Abstract
Previous work with spinach (Spinacia oleracea) has shown that the level of gibberellin (GA) 20-oxidase is strongly up-regulated by long days (LD). In the present work, the effect of photoperiod on expression of other GA dioxygenases was investigated and compared with that of GA 20-oxidase. Two GA 2-oxidases and one GA 3-oxidase were isolated from spinach by reverse transcription-polymerase chain reaction with degenerate primers and by 5′- and 3′-rapid amplification of cDNA ends. As determined by high-performance liquid chromatography with on-line radioactivity detection, the SoGA3ox1 gene product catalyzed 3β-hydroxylation of GA9 to GA4 and GA20 to GA1. The SoGA2ox1 and the SoGA2ox2 gene products catalyzed 2β-hydroxylation of GA9 to GA51 and GA20 to GA29. The product of GA20 metabolism by SoGA3ox1 was identified as GA1 by gas chromatography-mass spectrometry, whereas the products of GA1 and GA20 metabolism by SoGA2ox1 and SoGA2ox2 were identified as GA8 and GA29, respectively. SoGA2ox1 also metabolized GA53 to GA97. The levels of SoGA20ox1 transcripts were greatly increased in all organs tested in LD conditions, but the levels of SoGA3ox1 transcripts were only slightly increased in blades and petioles. A decrease in the levels of the SoGA2ox1 transcripts in young leaves and tips in LD conditions is opposite to the expression pattern of the SoGA20ox1. Expression of SoGA20ox1 in petioles and young leaves was strongly up-regulated by a supplementary 16 h of light, but the levels of SoGA3ox1 and SoGA2ox1 transcripts did not change. It is concluded that regulation and maintenance of GA1 concentration in spinach are primarily attributable to changes in expression of SoGA20ox1.
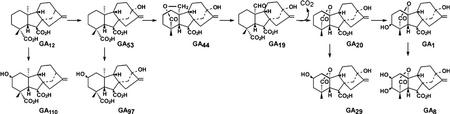
Long-day (LD) rosette plants, such as spinach (Spinacia oleracea), grow vegetatively and do not produce a stem when grown under short-day (SD) conditions. Upon transfer to LD conditions, stem elongation and flowering take place. In spinach, LD-induced stem elongation is dependent on gibberellin (GA)-regulated processes. There is considerable evidence that GA biosynthesis increases in rosette plants in LD conditions (Talon and Zeevaart, 1990; Talon et al., 1991). The major endogenous GAs of spinach belong to the early-13-hydroxylation pathway: GA53, GA44, GA19, GA20, GA1, GA8, and GA29 (Fig. 1; Talon et al., 1991), of which only GA1 is active per se (Zeevaart et al., 1993). Several steps of the GA biosynthetic pathway are regulated by photoperiod. For example, the biosynthesis of ent-kaurene, an early step in the GA biosynthetic pathway, is enhanced by LD (Zeevaart and Gage, 1993). The later steps of sequential oxidation and elimination of C-20 of C20-GAs (GA53→GA44→GA19→GA20), catalyzed by GA 20-oxidase, are also increased when spinach plants are transferred from SD to LD conditions (Wu et al., 1996).
Figure 1.
The early-13-hydroxylation pathway from GA12 to GA8 of GA biosynthesis and deactivation in spinach. GA53 is converted to GA20 by GA 20-oxidase via GA44 and GA19. GA20 can be converted to GA1 by GA 3-oxidase. GA12, GA53, GA20, and GA1 can be deactivated by 2β-hydroxylation to GA110, GA97, GA29, and GA8, respectively.
3β-Hydroxylation is the final step to active GAs, converting GA9 and GA20 to GA4 and GA1, respectively. Genes encoding 3-oxidases have been isolated from several species, and their expression and regulation have been studied (Chiang et al., 1995; Lester et al., 1997; Cowling et al., 1998; Yamaguchi et al., 1998; Itoh et al., 1999, 2001; Rebers et al., 1999). GA 2-oxidases introduce a 2β-hydroxyl group to biologically active GAs and deactivate C19-GAs, such as GA1, GA4, GA9, and GA20 (Lester et al., 1999; Thomas et al., 1999; Elliott et al., 2001). Both GA 3-oxidase and GA 2-oxidase can, therefore, directly regulate the levels of the active GAs, GA1 and GA4, and their immediate precursors, GA20 and GA9, respectively (for reviews, see Hedden and Phillips, 2000; Yamaguchi and Kamiya, 2000).
