Abstract
Understanding the genetic architecture of complex traits requires identification of the underlying genes and characterization of gene-by-gene and genotype-by-environment interactions. Behaviors that mediate interactions between organisms and their environment are complex traits expected to be especially sensitive to environmental conditions. Previous studies on the olfactory avoidance response of Drosophila melanogaster showed that the genetic architecture of this model behavior depends on epistatic networks of pleiotropic genes. We performed a screen of 1339 co-isogenic p[GT1]-element insertion lines to identify novel genes that contribute to odor-guided behavior and identified 55 candidate genes with known p[GT1]-element insertion sites. Characterization of the expression profiles of 10 p[GT1]-element insertion lines showed that the effects of the transposon insertions are often dependent on developmental stage and that hypomorphic mutations in developmental genes can elicit profound adult behavioral deficits. We assessed epistasis among these genes by constructing all possible double heterozygotes and measuring avoidance responses under two stimulus conditions. We observed enhancer and suppressor effects among subsets of these P-element-tagged genes, and surprisingly, epistatic interactions shifted with changes in the concentration of the olfactory stimulus. Our results show that the manifestation of epistatic networks dynamically changes with alterations in the environment.
BEHAVIORS are complex traits determined by many genes with allelic effects that are exquisitely sensitive to the environment (Anholt and Mackay 2004). We have used the olfactory avoidance response of Drosophila melanogaster as a model system to gain insights into the genetic architecture of behavior (Anholt et al. 1996). Olfactory avoidance responses to repellant odorants are essential for survival, and chemosensory behavior in general is critical for food localization, food intake, interactions with reproductive partners, and localization of oviposition sites.
The olfactory system of Drosophila is one of the best-characterized chemosensory systems, consisting of ∼1200 olfactory neurons compartmentalized in basiconic, coeloconic, and trichoid sensilla in the third antennal segments and of ∼120 olfactory neurons in basiconic sensilla in the maxillary palps (Shandbhag et al. 1999). Most olfactory neurons express a unique odorant receptor from a repertoire of 60 odorant receptor genes (Clyne et al. 1999; Gao and Chess 1999; Vosshall et al. 1999) together with the common Or83b receptor (Larsson et al. 2004), which is essential for transport and insertion of odorant receptors in the chemosensory dendritic membranes (Benton et al. 2006). Neurons that express the same receptor project bilaterally to 1 or 2 of ∼43 individually identifiable symmetrically located glomeruli in the antennal lobes (Laissue et al. 1999; Gao et al. 2000; Vosshall et al. 2000; Bhalerao et al. 2003), where olfactory information is encoded in a spatial and temporal pattern of glomerular activation (Wang et al. 2003). The glomerular map is decoded in central brain structures, the lateral horn of the protocerebrum, and the mushroom bodies, which receive chemosensory information from output neurons of the antennal lobes (Ng et al. 2002). Elegant single-unit electrophysiological recordings have characterized the molecular response profiles of many odorant receptors expressed in the maxillary palps (de Bruyne et al. 1999) and antennae (de Bruyne et al. 2001; Dobritsa et al. 2003; Hallem et al. 2004). Recently, this analysis has been extended to a comprehensive characterization of the molecular receptive fields and response characteristics of 24 odorant receptors with a panel of >100 odorants (Hallem and Carlson 2006).
Whereas the fly's odorant receptor repertoire determines its capacity for chemosensory stimulus recognition, processing and perception of the stimulus, assessment of its biological significance in the environmental context, and generation of an appropriate behavioral response depend on the recruitment of a vast array of interacting gene products. What are the properties of the genetic architecture that enable the nervous system to direct appropriate behavioral responses to chemical signals from the environment?
Understanding the genetic architecture of any complex trait requires first and foremost identifying the genes that contribute to manifestation of the trait, which can be achieved by mutagenesis screens. A second layer of analysis is determining how genes implicated in manifestation of the trait form functional ensembles through either additive or nonlinear interactions (Anholt and Mackay 2004; Anholt 2004).
Previous studies have identified several genes, other than chemoreceptors, that are essential for mediating chemosensory responses, e.g., scribble (Ganguly et al. 2003), the DSC1 sodium channel (Kulkarni et al. 2002), and Calreticulin (Stoltzfus et al. 2003). In addition, we identified 14 co-isogenic p[lArB] insertion lines with diminished olfactory avoidance responses to repellent odorants (Anholt et al. 1996) and showed that eight of these transposon-tagged smell-impaired (smi) loci formed a network of epistatic interactions (Fedorowicz et al. 1998). The success of these experiments motivated us to conduct a threefold larger P-element screen to extend the number of candidate genes that could be directly implicated in odor-guided behavior and might give additional insights into the extent of the plasticity of epistatic networks that govern behavioral phenotypes.
Here, we report the identification of 83 new P-element insertion lines in D. melanogaster with aberrant olfactory avoidance behavior from a screen of 1339 co-isogenic lines that contain a single marked p[GT1] gene-trap transposon (Lukacsovich et al. 2001; Bellen et al. 2004). The p[GT1] gene-trap element is a versatile transposable element designed to insert into or near target genes and contains an enhancer trap GAL4 cassette that can drive the expression of GAL4 under an endogenous promoter, thereby enabling potential transgene expression under UAS promoters for genetic manipulation of cells in which the target gene is expressed (Lukacsovich et al. 2001). Most of the p[GT1]-tagged candidate genes included in our study have been implicated in early development of the nervous system. Null mutants in these genes often result in developmental defects that will not allow the maturation of a healthy adult animal, thereby precluding assessment of the effects of such genes on adult behavior. Our hypomorphic P-element-induced mutations, however, allow ostensibly normal development, but have profound effects on adult olfactory avoidance behavior.
Since all p[GT1]-elements are in a co-isogenic background, we can assess epistatic interactions among them and ask whether epistatic networks are invariant or dynamic under different environmental conditions. We have performed this analysis with a set of 10 p[GT1]-element insertion lines and show that the manifestation of epistatic interactions among candidate genes depends on the stimulus concentration that elicits the avoidance response. Our results show that the composition of epistatic networks is more dynamic than generally appreciated. These studies lead us to propose a model in which the genetic architecture of olfactory behavior depends on genetic networks that comprise dynamic epistatic interactions with few stable hubs and with fluid enhancer/suppressor effects that are manifested under different environmental conditions.
MATERIALS AND METHODS
Fly stocks and mutagenesis screen:
p[GT1] insertion lines (Lukacsovich et al. 2001), constructed in co-isogenic Canton-S backgrounds (A, B, C, D, E, and F) as a resource for the Berkeley Drosophila Genome Project, were obtained from Hugo Bellen (Baylor College of Medicine, Houston; Bellen et al. 2004). Homozygous viable p[GT1] insertion lines were screened by quantifying olfactory avoidance behavior (exactly as described by Anholt et al. 1996) in single-sex groups of five flies/replicate and four replicates/sex at a stimulus concentration of 0.3% (v/v) benzaldehyde between 2:00 and 4:00 pm in a randomized design in which measurements on individual lines were collected over multiple days to average environmental variation. Avoidance scores of appropriate control lines were also determined on each day, with doubled sample size.
We assessed mutational variation in olfactory behavior by two-way mixed model analysis of variance (ANOVA) of replicate line means, expressed as deviation from their contemporaneous co-isogenic controls, according to the model Y = μ + S + L + S × L + ɛ, where μ is the overall mean, S is the fixed effect of sex, L is the random effect of the P-element insertion line, S × L is the sex-by-line interaction term, and ɛ is the environmental variance between replicates. We also ran reduced analyses for each sex separately and computed the variance components (σ2) for the random effects. The mutational broad sense heritability was computed as 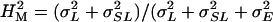 ) from the analysis pooled across sexes, where
) from the analysis pooled across sexes, where  , and
, and 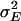 are, respectively, line, sex-by-line, and environmental variance components. The cross-sex genetic correlation was computed as
are, respectively, line, sex-by-line, and environmental variance components. The cross-sex genetic correlation was computed as 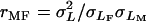 , where
, where 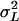 is the among-line variance component from the analysis pooled across sexes, and
is the among-line variance component from the analysis pooled across sexes, and 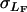 and
and 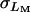 are the square roots of the variance components from the separate sex analyses of females and males, respectively.
are the square roots of the variance components from the separate sex analyses of females and males, respectively.
Approximately 10% of the lines with the lowest olfactory avoidance scores were retested by measuring 20 replicates for each sex to identify those lines that showed consistent statistically significant differences from the P-element-free Canton-S control. The data were analyzed by two-way fixed effects ANOVA, according to the model Y = μ + S + L + S × L + ɛ, where μ is the overall mean, S is the effect of sex, L is the effect of line (P-element insertion line and co-isogenic control), S × L is the sex-by-line interaction term, and ɛ is the environmental variance between replicates. All flies were reared on an agar yeast–molasses medium in vials maintained at 25° under a 12 hr light/dark cycle.
