Abstract
The FMR family of KH domain RNA-binding proteins is conserved from invertebrates to humans. In humans, inactivation of the X-linked FMR gene fragile X is the most common cause of mental retardation and leads to defects in neuronal architecture. While there are three FMR family members in humans, there is only a single gene, dfmr1, in flies. As in humans, inactivation of dfmr1 causes defects in neuronal architecture and in behavior. dfmr1 has other functions in the fly in addition to neurogenesis. Here we have analyzed its role during early embryonic development. We found that dfmr1 embryos display defects in the rapid nuclear division cycles that precede gastrulation in nuclear migration and in pole cell formation. While the aberrations in nuclear division are correlated with a defect in the assembly of centromeric/centric heterochromatin, the defects in pole cell formation are associated with alterations in the actin–myosin cytoskeleton.
MUTATIONS in the fragile X mental retardation gene (FMR1) are responsible for the most common form of hereditary mental retardation in humans known as the fragile X syndrome. While there are no gross lesions in the central nervous system (CNS), there is an excess density of dendritic spines and the spines are considerably longer than normal (for a detailed review, see Jin et al. 2004a,b). In addition to mental retardation, the fragile X syndrome is associated with a variety of other behavioral abnormalities, male sterility, and defects in oogenesis. In most cases the lesion in the FMR1 gene is caused by the expansion of a CGG trinucleotide repeat near the 5′-end, which results in hypermethylation and transcriptional silencing (Darnell et al. 2001). The X-linked FMR1 gene and its two autosomal homologs, FXR1P and FXR2P, encode KH domain RNA-binding proteins. These proteins exist in multi-component cytoplasmic RNP complexes and are associated with polysomes where they are believed to function in repressing the translation of target mRNAs. Biochemical studies indicate that FMR1 recognizes a G-quartet motif and this motif is present in many of the potential FMR1 regulatory target mRNAs that have been identified by microarray analysis in humans and mice (Brown et al. 2001).
Unlike mammals, Drosophila has only a single FMR gene, dfmr1. Although null dfmr1 mutant animals survive, viability is reduced and the mutant animals exhibit an array of defects in neuronal structure and function, behavior, and germline development that are broadly similar to those seen for FMR1 mutations in mice and humans (Morales et al. 2002; Zhang and Broadie 2005). Neurons in dfmr1 mutant flies have much more complex architectures with excess branching and synapse formation. Conversely, overexpression of dFMR1 simplifies neuronal structure and suppresses synapse formation. Recent studies have identified several important in vivo targets in the CNS for dfmr1 regulation. One of the targets in the nervous system is the futsch mRNA, which encodes the Drosophila microtubule-associated protein 1B (MAP1B) (Zhang et al. 2002). In dfmr1mutants, Futsch protein levels are elevated in neurons, while overexpression of dFMR1 protein reduces Fustch protein levels. Consistent with the idea that futsch overexpression contributes to the dfmr1 neuronal phenotypes, excess branching and synapse formation can be suppressed by mutations in futsch. Other targets in the nervous system include chickadee mRNA, which encodes profilin; dRac1 mRNA, which encodes a Rho GTPase; and pickpocket1 (ppk1), which encodes a sodium channel protein (Xu et al. 2004; Reeve et al. 2005). Both chickadee and dRac1 are involved in the remodeling of the actin cytoskeleton and have been implicated in synapse organization and structure.
Targets for dfmr1 regulation in fly ovaries have also been identified (Costa et al. 2005). One of the key targets in the ovary is orb, which encodes the fly germline-specific cytoplasmic polyadenylation element-binding protein. orb is required for oocyte specification and for the establishment of the anterior–posterior and dorsal–ventral axes of the developing egg. Orb functions in these critical morphogenetic processes by binding to the 3′-UTRs of localized mRNAs such as K(10), Bicaudal-D, oskar, and orb itself and by promoting their polyadenylation and translation activation. Costa et al. (2005) showed that dFMR1 is in an RNP complex with Orb and that it negatively regulates Orb activity. Interestingly, dFMR1 only appears to inhibit the expression of a subset of the Orb target mRNAs, downregulating K(10) and Orb protein expression but not Oskar. This specificity may be related to the presence or the absence of the dFMR1 G-quartet recognition motif in the different orb target mRNAs.
While it is not known how dFMR1 negatively regulates Orb activity, recent studies have suggested that dFMR1 may function as a component of the RNA interference (RNAi) machinery (for a review, see Carmell et al. 2002; Tomari and Zamore 2005). dFMR1 is found in complexes with four proteins known to be involved in RNAi-mediated translational repression: Argonaute-2 (Ago-2), a PAZ and Piwi domain protein; Dmp68, a fly homolog of the p68 RNA helicase; Dicer, a double-strand RNase; and VIG, an RNA-binding protein (Hammond et al. 2001; Caudy et al. 2002; Ishizuka et al. 2002). Moreover, Xu et al. (2004) have shown that both dFMR1 and Ago-2 are required to downregulate the level of pickpocket1 mRNA in the larval nervous system and have suggested that the two proteins collaborate in this process.
We have recently shown that ago-2 is required for the proper execution of the rapid nuclear division cycles in early fly embryos and for the proper formation of both the germline pole cells and the somatic cells (Deshpande et al. 2005). The connections among dFMR1, orb, and the RNAi machinery led us to wonder whether dfmr1 plays an equivalent role in early embryogenesis. In the studies reported here we have analyzed the function of dFMR1 protein during early embryonic development in various contexts.
MATERIALS AND METHODS
Strains and culturing:
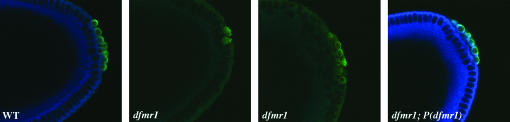
Flies were grown on a standard medium at 25° unless otherwise noted. Homozygous dfmr13 adults were obtained from a w; dfmr13/TM6 Sb stock. To generate dfmr13 embryos, dfmr13/dmr13 females were mated with dfmr13/dfmr13 males. The resulting dfmr13 embryos are completely devoid of the dFMR1 protein (see Figure 6). A fourth chromosome white insert (obtained from Lori Wallrath) was used to determine the effect of dfmr1 on heterochromatic silencing. A P(dfmr1);dfmr13 stock was used for rescue experiments (from T. Jongens).
Figure 6.—
P(dfmr1) transgene rescues the pole cell formation defects in dfmr1 embryos. Wild-type (left), dfmr13 (middle), and dfmr13; P(dfmr1) (right) embryos, 0–4 hr old, were fixed and stained with antibodies against dFMR1 (imaged in blue) and Vasa (imaged in green). The dfmr13 embryos were generated by mating homozygous dfmr13 females to homozygous dfmr13 males. The dfmr13; P(dfmr1) embryos were from a stock that is homozygous dfmr13 and is homozygous for the rescue P(dfmr1) transgene. The defects in pole cell formation evident in the dfmr13 embryos are rescued by the P(dfmr1) transgene.
Immunohistochemistry:
The embryo stainings were performed essentially as described in Deshpande et al. (1995). Embryos were usually fixed in paraformaldehyde/hepatane mix for 15–20 min and vitelline membranes were subsequently removed by shaking in methanol upon removal of paraformaldehyde. In the case of antitubulin antibodies, embryos were fixed only in methanol/heptane mix for 1 hr. The following antibodies were used at a prescribed dilution in a standard immunohistochemical analysis: anti-Peanut (mouse monoclonal from Developmental Hybridoma Bank used at 1:20; Neufeld and Rubin 1994); anti-Anillin (rabbit polyclonal used at 1:500; a gift from Chris Field; Oegema et al. 2000); anti-phospho-histone H3 (rabbit polyclonal used at 1:500 from Upstate Biotechnology); anti-CID (chicken polyclonal used at 1:100; a gift from Gary Karpen); anti-centrosomin (rabbit polyclonal used at 1:500; a gift from Thomas Kaufman); anti-HP1 (rabbit polyclonal used at 1:500; a gift from Rebecca Kellum); anti-Vasa (rabbit polyclonal antibody, which was preabsorbed against wild-type embryonic samples and was subsequently used at 1:500 dilution; a gift from Paul Lasko); and anti-β-galactosidase (rabbit) antibody purchased from Kappel and used at 1:1000 dilution after preabsorbing it against wild-type embryonic sample. The dfmr13 mutant embryos were stained with anti-dFMR1 antibodies (a gift from Tom Jongens) to confirm the protein null nature of the allele.
