Abstract
In higher plants, fat-storing seeds utilize storage lipids as a source of energy during germination. To enter the β-oxidation pathway, fatty acids need to be activated to acyl-coenzyme As (CoAs) by the enzyme acyl-CoA synthetase (ACS; EC 6.2.1.3). Here, we report the characterization of an Arabidopsis cDNA clone encoding for a glyoxysomal acyl-CoA synthetase designated AtLACS6. The cDNA sequence is 2,106 bp long and it encodes a polypeptide of 701 amino acids with a calculated molecular mass of 76,617 D. Analysis of the amino-terminal sequence indicates that acyl-CoA synthetase is synthesized as a larger precursor containing a cleavable amino-terminal presequence so that the mature polypeptide size is 663 amino acids. The presequence shows high similarity to the typical PTS2 (peroxisomal targeting signal 2). The AtLACS6 also shows high amino acid identity to prokaryotic and eukaryotic fatty acyl-CoA synthetases. Immunocytochemical and cell fractionation analyses indicated that the AtLACS6 is localized on glyoxysomal membranes. AtLACS6 was overexpressed in insect cells and purified to near homogeneity. The purified enzyme is particularly active on long-chain fatty acids (C16:0). Results from immunoblot analysis revealed that the expression of both AtLACS6 and β-oxidation enzymes coincide with fatty acid degradation. These data suggested that AtLACS6 might play a regulatory role both in fatty acid import into glyoxysomes by making a complex with other factors, e.g. PMP70, and in fatty acid β-oxidation activating the fatty acids.
Acyl-CoA synthetase (ACS; fatty acid:CoA ligase, AMP binding forming, EC 6.2.1.3) catalyzes the formation of acyl-CoA thioesters from free fatty acids in the presence of CoA, ATP, and Mg2+. This activation is a critical step in fatty acid metabolism in prokaryotes and eukaryotes. In fact, fatty acyl-CoAs represent important bioactive compounds, which are involved in many cellular processes in addition to serving as substrates for lipid biosynthesis and β-oxidation (Schulz, 1991).
Recent published papers have confirmed the importance of ACSs in various organisms. In Escherichia coli, ACS plays a pivotal role in the uptake of long-chain fatty acids and in regulating of the global transcriptional regulator FadR (Black et al., 1997). The E. coli ACS gene was cloned and its sequence is found to have a segment of 25 highly conserved amino acid residues that Black et al. (1997) proposed as a signature motif common to the family of fatty ACSs. In yeast (Saccharomyces cerevisiae), the activation of exogenous fatty acids before metabolic utilization proceeds through the ACSs Faa1p and Faa4p. It was also suggested that Faa1p and Faa4p function as components of the fatty acid intracellular utilization and signaling mechanisms (Færgeman et al., 2001). In mammals, both de novo-synthesized and dietary-derived fatty acids need to be metabolized via ACSs that are encoded by a multigene family characterized by a component number that is continuously increasing (Lewin et al., 2001).
In higher plants, oilseeds convert storage lipids to Suc after germination as energy sources. This unique type of gluconeogenesis occurs in the storage tissues of oilseeds, such as endosperm and cotyledons. The metabolic pathway involves many enzymes in several subcellular compartments, including lipid bodies, glyoxysomes (specialized peroxisomes), mitochondria, and cytosol. Within the entire gluconeo-genic pathway, fatty acids are converted to succinate in glyoxysomes, which contain enzymes for fatty acid β-oxidation spiral and the glyoxylate cycle. We have previously studied enzymes of β-oxidation and the glyoxylate cycle in pumpkin (Cucurbita sp. Kurokawa Amakuri) and Arabidopsis in detail (Mori and Nishimura, 1989; Kato et al., 1996b, 1998; Hayashi et al., 1998a, 1998b, 1999; De Bellis et al., 1999, 2000; Fukao et al., 2002). We extended our study to plant glyoxysomal ACSs.
Little is known about higher plant ACSs, although the corresponding enzymes, as indicated above, have been extensively characterized in various organisms. In 1997, Fulda et al. (1997) published the cloning of two ACS cDNAs from Brassica napus. They individuated five B. napus full-length clones encoding polypeptides of the AMP-binding families. After the expression of the clones in E. coli, ACS activity on oleic acid was confirmed for two of the clones. More recently, Shockey et al. (2000) identified and cloned 20 different genes bearing strong sequence similarity to known ACSs from other organisms. They divided the genes into two classes of 10 members: the ACS class and the general AMP-binding protein class. Some members of the ACS class were able to complement a yeast mutant with deletions in two of its ACSs that determine the inability to grow on media containing cerulenin to inhibit fatty acid synthetase and myristate as a carbon source. However, they did not mention the accession numbers of the deposited sequences. Schnurr et al. (2000) reported few data concerning the characterization of a plastidial ACS named ACS2, but the corresponding sequence was not also deposited in the public database. It was proposed to have a role as a putative plasma membrane Capsicum annuum ACS facilitating the fatty acid movement across the plasma membrane during plant defense responses to pathogen attacks (Lee et al., 2001). In another paper, B. napus ACS has been identified with a membrane-bound protein (Pongdontri and Hills, 2001). More recently, Shockey et al. (2002) identified nine long-chain ACS (LACS1–9) genes in Arabidopsis genome, and one of the ACSs, LACS9, was characterized as a plastidial ACS (Schnurr et al., 2002).
In this study, we report isolation and characterization of an Arabidopsis ACS from glyoxysomal membranes. The ACS corresponds to AtLACS6 in accordance with the proposed nomenclature (Shockey et al., 2002). We show here that the AtLACS6 is localized on the glyoxisomal membrane and catalyzes the activation of long-chain fatty acids to produce acyl CoA.
