Abstract
The ethylene receptor family of Arabidopsis consists of five members, one of these being ETR1. The effect of ethylene pathway mutations upon expression of ETR1 was examined. For this purpose, ETR1 levels were quantified in mutant backgrounds containing receptor loss-of-function mutations, ethylene-insensitive mutations, and constitutive ethylene response mutations. Ethylene-insensitive mutations of ETR1 resulted in a posttranscriptional increase in levels of the mutant receptor. Treatment of seedlings with silver, which leads to ethylene insensitivity, also resulted in an increase in levels of ETR1. Loss-of-function mutations of ETR1 resulted in both transcriptional and posttranscriptional changes in levels of the receptor. Most other ethylene pathway mutations, including a newly isolated T-DNA insertion mutation in the gene encoding the ethylene receptor ERS1, had relatively minor effects upon the expression of ETR1. Our results indicate that mutations in ETR1 can affect expression at the posttranscriptional level, and suggest that these posttranscriptional changes may contribute to the phenotypes observed in the mutants. Our results also refine the model on how mutations in ethylene receptors are able to confer dominant ethylene insensitivity upon plants.
Ethylene (C2H4) is a simple gaseous hydrocarbon that has profound effects upon plant growth and development. Ethylene regulates seed germination, seedling growth, leaf and petal abscission, organ senescence, ripening, stress responses, and pathogen responses (Mattoo and Suttle, 1991; Abeles et al., 1992). An important contribution to our understanding of ethylene signal transduction has come from the identification of mutants in Arabidopsis with altered ethylene sensitivity (Chang and Shockey, 1999; Stepanova and Ecker, 2000). These mutations fall into two main classes: (a) mutations that render a plant insensitive to ethylene, and (b) mutations that result in a constitutive ethylene response. Characterization of Arabidopsis mutants has led to the identification of ethylene receptors and additional components in the ethylene signal transduction pathway.
The ethylene receptor family of Arabidopsis contains five members (ETR1, ETR2, ERS1, ERS2, and EIN4; Schaller, 2000; Chang and Stadler, 2001), with ethylene binding confirmed for ETR1 and ERS1 (Schaller and Bleecker, 1995; Rodriguez et al., 1999; Hall et al., 2000). The receptors contain three N-terminal transmembrane domains that encompass the ethylene-binding site (Schaller and Bleecker, 1995; Rodriguez et al., 1999). The binding site contains a copper cofactor that is required for the high-affinity ethylene binding that receptors display (Rodriguez et al., 1999). In the C-terminal half, the receptors contain regions with similarity to His kinases and, in some cases, the receiver domains of response regulators (Schaller, 2000; Chang and Stadler, 2001), signaling elements originally identified as parts of bacterial two-component systems (Parkinson, 1993; Schaller, 2000). His kinase activity has been confirmed in vitro for ETR1 (Gamble et al., 1998), but the role of this activity in signal output is still unclear (Gamble et al., 2002).
Mutations in the ethylene receptors can result in ethylene insensitivity or constitutive ethylene responses, dependent on the nature of the mutation. Ethylene insensitivity can result from single amino acid changes within the region of the receptor involved in ethylene binding (Chang et al., 1993; Hua et al., 1995, 1998; Sakai et al., 1998). Evidence indicates that these gain-of-function mutations either disrupt ethylene binding or uncouple ethylene binding from signal output (Schaller and Bleecker, 1995; Hall et al., 1999; Rodriguez et al., 1999). For example, the etr1-1 mutation abolishes the ability of the receptor to coordinate the copper cofactor, and as a consequence, eliminates ethylene binding (Rodriguez et al., 1999). The ethylene-insensitive mutations are dominant and a single mutation in any one of the five family members can confer ethylene insensitivity upon the plant.
Loss-of-function mutations have been identified in four of five members of the ethylene receptor family (Hua and Meyerowitz, 1998). Single loss-of-function mutations have little or no effect upon ethylene signal transduction. However, in combination with the ETR1 loss-of-function mutation, the mutants show constitutive ethylene responses and this effect is most pronounced in triple and quadruple loss-of-function mutations (Hua and Meyerowitz, 1998). These results indicate that there is functional overlap among the receptor family members. These results also indicate that the receptors serve as negative regulators of the ethylene response pathway because elimination of receptors activates ethylene responses. According to this model for negative regulation, wild-type ethylene receptors actively repress ethylene responses in the air. In the presence of ethylene, wild-type receptors switch to a signaling inactive state that allows for induction of ethylene responses. Ethylene-insensitive mutant receptors, such as etr1-1, are apparently locked into the signaling state that they have in air, such that they repress ethylene responses even in the presence of ethylene (Bleecker, 1999).
Additional elements involved in ethylene signal transduction have also been identified by mutational analysis in Arabidopsis. RAN1 is a copper-transporting ATPase apparently required for addition of the copper cofactor to the ethylene receptors (Hirayama et al., 1999; Woeste and Kieber, 2000). Mutations in RAN1 alter ethylene signal transduction, a loss-of-function mutation resulting in a constitutive ethylene response. CTR1, EIN2, and EIN3 are all thought to act in the same primary response pathway and act downstream of the ethylene receptors. CTR1 belongs to the Raf family of protein Ser/Thr kinases that initiate mitogen-activated protein kinase cascades in eukaryotes (Kieber et al., 1993) and has been shown capable of physical interaction with the ethylene receptors ETR1 and ERS1 (Clark et al., 1998). Loss-of-function mutations in CTR1 result in constitutive ethylene responses (Kieber et al., 1993). EIN2 is an integral membrane protein with similarity to the Nramp family of metal ion transporters (Alonso et al., 1999). Loss-of-function mutations in EIN2 result in ethylene insensitivity. EIN3 belongs to a family of transcription factors that are directly activated by the ethylene signal transduction system and are required for ethylene-dependent gene induction (Chao et al., 1997). Loss-of-function mutations in EIN3 render a plant ethylene insensitive.
Here, we analyze the effect of ethylene pathway mutations upon expression of the ethylene receptor ETR1. This analysis was facilitated by the following: (a) the availability of a number of mutations within the receptor itself, thereby providing independent verification for effects of these mutations; (b) the availability of an antibody against ETR1, thereby allowing for analysis at the protein level; and (c) a detectable basal level of expression for ETR1, thereby allowing increases and decreases in expression to be determined. Our results lend insight into how ethylene receptor mutations affect expression and indicate that mutations within ETR1 can result in posttranscriptional changes in its own expression level. Our results also lend insight into the mechanism by which mutations within the receptors can lead to dominant ethylene insensitivity.