Assessment of gene expression levels:
We characterized 10 mutant lines, all derived in the Canton-S B isogenic background, in greater detail. We quantified mRNA levels in these lines by quantitative RT–PCR, using an ABI-7900 sequence detector with a SYBR green detection method, according to the protocol from Applied Biosystems (Foster City, CA) with glyceraldehyde-3-phosphate dehydrogenase as the internal standard. Independent triplicates of total RNA were isolated from female Canton-S control and mutant flies using the Trizol reagent (GIBCO-BRL, Gaithersburg, MD) and cDNA was generated from 150 to 200 ng of total RNA by reverse transcription. Transcript-specific primers were designed to amplify ∼60- to 100-bp regions of all 11 genes using the primer express program from Applied Biosystems. Primers were designed to encompass common regions of alternative transcripts. Negative controls without reverse transcriptase were used for all genes to exclude potential genomic DNA contamination. Statistical significance for differences in gene expression levels between P-element insertion lines and the control line was determined by two-tailed Student's t-tests.
To examine developmental stage-specific deficiencies in gene expression levels in mutant lines, relative levels of expression were analyzed in the same way after extraction of triplicate RNA samples from embryos between 13 and 16 hr after oviposition, third instar larvae, pupae, and adult female heads.
Diallel crosses and statistical analyses:
We analyzed epistasis among the 10 co-isogenic p[GT1] insertion lines with impaired olfactory behavior exactly as described previously (Fedorowicz et al. 1998) by crossing homozygous mutant parental strains to construct all 45 possible double heterozygous F1 genotypes with two P elements at different loci (excluding reciprocal crosses) in a half-diallel crossing design according to method 4, model 1 of Griffing (1956). Since 4 of the mutant lines had p[GT1] insertions on the X chromosome, we restricted our analysis of epistasis to females. Avoidance responses of trans-heterozygotes to 0.1% (v/v) and 0.3% (v/v) benzaldehyde were quantified with 20 replicate assays (100 flies/cross) between 8:00 and 11:00 am contemporaneously with the Canton-S control. Avoidance scores at this time interval were comparable to those recorded during the initial screen between 2:00 and 4:00 pm. Avoidance scores of the trans-heterozygote genotypes were analyzed by a two-way fixed effects ANOVA according to the model Y = μ + G + E + G × E + ɛ, where G denotes trans-heterozygote genotype, E is the benzaldehyde concentration (environment), and ɛ is the variance between individuals within each genotype and benzaldehyde concentration. We also ran reduced analyses separately for each concentration of benzaldehyde.
To analyze epistatic effects among the 10 loci, we first estimated the average heterozygous effect of each mutation in combination with all other mutations as the general combining ability (GCA), which reflects its average avoidance score as a trans-heterozygote when combined with all other mutations, expressed as the deviation from the overall mean (Sprague and Tatum 1942). Since the lines are co-isogenic, we can then estimate the expected phenotypic value of each trans-heterozygote on the basis of the GCA values of both parents under the null hypothesis that there is no epistasis between the two loci. Epistasis is inferred if the observed phenotypic value deviates significantly from the predicted value. Thus, the specific combining ability (SCA) of a trans-heterozygous genotype is defined as the difference between the observed avoidance score of the genotype, Xi,j (where i and j denote two different mutations), and the score expected from the sum of the corresponding GCAs of mutants i and j. The GCA for each mutant was estimated as
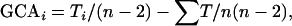 |
where Ti is the sum of mean avoidance score values (averaged over all replicates) of heterozygotes with the ith mutation, ∑T is twice the sum of mean avoidance score values of all heterozygotes, and n is the number of mutant lines (see also Falconer and Mackay 1996). The SCA effects were computed using the method of Griffing (1956) for each heterozygous genotype as
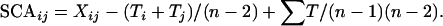 |
Standard errors of individual GCA and SCA effects were computed according to the formulas given by Griffing (1956) as
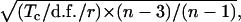 |
where Tc is the corrected total, d.f. is the degrees of freedom, and r is the number of replicates.
We used the Diallel-SAS05 program (Zang et al. 2005) to partition variance among trans-heterozygous genotypes into variance attributable to GCA and SCA; to partition the G × E interaction variance into variance attributable to GCA × E and SCA × E; and to estimate individual GCA and SCA effects and their standard errors.
RESULTS
Identification of co-isogenic p[GT1] insertion lines with aberrant olfactory avoidance behavior:
We measured olfactory avoidance responses of 1339 p[GT1] insertion lines for males and females separately. Two-way analysis of variance pooled over sexes revealed significant variation between the sexes among P-element insert lines and for the line-by-sex interaction term (Table 1, Figure 1). The broad-sense mutational heritability for olfactory behavior was high: 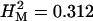 . Furthermore, the significant line-by-sex interaction term indicates that there was mutational variation in the difference in olfactory behavior between males and females; i.e., the effects of mutations on olfactory behavior were sex specific. The estimate of the cross-sex genetic correlation (±SE) was rMF = 0.587 ± 0.022.
. Furthermore, the significant line-by-sex interaction term indicates that there was mutational variation in the difference in olfactory behavior between males and females; i.e., the effects of mutations on olfactory behavior were sex specific. The estimate of the cross-sex genetic correlation (±SE) was rMF = 0.587 ± 0.022.
TABLE 1.
Analyses of variance of avoidance scores of 1339 p[GT1] insertion lines
| Analysis | Source | d.f. | SS | F | P | σ2a |
|---|---|---|---|---|---|---|
| Sexes pooled | Sex | 1 | 32.04 | 38.83 | <0.0001 | Fixed |
| Line | 1338 | 2397.79 | 3.85 | <0.0001 | 0.1209 | |
| Line × sex | 1338 | 1103.81 | 1.77 | <0.0001 | 0.0900 | |
| Error | 8034 | 3735.03 | 0.4649 | |||
| Males | Line | 1338 | 2044.94 | 3.02 | <0.0001 | 0.2557 |
| Error | 4017 | 2031.70 | 0.5058 | |||
| Females | Line | 1338 | 1456.65 | 2.57 | <0.0001 | 0.1662 |
| Error | 4017 | 1703.33 | 0.4240 |
SS, sum of squares.
Variance component.
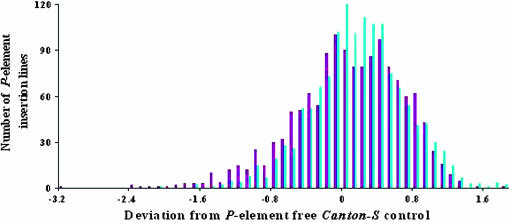
Figure 1.—
Distribution of avoidance scores of 1379 p[GT1] insertion lines from the initial behavioral screen. Blue and violet bars indicate the distributions of female and male avoidance scores, respectively. Data are standardized by calculating the deviation of the avoidance score for each p[GT1] insertion line from its co-isogenic control. Avoidance scores (±SEM) for males and females for the Canton S A line were 4.55 ± 0.12 (n = 20) and 4.03 ± 0.08 (n = 20), respectively; for the Canton S B control, 4.17 ± 0.07 (n = 152) and 4.03 ± 0.08 (n = 152), respectively; for the Canton S D line, 4.18 ± 0.41 (n = 4) and 4.25 ± 0.18 (n = 4), respectively; for the Canton S E line, 4.43 ± 0.16 (n = 16) and 4.38 ± 0.09 (n = 16), respectively; and, for the Canton S F control, 4.00 ± 0.15 (n = 52) and 3.90 ± 0.21 (n = 52), respectively.
Approximately 10% of the lines that showed the lowest avoidance scores were subjected to extensive retesting, which resulted in confirmation of aberrant behavioral responses for 83 p[GT1]-element insertion lines. Transposon insertion sites could be assigned to candidate genes for 55 of the mutant lines by identifying flanking sequences following inverse PCR (Table 2), whereas 28 did not yield amplification products. As the P-element insertion sites for these lines could not be determined, they were not considered further. The significance of the difference between 30 of the P-element insertion lines listed in Table 2 and their co-isogenic controls exceeds a conservative Bonferroni correction for multiple tests. The proportion of mutants represents ∼6% of the total lines screened, similar to a previous screen for smi mutants, which identified ∼4% of the p[lArB]-element insertion lines as hypomorpic behavioral mutants (Anholt et al. 1996).
TABLE 2.
