Abstract
The LIN-3–LET-23-mediated inductive signaling pathway plays a major role during vulval development in C. elegans. Studies on the components of this pathway have revealed positive as well as negative regulators that function to modulate the strength and specificity of the signal transduction cascade. We have carried out genetic screens to identify new regulators of this pathway by screening for suppressors of lin-3 vulvaless phenotype. The screens recovered three loci including alleles of gap-1 and a new gene represented by sli-3. Our genetic epistasis experiments suggest that sli-3 functions either downstream or in parallel to nuclear factors lin-1 and sur-2. sli-3 synergistically interacts with the previously identified negative regulators of the let-23 signaling pathway and causes excessive cell proliferation. However, in the absence of any other mutation sli-3 mutant animals display wild-type vulval induction and morphology. We propose that sli-3 functions as a negative regulator of vulval induction and defines a branch of the inductive signaling pathway. We provide evidence that sli-3 interacts with the EGF signaling pathway components during vulval induction but not during viability and ovulation processes. Thus, sli-3 helps define specificity of the EGF signaling to induce the vulva.
INDUCTION by intercellular signals mediated by growth factors play important roles in cell proliferation and fate specification. The intracellular signaling pathway mediated by epidermal growth factor (EGF) and its receptor (EGFR) has been extensively studied in many systems, and much is known about its components and how they are activated or inactivated (Moghal and Sternberg 2003b; Sundaram 2005). However, a signaling pathway is not a simply binary device, being either active or inactive. Once a pathway is activated, it can have many levels of activity. How these activities are regulated is not fully understood.
In Caenorhabditis elegans, the LIN-3–LET-23-mediated inductive signaling pathway is known to play multiple roles during development (Moghal and Sternberg 2003b). LIN-3 is a member of the EGF family that binds with the LET-23/EGFR to activate downstream pathway components including LET-60/Ras. Genetic analyses of the EGF signaling in C. elegans have revealed at least five different roles in regulating vulval formation, viability, ovulation, male spicule development, and posterior ectodermal P12 cell fate specification (Moghal and Sternberg 2003b). During vulval development and P12 fate specification activated LET-23 receptor transduces a signal through a conserved set of factors that includes LET-60/Ras, LIN-45/Raf, and MPK-1/MAP kinase. However, the ovulation process is mediated by a Ras-independent pathway and involves calcium signaling, which is regulated by inositol trisphosphate (IP3) and its receptor ITR-1 (Clandinin et al. 1998). Thus, different outcomes of the signaling depend upon tissue-specific effectors.
The molecular genetic studies of EGFR signaling during vulval development have revealed both positive and negative components that function to regulate the strength and specificity of the signal transduction cascade. The vulva is formed by the progeny of three (P5.p, P6.p, and P7.p) of six (Pn.p, n = 3–8) equipotential vulval precursor cells (VPCs) that are induced by the LIN-3/EGF produced by the gonadal anchor cell. Once induced, these three VPCs acquire 1° and 2° cell fates and undergo three rounds of cell divisions. Vulval progeny differentiate to form vulval tissue in adult animals.
Genetic analysis of the C. elegans vulva has revealed that in addition to LET-23-mediated inductive signaling two additional pathways, lateral signaling mediated by LIN-12/Notch and Wnt signaling mediated by BAR-1/β-Catenin, also participate in vulval development (Eisenmann 2005; Greenwald 2005; Sternberg 2005). Thus, vulval formation serves as a powerful system to investigate the underlying mechanisms regulating interactions between three evolutionarily conserved signaling pathways. Such studies have begun to reveal the components of these signaling pathways and their regulators, as well as target genes conferring specific developmental outcomes [e.g., lin-39/Hox, dpy-22/TRAP230, and lag-1/Su(H)/CBF1] (Christensen et al. 1996; Eisenmann et al. 1998; Moghal and Sternberg 2003a). To identify additional regulators of the EGF signaling pathway, we carried out a genetic screen to isolate suppressors of the lin-3(rf) Vulvaless (Vul) phenotype. Here, we report the isolation and characterizations of three mutations, one of which represents a new locus sli-3. The genetic analysis of sli-3 has revealed its function as a negative regulator of the EGF signaling pathway in vulval cells. Our epistasis experiments show that sli-3 functions either downstream or in parallel to transcriptional regulators lin-1 and sur-2. We also demonstrate that sli-3 specifically participates in the vulval function of EGF signaling but not in other developmental processes, i.e., viability, ovulation, and P12 cell fate specification. Furthermore, we find that sli-3 does not genetically interact with lin-12/Notch and Wnt pathway components bar-1/β-catenin and pry-1/axin. Taken together, our findings establish sli-3 as a tissue-specific regulator of the EGF signaling that helps establish proper signaling intensity during vulval development.
MATERIALS AND METHODS
General methods:
Worms were grown according to published methods (Brenner 1974). All experiments were performed at room temperature (20°) unless otherwise noted. Cell and tissue anatomy was observed under Nomarski DIC optics as described by Sulston and Horvitz (1977). Standard cellular and genetic nomenclature is as defined by Sulston and Horvitz (1977) and Horvitz et al. (1979).
Vulval induction was observed by scoring the number of VPCs that adopt vulval fates during the L4 stage. In wild-type animals (N2) vulval induction is three (one for each P5.p, P6.p, and P7.p). However, mutant animals have variable vulval induction (zero to six range). The vulval lineage was determined by direct observations of the cell-division patterns in animals between mid-L3 and early-L4 stages.
Strains and construction strategies:
The wild-type N2 and standard mutant strains are from Brenner (1974) and the Caenorhabditis Genetics Center. The mutants strains used in this study are as follows (references are given where appropriate):
LGI: pry-1(mu38) (Maloof et al. 1999), sur-2(ku9) (Singh and Han 1995), unc-13(e51) (Brenner 1974), unc-101(sy108) (Lee et al. 1994), and dpy-5(e61) (Brenner 1974).
LGII: dpy-10(e128), unc-4(e120), rol-1(e91), rol-6(e187) (Brenner 1974); let-23(sy15), let-23(sy97) (Aroian and Sternberg 1991); vab-9(e1744), let-239(mn93) (Sigurdson et al. 1984); mnC1[dpy-10(e128) unc-52(e444)] (Herman 1978); mnDf29, mnDf44, mnDf46, mnDf58, mnDf61, mnDf62, mnDf67, mnDf68, mnDf69, mnDf85, mnDf89, mnDf90, mnDf106 (all deficiencies are from Sigurdson et al. 1984); and mnDp34 (Herman et al. 1979).
LGIII: dpy-18(e364) (Brenner 1974), lin-39(n709) (Garriga et al. 1993), and lin-12(n137n460) (Greenwald et al. 1983).
LGIV: ark-1(sy187) (Hopper et al. 2000); let-59(s49), let-312(s1234) (Clark et al. 1988); itr-1(sy290) (Clandinin et al. 1998); dpy-20(e1282), unc-22(s7), unc-24(e138) (Moerman and Baillie 1979); let-60(n2034), unc-31(e169) (Beitel et al. 1990); lin-45(sy96) (Han et al. 1993); lin-3(n378), lin-3(n1058), lin-3(n1059), nT1[unc(n754dm) let] = DnT1 (Ferguson and Horvitz 1985); and sDf63, sDf67 (Clark and Baillie 1992).
LGV: dpy-11(e224), him-5(e1490), unc-34(e315) (Hodgkin et al. 1979); and lin-25(e1446), nT1[unc(n754dm) let](IV, V) (Ferguson and Horvitz 1985).
LGX: bar-1(ga80) (Eisenmann et al. 1998); dpy-6(e14), dpy-8(e120), lon-2(e678), unc-1(e719), unc-2(e55), unc-6(e78) (Brenner 1974); unc-97(su110) (Zengel and Epstein 1980); gap-1(n1691) (Hajnal et al. 1997); sli-1(sy143) (Jongeward et al. 1995); and mnDp31 (Herman et al. 1979).
To screen for the suppressors of lin-3 Vul phenotype, we constructed a strain PS1031 [let-312(s1234) lin-3(n378) unc-22(s7)/unc-24(e138) lin-3(n1059) dpy-20(e1282)] that carries two lin-3 hypomorphic alleles, n378 and n1059. PS1031 was constructed as follows: let-59 unc-22/nT1; +/nT1 hermaphrodites were mated with N2 males and the F1 males were mated with let-312 lin-3(n378)/DnT1; +/DnT1 hermaphrodites. Individual F1 non-Unc hermaphrodites were picked and let-312 lin-3(n378)/let-59 unc-22 progeny were identified on the basis of their segregation of both early and late larval lethals. The Unc progeny of these worms were crossed with N2 males, and F1 males were mated with unc-24 lin-3(n1059) dpy-20/DnT1; +/DnT1 hermaphrodites. Non-Unc cross-progeny were picked at L4 and those that became Vul adults were let-312 lin-3(n378) unc-22/unc-24 lin-3(n1059) dpy-20. In this strain vulval induction is severely defective (1% of wild type, see Table 1; also see Ferguson and Horvitz 1985) and animals exhibit fully penetrant egg-laying defective phenotype.