RESULTS
Cloning of the cDNA Encoding an AtLACS6
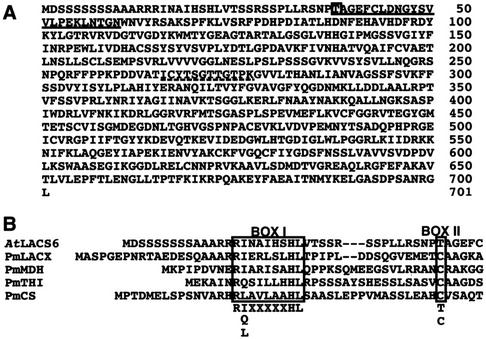
To analyze glyoxysomal membrane proteins, we isolated glyoxysomes from pumpkin seedlings that had been grown in darkness for 4 d. Isolated glyoxysomes were treated with 0.1 m NaCO3. The insoluble proteins by NaCO3 treatment were separated by SDS-PAGE and blotted on a polyvinylidene fluoride membrane. We have characterized the 31- and 38-kD proteins as peroxisomal ascorbate peroxidase (Nito et al., 2001) and ATP/ADP carrier protein (Fukao et al., 2001), respectively. In addition to these proteins, we started to analyze one of the major glyoxysomal membrane proteins with molecular mass of approximately 74 kD, which was cut out and subjected to protein sequencing by automated Edman degradation. The amino-terminal amino acid sequence is shown in Figure 1A (single underlined). A similarity search with this amino acid sequence in the GenBank database revealed that the terminal sequence of the 74-kD polypeptide is characterized by a high similarity with the N terminus of the B. napus AMP-binding protein (MF39, Z72152; Fulda et al., 1997). As the result of a similarity search with MF39 of its amino acid sequence, we received the cDNA clone (GenBank accession no. H76391) from the Arabidopsis Biological Resource Center (Ohio State University, Columbus) and fully sequenced it.
Figure 1.
Deduced amino acid sequences of the AtLACS6 cDNA and alignment of the AtLACS6 amino-terminal presequence with other presequences of glyoxysomal proteins that are synthesized as larger precursors. A, The amino-terminal amino acid presequence of the mature protein is single underlined. The amino acid residue of the processing site is marked with a box. The dotted line marks the putative AMP-binding motif. The GenBank accession number for AtLACS6 is AB030317. B, The amino acid sequences represent the N terminus of AtLACS6 and other glyoxysomal proteins that are synthesized as larger precursors. PmLACX, pumpkin long-chain acyl-CoA oxidase (LACX; Hayashi et al., 1998); PmMDH, pumpkin malate dehydrogenase (Kato et al., 1998); PmTHI, pumpkin 3-keto-acyl-CoA thiolase (Kato et al., 1996b); PmCS, pumpkin citrate synthase (Kato et al., 1995). BOX I indicates the PTS2 consensus. The processing sites of presequences determined by sequencing of the N terminal amino acids of mature proteins is shown by BOX II.
The full sequence of the clone indicated that the Arabidopsis cDNA is 2,295 bp. The predicted amino acid sequence of the clone, named AtLACS6 (accession no. AB030317), is presented in Figure 1A. The cDNA sequence contains a 2,106-bp open reading frame encoding a polypeptide of 701 amino acids that corresponds to a molecular mass of approximately 76 kD. The amino acid sequence is characterized by a typical peroxisomal targeting signal at the N terminus (PTS2). This suggests that the AtLACS6 protein might be synthesized as a large precursor with a presequence cleaved at the docking of glyoxysomes. We assume that the processing site is located between Thr and Ala because the N-terminal amino acid sequence obtained from the Edman sequencing of the pumpkin membrane 74-kD protein starts with AGEF, and because of the similarity with the N terminus of the B. napus AMP-binding protein MF39 (Fulda et al., 1997). A putative AMP-binding domain signature (PS00455, PROSITE, Swiss Institute of Bioinformatics, Geneva) appears to be present from amino acids 266 to 277: [L/I/V/M/F/Y]-x(2)-[S/T/G]-[S/T/A/G]-G-[S/T]-[S/T/E/I]-[S/G]-x-[P/A/S/L/I/V/M]-[K/R] (Fig. 1A, dotted line). Instead, the proposed ACS signature motif DGWLHTGDIGXWXPXGXLKIIDRKK (Black et al., 1997) is located between amino acids 526 and 550. The overall amino acids sequence shows high identity (94%) with the B. napus AMP-binding protein MP39 (Fulda et al., 1997) and a considerable identity (40%–45%) with mammalian ACSs.
Figure 1B shows the alignment of the amino-terminal presequence of AtLACS6 with other glyoxysomal proteins that are synthesized as larger precursors. These sequences share conserved substitutions with the identified consensus sequence R-[I/Q/L]-x(5)-H-L-x(15–22)-C (Kato et al., 1996a), with the exception that in the N terminus of AtLACS6, T substitutes C.