p[GT1] insertion lines with aberrant olfactory avoidance behavior
| Mutational effects
|
P-values from ANOVA
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MASa
|
a/σpb
|
Sexes pooledc
|
Sexes separate
|
|||||||||
| Line | Candidate gene | Cytological location | p[GT1] insertion site | ♀ | ♂ | ♀ | ♂ | S | L | L × S | L♀ | L♂ |
| BG01883 (A) | ftz transcription factor 1 | 75D8-E1 | In intron 1 | 3.6 | 3.71 | 1.01 | 0.72 | NS | **** | NS | *** | NS |
| BG01918 (A) | Phosphoglycerate kinase | 23A3 | 1.8 kb upstream of exon1 | 2.89 | 3.45 | 1.75 | 1.02 | NS | **** | NS | ** | * |
| BG00386 (B) | NMDA receptor 1 | 83A6-7 | 302 bp downstream of 3′ region | 4.15 | 3.82 | 0.21 | 0.34 | NS | * | NS | NS | * |
| BG01037 (B) | βν integrin | 39A1 | 133 bp in exon 1 | 3.93 | 3.92 | 0.44 | 0.27 | NS | ** | NS | * | NS |
| BG01047 (B) | frizzled | 70D4-5 | 100 bp in exon 1 | 3.84 | 4.53 | 0.53 | −0.17 | NS | NS | ** | ** | NS |
| BG01214 (B) | sugarless/CG10064 | 65D4-5 | 275 bp upstream of exon 1 | 3.91 | 3.86 | 0.45 | 0.30 | NS | *** | NS | ** | * |
| BG01244 (B) | couch potato | 90D1-E1 | In intron 2 | 3.88 | 4.06 | 0.49 | 0.17 | NS | * | NS | ** | NS |
| BG01245 (B) | Sema-5c | 68F2 | 197 bp upstream of exon 1 | 2.97 | 3.41 | 1.42 | 0.63 | NS | ***** | NS | *** | *** |
| BG01279 (B) | CG17836 | 91D4-5 | In intron 3 | 4.16 | 3.68 | 0.20 | 0.44 | NS | ** | NS | NS | ** |
| BG01295 (B) | SRY interacting protein 1 | 54B6-7 | 17 bp in exon 1 | 3.48 | 4.13 | 0.90 | 0.12 | NS | *** | * | *** | NS |
| BG01416 (B) | bicoid-interacting protein 3 | 42A13-14 | In intron 1 | 4.36 | 3.65 | −0.00 | 0.47 | NS | * | * | NS | ** |
| BG01533 (B) | No predicted gene | 67E7 | 4.32 | 3.56 | 0.04 | 0.53 | NS | ** | * | NS | ** | |
| BG01543 (B) | Merlin | 18E1 | In exon 1 | 3.22 | 3.19 | 1.17 | 0.79 | NS | ***** | NS | *** | *** |
| BG01563 (B) | CG16708 | 82F11-83A1 | 109 bp upstream of exon 1 | 3.71 | 3.88 | 0.66 | 0.30 | NS | ***** | NS | *** | *** |
| BG01564 (B) | CG14430 | 6E4 | 0.5 kb upstream of exon 1 | 2.97 | 3.14 | 1.42 | 0.83 | NS | ***** | NS | *** | *** |
| BG01596 (B) | CG13377 | 1A1 | 317 bp upstream of exon 1 | 4.24 | 3.70 | 0.12 | 0.42 | NS | * | NS | NS | ** |
| BG01672 (B) | CG14591/SCAP | 42A8 | 27 bp in exon 5 | 3.19 | 3.67 | 1.20 | 0.45 | NS | ***** | NS | *** | * |
| BG01686 (B) | 1.28 | 42B3 | 265 bp upstream of exon 1 | 3.48 | 3.60 | 0.90 | 0.49 | NS | ***** | NS | *** | ** |
| BG01693 (B) | CG10777/CG10778 | 7C3-4 | 25 bp in exon1/1.9 kb downstream of 3′ region | 3.48 | 3.85 | 0.90 | 0.32 | NS | **** | NS | *** | * |
| BG01803 (B) | 1.28 | 42B3 | 265 bp upstream of exon 1 | 3.89 | 3.43 | 0.48 | 0.62 | NS | **** | NS | ** | ** |
| BG01909 (B) | CG14035 | 25C6 | 3.7 kb downstream of 3′ region | 3.65 | 3.13 | 0.72 | 0.83 | NS | ***** | NS | *** | *** |
| BG02022 (B) | CG6301 | 53D11 | 1.4 kb downstream of 3′ region | 3.22 | 2.71 | 1.17 | 1.14 | NS | ***** | NS | *** | *** |
| BG02053 (B) | Sema-5c | 68F2 | 55 bp upstream of exon 1 | 1.07 | 1.66 | 3.01 | 1.73 | NS | ***** | NS | *** | *** |
| BG02077 (B) | Rtnl1 | 25B9-C1 | In intron 1 | 3.68 | 2.76 | 0.69 | 1.10 | NS | ***** | ** | *** | *** |
| BG02081 (B) | Rtnl1 | 25B9-C1 | In intron 1 | 3.23 | 2.78 | 1.16 | 1.09 | NS | ***** | NS | *** | *** |
| BG02115 (B) | Calreticulin | 85E | 87 bp in exon1 | 3.76 | 3.49 | 0.60 | 0.58 | NS | ***** | NS | ** | ** |
| BG02169 (B) | High mobility group protein D | 57F10 | 633 bp of transcription initiation site | 4.14 | 3.78 | 0.22 | 0.37 | NS | * | NS | NS | * |
| BG02181 (B) | Glutamate dehydrogenase | 95C13-D1 | 126 bp 5′ of transcription initiation site | 3.93 | 3.85 | 0.43 | 0.32 | NS | ** | NS | * | * |
| BG02200 (B) | CG6782/CG6783 | 86E10 | In intron 1 | 3.38 | 3.27 | 1.00 | 0.73 | NS | ***** | NS | *** | *** |
| BG02206 (B) | lilliputian | 23C1-C3 | In intron 2 | 3.40 | 4.32 | 0.98 | −0.01 | NS | ** | *** | *** | NS |
| BG02209 (B) | escargot | 35D2 | 321 bp upstream of exon 1 | 3.55 | 3.15 | 0.83 | 0.82 | NS | ***** | NS | *** | *** |
| BG02251 (B) | innexin 2 | 6E4 | In intron 1 | 3.23 | 3.06 | 1.16 | 0.88 | NS | ***** | NS | *** | *** |
| BG02314 (B) | 1.28 | 42B3 | 289 bp upstream of exon 1 | 3.82 | 3.61 | 0.55 | 0.49 | NS | **** | NS | ** | ** |
| BG02320 (B) | Toll | 97D2 | 44 bp upstream of exon 1 | 3.78 | 3.51 | 0.59 | 0.57 | NS | **** | NS | ** | *** |
| BG02326 (B) | CG13889 | 61D2 | 68 bp in exon 1 | 4.22 | 3.73 | 0.14 | 0.41 | NS | * | NS | NS | * |
| BG02327 (B) | pipsqueak | 47A13-B1 | In intron 2 | 3.29 | 3.39 | 1.09 | 0.65 | NS | ***** | NS | *** | *** |
| BG02348 (B) | Heat shock protein 23 | 67B3 | 298 bp upstream of exon 1 | 3.19 | 3.15 | 1.20 | 0.82 | NS | ***** | NS | *** | *** |
| BG02354 (B) | escargot | 35D2 | 350 bp upstream of exon 1 | 3.43 | 3.11 | 0.95 | 0.85 | NS | ***** | NS | *** | *** |
| BG02386 (B) | Sema-5c | 68F2 | 81 bp upstream of exon 1 | 3.55 | 2.81 | 0.83 | 1.06 | NS | ***** | * | *** | *** |
| BG02391 (B) | neuralized | 85C2-3 | In intron 1 | 3.97 | 3.78 | 0.39 | 0.37 | NS | **** | NS | ** | ** |
| BG02418 (B) | CG14509 | 98F13-99A1 | In intron 1, 6.8 kb from 5′ region | 3.99 | 3.93 | 0.38 | 0.26 | NS | * | NS | * | NS |
| BG02439 (B) | CG32556 | 16B12-C1 | 324 bp upstream of exon 1 | 3.29 | 2.93 | 1.09 | 0.98 | NS | ***** | NS | *** | *** |
| BG02470 (B) | CG8963 | 53 E4 | 80 bp in exon 1 | 3.37 | 3.20 | 1.01 | 0.79 | NS | ***** | NS | *** | *** |
| BG02491 (B) | Ras-related protein | 3E5-6 | 4 kb upstream of exon 1 | 3.78 | 4.03 | 0.59 | 0.19 | NS | ** | NS | ** | NS |
| BG02510 (B) | chameau | 27F3-4 | 2.9 kb upstream of exon 1 | 3.97 | 3.76 | 0.39 | 0.39 | NS | ** | NS | * | * |
| BG02522 (B) | CG15816 | 16C1-C8 | 876 bp upstream of of exon 1 | 3.98 | 3.78 | 0.39 | 0.37 | NS | ** | NS | * | * |
| BG02566 (B) | Calreticulin | 85E1 | 114 bp in exon 1 | 3.69 | 3.74 | 0.69 | 0.40 | NS | ***** | NS | *** | *** |
| BG02601 (B) | CG14782/CG14781 | 2B1 | In intron 2/exon 1 | 3.85 | 3.54 | 0.51 | 0.54 | NS | ***** | NS | *** | *** |
| BG02745 (B) | CG14411/CG14408 | 12F5 | In exon 1 | 3.96 | 3.88 | 0.40 | 0.30 | NS | *** | NS | ** | * |
| BG01015 (F) | Moesin | 8B4-6 | In intron 1 | 3.97 | 4.00 | 0.41 | 0.33 | NS | *** | NS | * | * |
| BG01707 (F) | CG6301 | 53D11 | 1.4 kb downstream of 3′ region | 3.76 | 4.39 | 0.73 | 0.09 | * | ** | * | *** | NS |
| BG01084 (F) | CG9238 | 70E1-2 | 49 bp upstream of exon 1 | 4.06 | 3.66 | 0.37 | 0.90 | NS | **** | NS | * | ** |
| BG01095 (F) | pointed/DNApolymarase E | 94E10-13 | 742 bp in exon 1 | 4.03 | 4.08 | 0.41 | 0.44 | NS | ** | NS | * | * |
| BG01574 (F) | Sema-5c | 68F2 | 82 bp upstream of exon 1 | 3.93 | 4.11 | 0.53 | 0.40 | NS | ** | NS | * | * |
| BG01796 (F) | Spinophilin | 62E4-5 | In intron 1 | 3.68 | 4.35 | 0.84 | 0.13 | * | ** | * | *** | NS |
Candidate genes used for analysis of epistasis are underlined. Letters in parentheses after the line name denote different co-isogenic Canton S host strains for the p[GT1]-element insertion. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, *****P < 0.00001. P-values < 0.0001 exceed Bonferroni correction for multiple tests.