As shown in earlier work with spinach, transfer of plants from SD to LD causes an increase in all GAs of the early-13-hydroxylation pathway, with GA20, GA1, and GA8 showing the largest increases (Wu et al., 1996). The increase in GA20 can be explained as being attributable to increased expression of the gene encoding GA 20-oxidase, SoGA20ox1. However, to what extent the levels of GA1 and GA8 are determined by photoperiodically induced changes in expression of GA 3-oxidase and GA 2-oxidase is not known. In this study, we describe the molecular cloning of a GA 3-oxidase and two GA 2-oxidases from spinach and the regulation of their expressions by photoperiod in comparison with that of GA 20-oxidase. Our results indicate that expression of GA 20-oxidase1 and GA 2-oxidase1 is affected more by the photoperiod than is expression of GA 3-oxidase1 and GA 2-oxidase2.
RESULTS
Cloning of GA 3-Oxidase and GA 2-Oxidase cDNAs
Reverse transcription (RT)-PCR reactions were performed with total RNA as a template isolated from spinach grown under SD conditions. All combinations of degenerate primers were used in the PCR reactions with first-strand cDNA as a template. The combination of degenerate primers JZ418 and JZ432 (Table I) yielded a 521-bp fragment of GA 3-oxidase (data not shown). The sequence of this RT-PCR product was homologous to the GA 3-oxidase genes of Arabidopsis (Chiang et al., 1995) and pea (Pisum sativum; Lester et al., 1997). 5′- and 3′-RACE were performed with sequence-specific primers (JZ445 and JZ444) using the SMART RACE cDNA Amplification Kit (BD Biosciences Clontech, Palo Alto). The coding region of the SoGA3ox1 gene was obtained with primers JZ509 and JZ510 (Table I). The full-length cDNA clone of SoGA3ox1 (1,612 bp; accession no. AF506280) has an open reading frame (ORF) of 1,149 bp, encoding a putative protein of 382 amino acids with a 223-bp 5′-untranslated sequence and a 240-bp 3′-untranslated sequence. The predicted protein molecular mass is 42 kD, with an pI of 8.39 (Fig. 2). Northern-blot analysis showed two sizes of transcripts of the SoGA3ox1 gene in shoot tips around 1.6 and 2.0 kb (data not shown). The longer transcript is immature because it contains one intron of 361 bp. This exon/intron structure is conserved at the same position as in other GA3ox genes (Lester et al., 1997). When compared with other GA 3-oxidases, the deduced amino acid sequence of SoGA3ox1 belongs to the same class of enzymes and contains certain conserved amino acid sequences, such as His-227, Asp-229, and His-284, that may bind iron (Thomas et al., 1999). It shares 33% to 56% identity with the GA 3-oxidases listed in Figure 2.
Table I.
Primers used for the amplification of gibberellin dioxygenase genes from spinach
| Purpose | Primer | Primer Sequence (5′–3′) | Gene Amplified | Orientation |
|---|---|---|---|---|
| RT-PCR | JZ254 | TYGGRGARCAYCANGAYCC | GA2ox1 and -2 | S |
| RT-PCR | JZ258 | CCDSCRAARTADATCATHGA | GA2ox1 | AS |
| 3′-RACE | JZ282 | TTTCATACTCGTTGGGGACTC | GA2ox1 | S |
| 5′-RACE | JZ298 | CACCTGCAAAGAGTCCCCAACGAGT | GA2ox1 | AS |
| Coding region | JZ300 | AAACCCGGGATGGTGGTGTTATCTCATGTT | GA2ox1 | S |
| Coding region | JZ301 | CATCTCGAGTTAGTTATGATTTCTCTTTAC | GA2ox1 | AS |
| QRT-PCR | JZ385 | ACCAAGGCCTGAGGCGTTTGAAG | GA2ox1 | S |
| QRT-PCR | JZ386 | AACCTAGATTTGTAAGCTGAT | GA2ox1 | AS |
| QRT-PCR | JZ389 | ATGGAAGCTSCDGGWATYCA | Actin | S |
| QRT-PCR | JZ390 | TTARAARCAYTTYCTGTGCAC | Actin | AS |
| RT-PCR | JZ342 | TCATCWGGWGGRACWGARAYCCA | GA2ox2 | AS |
| 5′-RACE | JZ375 | CAAGCCATCAGGCATACAAATC | GA2ox2 | AS |
| 3′-RACE | JZ574 | CAAACAATTCAACAACCAAAGCA | GA2ox2 | S |
| Coding region | JZ582 | CGCGCTCGAGTCATTGGTCATTAATACGTCTC | GA2ox2 | AS |
| Coding region | JZ619 | CGCGGATCCCCATGGTGGTAGCTTCTCCA | GA2ox2 | S |
| RT-PCR | JZ418 | AARCTYATGTGGTMYGARGGVTTYAC | GA3ox1 | S |
| RT-PCR | JZ432 | GAHGGKGGNCCATAKARATARGC | GA3ox1 | AS |
| 5′-RACE | JZ445 | TGTAATTATTAGGCCAAAGTTGACGAGCAT | GA3ox1 | AS |
| 3′-RACE | JZ444 | GCAATACGACGGGGCTCCAAGTGT | GA3ox1 | S |
| Coding region | JZ509 | GCGCCCCGGGATGCCATCTAGGCCATCTC | GA3ox1 | S |
| Coding region | JZ510 | GCGCGTCGACTTAACCAAATACAGCAAGG | GA3ox1 | AS |
Y (C/T), N (A/T/G/C), R (A/G), S (C/G), M (A/C), H (A/T/C), D (A/G/T), K (G/T), V (A/C/G), B (C/G/T), W (A/T).