TABLE 1.
Vulval induction in lin-3 hypomorphs and suppressor alleles
| Suppressor | lin-3 | VPC inductiona | nb |
|---|---|---|---|
| + | e1417 | 0.07 ± 0.18 | 20 |
| + | n378 | 1.0 ± 1.1 | 43 |
| + | n1058 | 1.4 ± 1.2 | 14 |
| + | n378/n1059 | 0.03 ± 0.19 | 29 |
| sy330/sy330 | + | 3.0 ± 0.0 | 15 |
| sy340/sy340 | + | 3.0 ± 0.0 | 23 |
| sy341/sy341 | + | 3.0 ± 0.0 | 50 |
| sy330/+ | n378/n1059 | 3.0 ± 0.0 | 30 |
| sy340/sy340 | n378/n1059 | 2.8 ± 0.5 | 12 |
| sy340/+ | n378 | 2.2 ± 0.9 | 14 |
| sy341/sy341 | n378/n1059 | 2.6 ± 0.4 | 8 |
The wild-type loci have been marked as “+.”
The average number of vulval precursors that acquire 1° and 2° cell fates and undergo cell divisions. The range is between 0 (no induction) and 6 (all VPCs induced). In wild-type animals 3.0 induction is observed. The values are shown as mean ± standard deviation.
Number of L4 stage hermaphrodites examined.
To construct a sli-3; lin-3(n1058) strain, unc-24 n1058/DnT1; +/DnT1 hermaphrodites were crossed with N2 males. The non-Unc F1 males (unc-24 n1058/+) were mated with dpy-10 sy341 hermaphrodites and dpy-10 sy341; n1058 dpy-20 animals were obtained in two generations.
To construct a partial triploid strain carrying extra copies of sli-3, we used a free duplication mnDp34. For this, mnC1/unc-4 unc-52; mnDp34 hermaphrodites were mated with unc-4 sy341/+; n378 males and F1 worms were individually cloned. In the next generation non-Unc (unc-4) non-Dpy Egl worms were picked from a clone that segregated Unc (unc-4 non-unc-52) animals to establish a line of unc-4 sy341/mnC1; n378; mnDp34 animals. The presence of mnDp34 was confirmed by segregation of the Dpy animals.
For deficiency mapping of sy341, we constructed sy341/Df strains by mating dpy-10 sy341; n378 hermaphrodites with Df/mnC1; n378 males. In the F1 generation non-Dpy worms were individually picked to establish clonal populations.
Mutagenesis:
Worms were mutagenized by ethyl methanesulfonate (EMS) (Brenner 1974). To isolate the suppressors of the lin-3 Vul phenotype, we used strain PS1031 (see above) and screened ∼30,000 haploid genomes. Single non-Egl F2 worms from each P0 plate were individually cloned. For those plates that carried <10 progeny, worms were allowed to grow two additional generations before picking a putative Egl suppressor. We scored vulval induction of the progeny to confirm the phenotype and obtained five true breeding non-Egl lines.
To screen for sli-4 revertants, we mutagenized animals carrying the sy330 allele in a PS1031 background. Single Egl hermaphrodites were picked in F1 and lines were established for those candidates that continued to produce Egl progeny in subsequent generations. In this way, we isolated five recessive mutations, four of which (sy561, sy594, sy595, and sy596) are linked to LGIV. The Egl phenotype of these alleles results from severe defects in vulval induction (average VPC induction in sy561, 1.4, n = 30; in sy594, 0.2, n = 16; in sy595, 0.2, n = 12; and in sy596, 0.1, n = 19). The mutant animals also exhibit embryonic and L1 stage lethality.
Complementation and mapping:
sli-3 was mapped to LGII on the basis of the following experiments. sli-3 is not linked to lin-3 since animals heterozygous for sli-3 (sy341/+) and lin-3 (n378/+) cosegregate only approximately one-quarter of the total progeny. Linkage tests with other markers were performed in the background of lin-3(n378) using vulval induction as an assay. Three-factor mapping using rol-6(e187) unc-4(e120) revealed that sli-3 is likely to the right of unc-4. Specifically, all 8 Rol non-Unc recombinants picked up sli-3(sy341) whereas none of 7 Unc non-Rol picked up sli-3(sy341). Futher mapping was done using unc-4 let-25 and unc-4 let-246 strains. dpy-10 sy341; n378 hermaphrodites were mated with unc-4 let-25/mnC1; n378 and unc-4 let-246/mnC1; n378 males separately and viable Unc (recombinant) hermaphrodites were picked in the F2 generation. All 19 Unc non-Let (let-25) recombinants picked up sli-3(sy341), suggesting that sli-3 is either to the right of let-25 or very close to its left. Using unc-4 let-246 we obtained 19 recombinants, 15 of which picked up sli-3(sy341). Thus, sli-3 is located between unc-4 and let-246, closer to let-246 than unc-4. We also used deficiencies to map the sli-3 locus. The deficiencies mnDf58 and mnDf62 that uncover let-25 and let-246 fail to complement sli-3(sy341). By contrast, deficiencies mnDf61 and mnDf29 that cover let-25 and let-246 complement sli-3(sy341). Finally, deficiency mnDf90 that uncovers let-246 but not let-25 also complements sli-3(sy341).
The three sli-4 alleles (sy315, sy330, and sy339) are considered allelic since they fail to complement each other when assayed for the suppression of lin-3 Vul defect and confer similar phenotypes. The linkage was determined as follows. let-312 lin-3(n378) unc-22/lin-3(n1059) sli-4 dpy-20 hermaphrodites were crossed to lin-3(n378) dpy-20(e1282); him-5 males. F1 Dpy progeny had wild-type vulval induction, and non-Dpy cross-progeny were all Vul, which indicates that sli-4 is linked to lin-3 and is dominant. For further mapping on LGIV, we crossed n1059 sli-4 dpy-20/n378 dpy-20 hermaphrodites to n378; him-5 males, and Dpy animals were picked out individually from n1059 sli-4 dpy-20/n378. When using sy330, 10 of 12 Dpy animals picked up sy330, and 8 of 10 picked up sy339 when using sy339. In these experiments the presence of sli-4 alleles was determined by the suppression of n1059 lethality phenotype. The mapping results suggested that sli-4 is between lin-3 and dpy-20 and close to dpy-20.
The sli-4 revertants were linked to the sli-4 chromosome (LGIV) on the basis of the following experiment. Hermaphrodites from the strain carrying sy330 and one of the revertant alleles [let-312(s1234) lin-3(n378) unc-22(s7)/lin-3(n1059) sli-4(sy330) dpy-20(e1282); revertant] were crossed to mec-3(e1338) him-8(e1489) dpy-20(e1282)/+ males. In the F1 generation, Dpy males were crossed to let-312(s1234) lin-3(n378)/DnT1; +/DnT1 hermaphrodites. Non-Unc F2 worms were cloned and the progeny of those that did not segregate Dpy worms were examined for the Egl phenotype. The clones for all four sli-4 revertants exhibited fully penetrant Egl phenotype, demonstrating their linkage to the sli-4 chromosome. The deficiency mapping was carried out to further refine the genetic intervals of mutant loci. All four mutations complemented two deficiencies sDf8 and sDf62 and gave rise to viable progeny but not the deficiency sDf63. In this manner sli-4 revertants were placed on LGIV very close to the lin-3 locus. In the case of sDf8 and sDf62, mutant/Df animals gave rise to viable progeny that exhibited wild-type vulval induction (data not shown). However, mutant/sDf63 animals were embryonic/early L1 larval lethal, a phenotype that resembles lin-3(n1059) mutant animals. It should be pointed out that sDf63 (but not sDf8 and sDf62) uncovers the lin-3 locus (Clark and Baillie 1992).
To determine linkage of sli-5, hermaphrodites of the genotype n1059 dpy-20/let-312 n378 unc-22; sli-5 were crossed with n378 dpy-20; him-5 males. Some animals of the resulting cross-progeny were non-Egl, suggesting that sli-5 is not linked to lin-3 and is semidominant for the suppression of the Vul phenotype of lin-3. We crossed lin-3(n378); sli-5 males to hermaphrodites that carry Dpy or Unc markers, and, in F2, Vul animals not displaying the marker phenotype were individually picked. In this manner, sli-5 was assigned to linkage group X. From a three-factor mapping sli-5 was mapped to the left of lon-2. Twenty-seven of 27 Unc non-Lon and 0 of 30 Lon non-Unc recombinant animals from n378; lon-2 unc-97/sli-5 picked up sli-5.