MAS, mean avoidance scores.
Standardized mutational effect (see text for explanation).
S and L denote the main cross-classified effects of sex and line, respectively, in the ANOVA of avoidance scores.
Consistent with the imperfect correlation between the behavior of males and females, we observed variation in sexual dimorphism for the effects of p[GT1]-element insertions on olfactory behavior, as indicated by a significant line-by-sex interaction term in the ANOVA (Table 2). Among the 55 mutants with known insertion sites, 9 were significantly sex specific: 2 mutations affected males only, 5 affected females only, and 2 mutations affected both sexes, but with a large difference in the magnitude of their effects in males and females (Table 2). Several p[GT1] elements inserted near or in the same candidate gene, yielding independent duplicate insertion lines for Rtl1, Crc, and esg and quadruplicate transposon insertions for 1.28 and Sema-5C. As our mutagenesis screen is far from saturation, these candidate genes likely represent hot spots for p[GT1]-element insertion.
Candidate genes, disrupted by the insertion of the transposon, that contribute to olfactory behavior include a wide range of gene ontology categories, including transcriptional regulators, neurodevelopmental genes, and signal transduction components. Gene families that are clustered in the genome are notably refractory to P-element insertion and, therefore, we did not identify transposons in or near odorant receptor genes or genes encoding odorant-binding proteins. We quantified the standardized mutational effects of the P-element mutations as a/σP, where a is one-half of the difference between the homozygous mutant and control line, and σP is the phenotypic standard deviation of the control (Falconer and Mackay 1996). The average standardized mutational effect of the significant P-element insertions was 0.83 in females and 0.64 in males, with a range of 0.37–3.01 in females and 0.30–1.73 in males. Thus the effects of P-element insertions on odor-guided behavior ranged in magnitude from moderate to large (an effect of three standard deviations is nearly Mendelian). The variation in sexual dimorphism, range of magnitude of phenotypic effects, diversity of candidate genes, and proportion of mutant lines among the initial test population resemble observations from our previous p[lArB]-element screen for smi mutants using the same behavioral assay (Anholt et al. 1996).
Effects of p[GT1] insertions on expression of candidate genes implicated in odor-guided behavior:
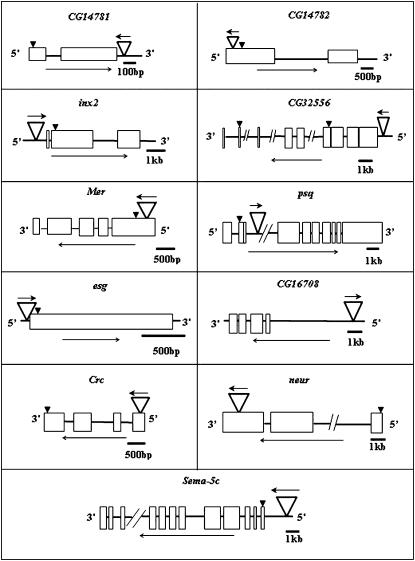
We selected 10 p[GT1] insertion lines with highly significant effects on olfactory behavior for further analysis; the average mutational effect of these lines is 1.06 and 0.77 phenotypic standard deviation in females and males, respectively (Table 2, Figure 2). In 6 of these lines, the transposon is located either within the immediate vicinity of the transcription initiation site or within an intron of the candidate gene (innexin2; CG32556, a predicted gene of unknown function; the transcriptional regulators pipsqueak and escargot; CG16708, which encodes a d-erythrosphingosine kinase; and Semaphorin-5C). In three lines, the p[GT1] element has inserted in an exon (Merlin, Calreticulin, and neuralized). In one case, the transposon has inserted in the vicinity of two predicted genes of unknown function—in the exon of CG14782, located immediately upstream of the 3′-end of the neighboring gene CG14781.
Figure 2.—
Diagram of p[GT1] insertion sites in candidate genes. Inverted open triangles indicate the P-element insertion sites. Boxes indicate exons. Orientation of the candidate gene and the P element are indicated by the long arrows below each diagram and the small arrows above the inverted triangles, respectively. Arrowheads indicate the position of the translation initiation ATG site of the coding sequence.
Among these candidate genes, pipsqueak (psq) is a transcriptional regulator associated with early development (Weber et al. 1995:Lehmann et al. 1998; Siegmund and Lehmann 2002), and escargot (esg) is a transcriptional regulator associated with development of the nervous system, including sensory bristles (Whiteley et al. 1992; Hayashi et al. 1993; Adelilah-Seyfried et al. 2000; Norga et al. 2003). Semaphorin 5C (Sema-5C) has been implicated in early embryogenesis (Khare et al. 2000). and neuralized (neur) encodes a ubiquitin ligase (Lai et al. 2001; Yeh et al. 2001), which regulates neurogenesis mediated by the Notch-Delta signaling pathway (Boulianne et al. 1991; Yeh et al. 2000) and has been implicated in the development of olfactory sensilla (Jhaveri et al. 2000). Merlin (Mer) is a homolog of the neurofibromatosis 2 tumor suppressor factor, implicated in axis specification during early embryonic development (MacDougall et al. 2001), and innexin2 (inx2) encodes a gap junction protein essential for epithelial morphogenesis (Bauer et al. 2002, 2004). The d-erythrosphingosine kinase, encoded by CG16708 (Renault et al. 2002), has been implicated in autophagic cell death (Gorski et al. 2003). Calreticulin (Crc) encodes a calcium-binding protein involved in intracellular protein transport and exocytosis, as well as in the development of the nervous system (Prokopenko et al. 2000), and Crc mutants are defective in olfactory avoidance behavior to 4-methylcyclohexanol, 3-octanol, and benzaldehyde (Stoltzfus et al. 2003). CG14872 has been implicated as a putative guanyl nucleotide exchange factor with a developmental function, whereas the functions of the gene products encoded by CG14781 and CG32556 remain unknown.
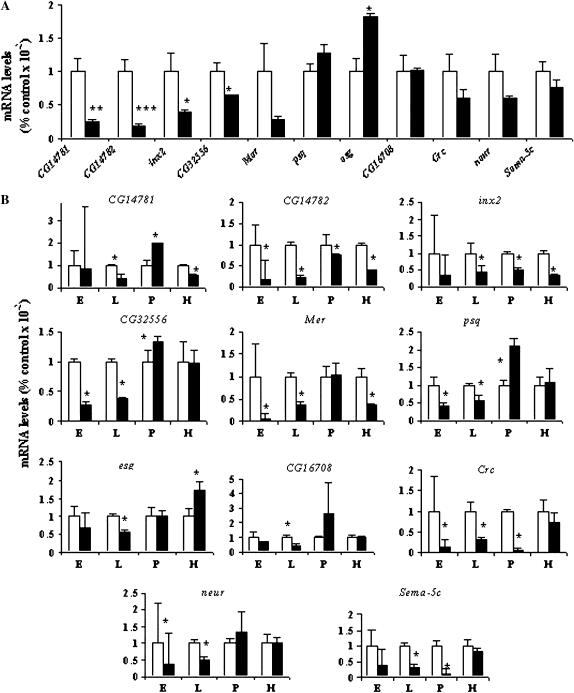
We performed quantitative RT–PCR to assess to what extent insertions of the p[GT1] elements disrupt expression of these candidate genes. First, we performed assays of control and mutant lines side-by-side on independent triplicate RNA samples extracted from whole flies (Figure 3A). Statistically significant effects on gene expression ranged from reductions in the cases of Mer, inx2, CG14781, CG14782, and CG32556 to significant increases in gene expression as in the case of esg. However, gene expression levels were not significantly different in the case of the remaining transposon-tagged genes such as Sema-5c, psq, neur, Crc, and CG16708 in whole bodies of adult female flies.
Figure 3.—
Quantitative RT–PCR analyses of candidate gene expression levels in whole female flies (A) and at four different developmental stages (B). mRNA expression levels are standardized against the Canton S B control (open bars). Solid bars show the relative expression levels in the p[GT1] insertion lines. E, embryos; L, larvae; P, pupae; H, adult female heads. In A, significance levels for differences between mutants and the control were estimated with two-tailed Student's t-test. *P < 0.05; **P < 0.01; ***P < 0.001. In B, differences in gene expression levels, indicated by the asterisks, were considered significant when 95% confidence intervals around the mean value of the mutant and that of the control did not overlap.