RESULTS
Effect of Ethylene Insensitivity Conferring Mutations upon Expression of ETR1
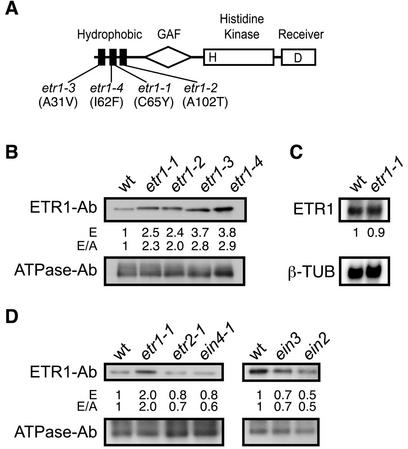
Four dominant mutations have been isolated in ETR1 that confer ethylene insensitivity on plants. These mutations, designated etr1-1, etr1-2, etr1-3, and etr1-4, all result in single amino acid changes within the hydrophobic domain of ETR1 that has been implicated in ethylene binding (Fig. 1A; Chang et al., 1993). The etr1-1, etr1-3, and etr1-4 mutations either reduce or eliminate ethylene binding (Hall et al., 1999). The etr1-2 mutation does not disrupt ethylene binding, but apparently uncouples ethylene binding from signal output (Hall et al., 1999). Based on immunoblot analysis, the protein levels of the mutant receptors etr1-1, etr1-2, etr1-3, and etr1-4 were all approximately 2- to 3-fold higher than that of the wild-type receptor ETR1 when analyzed in etiolated seedlings (Fig. 1B). To determine if the effect upon expression occurred at the transcriptional or posttranscriptional level, transcript levels of the receptor were determined by northern blot in both wild-type and etr1-1 backgrounds (Fig. 1C). No difference in transcript levels was found for the receptor between wild type and etr1-1. However, as previously observed (Fig. 1B), we did find that the etr1-1 protein was present at 2-fold higher levels than the ETR1 protein when analyzed by immunoblot using a portion of the same plant material examined by northern blot (results not shown). Thus, the increase in expression of ethylene-insensitive mutations of ETR1 occurs at the posttranscriptional level.
Figure 1.
Effect of ethylene-insensitive mutations upon expression of ETR1. A, Structure of ETR1 and position of ethylene-insensitive mutations. The hydrophobic ethylene-sensing domain, the GAF domain, the His kinase domain, and the receiver domain are indicated. The letters H and D indicate putative phosphorylation sites. B, Immunoblot analysis of wild-type and ethylene-insensitive mutants of ETR1. Etiolated seedlings were grown for 4 d, and the level of immunodetectable full-length receptor then determined from 10 μg of membrane proteins using an antibody directed against ETR1. Expression levels were quantified densitometrically (E) and also normalized against immunologically determined levels of the H+-ATPase (E/A) as an internal control. C, Northern-blot analysis of mRNA obtained from wild-type and etr1-1 seedlings. Blots were probed with an ETR1 probe and a β-tubulin gene probe as an internal control. The numbers represent the expression level of the ethylene receptor gene after normalization for the level of β-tubulin expression. D, Immunoblot analysis of ETR1 levels in additional ethylene-insensitive mutant backgrounds.
To determine if increased expression of the receptor was restricted to mutant lesions in ETR1 or was a general feature of ethylene insensitivity in Arabidopsis, we examined other ethylene-insensitive mutations. Seedlings were examined that contained dominant ethylene-insensitive mutations in other ethylene receptors (etr2-1 and ein4-1). Seedlings were also examined that contained ethylene-insensitive mutations in the downstream ethylene signaling components EIN2 and EIN3. The expression level of ETR1 based on immunoblot in these other mutant backgrounds was comparable with or less than that found in the wild-type background (Fig. 1D). Thus, the increased expression of ethylene-insensitive mutants of ETR1 is restricted to those lesions present in ETR1 itself, rather than being a general feature of ethylene-insensitive mutations.
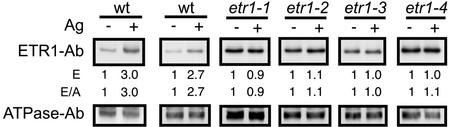
Some chemical compounds are able to induce ethylene insensitivity in plants by interacting with the ethylene receptors. Silver is thought to replace the copper cofactor present in the ethylene-binding site of the receptor. Receptors containing silver are still able to bind ethylene but the binding site is apparently perturbed such that ethylene binding is uncoupled from signal output (Rodriguez et al., 1999). We hypothesized that binding of silver by an ethylene receptor might mimic the effect of an ethylene-insensitive mutation in that receptor, and result in an increased expression level of the receptor. Consistent with this hypothesis, we observed that wild-type seedlings treated with 10 μg mL−1 silver nitrate had higher levels of ETR1 than control untreated seedlings based upon immunoblot analysis (Fig. 2). The stimulatory effect of silver upon expression was lacking with ethylene-insensitive mutations of ETR1 (Fig. 2). This supports the hypothesis that silver mimics the effect of the ethylene-insensitive mutation because there is no additive effect of silver on expression of the ethylene-insensitive mutants.
Figure 2.
Effect of silver treatment upon expression of ETR1. Wild-type and etr1 mutant seedlings were grown in the presence or absence of 10 μg mL−1 silver nitrate (Ag). Immunoblot analysis was then performed using antibodies directed against ETR1 and the H+-ATPase as an internal control on 10 μg of membrane protein. Expression levels are given based directly upon that determined with anti-ETR1 antibody (E) and normalized against the ATPase levels (E/A). For each plant background, expression level of the receptor in the presence of silver is given relative to that observed in the absence of silver. Results from two independent experimental treatments of wild-type plants with silver are shown.