On the basis of its map position, we tested whether the sli-5(sy340) mutation was a new allele of sli-1 or gap-1, two previously identified negative regulators of vulval development that map to the left arm of the X chromosome (Yoon et al. 1995; Hajnal et al. 1997). lin-3(n378); sli-5(sy340) males were crossed into lin-3(n378); unc-1 sli-1 or dpy-5; lin-3(n378); gap-1 unc-2 hermaphrodites. sli-5(sy340) failed to complement both sli-1 [2.78 cells induced (n = 20) for sli-1/sli-5(sy340) vs. 1.80 cells induced (n = 20) for sli-1/+] and gap-1 [2.95 cells induced (n = 20) for gap-1/sli-5(sy340) vs. 1.93 cells induced for gap-1/+ (n = 22)]. We have previously observed suppression of other let-23 pathway mutations [e.g., let-23(sy1)] by a trans-heterozygous combination of sli-1 and gap-1 alleles (data not shown). Thus, the complementation data are consistent with sli-5(sy340) being an allele of either locus.
Molecular analysis of sli-4:
The genetic linkage mapping experiments revealed that all four sli-4 revertants are tightly linked to sli-4, suggesting that they are either intragenic alleles or mutations in closely linked loci. Potential candidates in the lin-3 genetic region include let-60 (0.36 MU away from lin-3). Gain-of-function mutations in let-60 (e.g., n1046dn) are known to be epistatic to lin-3 (Han et al. 1990). Since let-60(dn) alleles have been shown to alter the coding region of the gene (Beitel et al. 1990), we sequenced let-60 RT–PCR products from each of the five sli-4 revertants. None of the cDNA clones showed any mutation in let-60 exons, suggesting that let-60 is unaffected in mutant animals. To test the possibility that sy330 revertants are allelic to lin-3, we sequenced the lin-3 genomic region in lin-3(n1059) sy330/lin-3(n378) animals and detected a molecular change corresponding to n378 (G61 to A) but none to n1059 (G564 to A). Hence sy330 represents a wild-type allele of lin-3. We also sequenced the lin-3 genomic region in three of the revertant alleles (sy561, sy594, and sy596) that revealed mutations in the lin-3 open reading frame. Two of these, sy594 and sy596, have a premature stop codon (C595 to T and G80 to A, respectively) whereas sy561 has a mutation identical to n378 (G61 to A).
Analyses of the survival rate and fertility:
Survival rate in different genotypes was calculated as described by Aroian and Sternberg (1991). Mutant hermaphrodites (for example, unc-24 n1058/DnT1) were mated with N2 males and F1 non-Unc hermaphrodites (unc-24 n1058/+ in this case) were individually picked and placed on plates. In F2 the numbers of Unc and wild-type progeny were counted to determine the percentage of survival. For the N2 strain, survival was the ratio of the number of fertilized eggs and hatched progeny.
sli-3(sy341) animals have significantly low brood size compare to the wild type and are partially sterile. To determine whether these phenotypes are caused by sy341 and not an unlinked mutation, we examined recombinant animals from a three-factor mapping cross with unc-4 let-246 (∼1.3 MU interval). All 15 recombinants that suppressed the lin-3(n378) Vul defect (see above) showed reduced brood size. The tight association of the two phenotypes and their location within a small genetic interval (1.3 MU) suggest that the fertility defect is most likely caused by sy341. However, we cannot rule out the involvement of an unlinked very closely located mutation.
To understand the cellular basis of sterility in sli-3(sy341) animals, we examined the morphology of adult gonad and developing oocytes. In contrast to wild-type animals where oocytes in diakinesis appear almost square and are aligned in a row, sy341 oocytes have abnormal morphology and do not align correctly. In addition, the number of oocytes in the gonad arms of 1- to 2-day-old sy341 adults is significantly lower compared to the control (one gonad arm, 4.3 ± 1.9, n = 20; wild type, 7.3 ± 2.5, n = 27). While these defects do provide a partial explanation of the sterility in sy341 animals, they do not rule out the possibility of an ovulation defect as an additional factor. To examine this possibility, we observed ovulation events in sy341 animals under Nomarski optics but did not find any defect (n = 4 ovulations). Thus ovulation does not appear to be visibly compromised in sli-3 mutants.
Molecular biology:
gap-1 and sli-1 open reading frames (including introns) from sli-5(sy340) and wild-type N2 animals were amplified by PCR using primer pairs GL92/93 (sli-1: 5.6 kb) and GL94/95 (gap-1: 2.8 kb). sli-1 DNA was sequenced using primers GL92, GL93, GL100, and GL101 and found to be wild type in sy340 animals. Primers GL94, GL95, GL98, and GL99 were used to sequence the gap-1 genomic DNA. We found one G-to-A mutation within a conserved 5′ splice site (GT) in intron 8 that prematurely introduces two in-frame stop codons (TGA and TAA). This was confirmed by sequencing both strands. Primer sequences are: GL92 (gccactggacttcacatcatatcacc), GL93 (cacaagtctactcccgctcactgttc), GL94 (atggttctatcttgcagagtcgtcgac), GL95 (cttctactcactttgttctccttctcg), GL98 (ggaaaccttcaacaagttgaccgaagc), GL99 (ctgacactacagttagacagcctttg), GL100 (gccaaattgcccaggtaattgaaac), and GL101 (gcaatgcaaagcatgcacattatctc).
RESULTS
Isolation of lin-3 pathway regulators:
To study mechanisms that regulate the LIN-3–LET-23-mediated vulval induction pathway in C. elegans, we carried out a novel genetic screen. The screen was designed to isolate suppressors of a severe, but nonnull lin-3 phenotype. We used a strain (PS1031) that carries the hypomorphic lin-3 allele n378 in trans to the null allele n1059 (see materials and methods). While n378 is a viable allele with significantly reduced vulval induction (Table 1), n1059 is an embryonic/early L1 stage lethal (Ferguson and Horvitz 1985). We chose this genotype since none of the known viable alleles of lin-3 (e.g., e1417, n378, and n1058) are completely penetrant for the vulval induction defect (see Table 1). In lin-3(n378)/lin-3(n1059) heterozygous animals, vulval induction is severely reduced (average induction 0.03, n = 29) and all animals are egg-laying defective (Egl) (100%, n = 244). The severity of the vulval induction and egg-laying defects in this strain facilitated isolation of suppressors simply on the basis of the egg-laying defect. We expected to obtain two kinds of mutants from the screen: reduction-of-function (rf) alleles of negative regulators and gain-of-function (gf) alleles of positive regulators.
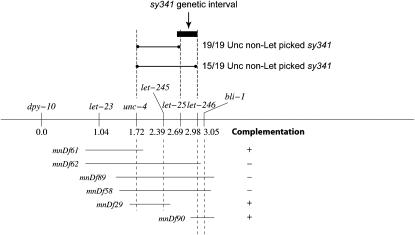
F2 progeny of EMS-mutagenized PS1031 animals were screened for suppressors that would revert the Egl phenotype. After screening nearly 30,000 haploid genomes, we isolated five mutations that suppress Egl and Vul defects of lin-3(n378)/lin-3(n1059) to almost wild-type levels (Table 1). Mutations were mapped to linkage groups using genetically marked strains and deficiencies (see materials and methods). Complementation and mapping experiments revealed that three of the suppressors, sy315, sy330, and sy339, are allelic and define a single locus (sli-4) on LGIV. We chose sy330 as a representative allele for further experiments. A combination of two- and three-factor mapping experiments helped localize sli-4 on LGIV close to dpy-20. Of the remaining two suppressors, sli-3(sy341) maps on LGII either very close to the left of let-25 (2.69 MU) or between let-25 and let-246 (2.98 MU) (Figure 1) and sli-5(sy340) maps on LGX to the left of lon-2 (−6.70 MU) (see materials and methods).
Figure 1.—
A partial genetic map of C. elegans showing some of the markers and deficiencies used to define the interval of sli-3. The map positions of markers and extents of various deficiencies are indicated. The results of complementation tests between sli-3(sy341) and deficiencies have been marked with either + (Df complements) or − (Df does not complement). The three-factor mapping experiments place sli-3 very close to the left of let-25 or between let-25 and let-246. In the case of the unc-4 let-246 double, 19 of 19 Unc non-Let recombinants picked up sli-3(sy341). Whereas, in the case of the unc-4 let-25 double, 15 of 19 Unc non-Let recombinants picked up sli-3(sy341) (see materials and methods for details).
Characterization of lin-3 suppressors:
We examined the three classes of lin-3 suppressor loci (sli-3, sli-4, and sli-5) by analyzing the ability of mutant alleles to suppress the lin-3(n378)/lin-3(n1059) Vul defect. sli-3(sy341) is a recessive loss-of-function allele by the following three criteria. First, sy341/+ animals do not suppress the vulval induction defect in lin-3(n378) animals (average induction 1.3 ± 1.0, n = 31, P = 0.2437). Second, a duplication of the sli-3 region mnDp34 can suppress the sy341 phenotype in the lin-3(n378) background (average induction 1.8 ± 1.2, n = 25 in sy341/sy341, mnDp34 animals; P = 0.1411 when compared to sy341/+). Third, sli-3(sy341)/Df animals exhibit vulval induction comparable to that of sli-3(sy341) homozygotes in the lin-3(n378) genetic background (Table 2). The sli-4 alleles (sy315, sy330, and sy339) are dominant suppressors of the lin-3(n378)/lin-3(n1059) Vul defect (Table 1). Finally, sy340, a single allele of sli-5, is semidominant as one copy of the mutation partially suppresses the vulval induction defect in lin-3(n378)/lin-3(n1059) animals (Table 1). All three classes of suppressor mutations are silent in an otherwise wild-type genetic background, suggesting that they are part of a redundantly acting regulatory network (Table 1).