Figure 2.
Alignment of the deduced amino acid sequences for the SoGA3ox1 gene from spinach with other GA 3-oxidases. Conserved regions are boxed in black. Asterisks indicate the putative Fe2+-binding motif at the active site of 2-oxoglutarate-dependent dioxygenases (Thomas et al., 1999). SoGA3ox1, GA 3-oxidase from spinach (GenBank accession no. AF506280); NtGA3ox, GA 3-oxidase from tobacco (Nicotiana tabacum; accession no. BAA89316); StGA3ox, GA 3-oxidase from potato (Solanum tuberosum; accession no. AAK91507); PsGA3ox, GA 3-oxidase from garden pea (GenBank accession no. AAC96015); AtGA3ox, GA 3-oxidase from Arabidopsis (accession no. T51691); CmGA3ox and CmGA23ox, GA 3-oxidases from pumpkin (Cucurbita pepo; accession nos. AAB64347 and CAB92914); and OsGA3ox, GA 3-oxidase from rice (Oryza sativa; accession no. BAB62072).
The primer combination JZ254 and JZ258 yielded a 236-bp fragment of GA 2-oxidase1, and JZ 254 and JZ 342 yielded a 119-bp fragment of GA 2-oxidase2 (data not shown). The sequence of each RT-PCR product was homologous to the GA 2-oxidase genes of runner bean and Arabidopsis (Thomas et al., 1999) and pea (Lester et al., 1999). 5′- and 3′-RACE were performed with sequence-specific primers (JZ282 and JZ298 for GA 2-oxidase1, and JZ375 and JZ574 for GA 2-oxidase2; Table I) using the SMART RACE cDNA amplification kit (BD Biosciences Clontech). The coding region of the SoGA2ox1 gene was obtained with primers JZ300 and JZ301 (Table I). The full-length SoGA2ox1 cDNA clone (1,229 bp, accession no. AF506281) has an ORF of 1,014 bp and encodes a protein of 337 amino acids with a 114-bp 5′-untranslated sequence and a 101-bp 3′-untranslated sequence. The molecular mass of the predicted protein is 38 kD, with an pI of 8.46. The SoGA2ox1 protein shares 40% to 61% identity with the other GA 2-oxidases listed in Figure 3. Northern-blot analysis showed that the size of the SoGA2ox1 transcript is 1.3 kb (data not shown).
Figure 3.
Alignment of the deduced amino acid sequences for the SoGA2ox1 and SoGA2ox2 genes from spinach with other GA 2-oxidases. Conserved regions are boxed in black. Asterisks indicate the putative Fe2+-binding motif at the active site of 2-oxoglutarate-dependent dioxygenases (Thomas et al., 1999). SoGA2ox1 and SoGA2ox2, GA 2-oxidases from spinach (accession nos. AF506281 and AF506282); PcGA2ox1, GA 2-oxidase from runner bean (accession no. AT132438); PsGA2ox1 and PsGA2ox2, GA 2-oxidases from garden pea (accession nos. AF100954 and AF100955); AtGA2ox1, AtGA2ox2, and AtGA2ox3, GA 2-oxidases from Arabidopsis (accession nos. AJ132435, AJ132436, and AJ132437); OsGA2ox1, GA 2-oxidase from rice (accession no. BAB40934).