Expression and Purification of the AtLACS6
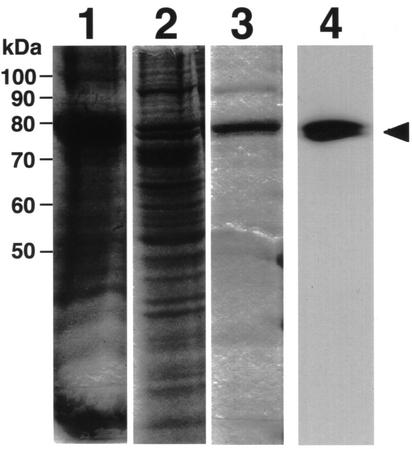
The full-length Arabidopsis cDNA was subcloned into an expression vector under the control of polyhedrin promoter (pBluebac 4.5) that was then used to transform insect cells. The transformed insect cells showed a high ACS activity on palmitic acid (data not shown). Therefore, we decided to purify the expressed protein. Crude homogenates were subjected to Blue Sepharose chromatography on a HiTrap Blue column. Fractions containing high ACS activity were pooled and concentrated by ultrafiltration. Then, the sample was loaded on a Resource S cation-exchange column. Fractions from each purification step were subjected to SDS-PAGE and immunoblotting employing an antiserum against AtLACS6 (C-terminal region, 601–701) fusion protein expressed in E. coli (Fig. 2). The immunoblot analysis of homogenates of Arabidopsis seedlings and pumpkin cotyledons using the antiserum showed only one band with molecular mass of 72 kD for Arabidopsis and 74 kD for pumpkin, respectively (data not shown). Immunogold and subcellular localization analyses showed that the immunoreactive protein is localized on glyoxysomal membranes (see below). These results suggest that an antiserum against AtLACS6 might be monospecific for AtLACS6, although many enzymes of ACSs are reported to be present in higher plant cells. This value, 72 kD for Arabidopsis, corresponds to the size of the AtLACS6 polypeptide without the presequence. Furthermore, the polypeptide was perfectly recognized by an antiserum against AtLACS6 (arrowhead). The Resource S chromatography failed to purify the enzyme to homogeneity, but only minor contaminants appeared to be present in the enzyme preparation (Fig. 2, lane 3) that was subsequently employed to test the enzyme substrate specificity.
Figure 2.
SDS-PAGE and immunoblot analyses of samples taken at various steps during the purification of AtLACS6 expressed in insect cells. Lane 1, Cell homogenate from insect cells infected with the recombinant baculovirus (harboring the cDNA of AtLACS6). Lane 2, HiTrap Blue column fraction. Lanes 3 and 4, Same Resource S column fraction (5 μg of purified protein). Lanes 1 through 3 represent lanes from SDS-PAGE (10% [w/v] acrylamide gel) stained with Coomassie Brilliant Blue R-250. Lane 4 represents an immunoblot analysis of the same sample used in lane 3. It was analyzed employing an antiserum against AtLACS6. The arrowhead marks the bands corresponding to the AtLACS6.
The ACS activity of the purified preparation was assayed using three different fatty acids, caprylic acid (C8:0), palmitic acid (C16:0), and lignoceric acid (C24:0), as substrates. The maximum activity, 236 × 10−3 units mg−1, was observed on palmitic acid (C16:0), a low level of activity was detected on caprylic acid (C8:0, 36.2 × 10−3 units mg−1), and on lignoceric acid (C24:0, 10.0 × 10−3 units mg−1), respectively. The results show that the ACS is predominantly active on long-chain fatty acids (C16:0).
Subcellular Localization of the Cloned AtLACS6
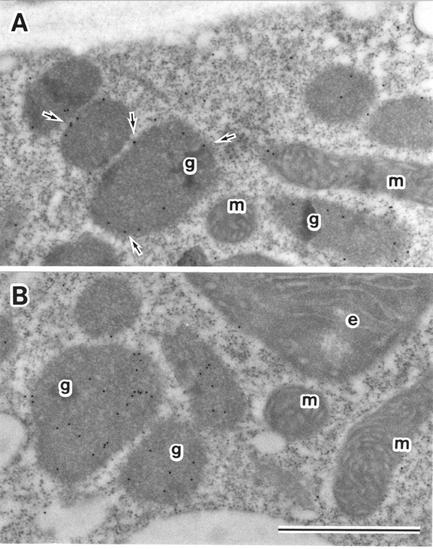
The presence of the PTS2 signal in the AtLACS6 presequence suggests that AtLACS6 is a glyoxysomal protein. Nevertheless, mammalian and yeast ACSs did not have a clear peroxisomal targeting signal. To confirm the subcellular localization of AtLACS6, we have performed an immuno-electron microscopy observation. Arabidopsis etiolated cotyledon from 5-d-old seedlings were fixed and thin sections were processed employing antisera against AtLACS6, pumpkin catalase, and nonimmune serum. The labeling shown in Figure 3A was exclusively localized in glyoxysomes, particularly on glyoxysomal membrane (arrow), whereas the catalase is localized in the glyoxysomal matrix. No gold particles were observed in other organelles such as mitochondria and etioplasts (Fig. 3A). No gold particles in any organelles were found in the case of nonimmune serum (data not shown). These data suggest that AtLACS6 is localized on glyoxysomal membranes.
Figure 3.
Immunoelectron microscope analysis of AtLACS6. Immunostaining of etiolated cotyledons from Arabidopsis seedlings grown for 5 d in the dark. Antisera against AtLACS6 (A) and catalase (B) were employed after IgG-purification. g, Glyoxysome; m, mitochondrion; e, etioplast. The arrowhead indicates the staining on the glyoxysomal membrane (15-nm gold particles). Bar = 1 μm.