TABLE 2.
Deficiency mapping of sli-3
| Genotype | lin-3 | VPC induction | n |
|---|---|---|---|
| + | + | 3.0 ± 0.0 | >100 |
| + | n378 | 1.0 ± 1.1 | 43 |
| sy341/sy341 | n378 | 3.0 ± 0.0 | 35 |
| mnDf62/+ | n378 | 1.3 ± 1.0 | 16 |
| sy341/mnDf62 | n378 | 2.4 ± 0.7 (P = 0.0023) | 14 |
| mnDf58/+ | n378 | 1.3 ± 1.2 | 39 |
| sy341/mnDf58 | n378 | 2.5 ± 0.9 (P < 0.0001) | 50 |
| mnDf61/+ | n378 | 1.4 ± 1.0 | 21 |
| sy341/mnDf61 | n378 | 1.0 ± 0.8 (P = 0.1867) | 27 |
| mnDf29/+ | n378 | 1.7 ± 1.1 | 14 |
| sy341/mnDf29 | n378 | 1.2 ± 1.3 (P = 0.3181) | 16 |
| mnDf90/+ | n378 | 1.2 ± 0.9 | 32 |
| sy341/mnDf90 | n378 | 1.0 ± 1.2 (P = 0.5554) | 31 |
sli-3 is uncovered by two deficiencies, mnDf62 and mnDf58. The other three deficiencies, mnDf61, mnDf29, and mnDf90, complement sli-3. VPC induction and “n” are defined in Table 1. The P-values are given in parentheses. The two groups of data are statistically significantly different if P < 0.05.
Epistasis experiments revealed that sli-4(sy330) suppresses the Vul defect associated with lin-3(rf) but not let-23(sy97) (Table 3). Since the dominant nature of sli-4 alleles (sy315, sy330, and sy339) did not allow further genetic studies, we sought to isolate recessive loss-of-function alleles by screening for sy330 suppressors that exhibit a Vul phenotype. From a screen of 50,000 F1's, we isolated four mutants—sy561, sy594, sy595, and sy596—that are phenotypically similar to lin-3(n378)/lin-3(n1059) and have almost zero vulval induction (see materials and methods). Three of these (sy594, sy595, and sy596) also exhibit embryonic/early L1 larval stage lethality. The results of our three-factor and deficiency mapping as well as allele sequencing experiments (see materials and methods) show that sli-4 alleles correspond to the lin-3 locus. Thus the dominant alleles of sli-4 are intragenic revertants of lin-3, whereas the revertant alleles define new lin-3 hypomorphs.
TABLE 3.
Epistasis test of lin-3 suppressor loci
| Suppressors | EGF pathway genes | VPC induction | n |
|---|---|---|---|
| + | let-23(sy97) | 0 | 21 |
| sy330 | sy97 | 0 | 27 |
| + | mpk-1(ku1) | 2.8 ± 0.7 | 22 |
| sy330 | ku1 | 2.3 ± 0.5 (P = 0.09) | 22 |
| sy340 | sy97 | 2.9 ± 0.2 | 10 |
| + | let-23(sa62) | 3.1 ± 0.4a | 14 |
| sy340 | sa62 | 4.3 ± 0.5b (P < 0.000004) | 12 |
| + | let-60(n2034) | 0 | 11 |
| sy340 | n2034 | 2.3 ± 0.7 | 19 |
| + | lin-45(sy96) | 1.6 ± 1.1 | 29 |
| sy340 | sy96 | 0.4 ± 1.3 | 13 |
Fourteen percent of animals exhibited a Muv phenotype. The genotype was let-23(sa62) unc-4/mnC1; lon-2.
sli-5(sy340) strongly suppresses the lin-3(rf) Vul phenotype, suggesting that sli-5 functions as a negative regulator of vulval induction. Consistent with this hypothesis, we found that sy340 enhances the Muv phenotype of let-23 gain-of-function allele sa62 (Table 3). To further study sli-5 in vulval development we carried out epistasis experiments with hypomorphs of the LET-23-mediated EGF signaling pathway. sy340 suppresses the Vul defect in let-23(sy97) and let-60(n2034) but not lin-45(sy96) animals (Table 3). Thus sli-5 appears to function at the level of let-60 during vulval induction. This property of sli-5 resembles that of gap-1, which encodes a GTPase-activating protein for LET-60/Ras (Hajnal et al. 1997), and sli-1, an ortholog of c-Cbl, both of which map to the same region as sli-5 (left of lon-2 on LGX, see materials and methods). We carried out complementation tests with both gap-1 and sli-1 and found that sli-5 fails to complement both loci. We interpreted these data to suggest that sli-5 was either gap-1 or sli-1 since we had previously found that a trans-heterozygous combination of a single sli-1 and gap-1 mutation can suppress hypomorphs in the let-23 pathway (data not shown). Therefore, we sequenced the gap-1 and sli-1 genomic coding regions (including introns) in sy340 animals. The sequence analysis identified a single G-to-A mutation in gap-1, while we failed to detect any mutations in sli-1. The G-to-A mutation is in the conserved splice donor site (GT) of intron 8 and is predicted to immediately add two consecutive STOP codons, which would prematurely truncate the protein after amino acid 497. This truncation would remove the last 12 amino acids of the PH domain, which might prevent GAP-1 from properly localizing with LET-60/Ras at the plasma membrane. On the basis of these results, we conclude that sli-5 is a new allele of gap-1.
sli-3 interacts with Ras-pathway genes in vulval cells:
The suppression of the lin-3 Vul defect by sy341 suggested that wild-type sli-3 functions as a negative regulator of vulval development. Apart from the lin-3 heteroallelic combination n378/n1059, we also tested the effect of sli-3(sy341) on two homozygous lin-3 hypomorphs, n378 and n1058. Vulval induction in these lin-3 mutant animals is significantly reduced compared to that in the wild type (Table 1) and, in addition, lin-3(n1058) but not lin-3(n378) animals exhibit a fully penetrant sterile phenotype (Ferguson and Horvitz 1985; Clandinin et al. 1998). sy341 suppresses the vulval defect in both lin-3 alleles (Table 4) but not the sterile phenotype of lin-3(n1058) (see below and Table 8). To determine whether sli-3-mediated Vul suppression was limited to lin-3, we examined the ability of sy341 to suppress vulval defects caused by mutations in other components of the LET-23 signaling pathway, including let-23, let-60, lin-45, and mpk-1. sy341 suppresses the Vul defect associated with viable loss-of-function mutations in all of these genes. Thus, severe hypomorphic alleles of let-23, let-60, and lin-45 are strongly suppressed by sy341 (Table 4). The weak hypomorphs of let-60 and mpk-1 (n2021 and ku1, respectively) are suppressed to wild-type levels (Table 4). These epistasis results suggest that sli-3 negatively regulates the LET-23 signaling pathway during vulval development.
TABLE 4.
Epistasis test of sli-3 with let-23-mediated EGF pathway genes
| Genotype
|
||||
|---|---|---|---|---|
| Vulval mutant | sli-3 | % Egl | VPC induction | n |
| + | + | 0 (>100) | 3.0 ± 0.0 | >100 |
| lin-3(n1058) | + | NA | 1.4 ± 1.2 | 14 |
| n1058 | sy341 | NA | 2.9 ± 0.3 (P = 0.0002) | 14 |
| lin-3(n378) | + | 83 (72) | 1.0 ± 1.1 | 43 |
| n378 | sy341a | 3 (72) | 3.0 ± 0.0 | 35 |
| n378 | sy341b | ND | 2.7 ± 0.7 | 31 |
| let-23(sy97) | + | 100 (50) | 0 | 20 |
| sy97 | sy341 | 46 (13) | 1.7 ± 1.2 | 15 |
| let-60(n1876) | + | 100 (16)c | 0 | 16 |
| n1876 | sy341 | 100 (6)c | 2.2 ± 0.5 | 6 |
| let-60(n2021) | + | 21 (19) | 2.6 ± 0.7 | 21 |
| n2021 | sy341 | ND | 3.0 ± 0.0 (P = 0.0240) | 30 |
| lin-45(sy96) | + | 100 (60) | 1.6 ± 1.1 | 29 |
| sy96 | sy341 | 100 (14) | 2.9 ± 0.2 (P = 0.0028) | 8 |
| mpk-1(ku1) | + | ND | 2.8 ± 0.7 | 22 |
| ku1 | sy341 | ND | 3.0 ± 0 (P = 0.0471) | 37 |
| lin-1(n1790gf) | + | 69 (52) | 2.9 ± 0.4d | 36 |
| n1790 | sy341 | 48 (64) | 3.3 ± 0.5d (P = 0.0019) | 29 |
| lin-1(n1761gf) | + | ND | 2.7 ± 0.4 | 16 |
| n1761 | sy341 | ND | 3.0 ± 0.1 (P = 0.0070) | 22 |
| lin-25(e1446) | + | 100 (136) | 1.4 ± 0.4 | 20 |
| e1446 | sy341 | 100 (88) | 1.6 ± 0.6 (P = 0.2039) | 18 |
| sur-2(ku9) | + | 100 (45) | 0.8 ± 0.6 | 25 |
| ku9 | sy341 | 100 (50) | 1.4 ± 0.7 (P = 0.0028) | 16 |
% Egl, percentage of egg-laying-defective animals; NA, not applicable; ND, not done.