We assessed whether differences in expression levels that occur during earlier developmental stages could account for the aberrant behavioral phenotypes observed in adults. Surprisingly, p[GT1]-induced disruptions of gene expression showed a marked dependence on developmental stage (Figure 3B). Only inx2 and CG14782 showed reduced expression from embryonic stages throughout development. Expression of Mer is reduced in all developmental stages, except the pupa stage. CG16708 shows a profound reduction in expression only in the larval stage, while CG32556 is reduced in both embryos and larvae. After puparium formation, CG16708 and CG32556 expression levels are indistinguishable from control levels. Effects of the p[GT1] insertion in esg are especially intriguing, showing reduction in embryos and larvae, but a significant increase in transcription in adult heads, consistent with the expression profile observed in adult whole bodies (Figure 3A). The expression profile of esg is mimicked by psq, albeit less extremely. Disruption of neur expression is especially pronounced in embryos and larvae. In the cases of Crc and Sema-5C, expression is decreased in embryos, larvae, and especially during puparium formation, but is restored to near control levels in adult flies (Figures 3, A and B). Both CG14781 and its neighboring gene CG14782 are affected by the insertion of the p[GT1] element. In CG14781, there is a reduction of expression in larvae and adults with an upregulation in pupae. The reduction in expression in whole bodies for these genes is much larger than that observed in heads only (compare A and B in Figure 3), suggesting that CG14781 and CG14782 are also expressed abundantly elsewhere in the body. Thus, the transcriptional effects exerted by the transposons are surprisingly diverse and heterogeneous during development, possibly reflecting effects on different promoter/enhancer elements that are recruited for regulation of gene expression at different developmental stages. Our results indicate that, at least in some cases, hypomorphic disruptions that occur in early development may account for the observed aberrant adult behavioral phenotype.
Dose-response relationships of olfactory avoidance behavior in p[GT1] insertion lines:
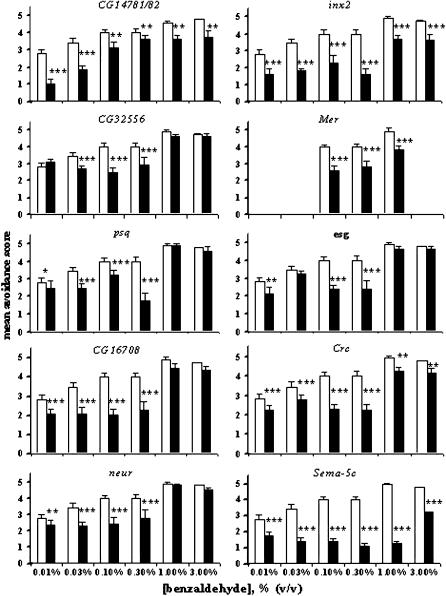
To further characterize the phenotypic effects of P-element insertions on the olfactory avoidance response to benzaldehyde and to establish a discriminating concentration range for detection of enhancer or suppressor effects in our subsequent analysis of epistasis, we measured dose-response relationships over a wide range of benzaldehyde concentrations from 0.01 to 3.0% (v/v) (Figure 4). There was no linear relationship between the magnitude of reduction of transcript abundance in adult flies (Figure 3) and reduction in the avoidance response to benzaldehyde. For example, the p[GT1] insertion near Sema-5C has no detectable effect on transcript abundance in the adult, but profoundly affects olfactory behavior. Whereas odor-guided behavior is restored to control levels at saturating concentrations of benzaldehyde in CG32556, psq, esg, and neur, in all other cases avoidance responses are statistically significantly reduced over the entire range of concentrations in mutants compared to the Canton-S control (Figure 4). On the basis of dose-response relationships, we selected 0.1 and 0.3% (v/v) benzaldehyde as stimulus concentrations for our subsequent analysis of epistatic interactions, as this concentration range elicited a significant behavioral response above background, yet was below saturation for the Canton-S control and discriminated responses of all mutant lines from the control.
Figure 4.—
Dose responses for olfactory avoidance behavior to benzaldehyde of female flies. Solid bars correspond to p[GT1]-element insertion lines and open bars to the Canton S B control. Significance levels for differences between mutants and the control were estimated with Student's t-test. *P < 0.05; **P < 0.01; ***P < 0.001.
Epistasis among 10 co-isogenic p[GT1]-insertion lines with transposon insertions at candidate genes implicated in odor-guided behavior:
Since the transposons in the 10 p[GT1] insertion lines were introduced in the same genetic background, we can examine epistasis among them by separating heterozygous effects from epistasis in double heterozygotes using a half-diallel crossing design (Table 3; Fedorowicz et al. 1998). The average dominance effect for each transposon-tagged gene as a heterozygote in combination with all other p[GT1] insertion lines can be estimated as its GCA. The GCA values of both parents allow us to predict the avoidance score expected for the trans-heterozygous offspring. Any statistically significant deviation of the observed value from the predicted value (SCA) in this scenario is attributable to epistasis. Enhancer effects will result in the trans-heterozygote showing an avoidance score that is more biased toward the mutant phenotype than predicted, whereas suppressor effects will reflect an avoidance score that is more wild type than predicted.
TABLE 3.
Diallel crosses among 10 p[GT1] insertion lines with aberrant olfactory avoidance behavior
| CG 32556 | CG 16708 | CG14781/CG14782 | Crc | inx2 | Mer | psq | neur | esg | Sema-5c | T | GCA | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | ||||||||||||
| CG32556 | 4.170 | 4.260 | 4.400 | 4.290 | 3.680 | 4.510 | 4.375 | 4.220 | 4.305 | 38.210 | 0.049 | |
| CG16708 | 4.325 | 4.195 | 4.215 | 3.620 | 4.165 | 4.240 | 4.270 | 4.060 | 37.260 | −0.070 | ||
| CG14781/CG14782 | 4.300 | 4.365 | 3.550 | 4.385 | 4.335 | 4.540 | 3.980 | 38.040 | 0.028 | |||
| Crc | 4.435 | 4.055 | 4.525 | 4.150 | 4.210 | 4.145 | 38.415 | 0.075 | ||||
| inx2 | 3.465 | 4.300 | 4.225 | 4.285 | 4.195 | 37.775 | −0.005 | |||||
| Mer | 3.840 | 4.260 | 4.030 | 3.590 | 34.090 | −0.466 | ||||||
| psq | 4.525 | 4.405 | 4.420 | 39.075 | 0.157 | |||||||
| neur | 4.595 | 4.195 | 38.900 | 0.135 | ||||||||
| esg | 4.485 | 39.040 | 0.153 | |||||||||
| Sema-5c | 37.375 | −0.055 | ||||||||||
| B | ||||||||||||
| CG32556 | 4.615 | 4.750 | 4.640 | 4.285 | 4.565 | 4.900 | 4.710 | 4.570 | 4.220 | 41.255 | −0.002 | |
| CG16708 | 4.450 | 4.660 | 4.645 | 4.525 | 4.755 | 4.775 | 4.600 | 4.590 | 41.615 | 0.043 | ||
| CG14781/CG14782 | 4.670 | 4.530 | 4.500 | 4.530 | 4.825 | 4.550 | 4.410 | 41.215 | −0.007 | |||
| Crc | 4.640 | 4.475 | 4.895 | 4.710 | 4.805 | 4.580 | 42.075 | 0.101 | ||||
| inx2 | 4.095 | 4.785 | 4.485 | 4.770 | 4.575 | 40.810 | −0.057 | |||||
| Mer | 4.525 | 4.410 | 4.180 | 4.075 | 39.350 | −0.240 | ||||||
| psq | 4.755 | 4.830 | 4.664 | 42.639 | 0.172 | |||||||
| neur | 4.750 | 4.640 | 42.060 | 0.099 | ||||||||
| esg | 4.425 | 41.480 | 0.027 | |||||||||
| Sema-5c | 40.179 | −0.136 | ||||||||||
Avoidance scores for trans-heterozygous females and calculated GCA values at 0.1% v/v (A) and 0.3% v/v (B) benzaldehyde are listed. Parental homozygous p[GT1] insertion lines are indicated on the top row and in the first column of A and B. The means of avoidance scores are derived from 20 replicate measurements for each hybrid cross. “T” is the sum of avoidance scores used to compute the GCA values. Significant GCA values are underlined (see supplemental Table 1 at http://www.genetics.org/supplemental/.)
Analysis of variance showed significant variation in avoidance scores between the two stimulus environments and among double-heterozygous genotypes as well as a significant genotype-by-environment interaction term (Table 4). The effect of genotype was also highly significant (P < 0.0001) when analyses were performed separately for each environment (Table 4). Next, we asked whether we could observe environment-dependent variation in GCA and SCA values. Analysis of variance revealed significant variation in GCA and SCA values when pooled over both concentrations of benzaldehyde (Table 5). In addition to variation due to the environment per se, both the GCA by environment and SCA by environment interaction terms were statistically significant (Table 5) and we also observed statistically significant variation in both GCA and SCA when analyses were performed separately for each concentration of benzaldehyde (Table 5). As expected from the steeper rise in the dose response curve at 0.1% (v/v) benzaldehyde than at 0.3% (v/v) benzaldehyde, which is near the inflection point that approaches the response maximum, variation in mean avoidance scores among the double heterozygotes was greater at the lower stimulus concentration (Table 4, Figure 5). The genotype-by-environment interaction is evident from both the analysis of variance (Table 4) and the crossing over of reaction norms (Figure 5).
TABLE 4.
Analyses of variance of avoidance response of double-heterozygote genotypes to two concentrations of benzaldehyde
| Analysis | Source | d.f. | SS | F | P |
|---|---|---|---|---|---|
| 0.1% (v/v) and 0.3% (v/v) (pooled) | Environment | 1 | 66.083 | 255.46 | <0.0001 |
| Genotype | 44 | 74.369 | 6.53 | <0.0001 | |
| Genotype × environment | 44 | 23.961 | 2.11 | <0.0001 | |
| Error | 1710 | 442.352 | |||
| 0.1% (v/v) | Genotype | 44 | 64.597 | 5.24 | <0.0001 |
| Error | 855 | 239.499 | |||
| 0.3% (v/v) | Genotype | 44 | 33.732 | 3.23 | <0.0001 |
| Error | 855 | 202.853 |
SS, sum of squares.