RESULTS
dfmr1 is required for pole cell formation:
After fertilization and fusion of the male and female pronuclei, there is a period of rapid synchronous nuclear division in the center of the embryo (Baker et al. 1993; Sullivan et al. 1993; Sullivan and Theurkauf 1995). The nuclei replicate their DNA and divide approximately every 5–8 min. After the fourth cycle the nuclei begin to spread out along the anterior–posterior axis, occupying discrete and roughly evenly spaced domains (Foe et al. 2000; Royou et al. 2002). During cycles 8 and 9, the nuclei begin migrating toward the cortex of the embryo and a few nuclei enter the specialized cytoplasm at the posterior pole, the pole plasm, and form polar buds, which protrude from the end of the embryo. The nuclei in the polar buds, together with the surrounding pole plasm, undergo cellularization to form the pole cells, the precursors of the germline (Williamson and Lehman 1996). Formation of pole cells is marked by the constriction of actin rings at the base of individual pole buds. Other cytoskeletal proteins such as Anillin, Peanut (Pnut), and Rhogef2 appear to localize to the contractile rings and are found along the membranes of the newly formed cells (Field et al. 2005; Padahs Barmchi et al. 2005). The localized accumulation of these proteins appears to be necessary for cellularization as embryos compromised for these cytoskeletal components display defective pole cell formation. The newly formed pole cells incorporate a number of maternally derived mRNAs and proteins that are specifically localized at the posterior pole of early embryos. These include osk, nanos, and orb mRNAs and Vasa protein. After the pole cells form they undergo one or two rounds of asynchronous division and then cease dividing. The remaining nuclei reach the periphery of the embryo during nuclear cycle 10 and, unlike the pole cells, continue to divide synchronously until the end of nuclear cycle 14, when the embryo cellularizes and commences gastrulation.
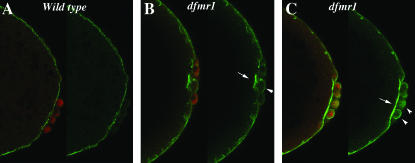
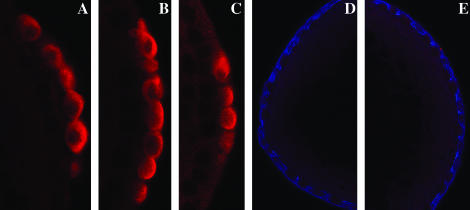
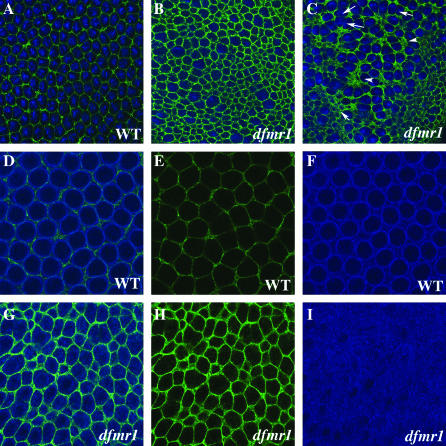
Since orb is the only maternal “pole cell” mRNA known to be subject to dfmr1 regulation, we examined the expression of Orb protein in pole cells of dfmr13 mutant embryos (generated by crossing homozygous dfmr13 mothers to homozygous dfmr13 fathers). Most of the dfmr1 transcription unit is deleted in dfmr13 and it is a protein null (Dockendorff et al. 2002). In contrast to what is seen in the ovary, we found that loss of dfmr1 activity had no apparent effect on the expression of Orb protein in pole cells. However, we noted that the process of pole cell formation is abnormal. Abnormalities are first evident when the polar buds begin to protrude from the posterior end. As shown in Figure 1A, when wild-type pole cells (visualized with Vasa antibody in red) complete the budding process and cellularize, they lie on the outside surface of the embryo. At this stage the surface of the embryo is marked by a thin band of Anillin (visualized with Anillin antibody in green). Anillin interacts with nonmuscle myosin II and is thought to regulate its contraction (Straight et al. 2005). It is also known to bind and bundle actin filaments and is thought to link actin to other components of the cytoskeleton (Field and Alberts l995; Kinoshita et al. 2002). The membranes of each newly formed pole cell are also outlined by a weak but readily discernible Anillin ring. Figure 1, B and C, shows pole cells in dfmr13 embryos that have completed or have just about completed the process of budding and cellularization. In contrast to wild-type, the newly formed pole cells in dfmr1 mutants are not fully extruded from the surface of the embryo. The pole cells in the embryo in Figure 1B remain partially embedded in the surface of the embryo, while in Figure 1C the posterior end of the embryo is abnormally indented and the pole cells lie along the indentation. dfmr13 embryos also differ from wild type in the pattern of Anillin staining. In some cases there are abnormal concentrations of Anillin protein in patches just underlying the newly formed pole cells (see arrow in Figure 1B), while in other cases the band of Anillin protein separating the pole cells from the soma is much thicker and more brightly stained than in the wild-type control. In addition, several pole cells appear to have higher-than-normal concentrations of anillin (see arrowheads in Figure 1, B and C).
Figure 1.—
Abnormalities in pole bud formation in dfmr13 embryos. (A) Wild-type and (B and C) dfmr13 embryos, 0–3 hr old, were fixed and stained with Vasa antibody (imaged in red) and Anillin antibody (imaged in green). (A) An accumulation of Anillin protein at the base of the pole buds in wild-type embryos. In dfmr13 embryos (B and C), the budding process is abnormal and the pole cells do not protrude properly from the posterior surface of the embryo. There are also abnormalities in Anillin accumulation. In B, Anillin is concentrated in irregular patches while in C there seem to be elevated levels of Anillin just underneath the pole buds.
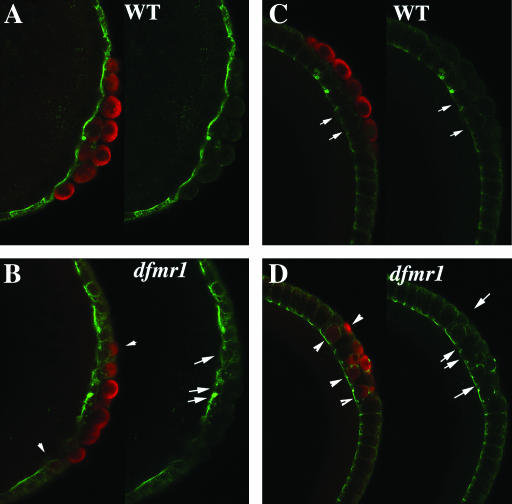
The abnormalities seen in newly formed pole cells in nuclear cycle 9–10 embryos persist through the cellular blastoderm stage. Figure 2, A and B, shows pole cells and Anillin protein in nuclear cycle 12–13 wild-type and dfmr13 embryos. In wild type, the cluster of pole cells is separated from the underlying soma by a band of Anillin that extends along the posterior surface of embryo. In the dfmr1 mutant embryo, the pole cells are not tightly clustered and several of them are separated from the main group (see arrowheads). The Anillin band underlying the pole cells is irregular in shape and the distribution of Anillin protein is nonuniform (see arrows). Figure 2, C and D, shows pole cells and Anillin in wild-type and dfmr13 embryos that are completing cellularization. The somatic cells in wild type are elongated along the apical–basal axis and the surface of the cells are outlined by an Anillin ring with the highest concentrations of protein at the basal surface of the cell. The pole cells lie on the outside of the embryo, while the underlying somatic cells are slightly less elongated than other somatic cells in the embryo (see arrows in Figure 2C). While more anterior somatic cells appear to be properly elongated in the dfmr1 mutant embryo (Figure 2D), somatic cells (arrows in Figure 2D) at the posterior are intermingled with pole cells (arrowheads in Figure 2D) and have irregular shapes and size. As was observed in earlier stages, many of the pole cells lie on the same plane as the somatic cells instead of on the outside surface of the embryo (arrowheads on the inside of embryo in Figure 2D).