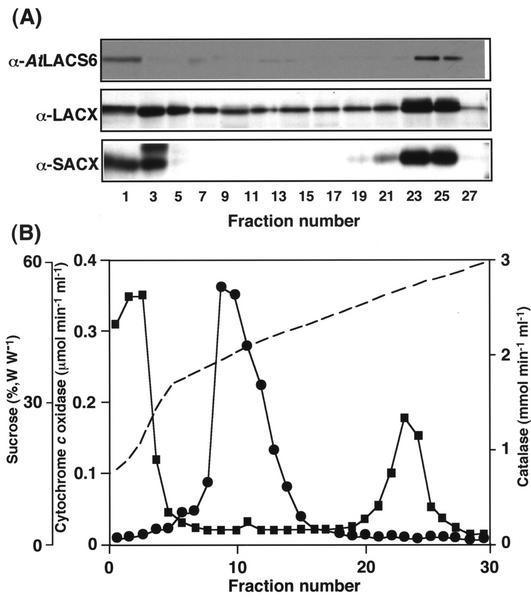
In addition, we analyzed the subcellular localization of pumpkin homolog of LACS6 in pumpkin cotyledons. Homogenates from 5-d-old etiolated pumpkin cotyledons were subjected to Suc density gradient centrifugation. It was shown from the marker enzyme assays that glyoxysomes are clearly separated from mitochondria in the Suc density gradient (Fig. 4B). Immunoblotting analyses were performed on fractions obtained from the Suc density gradient centrifugation employing antisera against AtLACS6 and other β-oxidation enzymes. Figure 4 indicates that the pumpkin homolog of LACS6 protein is mainly present in fractions 21 through 23, characterized by a Suc concentration of approximately 52% (w/w), the equilibrium density for glyoxysomes after Suc gradient fractionation. SACX and LACX, typical glyoxysomal enzymes, were also detected in large amounts in the same fractions. The presence of a signal in other fractions and particularly the first few fractions (corresponding to the top of the gradient) is because of the presence of enzymes leaked out from organelles damaged and broken during the homogenization procedure. These results clearly show that the pumpkin homolog of LACS6 is localized in glyoxysomes.
Figure 4.
Immunoblot analysis of Suc gradient fractions from pumpkin etiolated cotyledons. An extract from 5-d-old etiolated cotyledons was fractionated by Suc density gradient centrifugation. Each lane was loaded with 20 μL of samples from each odd-numbered fraction. Three different polyclonal antisera were employed. They were raised against AtLACS6, Arabidopsis LACX, and Arabidopsis short-chain acyl-CoA oxidase (SACX).
Developmental Change during Germination in the Level of AtLACS6
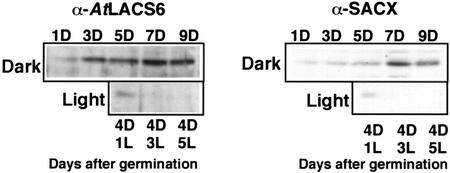
Figure 5 shows the comparison between the relative level of AtLACS6 and SACX, a typical enzyme for fatty acid β-oxidation, obtained by immunoblot analysis after SDS-PAGE of extracts from cotyledons of Arabidopsis seedlings grown for the time indicated in the dark or moved to the light after 4 d in the dark. The levels of both proteins increased in the dark, reaching a maximum level after 5 to 7 d and decreasing at d 9. A light treatment after 4 d in the dark determined a marked drop in the level of AtLACS6 and SACX so that both proteins were barely detectable after 3 d in the light. In conclusion, the protein levels of AtLACS6 and SACX showed a similar pattern during the development of Arabidopsis cotyledons.
Figure 5.
Developmental changes in the protein level of AtLACS6 and SACX during Arabidopsis germination. Each lane was loaded with an extract from Arabidopsis cotyledon extract containing approximately 10 μg of total proteins. Antisera against AtLACS6 and SACX were employed. Top, Immunoblots of samples from seedlings grown in the dark (D) for the time (days) indicated. Bottom, Immunoblots of samples from seedlings grown in the dark for 4 d (4D) plus 1, 3, or 5 d in the light (1L, 3L, and 5L).
DISCUSSION
In this report, we show that an Arabidopsis cDNA, designated as AtLACS6, encodes a glyoxysomal long-chain fatty ACS. The amino acid sequence presents a typical peroxisomal targeting signal (PTS2) represented by a cleavable presequence (Fig. 1). It is important to note that the AtLACS6 is synthesized as larger precursor containing a cleavable amino-terminal presequence, as is the case for some other plant glyoxysomal proteins, such as malate dehydrogenase (Gietl, 1990; Kato et al., 1998), citrate synthase (Kato et al., 1995), thiolase (Preisig-Müller and Kindle, 1993; Kato et al., 1996b), and LACX (Hayashi et al., 1998). This further confirms the importance of the PTS2 signal for protein import in plant glyoxysomes. As a novel finding, we propose that the processing site may follow a C or a T indifferently.
The glyoxysomal localization was confirmed by the immuno-electron microscope analysis of Arabidopsis etiolated cotyledons, the signal being localized on glyoxysomal membranes (Fig. 3). This indication is consistent with the difficulties for AtLACS6 purification by traditional methods. The strict association of AtLACS6 with glyoxysomal membranes suggests that AtLACS6 also may have an additional regulatory role in fatty acid transport in glyoxysomes by an unknown interaction with other factors. In fact, previous reports proposed that ACSs plays a functional role in activation of fatty acids and concomitant to transport (i.e. vectorial transport) of fatty acids in plant cells (Hayashi et al., 2001), E. coli (Black et al., 1997), and mammalian cells (Toke and Martin, 1996). In these data, AtLACS6 may interact with other membrane proteins (e.g. PMP70; Imanaka et al., 1999) or fatty acid transporters facilitating the import of fatty acids into glyoxysomes.
Figure 1 shows that the amino acid sequence of AtLACS6 presents a perfectly conserved 25-amino acid consensus sequence, DGWLHTGDIGXWXPXGXLKIIDRKK, common to mammalian ACSs, which was proposed as a signature motif for these enzymes (Black et al., 1997). A mutational analysis determining single amino acid substitutions within the FACS signature motif suggested that this region of ACS is specifically required for fatty acid binding and fatty acid substrate specificity. As indicated above, 20 AMP-binding protein genes appear to be present in the Arabidopsis genome but the FACS signature motif appears to be completely conserved only in the AtLACS6 protein and not in any other deduced proteins (data not shown). This suggests different substrate specificity for each member of the plant ACS family.
The level of the AtLACS6 protein appears to be controlled by light because the enzyme disappears during the transition from glyoxysomes to leaf peroxisomes after the exposure of the seedlings to light (Fig. 5). Similar patterns have been observed previously for other plant glyoxysomal enzymes such as malate synthase (Mori and Nishimura, 1989), citrate synthase (Kato et al., 1995), and SACX (Hayashi et al., 1999).
