Google Scholar]
- Nanda, I., M. Kondo, U. Hornung, S. Asakawa, C. Winkler et al., 2002. A duplicated copy of Dmrt1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc. Natl. Acad. Sci. USA 99: 11778–11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oreal, E., S. Mazaud, J. Y. Picard, S. Magre and D. Carre-Eusebe, 2002. Different patterns of anti-Mullerian hormone expression, as related to Dmrt1, Sf-1, Wt1, Gata4, Wnt4, and Lhx9 expression, in the chick differentiating gonads. Dev. Dyn. 225: 221–232. [DOI] [PubMed] [Google Scholar]
- Ottolenghi, C., M. Fellous, M. Barbieri and K. McElreavey, 2002. Novel paralogy relations among human chromosomes support a link between the phylogeny of doublesex-related genes and the evolution of sex determination. Genomics 79: 333–343. [DOI] [PubMed] [Google Scholar]
- Palti, Y., A. Shirak, G. Hulata, R. R. Avtalion and M. Ron, 2002. Detection of genes with deleterious alleles in an inbred line of tilapia (Oreochromis aureus). Aquaculture 206: 151–164. [DOI] [PubMed] [Google Scholar]
- Rao, C. B., and K. C. Majumdar, 1998. Multivariate map representation of phylogenetic relationships: application to tilapiine fish. J. Fish Biol. 52: 1199–1217. [Google Scholar]
- Rodriguez-Mari, A., Y. L. Yan, R. A. Bremiller, C. Wilson, C. Canestro et al., 2005. Characterization and expression pattern of zebrafish anti-Mullerian hormone (Amh) relative to Sox9a, Sox9b, and Cyp19a1a, during gonad development. Gene Expr. Patterns 5: 655–667. [DOI] [PubMed] [Google Scholar]
- Rosenstein, S., and G. Hulata, 1994. Sex reversal in the genus Oreochromis: optimization of feminization protocol. Aquac. Fish Manage. 25: 329–339. [Google Scholar]
- Sarre, S. D., A. Georges and A. Quinn, 2004. The ends of a continuum: genetic and temperature dependent sex determination in reptiles. BioEssays 26: 639–645. [DOI] [PubMed] [Google Scholar]
- Schartl, M., 2004. Sex chromosome evolution in non-mammalian vertebrates. Curr. Opin. Genet. Dev. 14: 634–641. [DOI] [PubMed] [Google Scholar]
- Schepers, G., M. Wilson, D. Wilhelm and P. Koopman, 2003. Sox8 is expressed during testis differentiation in mice and synergizes with Sf1 to activate the Amh promoter in vitro. J. Biol. Chem. 278: 28101–28108. [DOI] [PubMed] [Google Scholar]
- Shan, Z., I. Nanda, Y. Wang, M. Schmid, A. Vortkamp et al., 2000. Sex-specific expression of an evolutionarily conserved male regulatory gene, DMRT1, in birds. Cytogenet. Cell Genet. 89: 252–257. [DOI] [PubMed] [Google Scholar]
- Shirak, A., Y. Palti, A. Cnaani, A. Korol, G. Hulata et al., 2002. Association between loci with deleterious alleles and distorted sex ratios in an inbred line of tilapia (Oreochromis aureus). J. Hered. 93: 270–276. [DOI] [PubMed] [Google Scholar]
- Taniguchi, N., J. M. Macaranas and R. S. V. Pullin, 1985. Introgressive hybridization in culture tilapia stocks in the Philippines. Bull. Jpn. Soc. Fish. 51: 1219–1224. [Google Scholar]
- Uchida, D., M. Yamashita, T. Kitano and T. Iguchi, 2002. Oocyte apoptosis during the transition from ovary-like tissue to testes during sex differentiation of juvenile zebrafish. J. Exp. Zool. 205: 711–718. [DOI] [PubMed] [Google Scholar]
- Valenzuela, N., D. C. Adams and F. J. Janzen, 2003. Pattern does not equal process: Exactly when is sex environmentally determined? Am. Nat. 161: 676–683. [DOI] [PubMed] [Google Scholar]
- Vidal, V. P., M. C. Chaboissier, D. G. De Rooij and A. Schedl, 2001. Sox9 induces testis development in XX transgenic mice. Nat. Genet. 28: 216–217. [DOI] [PubMed] [Google Scholar]
- Volff, J.-N., and M. Schartl, 2001. Variability of sex determination in poeciliid fishes. Genetica 111: 101–110. [DOI] [PubMed] [Google Scholar]
- von Hofsten, J., and P.-E. Olsson, 2005. Zebrafish sex determination and differentiation: involvement of Ftz-F1 genes. Reprod. Biol. Endocrinol. 3: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western, P. S., and A. H. Sinclair, 2001. Sex, genes, and heat: triggers of diversity. J. Exp. Zool. 290: 624–631. [DOI] [PubMed] [Google Scholar]
- Wohlfarth, G. W., and H. Wedekind, 1991. The heredity of sex determination in tilapias. Aquaculture 92: 143–156. [Google Scholar]
- Yao, H.-C., and B. Capel, 2005. Temperature, genes, and sex: a comparative view of sex determination in Trachemys scripta and Mus musculus. J. Biochem. 138: 5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, W., and D. Zarkower, 1999. Similarity of DNA binding and transcriptional regulation by Caenorhabditis elegans Mab-3 and Drosophila melanogaster Dsx suggests conservation of sex determining mechanisms. Development 126: 873–881. [DOI] [PubMed] [Google Scholar]
- Zarkower, D., 2001. Establishing sexual dimorphism: Conservation amidst diversity? Nat. Rev. Genet. 2: 175–185. [DOI] [PubMed] [Google Scholar]
- Zhang, J., 2004. Evolution of DMY, a newly emergent male sex-determination gene of medaka fish. Genetics 166: 1887–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]