Figure 2.—
dfmr13 pole cells are unable to separate from the somatic cell monolayer. (A and C) Wild-type and (B and D) dfmr13 embryos, 0–4 hr old, were fixed and stained with Vasa (imaged in red) and Anillin antibody (imaged in green). (A and B) Late syncytial blastoderm-stage embryos. (C and D) Cellular blastoderm-stage embryos. Note the mispositioned pole cells in the dfmr13 embryos and the abnormal accumulation of Anillin protein.
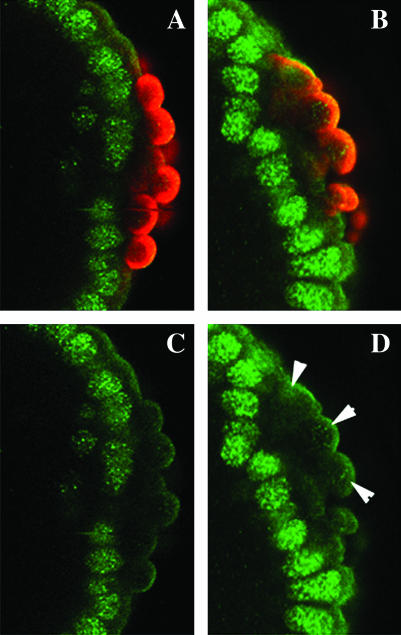
The profilin chickadee accumulation is altered in dfmr1 embryos:
Since defects in pole cell formation evident in dfmr1 embryos are associated with abnormalities in the organization and accumulation of the nonmuscle myosin binding protein Anillin, we wondered whether other components of the actin–myosin cytoskeleton are affected as well. Because previous studies of the fly nervous system indicated that dfmr1 regulates the translation of the profilin chickadee (chic), we examined the organization and accumulation of Chic during pole cell formation. In the pole buds of wild-type embryos, Chic (imaged in green in Figure 3) is organized in a ring (see carat) around the pole cell nucleus and is clearly separated from the Anillin ring (imaged in blue) that outlines the membrane of the budding cell. While similar Chic rings can also be detected at the pole bud stage in dfmr1 embryos, the rings are often disorganized (see arrow in Figure 3B) and displaced from the surface of the embryo (see arrow in Figure 3C). Moreover, as expected from the effects of dfmr1 on the expression of Chic in the nervous system, dfmr13 embryos appear to have significantly higher levels of protein as judged by antibody staining. Nascent pole buds outlined with Anillin can be also be seen in Figure 3B (arrowhead in inset) and in Figure 3C (arrow in inset). Note that the Anillin rings in these dfmr13 pole buds appear to be closing prematurely (see insets) and as a consequence the pole buds remain embedded in the soma instead of protruding from the end of the embryo.
Figure 3.—
Chickadee accumulation is altered in dfmr13 embryos. (A and D) Wild-type and (B, C, and E) dfmr1 embryos, 0–3 hr old, were fixed and stained with Vasa (imaged in red), Anillin (imaged in blue), and the profilin Chickadee (imaged in green) antibodies. (Top) All three labels are shown. (Bottom) Only the anti-Vasa- and anti-Chickadee-specific stainings are shown. (A–C) Presyncytial-stage embryos. (D and E) Syncytial blastoderm-stage embryos.The level of Chic protein and the pattern of accumulation in dfmr13 embryos is altered compared to wild type. (Insets in B and C) Abnormal “pole buds” in dfmr13 embryos labeled with Vasa, Anillin, and Chickadee antibodies shown at greater magnification.
Abnormalities in Chic organization are also evident in older syncytial blastoderm dfmr1 embryos (compare Figure 3, D and E). In some cases, the Chic rings surrounding the dfmr1 pole cell nuclei appear to be thicker and more intensely stained than wild type (arrows in Figure 3, D and E), while in other cases the rings have a somewhat irregular shape (arrowhead in Figure 3E). In addition, the overall level of Chic in these older embryos also appears to be higher than in wild type.
Number of pole cells is reduced in dfmr1 embryos:
Because of the defects in pole cell formation, we compared the number of pole cells in dfmr13 nuclear cycle 12–13 embryos with wild-type embryos at the same stage of development. While there were ∼23 pole cells in wild-type embryos (an average of 22.8 pole cells in 16 embryos), we found that dfmr13 embryos have only ∼15 pole cells (an average of 14.5 pole cells in 20 embryos). Strikingly, there was considerable variation in pole cell number from one dfmr13 embryo to the next as illustrated by the examples in Figure 4, where some dfmr13 embryos have near wild-type numbers of pole cells while in other embryos no pole cells can be detected. We also counted the number of germ cells in the coalesced gonad of stage 14–15 embryos. We found that the number of germ cells in the newly formed gonad of dfmr1 embryos is also reduced compared to the wild-type embryos at an equivalent stage. Although there were fewer germ cells in dfmr1 embryos, there was no difference in the number of somatic gonadal precursor cells.
Figure 4.—
Pole cell number is reduced in dfmr13 embryos. (A) Wild-type and (B–E) dfmr13 embryos, 0–4 hr old, were fixed and stained with either Vasa antibody alone (A–C) or with Vasa and Anillin antibodies (D and E). As described in the text, the number of pole cells in dfmr13 embryos is quite variable. In some dfmr13 embryos (B), the number of pole cells is equivalent to that seen in wild type, while in other embryos (C), pole cell number is greatly reduced. Some dfmr13 embryos completely lack pole cells. (D and E) An example of such an embryo: D is the anterior of the embryo, and E is the posterior.
A subset of dfmr1 pole cells are transcriptionally active:
One characteristic feature of pole cells that distinguishes them from the surrounding somatic nuclei is transcriptional quiescence. While RNA polymerase II transcription is activated in somatic nuclei soon after they migrate to the surface of the embryo, it remains inactive in pole cells until much later in development. Products from maternally active genes such as polar granule component (pgc), nanos (nos), and germ cell-less (gcl) have been shown to be required for the establishment and/or maintenance of transcriptional quiescence in pole cells (Asaoka et al. 1999; Deshpande et al. 1999, 2004; Leatherman and Jongens 2003; Schaner et al. 2003; Martinho et al. 2004). In embryos lacking these gene products, transcription is inappropriately activated in the pole cells at the syncytial blastoderm stage, while later in development the pole cells fail to migrate properly to the somatic gonad and are lost.
We wondered whether dfmr1 pole cells also failed to repress transcription. To test this possibility, we examined the phosphorylation status of the carboxyl terminal domain (CTD) of the largest RNA polymerase II subunit. When polymerase II is transcriptionally engaged, the heptad amino acid repeat of the CTD domain is phosphorylated on serine residue 2 (and also on serine residue 5). As reported by Seydoux and Dunn (l997), phospho-ser2 is detected in somatic but not in pole cell nuclei of wild-type syncytial blastoderm embryos (imaged in green in Figure 5, A and C). In contrast, the phospo-ser2 CTD modification could be detected in dfmr13 pole cells (Figure 5, B and D). While the number of pole cells in each embryo that had the CTD phospho-ser2 was variable, we found that overall ∼20% of the pole cells in dfmr13 embryos had readily detectable levels of the phospo-ser2. For comparison we quantitated the frequency of phospho-ser2-positive pole cells in embryos from nos and pgc mothers. For nos, ∼40% of the pole cells were phospho-ser-2 positive while nearly all pole cells were positive in pgc embryos.