The full-length sequence of the second GA 2-oxidase (1,130 bp), designated SoGA2ox2 (accession no. AF506282), contains an ORF of 990 bp encoding a protein of 329 amino acids (36 kD) with a 113-bp 5′-untranslated sequence, and a 27-bp 3′-untranslated sequence with an pI of 7.1. The coding region of the SoGA2ox2 gene was obtained by PCR with the sequence-specific primers JZ619 and JZ582 (Table I). SoGA2ox2 shares 37% to 55% identity with other GA 2-oxidases listed in Figure 3 and has a low identity (40%) with the SoGA2ox1 protein. We established by dot-blot analysis that there is no cross-reactivity between the SoGA2ox1 and SoGA2ox2 genes (data not shown). Figure 3 shows the conserved regions among various GA 2-oxidases. Alignment with GA 2-oxidases from other species indicates that the two putative GA 2-oxidases from spinach belong to the same class of proteins and contain highly conserved amino acid sequences that may bind iron at the active site.
Heterologous Expression in Escherichia coli
To determine that the SoGA3ox1 cDNA clone encodes a GA 3-oxidase and that the SoGA2ox1 and SoGA2ox2 clones encode GA 2-oxidases, each coding region was heterologously expressed as a fusion protein of the glutathione S-transferase fusion vector in E. coli, strain BL21p Lys21. Soluble protein extracts were used for assays of GA 3-oxidase and GA 2-oxidase activities with several radioactive GAs as substrates. The reaction products were separated by reverse phase HPLC with an on-line radioactivity detector. Retention times (Rts) of the products were compared with those of standard 14C-labeled GAs for tentative identification. The SoGA3ox1 protein converted radioactive GA9 and GA20 to products with Rts of 27.8 and 15.4 min, corresponding to those of GA4 and GA1, respectively. The product of [14C4]GA20 (specific activity 20 μCi μmol−1) incubated with SoGA3ox1 was identified as [14C4]GA1 (Table II), thus demonstrating conclusively that SoGA3ox1 encodes a GA 3-oxidase.
Table II.
Identification of products after incubation of recombinant GA 2-oxidases (SoGA2ox1 and SoGA2ox2) and GA 3-oxidase (SoGA3ox1) from spinach with different substrates
| Enzyme | Substrate | Product | Mass Spectra of Productsa |
|---|---|---|---|
| m/z (% relative abundance) | |||
| SoGA2ox1 | [14C4]GA20 | [14C4]GA29 | M+ 514 (7), 506 (100), 491 (12), 477 (4), 447 (6), 389 (7), 375 (9), 303 (14), 277 (3), 235 (5), 207 (17), 167 (5) |
| SoGA2ox1 | [2H2]GA1 | [2H2]GA8 | M+ 596 (100), 581 (7), 537 (4), 506 (2), 450 (10), 381 (6), 313 (2), 283 (3), 240 (7), 209 (10) |
| SoGA2ox1 | [2H2]GA53 | [2H2]GA97 | M+ 538 (35), 523 (11), 506 (11), 479 (14), 448 (10), 417 (8), 388 (11), 373 (9), 357 (6), 329 (21), 289 (4), 259 (5), 239 (47), 209 (100), 179 (15) |
| SoGA2ox2 | [2H2]GA20 | [2H2]GA29 | M+ 508 (100), 493 (11), 479 (4), 449 (6), 391 (8), 359 (4), 305 (18), 279 (3), 237 (5), 209 (23), 169 (6) |
| SoGA2ox2 | [2H2]GA1 | [2H2]GA8 | M+ 596 (100), 581 (12), 537 (10), 506 (9), 450 (16), 381 (14), 313 (4), 283 (7), 240 (12), 209 (16) |
| SoGA3ox1 | [14C4]GA20 | [14C4]GA1 | M+ 514 (7), 506 (100), 491 (12), 448 (16), 16 (3), 376 (13), 357 (3), 313 (7), 235 (6), 207 (20), 193 (8), 180 (5) |
As the methyl ester trimethylsilyl ethers.
Both SoGA2ox recombinant proteins converted radioactive GA9 and GA20 to putative GA51 (Rt = 25.8 min) and GA29 (Rt = 10.1 min), respectively. The products of [14C4]GA20 and [2H2]GA20 metabolism were identified as [14C4]GA29 and [2H2]GA29, respectively; [2H2]GA1 was converted to [2H2]GA8 (Table II), so that both GA20 and GA1 are substrates for these GA 2-oxidases. In addition to GA29, a second product (Rt = 15.0 min) was formed in incubations of GA20 with SoGA2ox1. This product was tentatively identified as GA29-catabolite, because [14C4]GA29 was converted by SoGA2ox1 to a product with the same Rt.