TABLE 5.
Analyses of variance of general and specific combining abilities of double-heterozygote genotypes at two concentrations of benzaldehyde
| Analysis | Source | d.f. | SS | F | P |
|---|---|---|---|---|---|
| 0.1% (v/v) and 0.3% (v/v) (pooled) | E | 1 | 66.083 | 255.46 | <0.0001 |
| GCA | 9 | 61.424 | 26.38 | <0.0001 | |
| SCA | 35 | 12.944 | 1.43 | 0.0501 | |
| GCA × E | 9 | 7.599 | 3.26 | 0.0006 | |
| SCA × E | 35 | 16.362 | 1.81 | 0.0027 | |
| Error | 1710 | 442.352 | |||
| 0.1% (v/v) | GCA | 9 | 48.027 | 19.05 | <0.0001 |
| SCA | 35 | 16.570 | 1.69 | 0.0080 | |
| Error | 855 | 239.499 | |||
| 0.3% (v/v) | GCA | 9 | 20.996 | 9.83 | <0.0001 |
| SCA | 35 | 12.737 | 1.53 | 0.0258 | |
| Error | 855 | 202.853 |
SS, sum of squares; E, environment.
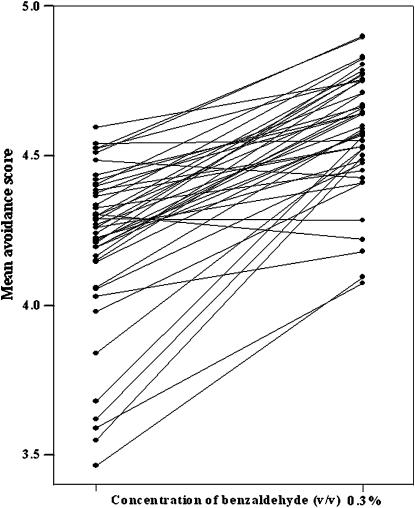
Figure 5.—
Variation in mean avoidance scores of trans-heterozygotes at two concentrations of benzaldehyde. Note the greater variation in scores at the lower stimulus concentration and the crossing over of reaction norms, which reflect genotype-by-environment interactions.
On the basis of previous observations (Fedorowicz et al. 1998), we predicted that we would uncover epistatic interactions for at least some of the trans-heterozygotes among the 10 P-element insertion lines. To assess to what extent the manifestation of such epistatic networks could be influenced by subtle changes in the chemosensory environment, we measured avoidance responses for trans-heterozygotes and established GCA values at the two stimulus concentrations of 0.1% (v/v) and 0.3% (v/v) benzaldehyde (Tables 3 and 5; supplemental Table 1 at http://www.genetics.org/supplemental/). We used the GCA values to estimate SCA values that showed statistically significant deviations from expected values on the basis of the GCA estimates (Table 5 and supplemental Table 2 at http://www.genetics.org/supplemental/).
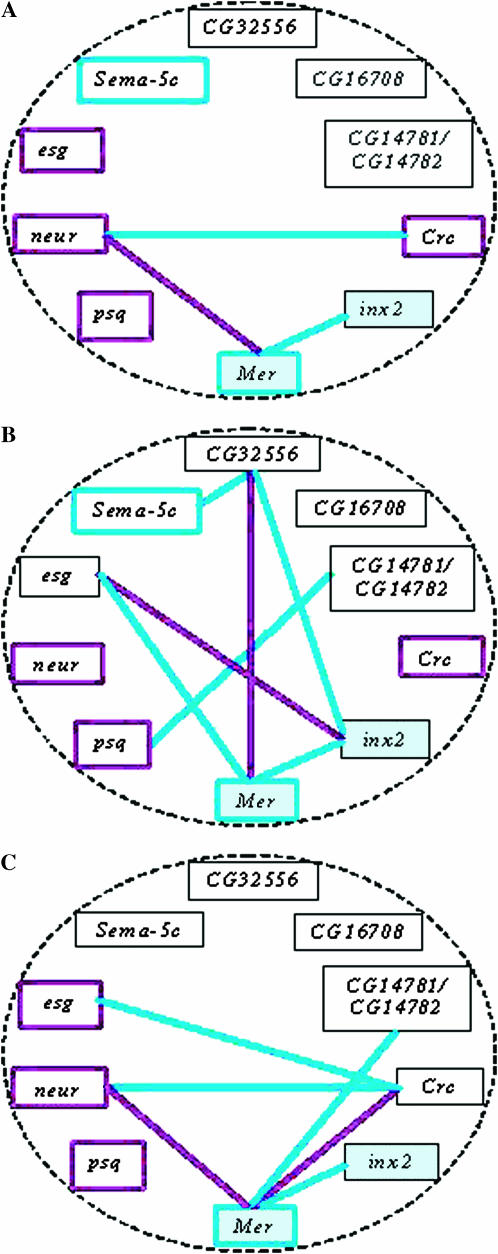
When the analysis was performed with the two stimulus concentrations pooled, we observed three epistatic interactions averaged over both stimulus concentrations: an enhancer effect between Mer and neur and suppressor effects between Mer and inx 2 and between Crc and neur (Figure 6A). However, the pattern of epistatic interactions was greatly enriched in the two individual chemosensory environments. We identified epistatic interactions in six trans-heterozygous genotypes when avoidance responses were quantified at 0.1% (v/v) benzaldehyde (Figure 6B). Enhancer effects were observed between Mer and Crc and between Mer and neur, whereas suppressor effects were evident between Crc and neur, Mer and inx2, Mer and CG14781/CG14782, and Crc and esg (Figure 6B). Surprisingly, the epistatic network shifted when avoidance responses were measured at a threefold higher benzaldehyde concentration (0.3% v/v). Under this condition, epistatic interactions comprised seven trans-heterozygous genotypes and included two enhancer effects, Mer–CG32556 and esg–inx2, and five suppressor effects, Sema-5C–CG32556, inx2–CG32556, psq–CG14781/CG14782, Mer–inx2, and Mer–esg (Figure 6C). Thus, a dynamic epistatic network is evident in which a small number of genes, such as Mer and inx2, emerge as nodes around which enhancer/suppressor effects are shaped, depending on the chemosensory environment.
Figure 6.—
Epistatic interactions between 10 p[GT1] insertion lines. (A) A diagram of enhancer (violet lines) and suppressor (blue lines) effects among transposon-tagged candidate genes when the analysis is performed with data pooled from both stimulus concentrations. Calculated SCA values for significant epistatic interactions are 0.1770 (Mer-neur), −0.2296 (Mer-inx2), and −0.1686 (Crc-neur). (B) A diagram of enhancer (violet lines) and suppressor (blue lines) effects among transposon-tagged candidate genes observed at 0.1% (v/v) benzaldehyde. Calculated SCA values for significant epistatic interactions are −0.2138 (Mer-CG14781/CG14782), 0.2444 (Mer-Crc), −0.2619 (Crc-neur), −0.2194 (Crc-esg), −0.2656 (Mer-inx2), and 0.3888 (Mer-neur). (C) A diagram of enhancer (violet lines) and suppressor (blue lines) effects among transposon-tagged candidate genes observed at 0.3% (v/v) benzaldehyde. Calculated SCA values for significant epistatic interactions are −0.2416 (CG32556-inx2), 0.2209 (CG32556-Mer), −0.2278 (CG32556-Sema-5C), −0.2197 (CG14781/CG14782-psq), −0.1935(Mer-inx2), 0.2153 (inx2-esg), and −0.1922 (Mer-esg). Mer and inx2 are highlighted against a blue background to emphasize their central positions and the invariant interaction between them in both environments. Violet- and blue-bordered boxes indicate significant positive and negative GCA values of the indicated candidate gene, respectively. A complete listing of all SCA values is available in supplemental Table 1 at http://www.genetics.org/supplemental/.
DISCUSSION
Previously, we described how co-isogenic P-element mutations have revealed extensive epistasis as a hallmark of the genetic architecture of odor-guided behavior (Anholt et al. 2003; Anholt 2004). Pervasive epistasis appears to be a characteristic feature of the genetic architecture of complex traits in Drosophila (Mackay 2004) and also influences sternopleural and abdominal bristle numbers (Dilda and Mackay 2002) and heat-stress-induced loss of locomotor coordination (van Swinderen and Greenspan 2005).
Since behaviors are complex traits that mediate interactions between an organism and its environment, behavioral phenotypes are expected to be especially susceptible to genotype-by-environment interactions. It was postulated previously that epistatic genetic networks for behavioral traits are modulated by sex, the physical and social environment, and developmental history (Anholt 2004). Here we provide direct evidence that confirms variation in sexual dimorphism among effects of P-element insertions on genes that contribute to olfactory avoidance behavior. Furthermore, we demonstrate that disruption of genes that act early in development can result in significant deficits in adult olfactory behavior, with effects ranging from subtle to large. Finally, we show that the manifestation of epistatic interactions between mutant alleles is exquisitely sensitive to the chemosensory environment.