Figure 5.—
Transcriptional quiescence in dfmr13 pole cells. (A and C) Wild-type and (B and D) dfmr13 embryos, 0–4 hr old, were fixed and stained with antibody against the polymerase II CTD domain phosphorylated on serine residue 2 (RP5) (imaged in green in A–D) and Vasa antibody (imaged in red in A and B). In wild-type CTD phospho-ser2 is found in somatic nuclei, but not in pole cell nuclei. In contrast, a subset of the dfmr13 pole cells have readily detectable levels of CTD phospho-ser2. Arrowheads in D indicate pole cells in the dfmr13 embryos with elevated levels of CTD phospho-ser2.
To provide additional evidence that a subset of the dfmr1 pole cells prematurely activated transcription, dfmr1 mutant females were crossed to males carrying two different transcription reporters, Sxl-Pe:LacZ and tailless:LacZ. While neither of these reporters is active in wild-type pole cells, we found that the tailless:LacZ reporter is activated in a small subset of dfmr1 pole cells (data not shown). We were not, however, able to detect any β-galactosidase expression from the Sxl-Pe:LacZ reporter. Interestingly, a similar promoter specificity was also observed for pgc; the tailless:LacZ reporter was activated in pgc pole cells, while the Sxl-Pe:LacZ reporter was not. Conversely, in nos pole cells Sxl-Pe:LacZ was activated while tailless:LacZ was not.
Defects in pole cell formation in dfmr13 embryos are rescued by a dfmr1 transgene:
To demonstrate that the abnormalities in pole cell formation evident in dfmr13 embryos are due to the loss of dfmr1 activity rather than to genetic background effects, we introduced a P(dfmr1) rescue construct into the dfmr13 stock (Dockendorf et al. 2002). As shown in Figure 6, dfmr13 embryos (generated by crossing homozygous dfmr13 females to homozygous dfmr13 males) have no detectable dFMR1 protein (imaged in blue) and exhibit the characteristic abnormalities in pole cell formation (imaged in green), including a failure in the budding process and a reduction in pole cell number. As can be seen in Figure 6, the P(dfmr1) transgene restores dFMR1 protein and rescues the abnormalities in pole cell formation evident in dfmr13 embryos. Note that the level of dFMR1 protein, as judged by staining, seems to be higher in the dfmr13; P(dfmr1) embryos than in wild type. Since the bulk of the dFMR1 protein in blastoderm-stage embryos is probably of maternal origin, the high levels of dFMR1 protein in these embryos is likely due to a chromosomal position effect that drives a high level of expression of mRNA from the P(dfmr1) transgene during oogenesis.
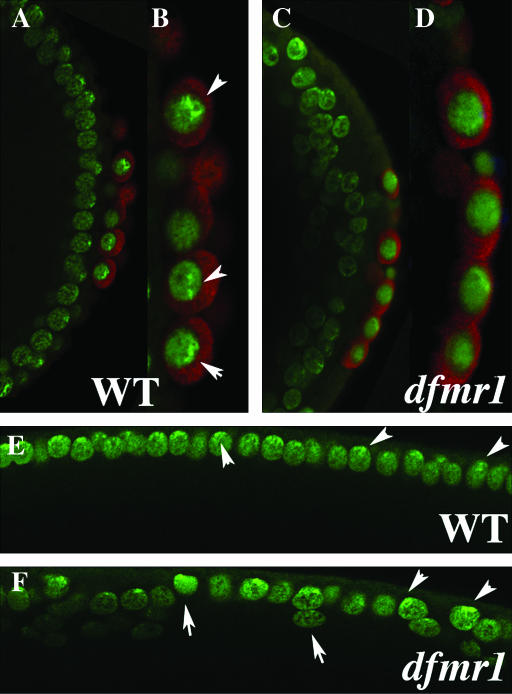
Heterochromatin protein HP-1 is mislocalized in dfmr1 embryos:
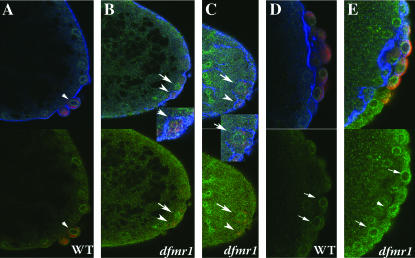
Although heterochromatin protein 1 (HP1) is present throughout much of the nucleus in pole cells of wild-type syncytial blastoderm-stage embryos, there are usually foci that have a much higher concentration of protein. This punctate localization pattern can be seen in Figure 7, A and B (arrowheads). To determine if HP1 is properly localized in dfmr1 pole cells, we probed dfmr13 embryos with HP1 antibody. As can be seen in Figure 7, C and D, the localization of HP1 in dfmr13 pole cell nuclei is more diffuse than in wild type, and sharp foci of protein are either not present or are less pronounced.
Figure 7.—
Localization of HP1 in dfmr13 embryos. (A, B, and E) Wild-type and (B, D, and F) dfmr1 syncytial blastoderm-stage embryos were labeled with HP1 (imaged in green) and Vasa antibody (imaged in red). (A–D) Merged images. (E and F) The HP1-specific staining. As described in the text, HP1 is localized in a punctate pattern in wild-type pole cells (see arrowheads) while its localization is much more diffuse in dfmr13 pole cells. In the case of somatic nuclei, HP1 is concentrated in a punctate pattern at the apical surface of wild-type nuclei. The distribution and level of HP1 in dfmr13 embryos varies from one nuclei to another. Also note that unlike wild-type nuclei, dfmr13 nuclei are not always elongated along the apical–basal axis and are often displaced from the surface (see arrows and arrowheads in F and somatic nuclei in C).
We also examined the distribution of HP1 protein in the somatic nuclei of wild-type and dfmr1 embryos (for a detailed description of HP1-specific staining in early embryos, see Kellum et al. 1995; Kellum and Alberts 1995). In wild-type cycle 10 embryos, HP1 protein is distributed almost uniformly throughout interphase nuclei, whereas by cycle 13 (see Figure 7E) it becomes enriched at the apical surface where centromeric heterochromatin is thought to be concentrated. Moreover, it is often concentrated in sharp foci that also contain high levels of DNA. Sharp foci of HP1 protein can also be seen near the apical surface of some dfmr13 nuclei (see arrowheads in Figure 7F); however, these foci are not present in all nuclei. Other differences in dfmr13 somatic nuclei are also evident. While somatic nuclei of wild-type nuclear cycle 13 embryos are elongated along the apical–basal axis and are distributed more or less evenly just underneath the surface of the embryo, this is not true for somatic nuclei in dfmr1 mutants. Many dfmr13 nuclei have an irregular shape or are elongated along the axis perpendicular to the apical–basal axis (arrows in Figure 7F). In addition, instead of a nearly uniform distribution along the exterior surface of the embryo, many of the dfmr13 nuclei do not appear to have reached the surface (see somatic nuclei in Figure 7, C and F).
Abnormalities in the nuclear division cycle:
The irregular positioning and shape of somatic nuclei at the surface of syncytial blastoderm and the alterations in the distribution of HP1 protein are reminiscent of defects seen in ago-2 mutant precellular blastoderm embryos. ago-2 mutants also exhibit a number of abnormalities in the rapid nuclear division cycles in presyncytial and syncytial blastoderm embryos and in the organization of the embryonic cytoskeleton. Since dFMR1 is found in a complex with Ago-2 and is thought to contribute to the activity of the RNAi machinery, we asked whether dfmr13 embryos exhibit any of the characteristic phenotypes of ago-2 mutant embryos.
