Abstract
Sex-ratio meiotic drive is the preferential transmission of the X chromosome by XY males, which occurs in several Drosophila species and results in female-biased progeny. Although the trait has long been known to exist, its molecular basis remains completely unknown. Here we report a fine-mapping experiment designed to characterize the major drive locus on a sex-ratio X chromosome of Drosophila simulans originating from the Seychelles (XSR6). This primary locus was found to contain two interacting elements at least, both of which are required for drive expression. One of them was genetically tracked to a tandem duplication containing six annotated genes (Trf2, CG32712, CG12125, CG1440, CG12123, org-1), and the other to a candidate region located ∼110 kb away and spanning seven annotated genes. RT–PCR showed that all but two of these genes were expressed in the testis of both sex-ratio and standard males. In situ hybridization to polytene chromosomes revealed a complete association of the duplication with the sex-ratio trait in random samples of X chromosomes from Madagascar and Reunion.
ACCORDING to Mendel's first law, each member of a pair of alleles or homologous chromosomes has an equal probability of being transmitted to the offspring. The physical basis of this principle is the balanced segregation of chromosomes during meiosis. However, a number of cases have been reported in which a particular allele or chromosome furthers its own transmission by altering some aspects of meiotic chromosome behavior or gamete formation in heterozygous individuals. Such infringements of the law, known as meiotic drive, have been observed in a wide range of organisms including fungi, plants, insects, and mammals (Lyttle 1991; Dawe and Hiatt 2004). These infringements could have a considerable impact on the natural history of organisms and on genome evolution (Orr and Presgraves 2000; Hurst and Werren 2001; Jaenike 2001; Pardo-Manuel de Villena and Sapienza 2001). However, there are very few cases in which any of the molecular players responsible for the drive have been identified and these all correspond to autosomal drive (Kusano et al. 2003; Lyon 2003). The work reported here provides the first identification of candidate genes for a case of meiotic drive involving the sex chromosomes.
The sex-ratio trait was originally reported in Drosophila obscura >75 years ago (Gershenson 1928). Since then it has been reported in a dozen Drosophila species and two other Diptera families (Jaenike 2001); however, its molecular basis remains completely unknown. Sex-ratio segregation distorters are typically linked to the X chromosome and prevent the production of Y-bearing sperm in heterogametic XY males, leading to the production of strongly female-biased progeny. In D. simulans, the object of this study, the loss of Y-bearing sperm is related to disjunction defects of the Y chromosome sister chromatids during the second meiotic division (Cazemajor et al. 2000) and results from the combined action of several loci, one of which has a major impact (Montchamp-Moreau and Cazemajor 2002). The genetic analysis of the trait in this species is facilitated by two facts: (i) it is not associated with complex chromosomal inversions, as is usually the case, and (ii) gene order is very well conserved between D. simulans and its close relative D. melanogaster, so that the D. melanogaster genome data can be used to perform fine genetic mapping in D. simulans. By this means, the major drive locus was recently shown to lie close to the Nrg gene, located in the cytological band 7E-F of the X chromosome (Derome et al. 2004).
Using the same sex-ratio (XSR6) and standard X chromosomes as in Derome et al. (2004) and additional polymorphic molecular markers, we have now more precisely mapped the candidate region for the major drive locus. We found that it contained a duplication, which was also studied by in situ hybridization to polytene chromosomes. Its association with the sex-ratio trait was then checked by examining X chromosomes sampled in two natural populations. Finally, we used RT–PCR to find out whether the candidate genes revealed by the mapping were expressed in the testis.
MATERIALS AND METHODS
Fly stocks used for genetic mapping and in situ hybridization:
To map the distorter elements on the X chromosome, we examined a collection of recombinants between the sex-ratio chromosome XSR6 (which produces 95% female progeny in a suppressor-free background) and the standard (nondriving) chromosome Xsn,lz carrying mutations at the singed (sn) and lozenge (lz) loci flanking the region of interest. The stocks carrying the XSR6 and Xsn,lz chromosomes, and the crossing scheme used to produce the recombinants, are described in Derome et al. (2004).