Effect of Loss-of-Function Mutations in ETR1 upon its Expression
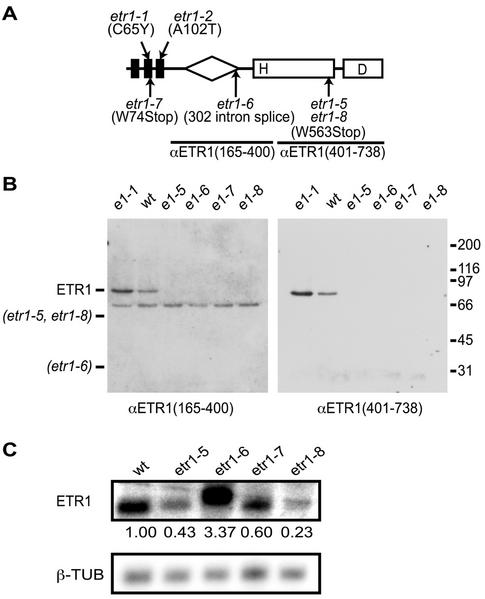
The mutations etr1-5, etr1-6, etr1-7, and etr1-8 are all loss-of-function mutations in ETR1 (Hua and Meyerowitz, 1998). The mutations etr1-5, etr1-6, and etr1-7 were isolated as intragenic suppressors of the ethylene insensitivity conferred by etr1-1, whereas etr1-8 was isolated as an intragenic suppressor of etr1-2 (Fig. 3A). The etr1-5, etr1-6, and etr1-8 mutations all introduce premature stop codons into the coding sequence. The etr1-6 mutation occurs at an intron splice site and retention of that intron would introduce a premature stop codon. All four mutants show similar ethylene responsiveness to that of wild-type plants (Hua and Meyerowitz, 1998). To determine whether the mutations result in the absence of the receptor or produce a truncated receptor incapable of signaling, we analyzed receptor expression by immunoblot. Two different antibodies, anti-ETR1(165–400) and anti-ETR1(401–738), were used that are targeted against different regions of the receptor (Fig. 3A). In initial experiments using recombinant fusion proteins expressed in bacteria, we confirmed that both antibodies recognized the etr1-5 and etr1-8 truncations as efficiently as full-length ETR1, and that they were incapable of detecting the etr1-6 truncation (results not shown). When Arabidopsis membranes were analyzed by immunoblot, no full-length protein was detected for any of the loss-of-function mutants (Fig. 3B). In addition, we did not detect any immunoreactive bands that would correspond to the truncated receptors. Note that the anti-ETR1(165–400) antibody does cross-react with a protein of 68 kD, but this is not derived from ETR1. A truncated protein for etr1-5 and etr1-8 would be detectable with both the anti-ETR1(165–400) and anti-ETR1(401–738) antibodies. Based on a control dilution series of the receptor, the anti-ETR1(165–400) antibody was capable of detecting a protein expressed at 10% of the level found with the wild-type receptor ETR1 or 5% of the level found with etr1-1. The anti-ETR1(401–738) antibody is even more sensitive and is capable of detecting proteins with at least 2-fold higher sensitivity than that of the anti-ETR1(165–400) antibody.
Figure 3.
Effect of loss-of-function mutations in ETR1 upon its expression. A, Positions of mutations in ETR1. The positions of ethylene-insensitive mutations are shown above the diagram of ETR1. The positions of intragenic suppressors of these mutations that result in loss of function are shown below the diagram of ETR1. Positions of regions used to generate the anti-ETR1(165–400) and anti-ETR1(401–738) antibodies are also indicated. B, Immunoblot analysis of ETR1 in different loss-of-function backgrounds. Membrane fractions (10 μg) from etiolated Arabidopsis seedlings were analyzed by immunoblot using the anti-ETR1(165–400) and anti-ETR1(401–738) antibodies. The migration position of ETR1 and predicted migration positions of the etr1-5, etr1-6, and etr1-8 truncated receptors are indicated on the left. Migration positions of molecular mass markers are indicated on the right in kilodaltons. C, Transcript levels of ETR1 in different loss-of-function backgrounds. Blots of mRNA were probed with an ETR1 probe and a β-tubulin gene probe as an internal control. The numbers represent the expression level of the ethylene receptor gene after normalization for the level of β-tubulin gene expression.
The lack of detectable protein for the etr1-5 and etr1-8 loss-of-function mutants could be because of instability of the truncated protein or of the mRNA. To differentiate between these possibilities, we performed northern-blot analysis. Transcripts were detected for all the loss-of-function mutations in ETR1 (Fig. 3C). The transcript for etr1-6 is slightly larger than the other transcripts, as predicted, because of the presence of an unspliced intron. Compared with wild type, the mRNA levels of etr1-5 and etr1-8 were reduced approximately 2- or 4-fold, respectively, whereas the mRNA level of etr1-6 was increased about 3-fold. The reduction in mRNA levels of etr1-5 and etr1-8 is significant but not sufficient to explain the lack of detectable protein, indicating that posttranscriptional mechanisms may also play a role in reducing the levels of the truncated proteins.
Effect of Loss-of-Function Mutations in Other Ethylene Receptors upon Expression of ETR1
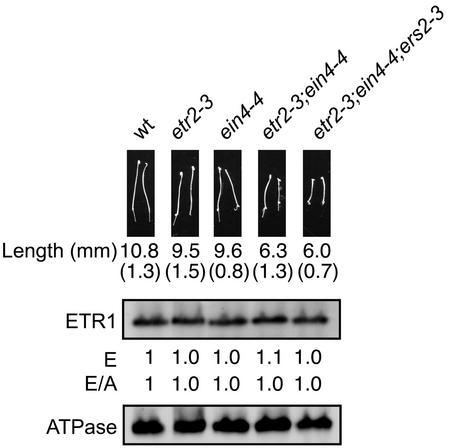
Loss of one member in a gene family can sometimes lead to functional compensation, whereby expression of another member of the same gene family is induced to compensate for activity of the missing family member (Bérard et al., 1997; Mulligan et al., 1998; Minkoff et al., 1999). An intriguing set of experiments suggests that functional compensation occurs within the ethylene receptor family of tomato (Tieman et al., 2000). Therefore, we examined the Arabidopsis ethylene receptor ETR1 to determine if its expression was affected by loss-of-function mutations in other ethylene receptor family members. Analysis was performed on single loss-of-function mutants (etr2-3 and ein4-4), a double mutant (etr2-3;ein4-4), and a triple mutant (etr2-3;ein4-4;ers2-3; Hua and Meyerowitz, 1998). The single mutants have little effect upon growth of etiolated Arabidopsis seedlings, but seedlings containing the double and triple mutant demonstrate partial induction of the triple-response phenotype, consistent with loss of receptors activating the ethylene response pathway (Fig. 4; Hua and Meyerowitz, 1998). The expression level of ETR1 protein in these mutant backgrounds was comparable with that found in the wild-type background (Fig. 4), indicating that ETR1 did not functionally compensate for the loss of these other members of the receptor family.