For Egl phenotype the numbers in parentheses represent animals examined. VPC induction and n are defined in Table 1. The P-values of average VPC induction are given in parentheses (the significance of data is defined in Table 2).
The genotype was unc-4(e120) sli-3(sy341); lin-3(n378).
The genotype was dpy-10(e128) sli-3(sy341); lin-3(n378).
The F1 progeny of homozygous let-60(n1876) hermaphrodites die during the L1 stage (Beitel et al. 1990). sy341 does not suppress larval lethality of n1876 animals.
n1790 animals exhibit weak Muv phenotype (8%). The penetrance is higher (44%) in sy341, n1790 double animals.
TABLE 8.
Phenotypic analysis of sli-3 mutants
| Genotype | % viabilitya | %fertilityb | Brood sizec |
|---|---|---|---|
| + | 96 (1226) | 100 (>100) | 252 ± 17 (7) |
| sli-3(sy341) | 95 (179) | 67 (70) | 50 ± 22 (21) |
| lin-3(n378) | 100 (100) | 100 (72) | 73 ± 20 (8) |
| sy341; n378 | 75 (138) | 83 (72) | 27 ± 13 (23) |
| lin-3(n1058) | ND | 6 (16) | 0.1 ± 0.5 (16) |
| sy341; n1058 | ND | 0 (14) | 0 (14) |
| let-23(sy97) | 21 (130) | 95 (22) | ND |
| sy97 sy341 | 29 (115) | 93 (14) | 10 ± 5 (13) |
| let-60(n1876) | 34 (80) | ND | 0 (12) |
| sy341; n1876 | 47 (69) | ND | 0 (6) |
| lin-45(sy96) | 52 (150) | 88 (8) | 3 ± 2 (8) |
| sy341; sy96 | 62 (88) | 85 (20) | 1 ± 1 (6) |
Numbers in parentheses represent animals examined for each phenotype. ND, not done.
Survival was calculated as described in materials and methods.
Calculated as the percentage of hermaphrodites laying fertilized eggs.
The number of progeny that survived to adulthood. Larvae that died during L1 and L2 stages were not counted.
To examine the possibility of sli-3 being a nuclear regulator of let-23-mediated signaling, we examined its interactions with known nuclear factors of the pathway. lin-1 encodes an ETS domain transcription factor that negatively regulates vulval induction (Beitel et al. 1990). We tested interactions of sy341 with two gain-of-function alleles of lin-1, n1761 and n1790, that cause an Egl phenotype due to reduced vulval induction and defective morphology (Beitel et al. 1990; Jacobs et al. 1998). The mutant lin-1 alleles disrupt a conserved MAP kinase docking site in LIN-1, thereby making the altered protein unresponsive to MPK-1-mediated negative regulation (Jacobs et al. 1998). Analysis of the vulval phenotype in double-mutant animals revealed that sli-3(sy341) suppresses the induction defect in both lin-1 alleles (Table 4). The Egl phenotype of n1790 animals is also suppressed by sy341 albeit only partially (Table 4). We also tested genetic interactions with mutations in two other nuclear regulators of the EGF signaling pathway, lin-25 (novel) and sur-2 (transcription mediator complex component). These gene products are essential for vulval induction and specify 1° and 2° cell fates (Singh and Han 1995; Tuck and Greenwald 1995; Stevens et al. 2002). Loss-of-function mutations in lin-25 and sur-2 result in the failure of VPCs to get induced, leading to a fully penetrant Egl phenotype. While sy341 showed no obvious suppression of the VPC induction defect in lin-25(e1446) animals, the suppression of sur-2(ku9) phenotype is statistically significant (Table 4). The enhancement in vulval induction in sur-2(ku9); sli-3(sy341) double-mutant animals is almost entirely due to the higher frequency of the P5.p and P7.p precursors adopting the 2° cell fate (37%, n = 32 in double-mutant animals compared to 14%, n = 50 in ku9 alone, where n stands for the total number of P5.p and P7.p VPCs scored). This is consistent with the analysis of the vulval cell lineages in mutant animals (Table 5). Taken together these results suggest that sli-3 functions most likely either downstream or in parallel to lin-1 and sur-2 in vulval cells.
TABLE 5.
Vulval cell lineages in sli-3 and sur-2 mutants
| VPC
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Genotype | P3.p | P4.p | P5.p | P6.p | P7.p | P8.p | n | ||
| sli-3(sy341) | S/SS | SS | LLTU | TTTT | UTLL | SS | 10 | ||
| sur-2(ku9) | SS | SS | SS | TTOO | UTS | SS | 1 | ||
| SS | SS | SOU | OOOO | SS | SS | 1 | |||
| SS | SS | SS | SS | SS | SS | 3 | |||
| SS | SS | SS | UOO | SS | SS | 2 | |||
| SS | SS | SOU | TTOO | SS | SS | 1 | |||
| SS | S | SS | SOU | TTOO | SS | S | SS | 1 | |
| ku9; sy341 | S | SS | SUUO | TOOT | UOLL | S | SS | 1 | |
| SS | SS | STTD | OOOT | SS | SS | 1 | |||
| SS | SS | LLTU | TTLT | UUU | SS | 1 | |||
| SS | S | SS | LDDU | OTTO | UUU | SS | 1 | ||
| SS | SS | LTUU | UOTO | SS | SS | 1 | |||
| S | SS | UUU | OOTO | UOS | SS | 1 | |||
| SS | SS | SS | SS | SS | SS | 1 | |||
| SS | S | SS | ssTU | ODOT | US | SS | 1 | ||
| S | SS | LUUU | TTTO | ssss | SS | 1 | |||
T, L, and O refer to transverse, longitudinal, and oblique axes of cell divisions of VPC granddaughters, respectively. D, cell division axis was not followed. U, undivided cell. S, syncytium fate after the first round of cell division of VPCs; s, syncytium fate after the second round of cell division of VPCs. sur-2(ku9) animals occasionally have extra VPCs. n, number of animals examined.
sli-3 interacts with negative regulators of the inductive signaling pathway:
Two properties of sli-3(sy341) with regard to vulval induction strongly resemble mutations of previously identified negative regulators sli-1, ark-1, unc-101, and gap-1 (Lee et al. 1994; Jongeward et al. 1995; Hajnal et al. 1997; Hopper et al. 2000). First, loss-of-function alleles of these genes have no effect on vulval induction and, second, mutations in each of these genes can suppress the Vul defect caused by mutations in the let-23 pathway. Given the genetic similarity between sli-3 and these negative regulators, we examined vulval phenotypes in double-mutant animals. sli-3(sy341) shows synergistic interactions with alleles of sli-1, ark-1, unc-101, and gap-1 by giving rise to a multivulva (Muv) phenotype at significantly high frequencies (Table 6). The strongest interaction was observed with gap-1 with 80% of the animals being Muv. The Muv phenotype was the result of ectopic induction in some or all of the P3.p, P4.p, and P8.p vulval precursors. Qualitative analysis of the Muv phenotypes, however, revealed some differences in the genetic interactions. We observed a high frequency of P8.p induction in sli-3; gap-1 (64%, n = 22) and unc-101; sli-3 (48%, n = 21) animals compared to sli-3; ark-1 (11%, n = 27) and sli-3; sli-1 (23%, n = 22) animals. gap-1 was previously shown to preferentially induce P8.p in the background of ark-1 mutation (Hopper et al. 2000). These results reveal functional differences between negative regulators in modulating the competence of different VPCs to respond to inductive signaling. This could help explain why there are so many different regulatory mechanisms.
TABLE 6.
Genetic interaction of sli-3 with other negative regulators
| Negative regulator | sli-3 | % Muv | VPC induction | n |
|---|---|---|---|---|
| + | + | 0 | 3.0 ± 0.0 | >100 |
| + | sy341 | 0 | 3.0 ± 0.0 | 50 |
| sli-1(sy143) | + | 0 | 3.0 ± 0.0 | 30 |
| sy143 | sy341 | 54 | 3.4 ± 0.4 | 22 |
| ark-1(sy247) | + | 6 | 3.0 ± 0.1 | 31 |
| sy247 | sy341 | 29 | 3.2 ± 0.4 | 51 |
| unc-101(sy108) | + | 3 | 3.0 ± 0.1 | 30 |
| sy108 | sy341 | 52 | 3.4 ± 0.4 | 21 |
| gap-1(n1691) | + | 0 | 3.0 ± 0.0 | 25 |
| n1691 | sy341 | 80 | 4.0 ± 0.7 | 15 |
The Muv phenotype and VPC induction were examined in L4 stage animals under a Nomarski microscope. Data for the ark-1(sy247); sli-3(sy341) double were pooled from two different genotypes, dpy-10(e128) sli-3(sy341); ark-1(sy247) unc-31(e169) (average induction = 3.2, Muv = 25%; n = 24) and unc-4(e120) sli-3(sy341); dpy-20(e1282) ark-1(sy247) (average induction = 3.2, Muv = 33%; n = 27). VPC induction and n are defined in Table 1.