ACSs occupy a pivotal role in lipid metabolism, being involved in lipid synthesis and modification or degradation of existing lipids. Therefore, the elucidation of the specific role of the different plant ACSs will contribute to the understanding of plant lipid metabolism. In addition, because the production of engineered triacylglycerols or novel biopolymers from fatty acids is a major goal of modern plant biotechnology, it is essential to know in detail the exact function of key enzymes such as ACS. AtLACS6, as an ACS specific for long-chain fatty acids localized on the glyoxysomal membrane, is a candidate for the control of the process of β-oxidation.
MATERIALS AND METHODS
Materials
[1-14C] octanoic acid and [1-14C] lignocerate were purchased from Muromachi Pharmacy Corp. (Tokyo). [1-14C]Palmitic acid was purchased from Moravek Biochemicals (Brea, CA). All other chemicals were purchased from Sigma (St. Louis).
Plant Materials
Pumpkin (Cucurbita sp. Kurokawa Amakuri) seeds were purchased from the Aisan Seed Company (Aichi, Japan). Seeds were soaked in running tap water overnight and germinated in rock fiber soil (66R, Nitto Boseki, Chiba, Japan) at 25°C in darkness. Seeds of Arabidopsis, ecotype Columbia, were surface sterilized in 2% (w/v) NaClO plus 0.02% (w/v) Triton X-100 and placed on agar-solidified medium in petri dishes. The growth medium contained 2.3 mg mL−1 Murashige and Skoog salts (Wako, Osaka), 1% (w/w) Suc, 100 μg mL−1 myoinositol, 1 μg mL−1 thiamine-HCl, 0.5 μg mL−1 pyridoxine, 0.5 μg mL−1 nicotinic acid, 0.5 mg mL−1 MES-KOH (pH 5.7), and 0.2% (w/v) Gellan gum (Wako). Petri dishes were placed at 22°C under continuous light to allow seed germination. Some seedlings, after 2 weeks in petri dishes under continuous light, were transferred to a 1:1 (w/v) mixture of perlite:vermiculite and kept under continuous light at 22°C. For some experiments, Arabidopsis seedlings were grown in darkness for 4 d and then transferred to continuous light at 22°C.
Plasmid
The cDNA clone (accession no. H76931) was obtained from the Arabidopsis Biological Resource Center. DNA sequencing was performed by the method of Sanger et al. (1977). DNA sequences were analyzed with GeneWorks Release 2.5 computer software (IntelliGenetics, Mountain View, CA). The BLAST server was utilized for the analysis of homologies among proteins. Alignment of several ACS adenylate-forming enzymes was performed using ClustalW software (Thompson et al., 1994).
Preparation of Glyoxysomes
Etiolated cotyledons (70 g fresh weight) were harvested from pumpkin seedlings that had been grown for 4 d in darkness. The cotyledons were homogenized twice for 3 s with 200 mL of grinding buffer (20 mm pyrophosphate-HCl [pH 7.5], 1 mm EDTA, and 0.3 m mannitol) in a chilled Waring blender (Dynamics Corporation of America, New Hartford, CT). The homogenate was squeezed through four layers of gauze, the filtrate was collected, and solid residues were further homogenized as before with 200 mL of grinding buffer. The two filtrates were combined and centrifuged at 1,500g for 15 min to remove plastids and cell debris. A second centrifugation at 10,000g for 20 min produced a pellet that was resuspended in 150 mL of grinding buffer. The differential centrifugation steps were repeated once and the final pellet was resuspended in 5 mL of buffer A (10 mm HEPES-KOH [pH 7.2], 1 mm EDTA, and 0.3 m mannitol). A 4-mL aliquot of the suspension was layered directly on top of 30 mL of a 28% (v/v) solution of Percoll in 10 mm HEPES-KOH (pH 7.2), 1 mm EDTA, and 0.3 mm raffinose and was centrifuged at 40,000g for 30 min with slow acceleration and deceleration. Glyoxysomes sedimented near the bottom of the self-generated gradient were collected with a Pasteur pipette. To remove the Percoll, glyoxysomes were washed by centrifugation at 5,000g for 10 min after the addition of 4 volumes of buffer A. Finally, the pellet was suspended carefully in 1 mL of buffer A and used as purified glyoxysome fraction for subsequent analysis.
Amino Acid Sequence Analysis
Determination of the amino-terminal sequence of the pumpkin 74-kD membrane protein was performed essentially as described by Matsudaira (1987). Isolated glyoxysomes were subjected to SDS-PAGE and proteins were transferred to a polyvinylidene difluoride membrane (Problot, Perkin-Elmer, Chiba, Japan). The membrane was stained with Coomassie Brilliant Blue R-250 and the band corresponding to 76 kD was cut out with a razor blade. Protein sequencing was performed by automated Edman degradation in a protein sequencer (model 473A, Applied Biosystems Japan, Tokyo).
Preparation of the Specific Antiserum
The partial sequence of Arabidopsis cDNA was inserted into a pET32 vector (Novagen, Madison, WI). A fusion protein between AtLACS6 (C-terminal region, 601–701) and a His tag was synthesized in Escherichia coli cells and purified employing a HiTrap chelating column (Amersham Pharmacia, Tokyo). The purified fusion protein (approximately 1 mg of protein) in 1 mL of phosphate buffered saline was emulsified with an equal volume of Freund's complete adjuvant (DIFCO, Detroit). The emulsion was injected subcutaneously on the back of a rabbit. Four weeks later, a booster injection (approximately 0.5 mg of protein) was similarly given to the first injection. The blood was taken from a vein in the ear 7 d after the second booster injection. The serum was used for immunoblotting.