Figure 4.
Effect of loss-of-function mutations in ETR2, EIN4, and ERS2 upon expression of ETR1. The phenotypes of 4-d-old dark-grown seedlings containing single, double, and triple mutant combinations of etr2-3, ein4-4, and ers2-3 are shown. The mean hypocotyl length is given in millimeters based on measurement of at least 25 seedlings with the sd in parentheses. Immunoblot analysis was performed using antibodies directed against ETR1 and the H+-ATPase as an internal control on 10 μg of membrane protein. Expression levels are given based directly upon that determined with anti-ETR1 antibody (E) and normalized against the ATPase levels (E/A).
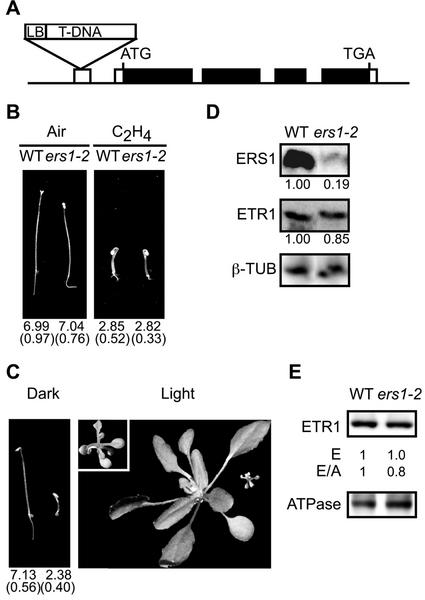
The ethylene receptor ERS1 is more closely related at the sequence level to ETR1 than are the other ethylene receptors of Arabidopsis (Chang and Stadler, 2001), but no loss-of-function mutations have been available for ERS1 (Hua and Meyerowitz, 1998) We isolated a T-DNA insertion in ERS1 by use of a PCR-based strategy, and determined by sequencing from the left-border junction that the T-DNA was inserted into the 5′-untranslated region of ERS1 (Fig. 5A). Sequence at the T-DNA junction with ERS1 was ATAACGCTCGGATCAATCAtactcga(atattcaattgtaaatggct), with capitals indicating ERS1 sequence and parentheses indicating T-DNA left border sequence. We named this mutant allele ers1-2 to differentiate it from the previously characterized ethylene-insensitive ers1-1 mutation (Hua et al., 1995). The responsiveness to ethylene of plants homozygous for the ers1-2 mutation was similar to that of wild-type plants (Fig. 5B). However, a double mutant of ers1-2 with the etr1-7 loss-of-function mutant displayed a strong ethylene response phenotype when grown in the absence of ethylene (Fig. 5C). Dark-grown ers1-2;etr1-7 seedlings displayed a triple-response phenotype in the air. Light-grown ers1-2;etr1-7 plants were dwarfed with compact and epinastic leaves in the air and died without bolting. Northern-blot analysis indicated a substantial reduction in mRNA levels of ers1-2 compared with that found in wild type, but low levels of transcript were detected (Fig. 5D). The significant reduction of ERS1 transcript levels in the ers1-2 mutant would contribute to the strong mutant phenotype observed when the ers1-2 mutant is combined with the etr1-7 mutant. The lack of a mutant phenotype in the ers1-2 mutant by itself could potentially be explained by functional compensation, ETR1 being a possible candidate because of its sequence similarity. However, the expression of ETR1 in the ers1-2 mutant background was comparable with that found in the wild-type background at both the mRNA and protein levels (Fig. 5, D and E), indicating that functional compensation was not because of changes in ETR1 expression.
Figure 5.
Analysis of the T-DNA insertional mutant ers1-2. A, Location of T-DNA insertion in the ERS1 gene. Black bars and white bars represent translated and untranslated regions of the ERS1 transcript, respectively. B, Phenotype of 3.5-d-old dark-grown seedlings containing the ers1-2 mutation grown in air or ethylene (50 μL L−1). Mean hypocotyl lengths are given in millimeters with sd in parentheses. C, Phenotype of the ers1-2;etr1-7 double mutant as compared with seedlings with wild-type phenotype segregating from the same population. Seedlings were grown in dark for 3.5 d or in the light for 4 weeks. The ers1-2;etr1-7 double mutant is on the right in each panel, and a 2-fold enlargement is also inset to reveal details of the light-grown seedling. D, Northern-blot analysis of ERS1 and ETR1 expression in the ers1-2 mutant line performed using 25 μg of total RNA. The numbers represent the expression level of the ethylene receptor genes after normalization for the level of β-tubulin expression. E, Effect of the ers1-2 mutation upon expression of ETR1 in etiolated seedlings. Immunoblot analysis was performed using antibodies directed against ETR1 and the H+-ATPase as an internal control on 15 μg of membrane protein. Expression levels are given based directly upon that determined with anti-ETR1 antibody (E) and normalized against the ATPase levels (E/A).
Effect of mutations in RAN1 and CTR1 upon Expression of ETR1
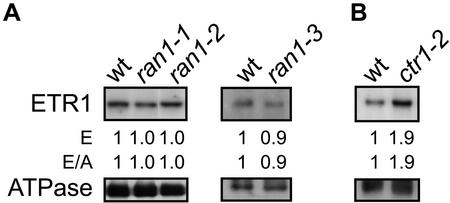
RAN1 is a copper-transporting ATPase implicated in the delivery of the copper cofactor to the ethylene receptors (Hirayama et al., 1999; Woeste and Kieber, 2000). The ran1-1 and ran1-2 mutations cause single amino acid changes in the RAN1 protein and are thought to alter rather than eliminate function (Hirayama et al., 1999). Plants containing these mutations demonstrate an induction of ethylene responses when treated with trans-cyclooctene, normally an antagonist of ethylene responses, but have no other discernible effect upon growth (Hirayama et al., 1999). Both ran1-1 and ran1-2 seedlings expressed ETR1 at levels similar to wild-type seedlings (Fig. 6A). The loss-of-function mutation ran1-3 results in constitutive activation of the ethylene response pathway. Because ran1-3 plants produce leaves but die without bolting (Woeste and Kieber, 2000), we identified homozygous ran1-3 plants based on phenotype from a segregating population of 4-week-old plants grown in the light. We observed no difference in ETR1 levels in ran1-3 plants compared with wild-type plants or members of the segregating population that lacked the ran1-3 phenotype (Fig. 6A). Loss-of-function mutations in the Ser/Thr kinase CTR1 also result in constitutive ethylene responses. We typically observed about a 2-fold increase in levels of ETR1 in the ctr1-2 mutant background relative to wild type (Fig. 6B). This could arise because of a low level of ethylene inducibility for the ETR1 transcript (Hua et al., 1998) or be an indirect effect of the phenotypic differences between ctr1-2 and wild-type plants (Kieber et al., 1993).