In addition to the negative regulators discussed above, synthetic multivulva (SynMuv) genes are also known to inhibit vulval induction (Ferguson and Horvitz 1989). These genes are divided into three functionally redundant classes (A, B, and C) that negatively regulate the fate of VPCs. We examined genetic interactions of sli-3(sy341) with class A and class B lin-15 alleles, n767 and n744, respectively. In each case >10 worms were examined but no Muv phenotype was observed. Thus sli-3 appears to function in a genetic pathway separate from that mediated by lin-15A and lin-15B SynMuv genes during vulval induction.
sli-3 does not interact with components of lin-12/Notch and Wnt signaling pathways:
In addition to LIN-3–LET-23-mediated inductive signaling, LIN-12-mediated lateral signaling also plays a crucial role in vulval induction (Greenwald 2005). Since our genetic experiments have demonstrated that sli-3 acts as a negative regulator in both 1° and 2° lineage cells, we examined its interaction with lin-12. We used lin-12(n137n460), a cold-sensitive allele of lin-12, which when grown at ≤20° gives rise to a fully penetrant Muv phenotype (Greenwald et al. 1983) (Table 7). The phenotype becomes progressively weaker at higher temperatures. Hence, while at 22° 63% (n = 155) of animals exhibit a Muv phenotype, at 25° lin-12(n137n460) animals are almost all wild type (3% Muv, see Table 7). We examined sli-3(sy341); lin-12(n137n460) double-mutant animals at 22° and 25° and found no enhancement in the Muv phenotype compared to the lin-12(n137n460) alone (Table 7). Thus sli-3 is not likely to play a major role in lin-12 signaling during establishment of the 2° lineage vulval fates.
TABLE 7.
VPC induction in sli-3 and lin-12 mutants
| Growth temperature | lin-12 | sli-3 | % Muv | VPC induction |
|---|---|---|---|---|
| 15° | n137n460 | + | 99 (167) | 5.0 ± 0.7 (23) |
| 20° | n137n460 | + | 99 (139) | 4.8 ± 0.7 (20) |
| 22° | n137n460 | + | 63 (155) | 3.4 ± 0.5 (24) |
| + | sy341 | 0 (20) | 3.0 ± 0.0 (20) | |
| n137n460 | sy341 | 50 (106) | 3.9 ± 0.6 (22), P = 0.0254 | |
| 25° | n137n460 | + | 3 (144) | 3.0 ± 0.0 (30) |
| + | sy341 | 0 (19) | 3.0 ± 0.0 (19) | |
| n137n460 | sy341 | 4 (57) | 3.2 ± 0.4 (18), P = 0.0206 |
The Muv phenotype of the lin-12 cold-sensitive allele, n137n460, varies with growth temperature. At 15° almost all animals exhibit Muv phenotype whereas at 25° rare animals do so. The phenotype was scored at plate level by looking for pseudovulvae (multiple ventral protrusions) in adult animals. Numbers in parentheses represent animals examined. The P-values are given where appropriate (the significance of data is defined in Table 2).
We also examined genetic interactions of sli-3 with two Wnt pathway components bar-1/β-catenin and pry-1/axin (Eisenmann et al. 1998; Eisenmann and Kim 2000; Gleason et al. 2002; Korswagen et al. 2002; Eisenmann 2005). Loss-of-function mutations in bar-1 frequently cause cell fusion and induction defects in VPCs due to decreased LIN-39 activity (Eisenmann et al. 1998; Gleason et al. 2002). We examined vulval cells in sli-3(sy341); bar-1(ga80) double-mutant animals and observed a statistically significant increase in the number of induced VPCs (average induction 2.8 ± 0.4, n = 39 compared to 2.2 ± 0.7, n = 41 in bar-1(ga80) animals alone, P < 0.0001). This increase was accompanied by a suppression of the cell fusion defect in P5.p, P6.p, and P7.p (31, 2, and 14% suppression, respectively), suggesting that once the presumptive precursors are prevented from fusing, they are likely to get induced. Since the cell fusion defect in the bar-1 mutant can be suppressed by elevated activity of the LET-60/Ras pathway (Eisenmann et al. 1998), we interpret this as an indirect effect of increased LET-60 pathway activity. In addition to reduced vulval induction, bar-1(ga80) animals also exhibit a P12 to P11 transformation defect (Eisenmann and Kim 2000). The penetrance of this phenotype is slightly reduced in a sli-3(sy341) genetic background [63%, n = 39 in double-mutant animals compared to 88%, n = 41 in bar-1(ga80) alone]. In contrast to bar-1(lf) animals that are Vul, hypomorphic alleles of pry-1 (e.g., mu38) exhibit a Muv phenotype (Gleason et al. 2002). We examined genetic interaction between sli-3(sy341) and pry-1(mu38) (at 22°) and found that Muv penetrance in sy341, mu38 double-mutant animals (36%, n = 39) is not significantly different from that in mu38 alone (28%, n = 46). Thus these results argue that sli-3 is not a component of the Wnt signaling pathway during vulval induction.
sli-3 does not participate in the IP3R-mediated let-23 fertility pathway:
Since sli-3 is a regulator of let-23-mediated EGF signaling in vulval cells, we examined whether the same regulation might also occur in ovulation. In wild-type animals, ovulation depends upon the contraction of the gonadal sheath and requires IP3-mediated calcium release (Clandinin et al. 1998; McCarter et al. 1999). The n1058 allele of lin-3 causes a fully penetrant sterile phenotype and partial defect in vulval induction (Tables 4 and 8). The sterility defect is due to the failure of the spermatheca to dilate correctly, thereby causing a defect in ovulation (Clandinin et al. 1998; Yin et al. 2004). We found that sli-3(sy341); lin-3(n1058) double-mutant animals are suppressed for the Vul defect but not the sterility (Tables 4 and 8). Hence, sli-3 appears to regulate only a subset of lin-3 functions. Alternatively, instead of being a negative regulator, sli-3 may act positively in the fertility pathway. This would still be consistent with the observation that sli-3(sy341) animals have a reduced brood size and exhibit defective morphology of the oocytes (Table 8 and Figure 2; also see materials and methods). We thus examined genetic interaction of sli-3 with gain-of-function mutations of itr-1, which encodes the IP3 receptor and functions as an effector of LET-23 to control ovulation through dilation of the adult spermatheca (Clandinin et al. 1998). Mutations in itr-1 suppress fertility defects in lin-3(n1058) animals, possibly by promoting ovulation (Clandinin et al. 1998). The brood size of sli-3(sy341); itr-1(sy290) double-mutant animals (32 ± 14; n = 12) is not significantly different from that of sy341 alone (50 ± 22; n = 21), suggesting that sli-3 does not function in the IP3-receptor-mediated let-23 fertility pathway.
Figure 2.—
sli-3(sy341) adult hermaphrodites exhibit defects in the morphology of oocytes and embryos. The arrows point to vulval opening. The spermatheca (Sp), embryos (E), and oocytes (Oo) are marked. In a wild-type animal (top), oocytes and embryos are aligned in a linear fashion. The sy341 animal (bottom), on the other hand, exhibits no such arrangement. Bar, 30 μm.
DISCUSSION
The lin-3–let-23 signaling pathway plays a central role in the development of the vulva in C. elegans hermaphrodites (Moghal and Sternberg 2003b; Sternberg 2005). To identify additional genes that interact with this pathway and control vulval development, we carried out a genetic screen using a heteroallelic combination of two lin-3 mutations, n378 and n1059. In this article, we report the isolation of three suppressor loci and present a detailed analysis of one of these, sli-3. Our results demonstrate that sli-3 is likely to function as a nuclear regulator of the LET-23/EGFR signaling pathway. The two other suppressors sli-4 and sli-5 are allelic to lin-3 and gap-1, respectively.
Overview of the suppressor screen:
Genetic screens in C. elegans have led to the successful identification of let-23 pathway components that control vulval induction (Sternberg 2005). Mutations in the core pathway genes were initially isolated on the basis of an Egl phenotype. These initial studies were followed by suppressor screens that identified additional pathway components as well as those that modulate pathway activity (e.g., positive and negative regulators). Our screening strategy was similar to many others carried out in the past, except that, in contrast to the previous screens that involved alleles of let-23, let-60, lin-10, and lin-15 (Beitel et al. 1990; Han et al. 1990, 1993; Hajnal et al. 1997; Moghal and Sternberg 2003b), we used alleles of lin-3.