Expression of Recombinant AtLACS6 from Insect Cells
The AtLACS6 was produced employing the baculovirus expression system of Invitrogen (San Diego) following the manufacturer's protocols. The system includes Spodeptera frugiperda (Sf 21) as the insect cell line, pBlueBac 4.5 (Luckow and Summers, 1988) as a transfer vector, and engineered baculovirus Autographa californica multiple polyhedrosis virus (Bac-N-Blue DNA) as an expression vector. In brief, the cDNA of the AtLACS6 was inserted into the pBlueBac 4.5 transfer vector and cotransfected together with linearized baculoviral Bac-N-Blue DNA in insect cells. Recombinant viruses were identified by plaque assay on a medium containing X-gal and recombinant plaques were confirmed by PCR. Afterward, a high-titer recombinant viral stock was generated and subsequently employed to determine the time course of protein expression. The optimization of protein expression in insect cells allowed a large-scale expression of recombinant AtLACS6.
Purification of Recombinant AtLACS6
Log phase-growing Sf 21 cells were seeded as half confluent in 10-cm petri dishes. The insect cells were infected with recombinant viral stock at a multiplicity of infection of 3. Sixty hours after the infection, the cells were dislodged from the bottom of the dishes and centrifuged at 500g for 5 min. All procedures were carried out at 4°C. The cell pellets were washed with phosphate-buffered saline and centrifuged at 500g for 5 min. The cell pellets were gently suspended in buffer A (50 mm Tris-HCl [pH 7.6], 1 mm EDTA, 1 mm dithiothreitol, 5 mm KCl, 1 mm phenylmethylsulfonyl fluoride, 0.5 m NaCl, 1% [w/v] octyl glucoside, proteinase inhibitor cocktail tablets [Complete, Boehringer Mannheim GmbH, Mannheim, Germany], and 10% [w/v] glycerol) and lysed by sonication (3 × 20 s at 10-min intervals on ice water). After centrifugation of the sample at 100,000g for 30 min, the supernatant was dialyzed against buffer B (50 mm Na-phosphate [pH 7.4], 20 mm NaCl, 1 mm EDTA, 1 mm dithiothreitol, 2 mm ATP, 0.2% [w/v] octyl glucoside, and 10% [w/v] glycerol). Afterward, the dialyzed sample was loaded on a HiTrap Blue column (Amersham Pharmacia). Proteins were eluted increasing step-wise the NaCl concentration in buffer B up to 1.8 m. Fractions of 0.5 mL were collected, pooled, and rapidly desalted using Centricon 30 concentrators (Amicon, Beverly, MA). The desalted sample was loaded on a Resource S column (Amersham Pharmacia) equilibrated with buffer C (50 mm MES-KOH [pH 6.4], 10 mm NaCl, 1 mm dithiothreitol, 0.1% [w/v] octyl glucoside, and 10% [w/v] glycerol) and proteins eluted with a liner a gradient of NaCl from 0 to 1 m in buffer C.
Subcellular Fractionation
Four-day-old pumpkin etiolated cotyledons (15 g fresh weight) were homogenized in a petri dish by chopping with a razor blade for 5 min in 10 mL of a medium that contained 150 mm Tricine-KOH (pH 7.5), 1 mm EDTA, and 0.5 m Suc. The homogenate was passed through four layers of cheesecloth. Three milliliters of the filtrate was layered onto a Suc gradient that consisted of a 1-mL cushion of 60% (w/w) Suc and 11 mL of a linear Suc gradient from 60% to 30% (w/w) without buffer. The gradient was centrifuged at 21,000 rpm for 3 h in an SW 28.1 rotor in an ultracentrifuge model XL-90 (Beckman, Palo Alto, CA). After centrifugation, fractions of 0.5 mL were collected with a gradient fractionator (model 185; ISCO, Lincoln, NE). All procedures were carried out at 4°C. Subcellular fractionation of Arabidopsis etiolated cotyledons was performed as follows. One hundred milligrams of seeds (approximately 5,000 seeds) was grown on growth medium for 5 d in darkness at 22°C. Etiolated cotyledons were harvested and chopped with a razor blade in a petri dish with 2 mL of chopping buffer (150 mm Tricine-KOH [pH 7.5], 1 mm EDTA, 0.5 m Suc, and 1% [w/v] bovine serum albumin). The extract was then filtered with a cell strainer (Becton-Dickinson, Franklin Lakes, NJ). Two milliliters of the homogenate was layered directly on top of a 16-mL linear Suc density gradient (30%–60% [w/w]) that contained 1 mm EDTA. Centrifugation was performed in an SW 28.1 rotor (Beckman) at 25,000 rpm for 2.5 h at 4°C. Fractions of 0.5 mL were collected with the gradient fractionator (model 185, ISCO).
Immunoelectron Microscopy
Arabidopsis etiolated cotyledons were harvested at 3 d in darkness. The samples were fixed, dehydrated, and embedded in LR white resin (London Resin, Basingstoke, UK) as described previously by Nishimura et al. (1993) and Hayashi et al. (1998). Ultrathin sections were cut with a Reichert ultramicrotome (Leica, Heidelberg) and mounted on uncoated nickel grids. The protein A-gold labeling procedure was essentially the same as that described by Nishimura et al. (1993) and Hayashi et al. (1998).