Figure 6.
Effect of mutations in RAN1 and CTR1 upon expression of ETR1. Immunoblot analysis was performed using antibodies directed against ETR1 and the H+-ATPase as an internal control. Expression levels are given based directly upon that determined with anti-ETR1 antibody (E) and normalized against the ATPase levels (E/A). A, Effect of ran1 mutations on expression of ETR1. For ran1-1 and ran1-2, etiolated seedlings were examined; for ran1-3, leaves of 4-week-old plants were examined. B, Effect of the ctr1-2 mutation upon expression of ETR1 in etiolated seedlings.
DISCUSSION
Expression of the ethylene receptor ETR1 was sensitive to mutations within its own coding sequence. Both gain-of-function mutations and loss-of-function mutations affected expression of ETR1 at the posttranscriptional level and, as discussed below, these posttranscriptional changes could contribute to the phenotypes observed in the mutants. Expression of ETR1 was affected to only a limited extent by mutations in other pathway components. For instance, loss-of-function mutations in other members of the ethylene receptor family had little effect upon expression of ETR1, indicating that ETR1 does not functionally compensate for the loss of these receptors by an increase in its own expression.
Expression analysis of ethylene pathway mutations refines the model shown in Figure 7 on how ethylene insensitivity is conferred by mutant forms of ETR1. Each of the four ethylene-insensitive mutations of ETR1 results in increased protein levels of the receptor, apparently through a posttranscriptional mechanism. The effect upon receptor expression can be phenocopied at the molecular level by treatment of plants with silver, which is also capable of generating ethylene insensitivity in plants. Both the ethylene-insensitive mutations (Hall et al., 1999) and silver (Rodriguez et al., 1999) are thought to perturb the ethylene-binding site (Fig. 7), and thus ethylene perception may play a role in regulating expression of the receptor. The ethylene-insensitive forms of the receptor could potentially have a slower rate of turnover than the wild-type receptors because turnover of animal hormone receptors is commonly regulated by ligand binding (Wiley, 1992). In such a case, endogenous ethylene levels within the plant would have to be sufficient to result in differing rates of turnover for the wild-type and mutant receptors.
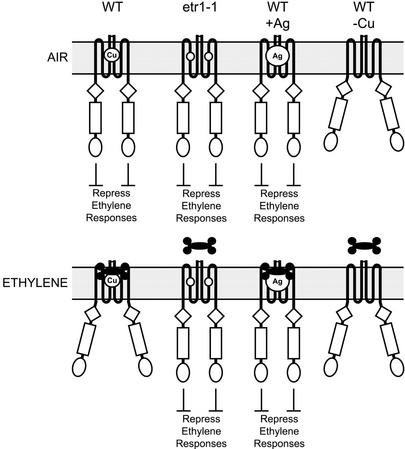
Figure 7.
A model for signaling by wild-type and mutant versions of the ethylene receptor ETR1. The ethylene receptor ETR1 contains one ethylene-binding site per homodimer, with ethylene binding mediated by a single copper ion (Cu) present in the ethylene-binding site. In air, wild-type (WT) receptors actively repress ethylene responses. In ethylene, wild-type receptors are inactivated, thereby relieving repression of the ethylene response pathway. The etr1-1 mutation (indicated by a white circle) eliminates binding of the copper cofactor and locks the receptor into a conformation such that the receptor represses ethylene responses even in the presence of ethylene. The replacement of the copper cofactor by silver (WT+Ag) also serves to lock the receptor into a conformation such that it continuously represses ethylene responses. In contrast, elimination of the copper cofactor (WT-Cu) results in the receptor adapting an inactive conformation in air and ethylene.
The discovery that ethylene-insensitive mutants of ETR1 have a higher expression level than wild-type receptors helps resolve an apparent paradox in our understanding of signaling by ethylene receptors (Hua and Meyerowitz, 1998; Bleecker, 1999; Chang and Stadler, 2001). An ethylene-insensitive mutation in one member of the five-member ethylene receptor family is sufficient to confer ethylene insensitivity, suggesting that signaling by one family member is enough to repress ethylene responses. On the other hand, loss-of-function mutations in three receptors simultaneously are sufficient to induce ethylene responses (Hua and Meyerowitz, 1998), a situation under which two family members would still theoretically be signaling to repress ethylene responses. Our data indicate that the signal output by an ethylene-insensitive receptor mutant is not equivalent to that of a wild-type receptor because of the difference in expression levels. The increase in expression of the ethylene-insensitive mutants of ETR1 would result in an increase in signal output and the ability to repress ethylene responses. Other mechanisms may also increase signal output of the ethylene-insensitive mutant receptors, such as their postulated ability to convert wild-type receptors to an ethylene-insensitive signaling state via heteromeric interactions (Chang and Stadler, 2001; Gamble et al., 2002).
Analysis of ETR1 expression in the ran1-3 background further clarifies the mechanism by which mutations in ethylene receptors confer ethylene insensitivity. The ran1-3 mutant eliminates a copper transporter required for delivery of the copper cofactor to the ethylene receptors (Hirayama et al., 1999; Woeste and Kieber, 2000). Plants containing the ran1-3 mutation display a constitutively active ethylene response (Woeste and Kieber, 2000). Interestingly, mutations like etr1-1 that produce a receptor unable to bind the copper cofactor result in the opposite phenotype: ethylene insensitivity (Rodriguez et al., 1999). This difference in phenotypes could be because of: (a) destabilization of the ethylene receptors in the ran1-3 background, or (b) functional differences between receptors lacking copper and the ethylene-insensitive receptor mutations. Our data support the second hypothesis. ETR1 protein was detected in the ran1-3 background at similar levels to that found in the wild-type background indicating that, although the receptor is present and lacking the copper cofactor, it does not confer ethylene insensitivity. Presumably, protein levels of the other members of the ethylene receptor family are similarly unaffected. Thus, wild-type ethylene receptors lacking the copper cofactor have a loss-of-function phenotype (i.e. the ran1-3 mutation produces the same constitutive ethylene response phenotype found in plant lines containing multiple loss-of-function mutations in the ethylene receptors). Wild-type receptors lacking the copper cofactor may adopt a signaling-inactive conformation similar to the conformation of wild-type receptors that have ethylene bound (Fig. 7). In contrast, the amino acid changes that result from mutations like etr1-1 (Cys-65-Tyr) result in a gain of function because they prevent not only copper binding but also lock the receptor into a signaling-active conformation such as it has in air (Fig. 7). The proposal that receptors in the ran1-3 background are not equivalent to receptors containing ethylene-insensitive mutations is consistent with the finding that the ethylene-insensitive etr1-3 mutant can suppress the ran1-3 constitutive ethylene phenotype (Woeste and Kieber, 2000). The finding that ETR1 is still present in the ran1-3 background also raises the possibility that not all mutations that eliminate ethylene binding will, as a consequence, confer ethylene insensitivity.