The p[GT1]-element insertional mutagenesis screen for genes affecting olfactory avoidance behavior reported here is more extensive than an earlier screen, which relied on insertions of the p[lArB] transposon (Anholt et al. 1996), and has generated approximately six times more candidate genes. The proportion of aberrant lines from among the total number of lines screened, however, is similar. The high broad-sense mutational heritability that we observed may be attributable to mutations with large effects on olfactory behavior and/or a large mutational target size (i.e., many genes potentially affect this trait). This is consistent with the high mutational heritability observed in our previous study of 379 P-element insert lines (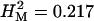 , averaged over second and third chromosomes; Anholt et al. 1996).
, averaged over second and third chromosomes; Anholt et al. 1996).
The p[GT1] dual gene-trap element was designed to insert into target genes (Lukacsovich et al. 2001). Indeed, in most instances, this transposon has inserted in exons, introns, or in close proximity to promoter/enhancer regions of the candidate genes (Figure 2). Rigorous evidence that the tagged gene is indeed responsible for the mutant phenotype is generally provided by demonstrating phenotypic rescue by P-element excision or introduction of a wild-type transgene into the mutant background. Transgenic rescue is technically challenging in the case of hypomorphic mutants where—as in this case—either under- or overexpression can result in aberrant behavioral phenotypes. The advantage of the p[GT1] transposon is that the intragenic location of the insertion itself and its corresponding effects on gene expression strongly implicate the candidate gene as causal to the phenotype, although additional effects on neighboring genes cannot be excluded. For example, one P-element insertion affects expression of both CG14781 and CG14782. Both genes encode transcripts of unknown function and it remains to be determined whether disruption of either one or both of these genes contributes to the observed aberrant olfactory avoidance responses.
Aberrant behavioral phenotypes in our P-element insertion lines could result from a reduction in mRNA of the mutant allele or from a change in the size of the message—which can result in changes in the amount of active protein produced—or from changes in mRNA splicing and ratios of alternative transcripts. Disruption of the open reading frame could conceivably occur when P elements insert into exons. Extensive studies at the protein level would be needed for a complete characterization of the molecular correlates associated with the P-element-induced mutations. A further complication is our previous observation that single P-element insertions result in transcriptional alterations at other loci in the genome (Anholt et al. 2003). Thus, indirect effects arising from altered expression elsewhere in the transcriptional network may contribute to the observed mutant phenotype.
One striking observation is how much the effects of single P-element insertions on gene expression depend on developmental stage. This phenomenon most likely reflects differential disruption by the transposon of distinct promoter elements that are active at different developmental stages. It should also be noted that several genes previously implicated only in early development, such as Sema-5C (Khare et al. 2000) and esg (Whiteley et al. 1992; Hayashi et al. 1993; Adelilah-Seyfried et al. 2000), are expressed also in adults and likely perform as yet unknown functions that may either recapitulate or be distinct from their functions in embryonic or larval stages. In cases in which gene expression in adult heads is similar in mutants and the wild-type control, adverse effects on odor-guided behavior are likely the consequence of early disruptions in the animal's developmental blueprint. Such disruptions are predicted to be subtle, as healthy adults emerge without apparent morphological defects. In cases in which gene expression is altered in adults as well as in prior developmental stages, we cannot discern whether aberrant olfactory behavior is a remnant of early developmental defects or directly related to functional disruption of the gene product in the adult brain. Future detailed neuroanatomical studies will be needed to determine whether alterations in neuronal connectivity or structure can be resolved or whether the precise spatial distribution of gene expression has changed in the mutant vs. its control at different developmental stages.
Availability of a co-isogenic collection of P-element insertion lines enables identification of epistatic effects by generating double heterozygotes according to the half-diallel crossing scheme of Griffing (1956) and by statistically separating heterozygous effects from epistasis. Previously, we conducted a screen of 379 co-isogenic lines with autosomal p[lArB]-element insertions and identified 14 smi loci (Anholt et al. 1996). Since all these lines were in the same genetic background, except for the differential insertion site of a single P element, we were able to assess the extent of epistasis among them. We accomplished this by generating all possible double heterozygotes of 12 smi lines in a half-diallel design and by separating average heterozygous dominance effects from epistatic enhancer and suppressor effects (Fedorowicz et al. 1998). The surprising result of this analysis revealed an extensive network of epistatic interactions among 8 of the 12 genes, which would not have been predicted a priori from a limited number of independently isolated mutants. Transcriptional profiling of 5 of these smi loci showed that disruption by a P element of its target gene results in changes in gene expression of, on average, ∼125 other genes. Quantitative complementation tests between mutants of genes with altered transcriptional regulation and the original smi mutants showed that transcriptional epistasis is significantly correlated with epistasis at the level of phenotype (Anholt et al. 2003).
Here, we analyzed epistatic interactions among 10 new p[GT1]-insertion lines. We used this new collection of mutants to extend our previous studies by asking whether epistatic networks among these loci are invariant or dynamic. It should be noted, however, that the half-diallel crossing design is limited in practice, as the number of measurements to be performed increases exponentially with each additional mutant (Anholt and Mackay 2004). Furthermore, the epistatic interactions that we observed are likely sensitive to the sex environment. However, we limited our analysis to females only, as four of the transposon-tagged candidate genes were located on the X chromosome. Despite the restricted scope of our experimental design, we were able to resolve epistatic interactions. Surprisingly, the nature of enhancer and suppressor effects was highly dynamic and depended strongly on the concentration of the stimulus used to elicit the olfactory avoidance response (Figure 6). Two transposon-tagged candidate genes, Mer and inx2, engage in epistatic interactions at both concentrations of benzaldehyde (Figure 6). It is tempting to hypothesize that these genes may represent focal points around which a dynamic epistatic network revolves. However, as the size of the networks considered here and the number of genes involved is small, this assessment at present is speculative. Nevertheless, the plasticity of genetic networks that mediate complex traits, illustrated here, suggests a dynamic continuum of epistatic partnerships that comprises the full spectrum from highly stable hubs to fragile, fleeting interactions.
Acknowledgments
We thank Christina Grozinger for helpful advice with the quantitative PCR experiments, Richard Lyman for assistance with statistical analyses, and Tatiana Morozova for helpful discussions. This work was supported by grants from the National Institutes of Health (GM59469 to R.R.H.A. and GM45146 to T.F.C.M.) and a Fogarty International Research Collaboration Award (TW007070 to J.J.F. and R.R.H.A.). D.S. was supported by a North Carolina State University W. M. Keck Center training fellowship from a grant from the University of North Carolina Office of the President (RA05-03). This is a publication of the W. M. Keck Center for Behavioral Biology at North Carolina State University.
References
- Abdelilah-Seyfried, S., Y. M. Chan, C. Zeng, N. J. Justice, S. Younger-Shepherd et al., 2000. A gain-of-function screen for genes that affect the development of the Drosophila adult external sensory organ. Genetics 155: 733–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anholt, R. R. H., 2004. Genetic modules and networks for behavior: lessons from Drosophila. BioEssays 26: 1299–1306. [DOI] [PubMed] [Google Scholar]
- Anholt, R. R. H., and T. F. C. Mackay, 2004. Quantitative genetic analyses of complex behaviours in Drosophila. Nat. Rev. Genet. 5: 838–849. [DOI] [PubMed] [Google Scholar]
- Anholt, R. R. H., R. F. Lyman and T. F. C. Mackay, 1996. Effects of single P-element insertions on olfactory behavior in Drosophila melanogaster. Genetics 143: 293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anholt, R. R. H., C. L. Dilda, S. Chang, J. J. Fanara, N. H. Kulkarni et al., 2003. The genetic architecture of odor-guided behavior in Drosophila: epistasis and the transcriptome. Nat. Genet. 35: 180–184. [DOI] [PubMed] [Google Scholar]
- Bauer, R., C. Lehmann, B. Fuss, F. Eckardt and M. Hoch, 2002. The Drosophila gap junction channel gene innexin 2 controls foregut development in response to Wingless signaling. J. Cell Sci. 115: 1859–1867. [DOI] [PubMed] [Google Scholar]
- Bauer, R., C. Lehmann, J. Martini, F. Eckardt and M. Hoch, 2004. Gap junction channel protein innexin2 is essential for epithelial morphogenesis in the Drosophila embryo. Mol. Biol. Cell 15: 2992–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen, H. J., R. W. Levis, G. Liao, Y. He, J. W. Carlson et al., 2004. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167: 761–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton, R., S. Sachse, S. W. Michnick and L. B. Vosshall, 2006. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 4(2): e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalerao, S., A. Sen, R. Stocker and V. Rodrigues, 2003. Olfactory neurons expressing identified receptor genes project to subsets of glomeruli within the antennal lobe of Drosophila melanogaster. J. Neurobiol. 54: 577–592. [DOI] [PubMed] [Google Scholar]
- Boulianne, G. L., A. De la Concha, J. A. Campos-Ortega, L. Y. Jan and Y. N. Jan, 1991. The Drosophila neurogenic gene neuralized encodes a novel protein and is expressed in precursors of larval and adult neurons. EMBO J. 10: 2975–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyne, P., C. Warr, M. Freeman, D. Lessing, J. Kim et al., 1999. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron 22: 327–338. [DOI] [PubMed] [Google Scholar]
- De Bruyne, M., P. Clyne and J. Carlson, 1999. Odor coding in a model olfactory organ: the Drosophila maxillary palp. J. Neurosci. 19: 4520–4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bruyne, M., K. Foster and J. Carlson, 2001. Odor coding in the Drosophila antenna. Neuron 30: 537–552. [DOI] [PubMed] [Google Scholar]
- Dilda, C. L., and T. F. C. Mackay, 2002. The genetic architecture of Drosophila sensory bristle number. Genetics 162: 1655–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa, A. A., W. van der Goes van Naters, C. G. Warr, R. A. Steinbrecht and J. R. Carlson, 2003. Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron 37: 827–841. [DOI] [PubMed] [Google Scholar]
- Falconer, D. S., and T. F. C. Mackay, 1996. Introduction to Quantitative Genetics, Ed. 4. Addison-Wesley/Longman, Harlow, UK.