Enzyme Assay
ACS activity was measured essentially as described previously (Singh et al., 1985; Choi and Martin, 1999). The fatty acid substrates, [1-14C] caprylic acid, [1-14C]palmitic acid, and [1-14C] lignoceric acid, were prepared as a 100 μm stock solution. Solubilized substrate was obtained by dissolving the dried fatty acid in 100 mm Tris-HCl (pH 8.5), containing 10 mg mL−1 α-cyclodextrin, and incubating for 30 min in a sonication at room temperature. Reaction mixture containing 50 mm Tris-HCl [pH 8.5], 10 mm ATP, 10 mm MgCl2, 0.01% [w/v] Triton X-100, 1 mm dithiothreitol, and 10 μm fatty acid dissolved in α-cyclodextrin was prepared to the final volume of 0.2 mL at purified protein concentrations of 200 ng/0.2 mL. After pre-incubation for 1 min at 25°C, reactions were started by adding 250 μm coenzyme A. Reactions terminated by adding 600 μL of Dole's reagent (isopropyl alcohol:heptane:1 m H2SO4, 40:10:1 [v/v]). After vigorous mixing, the lower (aqueous) layer by centrifugation was washed 6 times with 500 μL of heptane. The radioactivity in the final lower layer was measured by scintillation counting. Catalase and cytochrome c oxidase were assayed as previously (Hayashi et al., 1999).
Immunoblot Analysis
Arabidopsis and pumpkin cotyledons were homogenized in extraction buffer (0.1 m Tris-HCl [pH 8.0], 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, and 0.1% [w/v] SDS), the homogenate was centrifuged at 15,000g for 20 min, and the supernatant was applied to SDS-PAGE. Then, an immunoblot analysis was performed essentially following the method of Towbin et al. (1979). Immunodetection was realized monitoring the activity of horseradish peroxidase (ECL system, Amersham Pharmacia). Protein was quantitated with a protein assay kit (Nippon Bio-Rad Laboratories, Tokyo).
Footnotes
This work was supported in part by the Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists (Grant-in-Aid no. 12–2214 to H.H.) and by the Ministry of Education, Culture, Sports, Science and Technology of Japan (Grants-in-Aid for Scientific Research nos. 12440231 to M.N. and 12640625 to M.H.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.012955.
LITERATURE CITED
- Black PN, Zhang Q, Weimar JD, DiRusso CC. Mutational analysis of a fatty acyl-coenzyme A synthetase signature motif identifies seven amino acid residues that modulate fatty acid substrate specificity. J Biol Chem. 1997;272:4896–4903. doi: 10.1074/jbc.272.8.4896. [DOI] [PubMed] [Google Scholar]
- Choi J-Y, Martin CE. The Saccharomyces cerevisiae FAT1 gene encodes an acyl-CoA synthetase that is required for maintenance of very long chain fatty acid levels. J Biol Chem. 1999;274:4671–4683. doi: 10.1074/jbc.274.8.4671. [DOI] [PubMed] [Google Scholar]
- De Bellis L, Giuntini P, Hayashi H, Hayashi M, Nishimura M. Purification and characterization of pumpkin long-chain acyl-CoA oxidase. Physiol Plant. 1999;106:170–176. [Google Scholar]
- De Bellis L, Gonzali S, Alpi A, Hayashi H, Hayashi M, Nishimura M. Purification and characterization of a novel pumpkin short-chain acyl-coenzyme A oxidase with structural similarity to acyl-coenzyme A dehydrogenases. Plant Physiol. 2000;123:324–334. doi: 10.1104/pp.123.1.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Færgeman NJ, Black PN, Dan Zhao X, Knudsen J, DiRusso CC. The acyl-CoA synthetases encoded within FAA1 and FAA4 in Saccharomyces cerevisiae function as components of the fatty acid transport system linking import, activation, and intracellular utilization. J Biol Chem. 2001;276:37051–37059. doi: 10.1074/jbc.M100884200. [DOI] [PubMed] [Google Scholar]
- Fukao Y, Hayashi M, Nishimura M. Proteomic analysis of leaf peroxisomal proteins in greening cotyledons of Arabidopsis thaliana. Plant Cell Physiol. 2002;43:689–696. doi: 10.1093/pcp/pcf101. [DOI] [PubMed] [Google Scholar]
- Fukao Y, Hayashi Y, Mano S, Hayashi M, Nishimura M. Developmental analysis of a putative ATP/ADP carrier protein localized on glyoxysomal membranes during the peroxisome transition in pumpkin cotyledons. Plant Cell Physiol. 2001;42:835–841. doi: 10.1093/pcp/pce108. [DOI] [PubMed] [Google Scholar]
- Fulda M, Heinz E, Wolter FP. Brassica napus cDNAs encoding fatty acyl-CoA synthetase. Plant Mol Biol. 1997;33:911–922. doi: 10.1023/a:1005780529307. [DOI] [PubMed] [Google Scholar]
- Gietl C. Glyoxysomal malate dehydrogenase from watermelon is synthesized with an amino-terminal transit peptide. Proc Natl Acad Sci USA. 1990;87:5773–5777. doi: 10.1073/pnas.87.15.5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H, De Bellis L, Ciurli A, Kondo M, Hayashi M, Nishimura M. A novel acyl-CoA oxidase that can oxidize short-chain acyl-CoA in plant peroxisomes. J Biol Chem. 1999;274:12715–12721. doi: 10.1074/jbc.274.18.12715. [DOI] [PubMed] [Google Scholar]
- Hayashi H, De Bellis L, Yamaguchi K, Kato A, Hayashi M, Nishimura M. Molecular characterization of a glyoxysomal long-chain acyl-CoA oxidase that is synthesized as a precursor of higher molecular mass in pumpkin. J Biol Chem. 1998a;273:8301–8307. doi: 10.1074/jbc.273.14.8301. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Toriyama K, Kondo M, Nishimura M. 2,4-Dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid β-oxidation. Plant Cell. 1998b;10:183–195. doi: 10.1105/tpc.10.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Hayashi M, Hayashi H, Hara-Nishimura I, Nishimura M. Direct interaction between glyoxysomes and lipid bodies in cotyledons of the Arabidopsis thaliana ped1 mutant. Protoplasma. 2001;218:83–94. doi: 10.1007/BF01288364. [DOI] [PubMed] [Google Scholar]