The loss-of-function mutants etr1-5, etr1-6, etr1-7, and etr1-8 were isolated as intragenic suppressors of the ethylene insensitivity conferred by either etr1-1 or etr1-2, and are predicted to result in premature termination of the encoded protein (Hua and Meyerowitz, 1998). However, we have found that a truncated version of etr1-1 containing the first 349 amino acids is still capable of conferring ethylene insensitivity when transformed into Arabidopsis (Gamble et al., 2002). This raises the question as to why no ethylene insensitivity is observed with the loss-of-function mutants, in particular with etr1-5 and etr1-8, which are predicted to code for receptors containing 562 amino acids. Our data indicate that the loss-of-function mutants may reduce expression at the transcriptional and posttranscriptional levels. Transcript, but no protein, was detected for each of the ETR1 loss-of-function mutants. Examination of etr1-5 and etr1-8 indicated a reduction to 43% and 23%, respectively, of wild-type mRNA levels. This reduction in expression could be because of mechanisms for mRNA surveillance such as nonsense-mediated decay whereby mRNAs containing premature stop codons are targeted for degradation (van Hoof and Green, 1996). However, the reduction in mRNA expression levels of etr1-5 and etr1-8 is probably not sufficient to reduce protein levels below detection limits for the antibodies. Thus, the results obtained with the loss-of-function mutations suggest that premature termination of the protein may lead to an absence of receptor rather than a truncated receptor, presumably because of instability of the truncated protein. The genetic screen for intragenic suppressors may have favored the isolation of destabilizing mutations.
To facilitate our analysis of ethylene pathway mutations, we isolated a T-DNA insertion mutation in the ERS1 gene that, based on northern-blot analysis, substantially reduces expression of ERS1. As has been found in the analysis of loss-of-function genes in other ethylene receptors, the ers1-2 mutant by itself had little effect upon ethylene responses in the mutant seedlings. However, a double mutant of ers1-2 and etr1-7 exhibited a constitutive ethylene response. The phenotype observed with the ers1-2;etr1-7 double mutant was comparable with that previously reported for an etr1;etr2;ein4;ers2 quadruple loss-of-function mutant (Hua and Meyerowitz, 1998). These data suggest that ETR1 and ERS1 play more predominant roles in the regulation of ethylene signaling than the other three members of the ethylene receptor family. The relative importance of ETR1 and ERS1 could be because of the presence of His kinase activity (Gamble et al., 1998), the ability to interact with the downstream signaling component CTR1 (Clark et al., 1998), or possibly higher expression levels compared with those of the other ethylene receptors.
In summary, the results described here clarify the mode of action of ethylene pathway mutations previously identified in Arabidopsis. Mutations in the ethylene receptor ETR1 affected expression of the receptor at the posttranscriptional level. Similar mutations conferring ethylene insensitivity and intragenic suppressor mutations that result in premature stop codons have been identified in other members of the ethylene receptor family of Arabidopsis (Hua et al., 1995, 1998; Hua and Meyerowitz, 1998; Sakai et al., 1998). Thus, the mechanisms described here may be applicable to other ethylene receptors besides ETR1.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis mutants in the ecotype Columbia were used for all experiments except those involving the ers1-2 mutant, which was in the ecotype Wassilewskija. The ERS1 T-DNA insertion allele (ers1-2) was isolated from the 60,480 kanamycin-resistant T-DNA-tagged Arabidopsis lines of the University of Wisconsin Knockout Arabidopsis facility (http://www. biotech.wisc.edu/Arabidopsis). The mutant was identified with a PCR primer for the T-DNA left border (CATTTTATAATAACGCTGCGGACATCTAC) and an ERS1-specific primer (CAGAGAGTTCTGTCACTCCTGGAAATGGT). Plants containing the wild-type ERS1 gene were identified by use of PCR with the above ERS1 primer and a second ERS1-specific primer (CACAACCGCGCAAGAGACTTTAGCAATAGT). The ers1-2;etr1-7 double mutant was identified by crossing plants homozygous for the single mutations and subsequent PCR-based genotyping of F2 progeny according to Hua and Meyerowitz (1998). Upon request, the ers1-2 mutant and all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes.
Unless indicated otherwise, seedlings were grown on 0.8% (w/v) agar plates of one-half-strength Murashige and Skoog basal medium (pH 5.65) with Gamborg's vitamins (Murashige and Skoog media, Sigma, St. Louis). Seeds were stratified for 2 d at 4°C before growth at 22°C. Seeds were exposed to light for 12 h, then incubated in the dark. Seedlings were typically examined after 4 d, with time 0 corresponding to when the plates were removed from 4°C and brought to 22°C. For ethylene treatment, seedlings were grown in sealed chambers in the presence of 50 μL L−1 ethylene. Measurements of hypocotyl length were performed as described by Gamble et al. (2002). For analysis of the ran1-3 mutant, seedlings from a segregating population were grown for 4 weeks under an 8-h light cycle to allow for maximal rosette development before harvest. Homozygous ran1-3 seedlings were identified based on their readily distinguishable constitutive ethylene response phenotype (Woeste and Kieber, 2000).