We recovered a new locus sli-3, identified by a suppressor of the lin-3 Vul phenotype, that functions as a negative regulator of vulval induction. Several properties of sli-3 closely resemble those of previously identified negative regulators sli-1, ark-1, unc-101, and gap-1 (Moghal and Sternberg 2003b). Mutations in any of these genes alone do not exhibit a visible vulval defect but double-mutant combinations show synergistic interactions resulting in a Muv phenotype (Sternberg 2005; Sundaram 2005). Except for gap-1, our genetic screen did not recover alleles of known negative regulators, perhaps due to the smaller number of genomes screened. Compared to the previous genetic screens that were carried out at a larger scale (>100,000 haploid genome sets in the case of sli-1 and gap-1 in standard F2 generation screens and ∼25,000 in the case of the unc-101 synthetic enhancement screen), we screened fewer animals (∼30,000; see materials and methods). The fact that we recovered only one allele of sli-3 also supports this possibility. However, we cannot rule out an intrinsic bias in our screen toward recovering alleles of certain genes. Our genetic epistasis experiments have demonstrated that compared to other negative regulators that function either at the level of the LET-23 receptor (SLI-1, ARK-1, and UNC-101) or at that of LET-60/Ras (GAP-1) (Lee et al. 1994; Jongeward et al. 1995; Hopper et al. 2000), SLI-3 is the most downstream acting negative regulator of vulval induction identified thus far in C. elegans.
In addition to the alleles of sli-3 and sli-5, two extragenic suppressor loci, we also recovered sli-4 mutants that are allelic to lin-3. Our initial mapping experiments had suggested that sli-4 defines a new locus close to lin-3. This was due to the presence of an unidentified lethal mutation that, similar to lin-3(n1059), causes embryonic/early larval stage lethality. Since we relied upon the lin-3(n1059) larval lethality phenotype to map sli-4 (see materials and methods), the three-factor mapping experiments led us to conclude that sli-4 is genetically separate from lin-3. We do not know whether the lethal mutation arose spontaneously or due to the EMS treatment. The preliminary mapping experiments indicated tight linkage to lin-3 (∼1.8 MU to the left of lin-3).
SLI-3 is a tissue-specific regulator of the LET-23/EGFR signaling pathway:
The let-23 EGF receptor system is required for the development of multiple tissues in C. elegans (Moghal and Sternberg 2003b). Among at least five different roles of the pathway components identified so far, four (viability, vulval induction, P12 cell, and male tail spicule developments) are regulated by let-60/ras, whereas the fertility process utilizes IP3-mediated Ca2+ signaling. The presence of common let-23 signaling pathway components in multiple developmental processes suggests that functional specificity is likely to be mediated by tissue-specific downstream effectors/regulators. If so, carefully designed genetic screens should be able to identify such tissue-specific components. Our analysis of sli-3 function has revealed that sli-3 specifically functions in the let-23-mediated vulval induction pathway. Thus, the sli-3 mutation does not suppress nonvulval defects associated with let-23 pathway genes. The reduced viability and fertility defects in hypomorphic alleles of let-23, let-60, and lin-45 are significantly not altered in a sy341 background (Table 8). Furthermore, the P12 fate specification defect in let-23(sy97) animals is also not suppressed (data not shown).
The genetic analysis of let-23-mediated signaling in hermaphrodite fertility has revealed that some of the pathway components function in two distinct processes: germline development (cell cycle progression) and ovulation (spermathecal contraction). While let-60, mpk-1, and mek-1 play crucial roles in germline development (Church et al. 1995), genes such as lin-3, let-23, itr-1, and lfe-2 are involved in the ovulation process (Clandinin et al. 1998). Mutations in the former set of genes cause sterility due to the arrest of germ cell nuclei in the pachytene stage. On the other hand, mutations in the latter set of genes disrupt dilation of the spermatheca, leading to an ovulation defect. To determine whether sli-3 participates in any one or both of these processes, we tested its requirement in germline development and in ovulation by examining phenotypes of the mutant animals as well as by genetic interaction studies. Three experiments suggest a distinct role of sli-3 in hermaphrodite fertility. First, sy341 animals show neither an ovulation defect nor a diakinesis-stage arrest of germ cells (see materials and methods). Second, sli-3 shows no discernible genetic interaction with itr-1, suggesting that sli-3 is not a component of IP3 signaling during oocyte maturation and ovulation. Third, sli-3(sy341) does not enhance fertility defects of EGF pathway mutants (Table 8). Since sy341 hermaphrodites have morphologically defective oocytes and the sterility defect in mutant animals can be partially rescued by mating with wild-type males (data not shown), sli-3 is likely to play a role in gametogenesis.
sli-3 is a let-23/EGFR pathway-specific effector in vulval cells:
In C. elegans, vulval development is controlled by three evolutionarily conserved signaling pathways, namely LET-23/EGFR, LIN-12/Notch, and Wnt. Given that these pathways are also required for the development and patterning of other tissues, their specific responses are likely to be mediated by pathway components that possess cell- and/or tissue-specific activities. Our experiments have revealed that sli-3 negatively regulates the LET-23/EGFR signaling pathway in the vulva. In the absence of any other mutation, sli-3(sy341) animals exhibit wild-type vulval induction, suggesting that sli-3 is dispensable for normal development. The gene dosage studies reveal that the suppression of the lin-3(n378) VPC induction defect by sy341 can be ranked as follows (starting from the highest): sy341/sy341 > sy341/Df > sy341/sy341/+(mnDp34) ≈ sy341/+. This is most consistent with sy341 being a hypomorph.
Our epistasis experiments have revealed that the sli-3 mutation suppresses Vul defects caused by viable hypomorphic alleles of let-23, let-60, lin-45, and mpk-1. Similar epistasis experiments with nuclear targets of the let-23 signaling pathway have revealed that the sli-3 mutation can suppress the Vul defect caused by gain-of-function alleles of lin-1 and a severe hypomorphic allele of sur-2, genes that encode ETS domain protein and a component of the transcription mediator complex, respectively (Beitel et al. 1995; Singh and Han 1995; Stevens et al. 2002). These results indicate that sli-3 most likely functions in the nucleus either downstream or in parallel to lin-1 and sur-2 to specify 1° and 2° fates to VPCs. Furthermore, instead of being a major target of the let-23 pathway, sli-3 defines a regulatory branch of the signaling. The lineage analysis of sur-2 and sli-3 double-mutant animals has revealed a higher frequency of 2° cell fate specification (see Table 5). Since SUR-2 facilitates the crosstalk between the inductive and lateral signaling pathways to specify 2° fates to P5.p and P7.p (Shaye and Greenwald 2002), one possibility could be that SLI-3 mediates SUR-2 function during this specification process. In addition, SLI-3 may be regulated by LIN-1 to confer 1° fates on the VPCs. The molecular identity of sli-3 will help test these hypotheses and its precise mechanism of function in vulval cells.
To determine whether sli-3 is a pathway-specific effector, we examined its interactions with genes that encode components of LIN-12/Notch and Wnt signaling pathways. We found that sli-3 does not show strong genetic interactions with the lin-12, bar-1, and pry-1 alleles tested, suggesting that it may not function as a common regulator of multiple signaling pathways. The involvement of sli-3 in mediating let-23 pathway function in the vulva demonstrates that similar mechanisms are likely to exist in other tissues to provide specificity to let-60-mediated EGF signaling in C. elegans.
Acknowledgments
We thank Neil Hopper, Chieh Chang, Giovanni Lisa, Maureen Barr, and Minqin Wang for insights in this project. We also thank the anonymous reviewers for helpful comments on a draft of this article. This work was supported by the U.S. Public Health Service (USHS) grant HD23690 to P.W.S., the Canada Research Chair and McMaster research funds to B.P.G., and National Institutes of Health grant R01 GM073184 to N.M. Additional support was provided by the Howard Hughes Medical Institute, with which P.W.S. is an investigator and B.J.H. and B.P.G. were associates. J.L. was supported by a U.S.H.S. training grant. B.P.G. was a postdoctoral fellow of the Human Frontier Science Foundation. N.M. was a fellow of the California Breast Cancer Research Foundation. Some strains were provided by the Caenorhabditis Genetics Center.