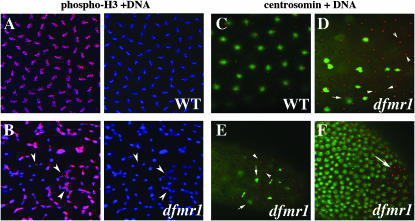
Unlike wild type, nuclear division in precellular blastoderm ago-2 embryos is often asynchronous. To test whether this is also true for dfmr1, we examined the distribution of phosphorylated histone H3, which is a marker for mitotic nuclei. In wild-type embryos, where the division cycles are highly synchronous, most nuclei enter mitosis synchronously and are labeled with the phospho-H3 antibody (Figure 8A). In contrast, the nuclear division cycles in 25–30% of the dfmr13 embryos are asynchronous, and while some nuclei are entering mitosis, others are at different stages of the nuclear division cycle (Figure 8B). Moreover, like ago-2, there is also evidence of abnormal mitotic figures, including lagging chromosomes, chromosome bridges, and chromosome breakage (see arrowheads in Figure 8B). Arguing that these nuclear division defects are due to the loss of dfmr1, rather than to some effect of genetic background, we found that the nuclear division defects in the dfmr13 mutant embryos were rescued by the P(dfmr1) transgene (data not shown).
Figure 8.—
Mitotic asynchrony in early dfmr13 embryos. Wild-type and dfmr13 embryos were stained with DNA dye, Hoechst (imaged in blue), and phospho-histone H3 (pH 3) antibody (imaged in red). (A) A close-up of a wild-type syncytial blastoderm embryo showing synchronously dividing, evenly spaced nuclei with pH 3. (B, left and right) A close-up of a similar age syncytial blastoderm dfmr1 embryo showing unevenly spaced and asynchronously dividing nuclei. (Arrowheads in B) Asynchronously dividing or incompletely separated nuclei. Note that only a subset of the nuclei in the dfmr1 embryo are positive for pH 3. (C–F) Defective centrosomal assembly in dfmr13 embryos. (C) Wild-type and (D–F) dfmr13 embryos stained with centrosomin (red) antibodies and DNA dye, Hoechst (green). In the wild-type embryo in C, there appear to be two centrosomes associated with each nucleus. (D and E) A subset of nuclei appear to be undergoing normal mitosis; however, a number of abnormalities are also evident. These include “orphan” centrosome pairs that are not associated with DNA/nuclei (arrowheads in D and E). Some of the duplicated centrosomes remain in close proximity (arrows in D and E). Similar abnormalities are seen in the stage 5 embryo in F.
A number of other nuclear division cycle defects are also evident in dfmr13 embryos. In wild-type nuclei, centrosomes duplicate early in the nuclear division cycle and then migrate around the nuclear membrane until they are on opposite sides of the nucleus (Figure 8C). While centrosome duplication and migration also take place in dfmr13 embryos, they can occur even in the apparent absence of a nucleus or DNA, and we observed many “free” centrosomes that seem to have migrated opposite each other even when there was no associated nucleus or chromosomes (see carats in Figure 8, D and E). There are also mitotic nuclei in which all of the chromosomes appear to be moving toward one of the centrosomes (see arrows in Figure 8E).
For ago-2, almost half of the embryos showed at least one of the different types of defects in the nuclear division cycles described above. Although dfmr13 embryos exhibited the same sorts of defects as seen in ago-2, the frequency is lower (25–30%) and the defects are typically somewhat less severe.
dfmr1 is required for assembly of centric heterochromatin:
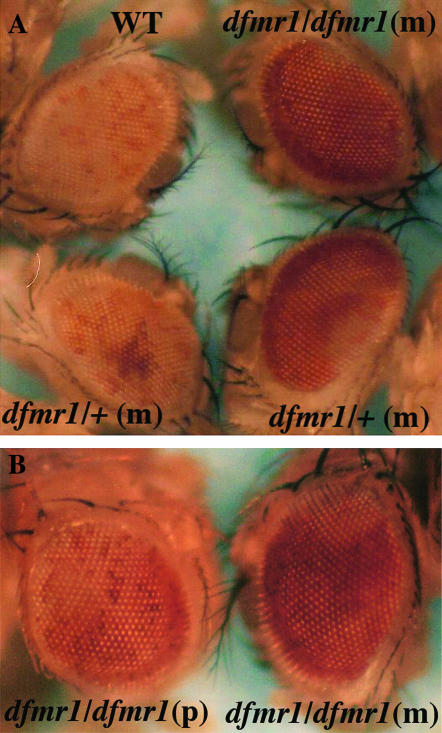
The various abnormalities in the nuclear division cycles seen in ago-2 mutant embryos were correlated with defects in the assembly and functioning of centric/centromeric heterochromatin. To ascertain if there are similar defects in the formation/maintenance of heterochromatin in dfmr1 embryos, we examined the silencing of a white transgene inserted into pericentric heterochromatin on the fourth chromosome (Cryderman et al. 1998) under conditions in which dfmr1 activity is reduced. In the first experiment, wild-type (w1) and dfrm13 homozygous females were crossed to males homozygous for the white insert on the fourth chromosome. As illustrated by the example shown in Figure 9, the eyes of the progeny from the cross with the wild-type control have only a low level of yellow pigmentation with a few scattered ommatidia that have a light red color. By contrast, all progeny from the dfrm13/dfmr13 females have much darker eyes. The eye color is not completely uniform with large blotches of relatively dark red over a generally orange background.
Figure 9.—
dfmr1 is required for heterochromatic silencing. (A) Males carrying a white transgene inserted into a heterochromatic region of the fourth chromosome (118E-10) (Cryderman et al. 1998) were crossed to w1/w1, 1/w; dfmr13/dfmr13, or w/w; dfmr13/+ females and the eye phenotype was examined in the adult offspring. Strong suppression was observed in all offspring of homozygous dfmr13 females. In the case of females that were heterozygous for the dfmr13 mutation, the progeny could be divided into two equal classes on the basis of their eye-color phenotype. One class showed relatively strong suppression as illustrated by the example at the bottom right of A. This class is presumed to correspond to flies that are dfmr13/+. The second class showed weak, but readily discernible, suppression as illustrated by the example at the bottom right of A. Flies in this class are presumed to be wild type for dfmr1. (B) Suppression of fourth chromosomal PEV when the father (left) or mother (right) is homozygous for the dfmr13 mutation. Note that no suppression was observed in ∼15% of the offspring from homozygous dfmr13 males. In contrast, suppression was observed in all progeny of homozygous dfmr13 females.
To determine whether the effects of dfmr1 on fourth chromosome position effect variegation (PEV) depend upon maternal or zygotic dfmr1 or both, we crossed dfmr13/dfmr13 males to females carrying the fourth chromosomal white insert. As shown in Figure 9B, suppression of heterochromatic silencing is observed when the father carries the dfmr13 mutation. This finding indicates that a reduction in the dose of the dfmr1 gene in the zygote is sufficient to compromise the heterochromatic silencing of a transgene inserted on the fourth chromosome. However, the extent of suppression was not equivalent to that observed when the mothers where homozygous for the dfmr13 mutation. First, while suppression was observed in all progeny of dfmr13 mutant mothers, no suppression was evident in ∼15% of the progeny of dfmr13 mutant fathers. Second, suppression was stronger when the mother rather than the father lacked dfmr1 activity. This is illustrated by the examples in Figure 9B. These findings indicate that the loss of maternally derived dfmr1 activity also perturbs heterochromatin silencing. To confirm this suggestion, we mated dfmr13/+ females (generated by mating homozygous dfmr13 females to w1 males) to males carrying the fourth chromosomal white transgene insert. As shown at the bottom of Figure 9A, the progeny from this cross could be divided into two equal classes. The first class showed a relatively strong suppression of white silencing (bottom right). The suppression was stronger than that observed when the father was homozygous dfmr13, but it was not as strong as that observed in the progeny of homozygous dfrm13 females. This class likely corresponds to flies that are heterozygous for the dfmr13 mutation. Suppression was also observed in the second class (bottom left); however, it was weaker, resembling that seen in progeny of homozygous dfmr13 fathers. Since flies in this class are likely to have two wild-type copies of the dfrm1gene, the suppression observed in these flies must be due to the fact that their mothers were heterozygous for dfmr13 and thus did not contribute a full complement of dfmr1 to the egg.