In situ hybridization was performed on the XSR6 and the Xsn,lz chromosomes. We also examined X chromosomes randomly sampled in the wild. A first sample of 10 sex-ratio and five standard X chromosomes was from Antananarivo (Madagascar), collected in 2000 and described in Derome et al. (2004). The second sample was collected in 1999, in La Saline (Reunion). The mean percentages of females produced in a suppressor-free background by the X chromosomes from La Saline were measured as in Derome et al. (2004). Seven were found to be standard [XSA138 (49% of females), XSA37 (50%), XSA8 (51%), XSA131 (51%), XSA10 (52%), XSA125 (52%), XSA40 (52%)] and six sex-ratio [XSA123 (72%), XSA130 (78%), XSA141 (82%), XSA13 (84%), XSA3 (92%), XSA22 (93%)].
DNA and RNA extraction and amplification, cloning, and sequencing:
Extraction of single male genomic DNA and PCR amplification were carried out using conventional protocols (see Derome et al. 2004). Total RNA extraction was performed with the SV total RNA isolation system (Promega, Madison, WI), according to the manufacturer's instructions. We used samples of 10 whole males (5 days old) and of the testes of 50 males dissected in PBS and then kept in RNAlater (Ambion, Austin, TX). RT–PCR was performed using the M-MLV kit (Promega). To prevent any confusion with amplifications of any residual DNA that could have been present in the RNA extracts, the markers were chosen so that they spanned at least one intron (with the exception of CG12081, which had no introns).
The PCR and RT–PCR amplification primers are shown in Table S1 (supplemental data at http://www.genetics.org/supplemental/). There were designed using the D. melanogaster genomic sequences from GenBank (accession nos. AE003443 and AE003444). They span a region ∼240 kb long surrounding the Nrg gene on the X chromosome of D. melanogaster (according to genome annotation Release 4.0). When necessary, they were corrected after BLAST on D. simulans sequences available at ftp://genome.wustl.edu/. The amplification products were cloned in the pGEM-T Easy Vector (Promega) according to the manufacturer's instructions. Sequencing was carried out using an ABI-3100 sequencer.
In situ hybridization:
Polytene chromosome spreads were prepared from salivary glands of male larvae, which had been fixed for 1 min in 45% acetic acid and then for 3 min in lactic acid/H2O/acetic acid (1:2:3, v:v:v). The clones used as probes were labeled with biotin, using the nick translation system (Invitrogen, San Diego), or with digoxygenin (DIG)-11-dUTP, using the DIG nick translation mix (Roche). The probe solution (∼0.01 μg DNA/μl for each probe) was prepared in 50% formamide/4× SSC/10% dextran sulfate, deposited on the slides (10 μl/slide), and then covered with a coverslip. Chromosome and probe DNA were simultaneously denatured for 5 min at 72° and incubated overnight at 37°. The slides were then washed for 15 min at 38° in 1× SSC/50% formamide, for 8 min at 38° in 2× SSC, and for 5 min at room temperature in 4× SSC/0.5% Tween. Detection was performed using fluorescein-labeled avidin and rhodamin-labeled antidigoxygenin (chromosome in situ system, Oncor-Appligene), following the manufacturer's instructions. After three washes in 4× SSC/0.5% Tween at room temperature, the preparations were counterstained and mounted in DAPI/Antifade (Oncor-Appligene).
Slides were observed under a Zeiss Axioplan 2 epifluorescence microscope. Chromosome images were generated using a highly sensitive CCD camera (Princeton Instruments, Evry, France) and the Metaview 4.1.7 image analysis system (Universal Imaging, West Chester, PA).