- Fedorowicz, G. M., J. D. Fry, R. R. H. Anholt and T. F. C. Mackay, 1998. Epistatic interactions between smell-impaired loci in Drosophila melanogaster. Genetics 148: 1885–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly, I., T. F. C. Mackay and R. R. H. Anholt, 2003. scribble is essential for olfactory behavior in Drosophila. Genetics 164: 1447–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Q., and A. Chess, 1999. Identification of candidate Drosophila olfactory receptors from genomic DNA sequence. Genomics 60: 31–39. [DOI] [PubMed] [Google Scholar]
- Gao, Q., B. Yuan and A. Chess, 2000. Convergent projections of Drosophila olfactory neurons to specific glomeruli in the antennal lobe. Nat. Neurosci. 3: 780–785. [DOI] [PubMed] [Google Scholar]
- Gorski, S. M., S. Chittaranjan, E. D. Pleasance, J. D. Freeman, C. L. Anderson et al., 2003. A SAGE approach to discovery of genes involved in autophagic cell death. Curr. Biol. 13: 358–363. [DOI] [PubMed] [Google Scholar]
- Griffing, B., 1956. Concept of general and specific combining ability in relation to diallel crossing systems. Aust. J. Biol. Sci. 9: 463–493. [Google Scholar]
- Hallem, E., M. Ho and J. Carlson, 2004. The molecular basis of odor coding in the Drosophila antenna. Cell 117: 965–979. [DOI] [PubMed] [Google Scholar]
- Hallem, E. A., and J. R. Carlson, 2006. Coding of odors by a receptor repertoire. Cell 125: 143–160. [DOI] [PubMed] [Google Scholar]
- Hayashi, S., S. Hirose, T. Metcalfe and A. D. Shirras, 1993. Control of imaginal cell development by the escargot gene of Drosophila. Development 118: 105–115. [DOI] [PubMed] [Google Scholar]
- Jhaveri, D., A. Sen and V. Rodrigues, 2000. Mechanisms underlying olfactory neuronal connectivity in Drosophila: the atonal lineage organizes the periphery while sensory neurons and glia pattern the olfactory lobe. Dev. Biol. 226: 73–87. [DOI] [PubMed] [Google Scholar]
- Khare, N., N. Fascetti, S. DaRocha, R. Chiquet-Ehrismann and S. Baumgartner, 2000. Expression patterns of two new members of the semaphorin family in Drosophila suggest early functions during embryogenesis. Mech. Dev. 91: 393–397. [DOI] [PubMed] [Google Scholar]
- Kulkarni, N. H., A. Yamamoto, K. O. Robinson, T. F. C. Mackay and R. R. H. Anholt, 2002. The DSC1 channel, encoded by the smi60E locus, contributes to odor-guided behavior in Drosophila melanogaster. Genetics 161: 1507–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, E. C., G. A. Deblandre, C. Kitner and G. M. Rubin, 2001. Drosophila neuralized is a ubiquitin ligase that promotes the internalization and degradation of delta. Dev. Cell. 1: 783–794. [DOI] [PubMed] [Google Scholar]
- Laissue, P., C. Reiter, P. Hiesinger, S. Halter, K. Fischbach et al., 1999. Three-dimensional reconstruction of the antennal lobe in Drosophila melanogaster. J. Comp. Neurol. 405: 543–552. [PubMed] [Google Scholar]
- Larsson, M., A. Domingos, W. Jones, M. Chiappe, H. Amrein et al., 2004. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron 43: 703–714. [DOI] [PubMed] [Google Scholar]
- Lehmann, M., T. Siegmund, K. G. Lintermann and G. Korge, 1998. The pipsqueak protein of Drosophila melanogaster binds to GAGA sequences through a novel DNA-binding domain. J. Biol. Chem. 273: 28504–28509. [DOI] [PubMed] [Google Scholar]
- Lukacsovich, T., Z. Asztalos, W. Awano, K. Baba, S. Kondo et al., 2001. Dual-tagging gene trap of novel genes in Drosophila melanogaster. Genetics 157: 727–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDougall, N., Y. Lad, G. S. Wilkie, H. Francis-Lang, W. Sullivan et al., 2001. Merlin, the Drosophila homologue of neurofibromatosis-2, is specifically required in posterior follicle cells for axis formation in the oocyte. Development 128: 665–673. [DOI] [PubMed] [Google Scholar]
- Mackay, T. F. C., 2004. The genetic architecture of quantitative traits: lessons from Drosophila. Curr. Opin. Genet. Dev. 14: 253–257. [DOI] [PubMed] [Google Scholar]
- Ng, M., R. Roorda, S. Lima, B. Zemelman, P. Morcillo et al., 2002. Transmission of olfactory information between three populations of neurons in the antennal lobe of the fly. Neuron 36: 463–474. [DOI] [PubMed] [Google Scholar]
- Norga, K. K., M. C. Gurganus, C. L. Dilda, A. Yamamoto, R. F. Lyman et al., 2003. Quantitative analysis of bristle number in Drosophila mutants identifies genes involved in neural development. Curr. Biol. 13: 1388–1397. [DOI] [PubMed] [Google Scholar]
- Prokopenko, S. N., Y. He, Y. Lu and H. J. Bellen, 2000. Mutations affecting the development of the peripheral nervous system in Drosophila: a molecular screen for novel proteins. Genetics 156: 1691–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault, A. D., M. Starz-Gaiano and R. Lehmann, 2002. Metabolism of sphingosine 1-phosphate and lysophosphatidic acid: a genome wide analysis of gene expression in Drosophila. Mech. Dev. 119: S293–S301. [DOI] [PubMed] [Google Scholar]
- Shandbhag, S., B. Muller and A. Steinbrecht, 1999. Atlas of olfactory organs of Drosophila melanogaster. 1. Types, external organization, innervation and distribution of olfactory sensilla. Int. J. Insect Morphol. Embryol. 28: 377–397. [Google Scholar]
- Siegmund, T., and M. Lehmann, 2002. The Drosophila Pipsqueak protein defines a new family of helix-turn-helix DNA-binding proteins. Dev. Genes Evol. 212: 152–157. [DOI] [PubMed] [Google Scholar]
- Sprague, G. F., and L. A. Tatum, 1942. General vs. specific combining ability in single crosses of corn. J. Am. Soc. Agron. 34: 923–932. [Google Scholar]
- Stoltzfus, J. R., W. J. Horton and M. S. Grotewiel, 2003. Odor-guided behavior in Drosophila requires calreticulin. J. Comp. Physiol. A 189: 471–483. [DOI] [PubMed] [Google Scholar]
- van Swinderen, B., and R. J. Greenspan, 2005. Flexibility in a gene network affecting a simple behavior in Drosophila melanogaster. Genetics 169: 2151–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosshall, L., H. Amrein, P. Morozov, A. Rzhetsky and R. Axel, 1999. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell 96: 725–736. [DOI] [PubMed] [Google Scholar]
- Vosshall, L., A. Wong and R. Axel, 2000. An olfactory sensory map in the fly brain. Cell 102: 147–159. [DOI] [PubMed] [Google Scholar]
- Wang, J., A. Wong, J. Flores, L. Vosshall and R. Axel, 2003. Two-photon calcium imaging reveals an odor-evoked map of activity in the fly brain. Cell 112: 271–282. [DOI] [PubMed] [Google Scholar]
- Weber, U., V. Siegel and M. Mlodzik, 1995. pipsqueak encodes a novel nuclear protein required downstream of seven-up for the development of photoreceptor R3 and R4. EMBO J. 14: 6247–6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley, M., P. D. Noguchi, S. M. Sensabaugh, W. F. Odenwald and J. A. Kassis, 1992. The Drosophila gene escargot encodes a zinc finger motif found in snail-related genes. Mech. Dev. 36: 117–127. [DOI] [PubMed] [Google Scholar]
- Yeh, E., L. Zhou, N. Rudzik and G. L. Boulianne, 2000. neuralized functions cell autonomously to regulate Drosophila sense organ development. EMBO J. 19: 4827–4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh, E., M. Dermer, C. Commisso, L. Zhou, C. J. McGlade et al., 2001. Neuralized functions as an E3 ubiquitin ligase during Drosophila development. Curr. Biol. 11: 1675–1679. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., M. S. Kang and K. R. Lamkey, 2005. DIALLEL-SAS05: a comprehensive program for Griffing's and Gardner–Eberhart analyses. Agron. J. 97: 1097–1106. [Google Scholar]