Localization of cytoskeletal proteins Peanut, Anillin, and Chickadee is disrupted in the soma:
In addition to abnormalities in the nuclear division cycle, dfmr1 embryos also exhibit defects in the migration of nuclei to the cortex of the embryo. As a result, an insufficient number of nuclei reached the periphery and there are gaps in the regular nuclear array of variable size in mutant syncytial blastoderm embryos (see arrow in Figure 7F). This may result from partial and/or defective cortical nuclear migration. Alternatively, these gaps might be a result of nuclei having fallen back inside the embryo after reaching the nuclear periphery. Since cortical nuclear migration and cytokinesis are thought to be dependent on the actin and nonmuscle myosin network, we used antibodies against the Drosophila homolog of septin, the Peanut (Pnut) protein, and against Anillin and Chickadee to determine if the cytoskeletal architecture in the soma of dfmr1 syncytial blastoderm embryos is correctly established.
Pnut is required for a late step in cellularization, and in wild-type embryos it localizes to the leading edge of the furrow during cellularization (Neufeld and Rubin 1994; Fares et al. l995). In surface views the Pnut protein is organized into a regular net- or mesh-like lattice pattern around each nucleus (Figure 10A). This regular hexagonal lattice pattern is disrupted in dfmr13 embryos. In some embryos, the size of the individual hexagons is much more variable than in wild type (compare A and B in Figure 10). In other embryos, regions of the lattice have abnormally high concentrations of Pnut protein (see arrowheads in Figure 10C), while in other regions there appears to be insufficient Pnut (see arrows in Figure 10C).
Figure 10.—
Localization of Peanut, Anillin, and Chickadee in the soma of wild-type and dfmr13 embryos. (A) Wild-type and (B and C) dfmr13 embryos, 0–4 hr old, were stained with Hoechst (blue) and Peanut antibody (green). The regular Pnut network in wild-type embryos is disrupted in dfmr embryos (compare A with B and C). (D–F) Wild-type and (G–I) dfmr13 embryos were stained with Anillin (green) and Chickadee (blue) antibodies. (D and G) The merged image. (E and H) Anillin. (F and I) Chickadee.
We also examined the organization of Anillin and Chic in syncytial blastoderm embryos. In wild-type, Anillin (Figure 10E) is arranged in a regular net-like hexagonal lattice much like that seen for Pnut (Figure 10A) (Field and Alberts 1995). As was observed for Pnut, this regular Anillin net-like lattice is disorganized in dfmr13 embryos, and the Anillin “hexagons” are quite irregular in size and shape. In surface views of wild-type embryos, most of the Chic protein is localized in a ring around each somatic nuclei (Figure 10F). The Chic “rings” appear to be located just inside of the Anillin “hexons” so that rings are separated from each other by a band of Anillin (Figure 10D). As can be seen in Figure 10, G and I, the regular array of Chic rings seen in wild-type embryos is disrupted in dfmr13 mutants. In these experiments, the wild-type and dfmr13 embryos were antibody stained and imaged under the same conditions. As was noted above, we found that the overall level of Anillin and also of Chic antibody staining is clearly elevated in the dfmr1 mutant.
DISCUSSION
dFMR1 and the nuclear division cycles:
Recent studies have shown that dFMR1 is physically associated with Ago-2 and other components of the RNAi machinery and have suggested that this association may be important for the regulatory functions of the dFMR1 protein (Ishizuha et al. 2002; Xu et al. 2004). Our analysis of early embryogenesis in dfmr1 mutant embryos provides additional evidence for a connection between dfmr1 and the RNAi machinery. We have previously found that Ago-2 activity is required for the proper execution of the rapid nuclear division cycles that precede the formation of the cellular blastoderm and a variety of defects are evident in ago-2 mutant embryos (Deshpande et al. 2005). These include asynchronous nuclear division, incomplete chromosome condensation, defects in chromosome segregation, and chromosome fragmentation. Significantly, the same defects in the nuclear division cycles are evident in dfmr1 embryos: nuclear division is asynchronous, mitotic figures have incompletely condensed and lagging chromosomes, and there is evidence of chromosome fragmentation. In addition, the assembly/functioning of the spindle apparatus is abnormal, and like ago-2, many dfmr1 embryos have “orphaned” centrosomes that have duplicated and migrated to “opposite poles” even though no nucleus is apparent. In the case of ago-2, around half of the embryos exhibit at least one of these nuclear division phenotypes, while for dfmr1 25–30% of the embryos show at least one of these phenotypes.
Many of the defects in nuclear division seen in ago-2 embryos can be attributed to a failure in the assembly of centromeric and centric heterochromatin (Deshpande et al. 2005). The centromeric-specific histone H3 variant CID (Blower and Karpen 2001) is often present in greatly reduced amounts or completely absent from the centromeric regions of ago-2 mitotic chromosomes. There are also abnormalities in the localization of the centric heterochromatin protein HP1. Although we have not examined CID in dfmr1 embryos, we did find that the centric heterochromatin protein HP1 is mislocalized in dfmr1 embryos much like that observed in ago-2 mutants. We also tested for the establishment/maintenance of functional heterochromatin by examining the effects of dfrm1 mutations on the silencing of a white transgene inserted on the fourth chromosome (Cryderman et al. 1998). We found that a reduction in dfmr1 activity was quite effective in suppressing the silencing of a white transgene inserted into a heterochromatic region of the fourth chromosome. Although the strongest suppression was observed when the mothers were homozygous for dfmr13, suppression was also evident in the reciprocal cross in which fathers were homozygous for dfmr13. These findings indicate that dfmr1 is required to maintain the silenced state as the fly develops. In addition, our results point to a role for dfmr1 in the initial establishment of the silenced state in the embryo. First, suppression is stronger when the mother is homozygous mutant than when the father is. Second, suppression is also seen in all progeny of heterozygous dfrm13 females. In this case, two equal classes were observed. In the first class, suppression is relatively strong, and these flies are presumed to be heterozygous for the dfmr13 mutation. Weak suppression is observed in the second class and these flies are presumed to be wild type for dfmr1. These findings indicate that homochromatic silencing is dependent upon maternally contributed dFMR1 and would be consistent with the idea that dFMR1 functions in the establishment of the silenced state at a point early in embryogenesis.
Taken together with the physical association between dFMR1 and the components of the RNAi machinery, our results would support the idea that dFMR1 functions as a cofactor in an RNAi pathway required in early embryos for the proper execution of the nuclear division cycles and for the assembly of functional centric/centromeric heterochromatin. On the other hand, there remains the question of why the defects in the nuclear division cycles in dfmr1 and also in ago-2 embryos are not fully penetrant. In the case of ago-2, there are other Argonaute-related genes in the fly that could potentially perform partially redundant or overlapping functions that compensate for the loss of Ago-2 (Carmell et al. 2002; Tomari and Zamore 2005). Consistent with this idea, a similar spectrum of nuclear division defects is evident in embryos from mothers deficient in piwi activity (G. Deshpande, unpublished data). Moreover, like ago-2, the nuclear division phenotypes in piwi embryos are not fully penetrant. However, this explanation would not account for the incomplete penetrance of dfmr1 as it is the only fragile X family member in the fly. One plausible idea is that dfmr1 functions as a facilitator in the nuclear division cycle RNAi pathway, but is not absolutely essential for the operation of this pathway. Alternatively, there may be other RNA-binding proteins that can substitute for dFMR1.
dfmr1 and pole cell formation:
While the various nuclear division cycle abnormalities in dfmr1 embryos closely resemble those observed in ago-2 (or in piwi), this is only partially true for the pole cell formation phenotypes. Like ago-2, there is a small but significant reduction in the number of pole cells in dfrmr1 embryos. Like ago-2, this could be due at least in part to defects in the migration of nuclei into the posterior pole, in which case it probably arises from the disruptions in the nuclear division cycles during the presyncytial blastoderm stages. However, this is not the most striking phenotype in dfmr1 embryos. In contrast to either ago-2 or wild-type embryos, the pole cells in dfmr1 embryos fail to properly segregate from the surrounding somatic nuclei/cells and instead remain intermingled with somatic nuclei in syncytial blastoderm embryos and somatic cells in cellular blastoderm embryos. Unlike the various nuclear division cycle phenotypes, the pole cell segregation phenotype is fully penetrant and is seen in virtually every appropriately staged dfmr1 embryo. dfmr1 also differs from ago-2 in that transcriptional quiescence is not properly established in all pole cells.