RESULTS
Characterization of the sex-ratio duplication:
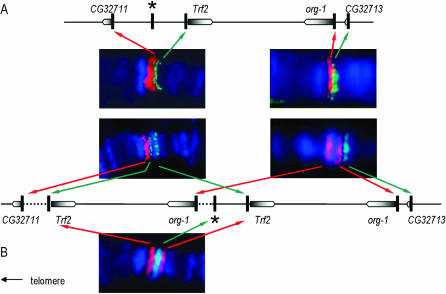
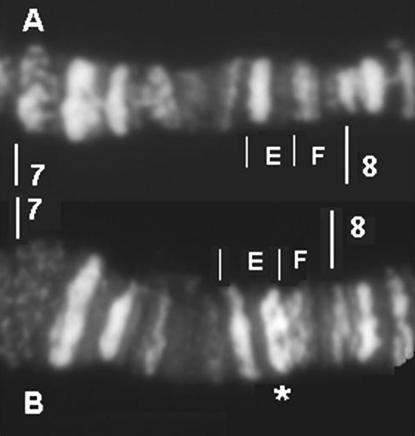
While fluorescent in situ hybridization to polytene chromosomes indicated that gene arrangement in the region was conserved between D. melanogaster and the standard chromosome of D. simulans, we did detect a duplication on XSR6, extending from gene Trf2 to gene org-1 (Figure 1). The four genes located between Trf2 and org-1 in D. melanogaster (CG32712, CG12125, CG1440, and CG12123) were present in the duplication (data not shown), whereas the genes located just outside the Trf2/org-1 fragment (CG32711 and CG32713) were not detected. The duplication appeared to have been inserted in a direct orientation (supplemental Figure S1 at http://www.genetics.org/supplemental/), distally from the original fragment, and within the intergenic region between the CG32711 and Trf2 genes. The duplication seemed to contain a complete copy of Trf2 since, on its distal (telomeric) side, a hybridization signal was obtained with a probe homologous to the noncoding region adjacent to the 5′-end of the gene. It seemed to end close to the 5′-end of org-1 on its proximal side. Indeed, there was no hybridization signal with a probe spanning the noncoding region adjacent to the 5′-end and the start of exon 1, whereas there was a hybridization signal with a probe straddling exon 1 and intron 1. According to genome data for D. melanogaster, the duplicate fragment is expected to be ∼40 kb in length. Repeated and independent sequencing of PCR products revealed sequence differences between the original (SRo) and duplicated (SRd) copies of Trf2, CG12125, and org-1 markers on the XSR6 chromosome. The divergence between the SRo and SRd copies was in the same range (0.0096 < D < 0.0152, depending on the marker) as between either copy and the sequence of the standard chromosome (0.0099 < D < 0.0258). In contrast, we obtained only one sequence for the markers within CG32712, CG1440, and CG12123, and this was different from that carried by the standard chromosome (0.0116 < D < 0.0150, depending on the marker). This implies either that we failed to amplify one copy of these markers or that the SRo and SRd copies are identical. Comparison with the banding pattern of standard X chromosomes of D. melanogaster (Canton-S) and D. simulans showed that the duplication is associated with an extra DAPI-positive band located in the 7E subdivision on XSR6, which suggested that AT-rich material had also been added (Figure 2).
Figure 1.—
Visualization of the sex-ratio duplication by in situ hybridization. The gene organization on (A) D. simulans standard X and (B) sex-ratio XSR6 chromosomes was deduced from the relative position of hybridization signals obtained with the corresponding markers in a double-labeling experiment with probes marked with fluorescein (green) or rhodamin (red) on polytene chromosomes counterstained with DAPI. Genes are represented by boxes (pointed end is the 3′-end); vertical bars indicate the position of molecular markers. The star corresponds to a marker located in the intergenic region between the CG32711 and Trf2 genes.
Figure 2.—
Banding pattern in the 7A–8A division of polytene chromosome X stained with DAPI: (A) D. simulans standard. (B) D. simulans sex-ratio (XSR6). The additional band on XSR6 is indicated by the star.
The sex-ratio XSR6 chromosome was extracted from laboratory stock descended from flies collected in Mahé (Seychelles) in 1981. All 16 sex-ratio chromosomes from Madagascar and Reunion that we examined carried both the duplication (in situ hybridization with Trf2 and org-1 markers) and the extra DAPI-positive band observed on XSR6, whereas the 12 standard chromosomes from these geographical locations did not have either. We therefore propose that the duplication, or a part of it, is a component of the major sex-ratio locus.