Antibodies
The anti-ETR1(401–738) antibody was prepared against a glutathione S-transferase (GST) fusion protein with amino acids 401 to 738 of ETR1 (Schaller et al., 1995) and was used for detection of ETR1 in all cases except where indicated in Figure 3. The serum was depleted of antibodies that cross-react with GST by passing through a column of Affigel-10 (Bio-Rad Laboratories, Hercules, CA) cross-linked to GST. The antibody was affinity purified by binding to an Affigel column cross-linked to GST-ETR1(401–738) (Schaller et al., 1995), then eluted with 0.1 m Gly (pH 2.5). The anti-ETR1(165–400) antibody used for Figure 3 was prepared against a GST fusion protein with amino acids 165 to 400 of ETR1 (Schaller et al., 1995) and was affinity purified as described (Gamble et al., 2002). The anti-(H+-ATPase) antibody (DeWitt et al., 1996) used as an internal loading control was provided by Dr. Michael Sussman (University of Wisconsin, Madison).
Protein Isolation and Immunoblot Analysis
For isolation of Arabidopsis membranes, plant material was homogenized at 4°C in extraction buffer (50 mm Tris [pH 8.5], 150 mm NaCl, 10 mm EDTA, and 20% [v/v] glycerol) containing 1 mm phenylmethylsulfonyl fluoride, 1 μg mL−1 pepstatin, 10 μg mL−1 aprotinin, and 10 μg mL−1 leupeptin as protease inhibitors. The homogenate was strained through Miracloth (Calbiochem-Novobiochem, San Diego) and centrifuged at 8,000g for 15 min. The supernatant was centrifuged at 100,000g for 30 min, and the membrane pellet was resuspended in 10 mm Tris (pH 7.5), 150 mm NaCl, 1 mm EDTA, and 10% (v/v) glycerol with protease inhibitors. Protein concentration was determined by a modification of the Lowry assay (Lowry et al., 1951) in which samples were treated with 0.4% (w/v) sodium deoxycholate (Schaller and DeWitt, 1995). Bovine serum albumin was used as a standard for protein assays.
For immunoblot analysis, membranes were mixed with SDS-PAGE loading buffer and incubated at 37°C for 1 h. Proteins were fractionated by SDS-PAGE using 8% (w/v) polyacrylamide gels (Laemmli, 1970). After electrophoresis, proteins were either stained with Coomassie Blue or electrotransferred to Immobilon nylon membrane (Millipore, Bedford, MA). Immunoblotting was performed by using anti-ETR1(165–400), anti-ETR1(401–738), or anti-(H+-ATPase) polyclonal antibodies. Immunodecorated proteins were visualized by enhanced chemiluminescence detection according to the manufacturer (Pierce Chemical, Rockford, IL). Densitometric analysis was performed by using the NIH Image program (http://rsb.info.nih.gov/nih-image) after first scanning the exposed film and then capturing the images with Photoshop (Adobe Systems, San Jose, CA). The relative expression level for ETR1 was quantified by comparison to a dilution series of ETR1.
Northern-Blot Analysis
Total RNA was extracted from Arabidopsis tissue according to the method of Carpenter and Simon (1998). For Figures 1 and 3, RNA was isolated from etiolated seedlings; and for Figure 5, RNA was isolated from 15-d-old leaf tissue of plants grown in liquid culture as described by Chang et al. (1992). mRNA was isolated from total RNA using the PolyATract mRNA isolation system (Promega, Madison, WI). For northern-blot analysis, RNA was separated on 1% (w/v) agarose gels using the NorthernMax-Gly kit (Ambion, Austin, TX) according to the manufacturer's instructions. RNA was transferred to nylon membrane by the capillary method and fixed by UV cross-linking. Hybridizations were performed using buffers supplied with the NorthernMax-Gly kit. Single-stranded DNA antisense probes were made using primers designed to anneal at the 3′ end of the selected genes. Radiolabeled probes were made and the blot stripped between hybridizations by using the Strip-EZ PCR kit (Ambion) according to the manufacturer's instructions. Radioactivity was imaged and quantitated by phosphor imaging with a Molecular Imager FX (Bio-Rad Laboratories), using accompanying Quantity One software.
ACKNOWLEDGMENTS
We thank Michael Sussman for providing the anti-(H+-ATPase) antibody, Jian Hua and Elliot Meyerowitz for the ethylene receptor loss-of-function seed lines, Yi-Feng Chen for assistance with affinity purification of antibodies, and Anita Klein and Estelle Hrabak for critical reading of the manuscript.
Footnotes
This work was supported by the National Science Foundation (grant nos. MCB–9982510 and DBI–9975908 to G.E.S.). This is scientific contribution no. 2,138 from the New Hampshire Agricultural Experiment Station.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.011635.
LITERATURE CITED
- Abeles FB, Morgan PW, Saltveit ME., Jr . Ethylene in Plant Biology. Ed 2. San Diego: Academic Press; 1992. [Google Scholar]
- Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science. 1999;284:2148–2152. doi: 10.1126/science.284.5423.2148. [DOI] [PubMed] [Google Scholar]
- Bérard J, Luo H, Chen H, Mukuna M, Bradley WE, Wu J. Abnormal regulation of retinoic acid receptor beta2 expression and compromised allograft rejection in transgenic mice expressing antisense sequences to retinoic acid receptor beta1 and beta3. J Immunol. 1997;159:2586–2598. [PubMed] [Google Scholar]
- Bleecker AB. Ethylene perception and signaling: an evolutionary perspective. Trends Plant Sci. 1999;4:269–274. doi: 10.1016/s1360-1385(99)01427-2. [DOI] [PubMed] [Google Scholar]
- Carpenter CD, Simon AE. Preparation of RNA. In: Martinez-Zapater JM, Salinas J, editors. Methods in Molecular Biology. 82: Arabidopsis Protocols. Totowa, NJ: Humana Press; 1998. pp. 85–89. [DOI] [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, Meyerowitz EM. Arabidopsis ethylene response gene ETR1: similarity of product to two-component regulators. Science. 1993;262:539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- Chang C, Schaller GE, Patterson SE, Kwok SF, Meyerowitz EM, Bleecker AB. The TMK1 gene from Arabidopsis codes for a protein with structural and biochemical characteristics of a receptor protein kinase. Plant Cell. 1992;4:1263–1271. doi: 10.1105/tpc.4.10.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Shockey JA. The ethylene-response pathway: signal perception to gene regulation. Curr Opin Plant Biol. 1999;2:352–358. doi: 10.1016/s1369-5266(99)00004-7. [DOI] [PubMed] [Google Scholar]