The pole cell segregation phenotype in dfmr1 embryos can be traced back to the budding stage. In wild-type embryos, nuclei migrating into the pole plasm induce the formation of buds. The outside surface of the buds is marked by actin and Anillin, while there is a Chickadee ring surrounding the pole bud nucleus. Once the buds, including the pole cell nuclei, have been fully extruded from the surface of the embryo, the actin/Anillin ring at the base of the bud contracts, completing the cellularization process. With the completion of cellularization, the exterior surface of the embryo is redefined with the pole lying outside of the embryo. In dfmr1 embryos, the pole buds fail to emerge from the surface of the embryo or, if they do, they do not enlarge properly. The process of pole bud emergence and/or enlargement appears to be short circuited by “precocious” cellularization. In our experiments, this is marked by the formation of partially closed and closed Anillin rings around unbudded or incompletely budded pole cells. Once the pole cells form, many remain embedded in the somatic layer instead of locating on the exterior of the embryo and, when the blastoderm cellularizes, the pole cells are often intermingled with somatic cells.
The dfmr1 pole cell segregation phenotype resembles that reported for drhogef2 mutants (Padash Barmchi et al. 2005). In drhogef2 mutant embryos, the pole buds fail to form cortical actin networks that are separated from the somatic layer of nuclei and, when the buds cellularize, they remain embedded in the soma. Because of the apparent similarities in phenotypes, we wondered whether the expression of dRhogef2 was impaired in dfmr13 embryos. Although there was no reduction in expression, we did find that the localization of Drhogef2 in the pole buds/cells and in the soma of dfmr13 embryos was abnormal much like that observed for Anillin (G. Deshpande, unpublished data). Taken together with the effects of dfmr1 on Profilin and Pnut, these findings would suggest that dfrm1 activity is required for the proper assembly and/or functioning of the actin–myosin cytoskeleton during pole cell formation (and later during the cellularization of the somatic nuclei).
Two different but not mutually exclusive models could potentially account for the disruptions in the actin–myosin cytoskeleton in dfmr1 embryos. In the first, dFMR1 would play a more or less direct role in organizing the cytoskeleton through protein–protein interactions with effector molecules. In this view, the organization of the cytoskeleton in dfmr1mutants would be perturbed because dFMR1 is not available to influence the activity of these effector molecules. This possibility is supported by a number of findings. First, dFMR1 interacts with the Rac1-GTP-binding protein Sra-1/CYFIP (Schenck et al. 2001). Sra-1/CYFIP is part of a multi-component complex (SCAR/WAVE) that regulates actin nucleation through the activation of Arp2/3 (Kunda et al. 2003). The complex is activated when Sra-1/CYFIP is displaced from the complex by the GTP-bound form of Rac1 (Kunda et al. 2003). Second, dFMR1 has also been shown to interact with the lethal-(2)-giant-larvae (LGL) protein (Zarnescu et al. 2005). LGL is a component of the PAR complex, which is involved in establishing cellular asymmetries. Additional support for this idea comes from the association of mammalian FMR proteins with the actin cytoskeleton (Castets et al. 2005). The second model is that dfmr1 influences the organization of the cytoskeleton indirectly through its ability to downregulate the translation of target mRNAs. In this case, the cytoskeleton would be perturbed in dfmr1 embryos because the stoichiometric balance between components of the cytoskeletal that are dfmr1 targets and those that are not would be altered. Consistent with this idea, several of the known mRNA targets for dfmr1 regulation in the nervous system encode components of both the actin–myosin and the microtubule cytoskeleton (rac1, chic, and futsch). Since our antibody-staining experiments suggest that the profilin Chic may be overexpressed in dfmr1 mutant embryos, it would be reasonable to think that the translation of chic mRNA may also be regulated by dFMR1 during early embryogenesis. Chic is probably not the only cytoskeletal target for dfmr1 that is misregulated in mutant embryos. While we did not examine either Rac1 or Futsch expression, our antibody-staining experiments indicate that Anillin is present at higher levels in dfmr1 embryos than in similarly staged wild-type embryos. Like many of the other known targets for dFMR1, Drosophila Anillin mRNA has several potential dFMR1 recognition motifs. In this respect, it is interesting to note that Orb protein does not seem to be overexpressed in dfmr1 pole cells, while it is overexpressed in dfmr1 ovaries (in both nurse cells and the oocyte). This finding suggests that the mRNA targets for dfmr1 regulation may vary with different tissues or stages of development. Presumably, this depends upon what other regulatory cofactors are present or absent.
Acknowledgments
We thank F. B. Gao, A. Muller, T. Schüpbach, H. Siomi, L. Wallrath, T. Jongens, and the Bloomington Stock Center for flies; and C. Field, G. Karpen, T. Kaufman, P. Lasko, R. Kellum, T. Jongens, and E. Wieschaus for various antibodies. We are grateful to D. Gohl and G. Shanower for advice and helpful discussions during the course of this work. We thank J. Goodhouse for help with confocal microscopy and Gordon Gray for fly food. This work was supported by a National Institutes of Health grant to P.S.
References
- Asaoka, M., M. Yamada, A. Nakamura, K. Hanyu and S. Kobayashi, 1999. Maternal pumilio acts togother with Nanos in germline development in Drosophila embryos. Nat. Cell Biol. 1 431–437. [DOI] [PubMed] [Google Scholar]
- Baker, J., W. E. Theurkauf and G. Schubiger, 1993. Dynamic changes in microtubule configuration correlate with nuclear migration in the preblastoderm Drosophila embryo. J. Cell Biol. 122 113–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower, M. D., and G. H. Karpen, 2001. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat. Cell Biol. 3 730–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, V., P. Jin, S. Ceman, J. C. Darnell, W. T. O'Donnell et al., 2001. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell 107 477–87. [DOI] [PubMed] [Google Scholar]
- Carmell, M., Z. Xuan, M. Zhang and G. J. Hannon, 2002. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 16 2733–2742. [DOI] [PubMed] [Google Scholar]
- Castets, M., C. Schaeffer, E. Bechara, A. Schenck, E. W. Khandjian et al., 2005. FMRP interferes with the Rac1 pathway and controls actin cytoskeleton dynamics in murine fibroblasts. Hum. Mol. Genet. 14 835–844. [DOI] [PubMed] [Google Scholar]
- Caudy, A. A., M. Myers, G. J. Hannon and S. M. Hammond, 2002. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 16 2491–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, A., Y. Wang, T. C. Dockendorff, H. Erdjument-Bromage, P. Tempest et al., 2005. The Drosophila fragile X protein functions as a negative regulator in the orb autoregulatory pathway. Dev. Cell 3 331–342. [DOI] [PubMed] [Google Scholar]
- Cryderman, D. E., M. H. Cuaycong, S. C. R. Elgin and L. L. Wallrath, 1998. Characterization of sequences associated with position-effect variegation at pericentric sites in Drosophila heterochromatin. Chromosoma 107 277–285. [DOI] [PubMed] [Google Scholar]
- Darnell, J. C., K. B. Jensen, P. Jin, V. Brown, S. T. Warren et al., 2001. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell 107 489–499. [DOI] [PubMed] [Google Scholar]