Mapping of the distorter elements:
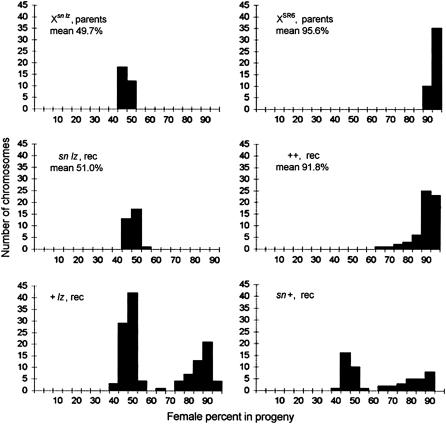
The segregation ratios of the parental sex-ratio XSR6 and standard Xsn,lz chromosomes as well as those of the recombinant chromosomes are given in Figure 3. All the + + recombinants were sex-ratio, although their drive ability was significantly lower on average than that of the parental XSR6 (91.8 and 95.6% of females in the progeny, respectively; Mann–Whitney rang test P < 10−4). The mean percentage of females produced by males carrying the sn lz recombinant chromosomes (51%) was slightly but not significantly higher than the values obtained with the parental standard chromosome (49.7%). The segregation ratios of the + lz and sn + recombinants followed a bimodal distribution, with a group of nondriving chromosomes (<60% of females) and a group of sex-ratio chromosomes (>70% of females). In both cases, the recombinant sex-ratio chromosomes had very variable segregation ratios and produced a segregation bias that was lower on average than that of the parental XSR6 (88.7 and 83.4% of females, respectively; Mann–Whitney rang tests P < 10−4). The data thus supported the previous conclusion that an additional locus with little or no intrinsic effect did enhance the sex-ratio bias induced by the major locus (Montchamp-Moreau and Cazemajor 2002).
Figure 3.—
Percentages of females in the progeny of males carrying the parental chromosomes Xsn,lz (standard) and XSR6 (sex-ratio), and the recombinant chromosomes sn lz, + +, sn +, and + lz. Each value plotted on the histograms corresponds to the mean calculated from the individual progeny (≥50 flies each) of five males carrying a given X chromosome in a drive-sensitive reference background (details on the protocol in Montchamp-Moreau and Cazemajor 2002).
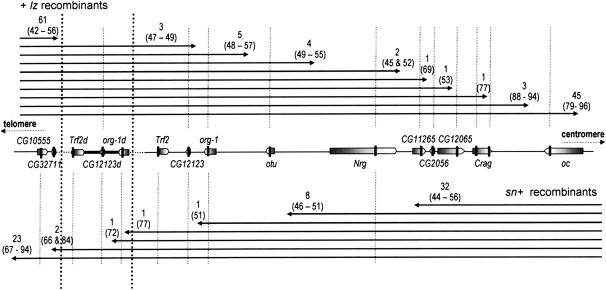
The genetic analysis of sn + and + lz recombinants was performed using the markers listed in supplemental Table S1 at http://www.genetics.org/supplemental/. The results were consistent with gene arrangement conservation in the surveyed region between D. melanogaster and the standard chromosomes of D. simulans and the direct orientation of the sex-ratio duplication. Several of the markers included in the duplication had polymorphic sites (see above), which made it possible to distinguish breakpoints within the duplicated fragment from those within the original fragment, and thus to find out whether the recombinant chromosomes retained a duplication. The results are shown in Figure 4.
Figure 4.—
Phenotype and genotype data on recombinant X chromosomes. The chromosomal region, from CG10555 to oceliless (oc), is represented according to its deduced organization on the sex-ratio XSR6 chromosome. The boxes represent the genes, the dotted lines aligned with the solid vertical bars show the position of the markers, and the two vertical dotted lines mark off the duplication. The arrows represent the part of the + lz (top) and sn + (bottom) recombinant chromosomes that comes from XSR6. The number of chromosomes with a breakpoint falling between two markers is given, together with the corresponding range of the mean percentage of females that they induce (within parentheses).
The examination of sn + recombinants showed that (i) none of the 41 chromosomes that had lost the duplication were able to induce drive, even when they retained part of the original Trf2/org-1 region from XSR6 (from 44 to 56% of females on average within the progeny), and (ii) the 27 chromosomes that still carried the duplication produced female-biased progeny, even when the breakpoint occurred within the duplicated fragment and replaced part of it with a fragment from the standard chromosome (in one case extending from its distal end to the org-1 marker inclusive). The mean female ratios observed varied widely (66–94%) among these chromosomes. Using a marker located within the sniffer gene (∼200 kb distal to CG10555), we showed that the source of this variation was located far from the region studied, since it was found in the same range regardless of whether the breakpoint was proximal or distal to sniffer (67–94 and 69–94%, respectively; Mann–Whitney test P = 0.81).