References
- Aroian, R. V., and P. W. Sternberg, 1991. Multiple functions of let-23, a Caenorhabditis elegans receptor tyrosine kinase gene required for vulval induction. Genetics 128: 251–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beitel, G. J., S. G. Clark and H. R. Horvitz, 1990. Caenorhabditis elegans ras gene let-60 acts as a switch in the pathway of vulval induction. Nature 348: 503–509. [DOI] [PubMed] [Google Scholar]
- Beitel, G. J., S. Tuck, I. Greenwald and H. R. Horvitz, 1995. The Caenorhabditis elegans gene lin-1 encodes an ETS-domain protein and defines a branch of the vulval induction pathway. Genes Dev. 9: 3149–3162. [DOI] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, S., V. Kodoyianni, M. Bosenberg, L. Friedman and J. Kimble, 1996. lag-1, a gene required for lin-12 and glp-1 signaling in Caenorhabditis elegans, is homologous to human CBF1 and Drosophila Su(H). Development 122: 1373–1383. [DOI] [PubMed] [Google Scholar]
- Church, D. L., K. L. Guan and E. J. Lambie, 1995. Three genes of the MAP kinase cascade, mek-2, mpk-1/sur-1 and let-60 ras, are required for meiotic cell cycle progression in Caenorhabditis elegans. Development 121: 2525–2535. [DOI] [PubMed] [Google Scholar]
- Clandinin, T. R., J. A. DeModena and P. W. Sternberg, 1998. Inositol trisphosphate mediates a RAS-independent response to LET-23 receptor tyrosine kinase activation in C. elegans. Cell 92: 523–533. [DOI] [PubMed] [Google Scholar]
- Clark, D. V., and D. L. Baillie, 1992. Genetic analysis and complementation by germ-line transformation of lethal mutations in the unc-22 IV region of Caenorhabditis elegans. Mol. Gen. Genet. 232: 97–105. [DOI] [PubMed] [Google Scholar]
- Clark, D. V., T. M. Rogalski, L. M. Donati and D. L. Baillie, 1988. The unc-22(IV) region of Caenorhabditis elegans: genetic analysis of lethal mutations. Genetics 119: 345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann, D. M., 2005. Wnt signaling (January 1, 2006), pp. 1–17 in WormBook, edited by I. Greenwald (http://www.wormbook.org).
- Eisenmann, D. M., and S. K. Kim, 2000. Protruding vulva mutants identify novel loci and Wnt signaling factors that function during Caenorhabditis elegans vulva development. Genetics 156: 1097–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann, D. M., J. N. Maloof, J. S. Simske, C. Kenyon and S. K. Kim, 1998. The β-catenin homolog BAR-1 and LET-60 Ras coordinately regulate the Hox gene lin-39 during Caenorhabditis elegans vulval development. Development 125: 3667–3680. [DOI] [PubMed] [Google Scholar]
- Ferguson, E. L., and H. R. Horvitz, 1985. Identification and characterization of 22 genes that affect the vulval cell lineages of the nematode Caenorhabditis elegans. Genetics 110: 17–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson, E. L., and H. R. Horvitz, 1989. The multivulva phenotype of certain Caenorhabditis elegans mutants results from defects in two functionally redundant pathways. Genetics 123: 109–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garriga, G., C. Desai and H. R. Horvitz, 1993. Cell interactions control the direction of outgrowth, branching and fasciculation of the HSN axons of Caenorhabditis elegans. Development 117: 1071–1087. [DOI] [PubMed] [Google Scholar]
- Gleason, J. E., H. C. Korswagen and D. M. Eisenmann, 2002. Activation of Wnt signaling bypasses the requirement for RTK/Ras signaling during C. elegans vulval induction. Genes Dev. 16: 1281–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald, I., 2005. LIN-12/Notch signaling in C. elegans (August 4, 2005), pp. 1–15 in WormBook, edited by L. R. Girard (www.wormbook.org).
- Greenwald, I. S., P. W. Sternberg and H. R. Horvitz, 1983. The lin-12 locus specifies cell fates in Caenorhabditis elegans. Cell 34: 435–444. [DOI] [PubMed] [Google Scholar]
- Hajnal, A., C. W. Whitfield and S. K. Kim, 1997. Inhibition of Caenorhabditis elegans vulval induction by gap-1 and by let-23 receptor tyrosine kinase. Genes Dev. 11: 2715–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, M., R. V. Aroian and P. W. Sternberg, 1990. The let-60 locus controls the switch between vulval and nonvulval cell fates in Caenorhabditis elegans. Genetics 126: 899–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, M., A. Golden, Y. Han and P. W. Sternberg, 1993. C. elegans lin-45 raf gene participates in let-60 ras-stimulated vulval differentiation. Nature 363: 133–140. [DOI] [PubMed] [Google Scholar]
- Herman, R. K., 1978. Crossover suppressors and balanced recessive lethals in Caenorhabditis elegans. Genetics 88: 49–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman, R. K., J. E. Madl and C. K. Kari, 1979. Duplications in Caenorhabditis elegans. Genetics 92: 419–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin, J., H. R. Horvitz and S. Brenner, 1979. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics 91: 67–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper, N. A., J. Lee and P. W. Sternberg, 2000. ARK-1 inhibits EGFR signaling in C. elegans. Mol. Cell 6: 65–75. [PubMed] [Google Scholar]
- Horvitz, H. R., S. Brenner, J. Hodgkin and R. K. Herman, 1979. A uniform genetic nomenclature for the nematode Caenorhabditis elegans. Mol. Gen. Genet. 175: 129–133. [DOI] [PubMed] [Google Scholar]
- Jacobs, D., G. J. Beitel, S. G. Clark, H. R. Horvitz and K. Kornfeld, 1998. Gain-of-function mutations in the Caenorhabditis elegans lin-1 ETS gene identify a C-terminal regulatory domain phosphorylated by ERK MAP kinase. Genetics 149: 1809–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongeward, G. D., T. R. Clandinin and P. W. Sternberg, 1995. sli-1, a negative regulator of let-23-mediated signaling in C. elegans. Genetics 139: 1553–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korswagen, H. C., D. Y. Coudreuse, M. C. Betist, S. van de Water, D. Zivkovic et al., 2002. The Axin-like protein PRY-1 is a negative regulator of a canonical Wnt pathway in C. elegans. Genes Dev. 16: 1291–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J., G. D. Jongeward and P. W. Sternberg, 1994. unc-101, a gene required for many aspects of Caenorhabditis elegans development and behavior, encodes a clathrin-associated protein. Genes Dev. 8: 60–73. [DOI] [PubMed] [Google Scholar]
- Maloof, J. N., J. Whangbo, J. M. Harris, G. D. Jongeward and C. Kenyon, 1999. A Wnt signaling pathway controls hox gene expression and neuroblast migration in C. elegans. Development 126: 37–49. [DOI] [PubMed] [Google Scholar]
- McCarter, J., B. Bartlett, T. Dang and T. Schedl, 1999. On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev. Biol. 205: 111–128. [DOI] [PubMed] [Google Scholar]
- Moerman, D. G., and D. L. Baillie, 1979. Genetic organization in Caenorhabditis elegans: fine-structure analysis of the unc-22 gene. Genetics 91: 95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghal, N., and P. W. Sternberg, 2003. a A component of the transcriptional mediator complex inhibits RAS-dependent vulval fate specification in C. elegans. Development 130: 57–69. [DOI] [PubMed] [Google Scholar]
- Moghal, N., and P. W. Sternberg, 2003. b The epidermal growth factor system in Caenorhabditis elegans. Exp. Cell Res. 284: 150–159. [DOI] [PubMed] [Google Scholar]
- Shaye, D. D., and I. Greenwald, 2002. Endocytosis-mediated downregulation of LIN-12/Notch upon Ras activation in Caenorhabditis elegans. Nature 420: 686–690. [DOI] [PubMed] [Google Scholar]
- Sigurdson, D. C., G. J. Spanier and R. K. Herman, 1984. Caenorhabditis elegans deficiency mapping. Genetics 108: 331–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, N., and M. Han, 1995. sur-2, a novel gene, functions late in the let-60 ras-mediated signaling pathway during Caenorhabditis elegans vulval induction. Genes Dev. 9: 2251–2265. [DOI] [PubMed] [Google Scholar]
- Sternberg, P. W., 2005. Vulval development (June 25, 2005), pp. 1–28 in Wormbook, edited by B. J. Meyer (http://www.wormbook.org). [DOI] [PMC free article] [PubMed]
- Stevens, J. L., G. T. Cantin, G. Wang, A. Shevchenko, A. Shevchenko et al., 2002. Transcription control by E1A and MAP kinase pathway via Sur2 mediator subunit. Science 296: 755–758. [DOI] [PubMed] [Google Scholar]
- Sulston, J. E., and H. R. Horvitz, 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56: 110–156. [DOI] [PubMed] [Google Scholar]
- Sundaram, M., 2005. RTKRas/MAP kinase signaling, pp. 1–19 in Wormbook (http://www.wormbook.org).
- Tuck, S., and I. Greenwald, 1995. lin-25, a gene required for vulval induction in Caenorhabditis elegans. Genes Dev. 9: 341–357. [DOI] [PubMed] [Google Scholar]
- Yin, X., N. J. Gower, H. A. Baylis and K. Strange, 2004. Inositol 1,4,5-trisphosphate signaling regulates rhythmic contractile activity of myoepithelial sheath cells in Caenorhabditis elegans. Mol. Biol. Cell 15: 3938–3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, C. H., J. Lee, G. D. Jongeward and P. W. Sternberg, 1995. Similarity of sli-1, a regulator of vulval development in C. elegans, to the mammalian proto-oncogene c–cbl. Science 269: 1102–1105. [DOI] [PubMed] [Google Scholar]
- Zengel, J. M., and H. F. Epstein, 1980. Identification of genetic elements associated with muscle structure in the nematode Caenorhabditis elegans. Cell Motil. 1: 73–97. [DOI] [PubMed] [Google Scholar]