- Deshpande, G., G. Calhoun, J. Yanowitz and P. Schedl, 1999. Novel functions of nanos in downregulating mitosis and transcription during the development of Drosophila germline. Cell 99 271–281. [DOI] [PubMed] [Google Scholar]
- Deshpande, G., G. Calhoun and P. D. Schedl, 2004. Overlapping mechanisms function to establish transcriptional quiescence in the embryonic Drosophila germline. Development 131 1247–1257. [DOI] [PubMed] [Google Scholar]
- Deshpande, G., G. Calhoun and P. Schedl, 2005. Drosophila argonaute-2 is required early in embryogenesis for the assembly of centric/centromeric heterochromatin, nuclear division, nuclear migration, and germ-cell formation. Genes Dev. 19 1680–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dockendorf, T. C., H. S. Su, S. M. J. Mcbride, Z. Yang, C. H. Choi et al., 2002. Drosophila lacking dfmr1 activity show defects in circadian output and fail to maintain courtship interest. Neuron 34 973–984. [DOI] [PubMed] [Google Scholar]
- Fares, H., M. Peifer and J. R. Pringle, 1995. Localization and possible functions of Drosophila septins. Mol. Biol. Cell 6 1843–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field, C. M., and B. M. Alberts, 1995. Anillin, a contractile ring protein that cycles from the nucleus to the cell cortex. J Cell Biol. 131 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field, C. M., M. Coughlin, S. Doberstein, T. Marty and W. Sullivan, 2005. Characterization of Anillin mutants reveals essential roles in septin localization and plasma membrane integrity. Development 132 2849–2860. [DOI] [PubMed] [Google Scholar]
- Foe, V. E., C. M. Field and G. M. Odell, 2000. Microtubules and mitotic cycle phase modulate spatiotemporal distributions of F-actin and myosin II in Drosophila syncytial blastoderm embryos. Development 127 1767–1787. [DOI] [PubMed] [Google Scholar]
- Hammond, S. M., S. Boettcher, A. A. Caudy, R. Kobayashi and G. J. Hannon, 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293 1146–1150. [DOI] [PubMed] [Google Scholar]
- Ishizuka, A., M. C. Siomi and H. Siomi, 2002. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 16 2497–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, P., R. S. Alisch and S. T. Warren, 2004. a RNA and microRNAs in fragile X mental retardation. Nat. Cell Biol. 11 1048–1053. [DOI] [PubMed] [Google Scholar]
- Jin, P., D. C. Zarnescu, S. Ceman, M. Nakamoto, J. Mowrey et al., 2004. b Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat. Neurosci. 2 113–117. [DOI] [PubMed] [Google Scholar]
- Kellum, R., and B. M. Alberts, 1995. Heterochromatin protein 1 is required for correct chromosome segregation in Drosophila embryos. J. Cell Sci. 108 1419–1431. [DOI] [PubMed] [Google Scholar]
- Kellum, R., J. W. Raff and B. M. Alberts, 1995. Heterochromatin protein 1 distribution during development and during the cell cycle in Drosophila embryos. J. Cell Sci. 108 1407–1418. [DOI] [PubMed] [Google Scholar]
- Kinoshita, K. B., C. M. Field, M. L. Coughlin, A. F. Straight, and T. J. Mitchison 2002. Self- and actin-templated assembly of mammalian septins. Dev. Cell 3 791–802. [DOI] [PubMed] [Google Scholar]
- Kunda, P., G. Craig, V. Dominguez and B. Baum, 2003. Abi, Sra1, and Kette control the stability and localization of SCAR/WAVE to regulate the formation of actin-based protrusions. Curr. Biol. 13 1867–1875. [DOI] [PubMed] [Google Scholar]
- Leatherman, J. L., and T. A. Jongens, 2003. Transcriptional silencing and translational control: key features of early germline development. BioEssays 25 326–335. [DOI] [PubMed] [Google Scholar]
- Martinho, R. G., P. S. Kunwar, J. Casanova and R. Lehmann, 2004. A non-coding RNA is required for the repression of RNApolII-dependent transcription in primordial germ cells. Curr. Biol. 14 159–165. [DOI] [PubMed] [Google Scholar]
- Morales, J., P. R. Hiesinger, A. J. Schroeder, K. Kume, P. Verstreken et al., 2002. Drosophila fragile X protein, DFXR, regulates neuronal morphology and function in the brain. Neuron 34 961–972. [DOI] [PubMed] [Google Scholar]
- Neufeld, T. P., and G. M. Rubin, 1994. The Drosophila peanut gene is required for cytokinesis and encodes a protein similar to yeast putative bud neck filament proteins. Cell 77 371–379. [DOI] [PubMed] [Google Scholar]
- Oegema, K., M. S. Savoian, T. J. Mitchison and C. M. Field, 2000. Functional analysis of a human homologue of the Drosophila actin binding protein Anillin suggests a role in cytokinesis. J. Cell Biol. 150 539–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padash Barmchi, M., S. Rogers and U. Hacker, 2005. DRhoGEF2 regulates actin organization and contractility in the Drosophila blastoderm embryo. J Cell Biol. 168 575–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve, S. P., L. Bassetto, G. K. Genova, Y. Kleyner, M. Leyssen et al., 2005. The Drosophila fragile X mental retardation protein controls actin dynamics by directly regulating profilin in the brain. Curr. Biol. 15 1156–1163. [DOI] [PubMed] [Google Scholar]
- Royou, A., W. Sullivan and R. Karess, 2002. Cortical recruitment of nonmuscle myosin II in early syncytial Drosophila embryos: its role in nuclear axial expansion and its regulation by Cdc2 activity. J. Cell Biol. 158 127–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaner, C. E., G. Deshpande, P. D. Schedl and W. G. Kelly, 2003. A conserved chromatin architecture marks and maintains the restricted germ cell lineage in worms and flies. Dev. Cell 5 747–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenck, A., B. Bardoni, C. Langmann, N. Harden, J. L. Mandel et al., 2003. CYFIP/Sra-1 controls neuronal connectivity in Drosophila and links the Rac1 GTPase pathway to the fragile X protein. Neuron 38 887–898. [DOI] [PubMed] [Google Scholar]
- Seydoux, G., and M. A. Dunn, 1997. Transcriptionally repressed germ cells lack a subpopulation of phosphorylated RNA polymerase II in early embryos of Caenorhabditis elegans and Drosophila melanogaster. Development 124 2191–2201. [DOI] [PubMed] [Google Scholar]
- Straight, A. F., C. M. Field and T. J. Mitchison, 2005. Anillin binds nonmuscle myosin II and regulates the contractile ring. Mol. Biol. Cell 16 193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, W., and W. E. Theurkauf, 1995. The cytoskeleton and morphogenesis of the early Drosophila embryo. Curr. Opin. Cell Biol. 7 18–22. [DOI] [PubMed] [Google Scholar]
- Sullivan, W., P. Fogarty and W. Theurkauf, 1993. Mutations affecting the cytoskeletal organization of syncytial Drosophila embryos. Development 118 1245–1254. [DOI] [PubMed] [Google Scholar]
- Tomari, Y., and P. D. Zamore, 2005. Perspective: machines for RNAi. Genes Dev. 19 517–529. [DOI] [PubMed] [Google Scholar]
- Williamson, A., and R. Lehmann, 1996. Germ cell development in Drosophila. Annu. Rev. Cell Dev. Biol. 12 365–391. [DOI] [PubMed] [Google Scholar]
- Xu, K., B. A. Bogert, W. Li, K. Su, A. Lee et al., 2004. The fragile X-related gene affects the crawling behavior of Drosophila larvae by regulating the mRNA level of the DEG/ENaC protein pickpocket1. Curr. Biol. 14 1025–1034. [DOI] [PubMed] [Google Scholar]
- Zarnescu, D. C., P. Jin, J. Betschinger, M. Nakamoto, Y. Wang et al., 2005. Fragile X protein functions with lgl and the par complex in flies and mice. Dev. Cell 8 43–52. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. Q., and K. Broadie, 2005. Fathoming fragile X in fruit flies. Trends Genet. 1 37–45. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. Q., A. M. Bailey, H .J. Matthies, R. B. Renden, M. A. Smith et al., 2001. Drosophila fragile X-related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell 107 591–603. [DOI] [PubMed] [Google Scholar]