Among the + lz recombinant chromosomes, the phenotypic shift occurred at some distance from the duplication, beyond the Nrg marker. Further molecular analysis confirmed that the duplication was not sufficient per se for drive to be expressed. Indeed, the six chromosomes with a breakpoint located between the otu and CG11265 markers, which all induced equal-progeny sex ratios (45–55% of females), carried the duplication (control with Trf2 and org-1 markers). A slight bias toward females (69%) was obtained with a chromosome that had a breakpoint between the CG11265 and CG2056 markers. A chromosome with its breakpoint between CG2056 and CG12065 produced an equal sex ratio, whereas all the chromosomes with more distant breakpoints, the closest of which was within the Crag gene, produced a marked female bias (≥77%). Moderate distorters are known to have very unstable drive expression (Montchamp-Moreau and Cazemajor 2002), and so we could not be sure that the apparently nondriving chromosome was totally devoid of drive activity. Unfortunately, we had not kept the corresponding iso-X line and so could not check its phenotype. Whatever it is, some other element, located between the Nrg and Crag markers, is required for drive expression. As had already been found for the sn + recombinants, a large range of segregation ratios was observed among the 48 + lz chromosomes with a breakpoint beyond Crag. The source of this variation was probably remote from the region studied since the range of variation was similar regardless of whether the recombination occurred proximal or distal to an additional marker designed within the gene Lim1, located ∼200 kb from Crag (80–96% of females and 77–96%, respectively, Mann–Whitney test P = 0.99).
Testicular expression of genes located within the candidate regions:
The expression in the testis of the six duplicated genes (Trf2, CG32712, CG12125, CG1440, CG12123, and org-1) and the seven genes located in the second candidate region (CG11265, CG12111, CG2056, CG12065, CG12081, CG12659, and Crag) was tested by RT–PCR of appropriate markers (supplemental Table S1 at http://www.genetics.org/supplemental), followed by cloning and sequencing (sequence accession nos. AM040157–AM040171). All the genes apart from CG12081 and CG12659 were found to be expressed in the testes of sex-ratio XSR6 and standard males. The failure of RT–PCR, with regard to the CG12081 and CG12659 markers, probably resulted from the absence or a very low level of transcripts in the testes of both types of males. Indeed, we easily obtained amplification products using RNA extracts from entire flies or genomic DNA (for CG12659) or genomic DNA only (for CG12081). Regarding the genes included in the duplication, sex-ratio males provided two different cDNA sequences for Trf2, CG12125, and org-1 markers (four polymorphic positions among 764, one among 929 and six among 926, respectively), which were consistent with those obtained from genomic DNA, indicating that both copies of these genes were expressed. We failed to detect any trace of a second sequence for CG32712, CG1440, or CG12123, but this was to be expected from the genomic DNA data (no polymorphism was detected between SRo and SRd markers, or polymorphic sites were restricted to introns). Therefore, we cannot draw any conclusion regarding these three genes.
DISCUSSION
We have shown that the primary sex-ratio locus on the chromosome XSR6 of D. simulans contains two interacting elements. One of these elements was genetically tracked to a tandem duplication, containing six annotated genes, at least three of which are transcriptionally active in the testis. The observation that drive persists after recombination has changed the alleles within a very large proportion of the duplication suggests that it does not involve specific, functionally divergent alleles from XSR6, but rather a difference in gene dosage where two copies confer drive. This possibility is worth noting, because it may be a common feature of meiotic drive systems. Indeed, as far as we are aware, the only other driver for which we have sufficient genetic resolution, the autosomal Segregation distorter of D. melanogaster, also displays a duplication. In that case, it has been demonstrated that gene dosage alone, either of the key distorter locus or of a modifier locus, can confer drive (Kusano et al. 2002).