- Imanaka T, Aihara K, Takano T, Yamashita A, Sato R, Suzuki Y, Yokota S, Osumi T. Characterization of the 70-kDa peroxisomal membrane protein, an ATP binding cassette transporter. J Biol Chem. 1999;274:11968–11976. doi: 10.1074/jbc.274.17.11968. [DOI] [PubMed] [Google Scholar]
- Kato A, Hayashi M, Kondo M, Nishimura M. Targeting and processing of a chimeric protein with the N-terminal presequence of the precursor to glyoxysomal citrate synthase. Plant Cell. 1996a;8:1601–1611. doi: 10.1105/tpc.8.9.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A, Hayashi M, Mori H, Nishimura M. Molecular characterization of a glyoxysomal citrate synthase that is synthesized as a precursor of higher molecular mass in pumpkin. Plant Mol Biol. 1995;27:377–390. doi: 10.1007/BF00020191. [DOI] [PubMed] [Google Scholar]
- Kato A, Hayashi M, Takeuchi Y, Nishimura M. cDNA cloning and expression of a gene for 3-ketoacyl-CoA thiolase in pumpkin cotyledons. Plant Mol Biol. 1996b;22:843–852. doi: 10.1007/BF00019471. [DOI] [PubMed] [Google Scholar]
- Kato A, Takeda-Yoshikawa Y, Hayashi M, Kondo M, Hara-Nishimura I, Nishimura M. Glyoxysomal malate dehydrogenase in pumpkin: cloning of a cDNA and functional analysis of its presequence. Plant Cell Physiol. 1998;39:186–195. doi: 10.1093/oxfordjournals.pcp.a029356. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Suh MC, Kim S, Kwon JK, Kim M, Paek KH, Choi D, Kim BD. Molecular cloning of a novel pathogen-inducible cDNA encoding a putative acyl-CoA synthetase from Capsicum annuum L. Plant Mol Biol. 2001;46:661–671. doi: 10.1023/a:1011677028605. [DOI] [PubMed] [Google Scholar]
- Lewin TM, Kim J-H, Granger DA, Vance JE, Coleman RA. Acyl-CoA synthetase isoforms 1, 4, and 5 are present in different subcellular membranes in rat liver and can be inhibited independently. J Biol Chem. 2001;276:24674–24679. doi: 10.1074/jbc.M102036200. [DOI] [PubMed] [Google Scholar]
- Luckow VA, Summers MD. Signals important for high-level expression of foreign genes in Autographa californica nuclear polyhedrosis virus expression vectors. Virology. 1988;167:56–71. doi: 10.1016/0042-6822(88)90054-2. [DOI] [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- Mori H, Nishimura M. Glyoxysomal malate synthetase is specifically degraded in microbodies during greening of pumpkin cotyledons. FEBS Lett. 1989;244:163–166. [Google Scholar]
- Nishimura M, Takeuchi Y, De Bellis L, Hara-Nishimura I. Leaf peroxisomes are directly transformed to glyoxysomes during senescence of pumpkin cotyledons. Protoplasma. 1993;175:131–137. [Google Scholar]
- Nito K, Yamaguchi K, Kondo M, Hayashi M, Nishimura M. Pumpkin peroxisomal ascorbate peroxidase is localized on peroxisomal membranes and unknown membranous structures. Plant Cell Physiol. 2001;42:20–27. doi: 10.1093/pcp/pce003. [DOI] [PubMed] [Google Scholar]
- Pongdontri P, Hills M. Characterization of a novel plant acyl-CoA synthetase that is expressed in lipogenic tissues of Brassica napus L. Plant Mol Biol. 2001;47:717–726. doi: 10.1023/a:1013652014744. [DOI] [PubMed] [Google Scholar]
- Preisig-Müller R, Kindle H. Thiolase mRNA translated in vivo yields a peptide with a putative N-terminal presequence. Plant Mol Biol. 1993;22:59–66. doi: 10.1007/BF00038995. [DOI] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnurr JA, Shockey JM, Browse JA. Characterization of an acyl-CoA synthetase from Arabidopsis thaliana. Biochem Soc Trans. 2000;28:957–958. [PubMed] [Google Scholar]
- Schnurr JA, Shockey JM, de Boer G-J, Browse JA. Fatty acid export from the chloroplast. Molecular characterization of a major plastidial acyl-coenzyme A synthetase from Arabidopsis. Plant Physiol. 2002;129:1700–1709. doi: 10.1104/pp.003251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz H. Beta oxidation of fatty acids. Biochim Biophys Acta. 1991;1081:109–120. doi: 10.1016/0005-2760(91)90015-a. [DOI] [PubMed] [Google Scholar]
- Shockey JM, Fulda MS, Browse JA. Arabidopsis contains nine long-chain acyl-coenzyme A synthetase genes that participate in fatty acid and glycerolipid metabolism. Plant Physiol. 2002;129:1710–1722. doi: 10.1104/pp.003269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shockey JM, Schnurr JA, Browse JA. Characterization of the AMP-binding protein gene family in Arabidopsis thaliana: will the real acyl-CoA synthetases please stand up? Biochem Soc Trans. 2000;28:955–957. [PubMed] [Google Scholar]
- Singh I, Singh R, Bhushan A, Singh AK. Lignoceroyl-CoA ligase activity in rat brain microsomal fraction: topographical localization and effect of detergents and α-cyclodextrin. Arch Biochem Biophys. 1985;236:418–426. doi: 10.1016/0003-9861(85)90642-3. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;8:4321–4325. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toke DA, Martin CE. Isolation and characterization of a gene affecting fatty acid elongation in Saccharomyces cerevisiae. J Biol Chem. 1996;271:18413–18422. doi: 10.1074/jbc.271.31.18413. [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacryl amide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]