- Chang C, Stadler R. Ethylene hormone receptor action in Arabidopsis. BioEssays. 2001;23:619–627. doi: 10.1002/bies.1087. [DOI] [PubMed] [Google Scholar]
- Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR. Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell. 1997;89:1133–1144. doi: 10.1016/s0092-8674(00)80300-1. [DOI] [PubMed] [Google Scholar]
- Clark KL, Larsen PB, Wang X, Chang C. Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS1 ethylene receptors. Proc Natl Acad Sci USA. 1998;95:5401–5406. doi: 10.1073/pnas.95.9.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWitt ND, Hong B, Sussman MR, Harper JF. Targeting of two Arabidopsis H+-ATPase isoforms to the plasma membrane. Plant Physiol. 1996;112:833–844. doi: 10.1104/pp.112.2.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble RL, Coonfield ML, Schaller GE. Histidine kinase activity of the ETR1 ethylene receptor from Arabidopsis. Proc Natl Acad Sci USA. 1998;95:7825–7829. doi: 10.1073/pnas.95.13.7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble RL, Qu X, Schaller GE. Mutational analysis of the ethylene receptor ETR1. Role of the histidine kinase domain in dominant ethylene insensitivity. Plant Physiol. 2002;128:1428–1438. doi: 10.1104/pp.010777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AE, Chen QG, Findell JL, Schaller GE, Bleecker AB. The relationship between ethylene binding and dominant insensitivity conferred by mutant forms of the ETR1 ethylene receptor. Plant Physiol. 1999;121:291–299. doi: 10.1104/pp.121.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AE, Findell JL, Schaller GE, Sisler EC, Bleecker AB. Ethylene perception by the ERS1 protein in Arabidopsis. Plant Physiol. 2000;123:1449–1458. doi: 10.1104/pp.123.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T, Kieber JJ, Hirayama N, Kogan M, Guzman P, Nourizadeh S, Alonso JM, Dailey WP, Dancis A, Ecker JR. RESPONSIVE-TO-ANTAGONIST1, a Menkes/Wilson disease-related copper transporter, is required for ethylene signaling in Arabidopsis. Cell. 1999;97:383–393. doi: 10.1016/s0092-8674(00)80747-3. [DOI] [PubMed] [Google Scholar]
- Hua J, Chang C, Sun Q, Meyerowitz EM. Ethylene insensitivity conferred by Arabidopsis ERS gene. Science. 1995;269:1712–1714. doi: 10.1126/science.7569898. [DOI] [PubMed] [Google Scholar]
- Hua J, Meyerowitz EM. Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell. 1998;94:261–271. doi: 10.1016/s0092-8674(00)81425-7. [DOI] [PubMed] [Google Scholar]
- Hua J, Sakai H, Nourizadeh S, Chen QG, Bleecker AB, Ecker JR, Meyerowitz EM. EIN4 and ERS2 are members of the putative ethylene receptor family in Arabidopsis. Plant Cell. 1998;10:1321–1332. doi: 10.1105/tpc.10.8.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldman KA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Mattoo AK, Suttle JC, editors. The Plant Hormone Ethylene. Boca Raton, FL: CRC Press, Inc.; 1991. [Google Scholar]
- Minkoff R, Bales ES, Kerr CA, Struss WE. Antisense oligonucleotide blockade of connexin expression during embryonic bone formation: evidence of functional compensation within a multigene family. Dev Genet. 1999;24:43–56. doi: 10.1002/(SICI)1520-6408(1999)24:1/2<43::AID-DVG6>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Mulligan GJ, Wong J, Jacks T. p130 is dispensable in peripheral T lymphocytes: evidence for functional compensation by p107 and pRB. Mol Cell Biol. 1998;18:206–220. doi: 10.1128/mcb.18.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson JS. Signal transduction schemes of bacteria. Cell. 1993;73:857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- Rodriguez FI, Esch JJ, Hall AE, Binder BM, Schaller GE, Bleecker AB. A copper cofactor for the ethylene receptor ETR1 from Arabidopsis. Science. 1999;283:996–998. doi: 10.1126/science.283.5404.996. [DOI] [PubMed] [Google Scholar]
- Sakai H, Hua J, Chen QG, Chang C, Medrano LJ, Bleecker AB, Meyerowitz EM. ETR2 is an ETR1-like gene involved in ethylene signaling in Arabidopsis. Proc Natl Acad Sci USA. 1998;95:5812–5817. doi: 10.1073/pnas.95.10.5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE. Histidine kinases and the role of two-component systems in plants. Adv Bot Res. 2000;32:109–148. [Google Scholar]
- Schaller GE, Bleecker AB. Ethylene-binding sites generated in yeast expressing the Arabidopsis ETR1 gene. Science. 1995;270:1809–1811. doi: 10.1126/science.270.5243.1809. [DOI] [PubMed] [Google Scholar]
- Schaller GE, DeWitt ND. Analysis of the H+-ATPase and other proteins of the Arabidopsis plasma membrane. Methods Cell Biol. 1995;50:129–148. [PubMed] [Google Scholar]
- Schaller GE, Ladd AN, Lanahan MB, Spanbauer JM, Bleecker AB. The ethylene response mediator ETR1 from Arabidopsis forms a disulfide-linked dimer. J Biol Chem. 1995;270:12526–12530. doi: 10.1074/jbc.270.21.12526. [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Ecker JR. Ethylene signaling: from mutants to molecules. Curr Opin Plant Biol. 2000;3:353–360. doi: 10.1016/s1369-5266(00)00096-0. [DOI] [PubMed] [Google Scholar]
- Tieman DM, Taylor MG, Ciardi JA, Klee HJ. The tomato ethylene receptors NR and LeETR4 are negative regulators of ethylene response and exhibit functional compensation within a multigene family. Proc Natl Acad Sci USA. 2000;97:5663–5668. doi: 10.1073/pnas.090550597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Green PJ. Premature nonsense codons decrease the stability of phytohemagglutinin mRNA in a position-dependent manner. Plant J. 1996;10:415–424. doi: 10.1046/j.1365-313x.1996.10030415.x. [DOI] [PubMed] [Google Scholar]
- Wiley HS. Receptors: topology, dynamics, and regulation. In: Bittar EE, editor. Fundamentals of Medical Cell Biology. 5A. Greenwich, CT: JAI Press Inc.; 1992. pp. 113–142. [Google Scholar]
- Woeste KE, Kieber JJ. A strong loss-of-function mutation in RAN1 results in constitutive activation of the ethylene response pathway as well as a rosette-lethal phenotype. Plant Cell. 2000;12:443–455. doi: 10.1105/tpc.12.3.443. [DOI] [PMC free article] [PubMed] [Google Scholar]