Sex-ratio drive in D. simulans is linked to nondisjunction of the Y chromosome sister chromatids during meiosis II (Cazemajor et al. 2000). Apart from Trf2, none of the genes included in the duplication are known to be involved in meiosis. CG32712, CG12125, CG1440, and CG12123 all encode products with an unknown or only putatively assigned function (Drysdale and Crosby 2005), whereas the product of org-1 is a member of a family of transcription factors that plays a wide range of roles during development (Lee et al. 2003). Trf2 encodes a member of the TATA box-binding protein family (Rabenstein et al. 1999). The complex utilizing Trf2 probably has numerous targets (Ohler et al. 2002), but interestingly these include genes involved in DNA replication and chromatin remodeling (Hochheimer et al. 2002). It has recently been shown that in D. melanogaster certain defects in Trf2 cause chromosome nondisjunction (Simonova et al. 2005). In addition, its human and murine homologs are important components of gene expression in the testis (Rabenstein et al. 1999; Jiao et al. 2002). Both copies of Trf2 are expressed in the testes of sex-ratio males, which can be expected to change the transcript level relative to that of standard males. Among the testis-expressed genes located within the second mapped region, CG12111, CG2056, and CG12065 encode products with unknown or putative molecular functions, lacking any apparent relationship with meiosis (Drysdale and Crosby 2005). So does the gene Crag, which encodes a calmodulin-binding protein that is related to a Rab3 GDP/GTP exchange protein and putatively involved in synaptic transmission (Xu et al. 1998). In contrast, the product of CG11265 is a more likely candidate as its yeast homolog is a DNA polymerase required for sister-chromatid cohesion (Wang et al. 2000). In planning further studies, however, three points must be borne in mind: (i) the two groups of genes mapped here mark off the chromosomal region required for drive expression, and so other element(s) located between them may also be involved; (ii) it is theoretically possible that the additional AT-rich material accompanying the duplication, rather than the duplication itself, causes drive; and (iii) the function of a segregation distorter is not necessarily directly related to the mechanism of maintaining Mendelian fairness (Hawley 2001).
The identification of candidate genes constitutes a major step toward unraveling the molecular mechanism underlying sex-ratio drive in D. simulans, and beyond it in other Drosophila or dipteran species, where a genetic analysis of the trait is difficult or even impossible to perform. This could make it possible to find out whether sex-ratio drive is related to a common molecular pathway in all these species. Given their lack of a homologous region, segregation distorters acting against the Y chromosome could theoretically evolve anywhere on the Drosophila X chromosome.
Sex-ratio drive in D. simulans exhibits the genetic complexity shared by all the drive systems that have been investigated in this regard (Wu and Beckenbach 1983; Temin et al. 1991; Hiatt and Dawe 2003; Lyon 2003). This supports the view that there is an endless arms race between the X chromosome and the rest of the nuclear genome during which the X chromosome would tend to accumulate distorters, whereas the Y chromosome and the autosomes would accumulate drive suppressors. However, we have no definite evidence so far that cycles of distortion and suppression can happen repeatedly in a given species. The identification of these two interacting elements on the XSR6 chromosome will give an opportunity to test this hypothesis in D. simulans. Indeed, it has been demonstrated that distorter alleles have recently spread in this species, causing a selective sweep at the neighboring locus Nrg (Derome et al. 2004). Surveying the pattern of DNA sequence polymorphism and linkage disequilibrium along the two candidate regions mapped here and in the surrounding area should help to identify the distorter elements and also yield information about where and under what conditions each of them became established in the populations. The challenge lying ahead will be to trace the evolutionary history of the sex-ratio system.
Acknowledgments
We thank Linda Louis and Caroline Bragé for their technical assistance. This work was funded by the Centre National de la Recherche Scientifique.
References
- Cazemajor, M., D. Joly and C. Montchamp-Moreau, 2000. Sex-ratio meiotic drive in Drosophila simulans is related to equational nondisjunction of the Y chromosome. Genetics 154: 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe, R. K., and E. N. Hiatt, 2004. Plant neocentromeres: fast, focused, and driven. Chromosome Res. 12: 655–669. [DOI] [PubMed] [Google Scholar]
- Derome, N., K. Métayer, C. Montchamp-Moreau and M. Veuille, 2004. Signature of selective sweep associated with the evolution of sex-ratio drive in Drosophila simulans. Genetics 166: 1357–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale, R. A., and M. A. Crosby, 2005. The FlyBase Consortium. Nucleic Acids Res. 33: D390–D395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershenson, S., 1928. A new sex-ratio abnormality in Drosophila obscura. Genetics 13: 488–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley, R. S., 2001. Altered nuclear transport and the most selfish of genes. Dev. Cell 1: 311–313. [DOI] [PubMed] [Google Scholar]
- Hiatt, E. N., and R. K. Dawe, 2003. Four loci on abnormal chromosome 10 contribute to meiotic drive in maize. Genetics 164: 699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochheimer, A., S. Zhou, S. Zheng, M. C. Holmes and R. Tjian, 2002. TRF2 associates with DREF and directs promoter-selective gene expression in Drosophila. Nature 420: 439–445. [DOI] [PubMed] [Google Scholar]
- Hurst, G. D., and J. H. Werren, 2001. The role of selfish genetic elements in eukaryotic evolution. Nat. Rev. Genet. 2: 597–606. [DOI] [PubMed] [Google Scholar]
- Jaenike, J., 2001. Sex chromosome meiotic drive. Annu. Rev. Ecol. Syst. 32: 25–49. [Google Scholar]
- Jiao, X., P. Trifillis and M. Kiledjian, 2002. Identification of target messenger RNA substrates for the murine deleted in azoospermia-like RNA-binding protein. Biol. Reprod. 66: 475–485. [DOI] [PubMed] [Google Scholar]
- Kusano, A., C. Staber and B. Ganetzky, 2002. Segregation distortion induced by wild-type RanGAP in Drosophila. Proc. Natl. Acad. Sci. USA 99: 6866–6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano, A., C. Staber, H. Y. Chan and B. Ganetzky, 2003. Closing the (Ran)GAP on segregation distortion in Drosophila. BioEssays 25: 108–115. [DOI] [PubMed] [Google Scholar]
- Lee, H. H., A. Norris, J. B. Weiss and M. Frasch, 2003. Jelly belly protein activates the receptor tyrosine kinase Alk to specify visceral muscle pioneers. Nature 425: 507–512. [DOI] [PubMed] [Google Scholar]
- Lyon, M. F., 2003. Transmission ratio distortion in mice. Annu. Rev. Genet. 37: 393–408. [DOI] [PubMed] [Google Scholar]
- Lyttle, T. W., 1991. Segregation distorters. Annu. Rev. Genet. 25: 511–557. [DOI] [PubMed] [Google Scholar]
- Montchamp-Moreau, C., and M. Cazemajor, 2002. Sex-ratio drive in Drosophila simulans: variation in segregation ratio of X chromosomes from a natural population. Genetics 162: 1221–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohler, U., G. C. Liao, H. Niemann and G. M. Rubin, 2002. Computational analysis of core promoters in the Drosophila genome. Genome Biol. 3: RESEARCH0087. [DOI] [PMC free article] [PubMed]
- Orr, H. A., and D. C. Presgraves, 2000. Speciation by postzygotic isolation: forces, genes and molecules. BioEssays 22: 1085–1094. [DOI] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena, F., and C. Sapienza, 2001. Female meiosis drives karyotypic evolution in mammals. Genetics 159: 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabenstein, M. D., S. Zhou, J. T. Lis and R. Tjian, 1999. TATA box-binding protein (TBP)-related factor 2 (TRF2), a third member of the TBP family. Proc. Natl. Acad. Sci. USA 96: 4791–4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonova, O. B., E. A. Modestova, J. E. Vorontsova and L. I. Korochkin, 2005. Leg-arista-wing complex mutations cause not only morphological abnormalities, but also chromosome non-disjunction. Annual Drosophila Research Conference, Vol. 46. The Genetics Society of America, Bethesda, MD.
- Temin, R. G., B. Ganetzky, P. A. Powers, T. W. Lyttle, S. Pimpinelli et al., 1991. Segregation distortion in Drosophila melanogaster: genetic and molecular analyses. Am. Nat. 137: 287–331. [Google Scholar]
- Wang, Z., I. B. Castano, A. De Las Penas, C. Adams and M. F. Christman, 2000. Pol kappa: a DNA polymerase required for sister chromatid cohesion. Science 289: 774–779. [DOI] [PubMed] [Google Scholar]
- Wu, C. I., and A. T. Beckenbach, 1983. Evidence for extensive genetic differentiation between the sex-ratio and the standard arrangement of Drosophila pseudoobscura and D. persimilis and identification of hybrid sterility factors. Genetics 105: 71–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X. Z., P. D. Wes, H. Chen, H. S. Li, M. Yu et al., 1998. Retinal targets for calmodulin include proteins implicated in synaptic transmission. J. Biol. Chem. 273: 31297–31307. [DOI] [PubMed] [Google Scholar]