SoGA2ox1, but not SoGA2ox2, also converted GA53 (Rt = 29.3 min) to GA97 (Rt = 12.4 min; Table II) and a second metabolite (Rt = 17.0 min), which was not identified. Thus, it appears from these results that the substrate specificity of the two GA 2-oxidases isolated from spinach is similar to that of other GA 2-oxidases (Thomas et al., 1999; Elliott et al., 2001) in that they all catalyze 2β-hydroxylation of C19-GAs, but that SoGA2ox1 is also capable of 2β-hydroxylation of the C20-GA, GA53.
Gene Expression in SD and LD Conditions
SoGA2ox1 transcripts were barely detectable on northern blots, so SoGA2ox1 gene expression was measured by the quantitative RT-PCR method (Cowling et al., 1998). For the PCR reaction, 29 cycles were chosen as the point where PCR products of the SoGA2ox1 and SoActin genes were increasing exponentially (data not shown).
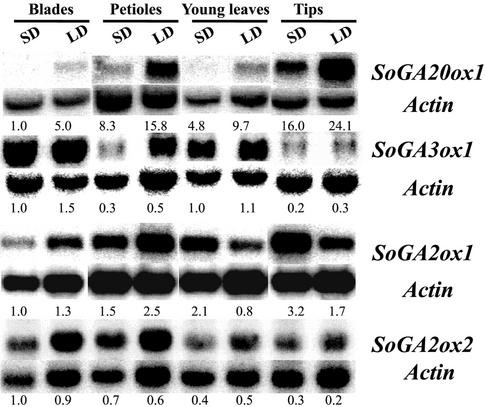
To investigate how the photoperiod regulates expression of the GA dioxygenases isolated, spinach plants grown in SD conditions were transferred to LD conditions for 8 d. Figure 4 shows the expression patterns of four GA dioxygenases in several organs. In SD conditions, the levels of the SoGA20ox1 transcripts were low in blades, petioles, and young leaves, but the SoGA20ox1 transcripts accumulated in all organs tested, especially in petioles and tips, when spinach plants were transferred from SD to LD conditions.
Figure 4.
Expression patterns of the SoGA20ox1, SoGA3ox1, SoGA2ox1, and SoGA2ox2 genes in various organs of spinach plants grown in SD and after 8 LD. Northern blots were prepared by separating 30 μg of total RNA of each organ through 1.2% (w/v) agarose gels containing formaldehyde, followed by transfer to Hybond-N+ membranes. The blots were separately hybridized to 32P-labeled cDNAs of SoGA20ox1, SoGA3ox1, SoGA2ox2, or SoActin. After RT-PCR of SoGA2ox1 and SoActin, 10 μL of the PCR reaction products were separated through 2.0% (w/v) agarose gel by electrophoresis and hybridized to a 32P-labeled cDNA of SoGA2ox1 or SoActin. The numbers under the blots indicate the relative amount of each transcript after standardization using SoActin as a loading control. The value for each gene transcript in blades of spinach grown in SD conditions was arbitrarily set at 1.0.
The levels of the SoGA3ox1 transcripts of plants in SD conditions were lower in petioles and tips than in leaf blades and young leaves. There was a slight increase in SoGA3ox1 transcript levels in leaf blades and petioles when spinach plants were transferred from SD to LD conditions, but the change in photoperiod had very little effect on transcript level in young leaves and tips (Fig. 4), the organs where growth takes place.
The levels of the SoGA2ox1 transcripts in SD-grown plants were lower in leaf blades than in petioles, young leaves, and tips. But in LD conditions, the levels of the SoGA2ox1 transcripts were lower in young leaves than in other organs. Thus, the levels of the SoGA2ox1 transcripts increased in blades and petioles when spinach plants were transferred from SD to LD conditions. By contrast, the level of SoGA2ox1 transcripts decreased in young leaves and tips. This decrease in the level of SoGA2ox1 transcripts in young leaves and tips in LD conditions is opposite to the increase in the level of SoGA20ox1 transcripts.
The SoGA2ox2 gene was highly expressed in leaf blades and petioles. However, there were no significant differences between transcript levels in SD and LD, so that expression of SoGA2ox2 was not further investigated in subsequent experiments.
Time Course of LD Treatment
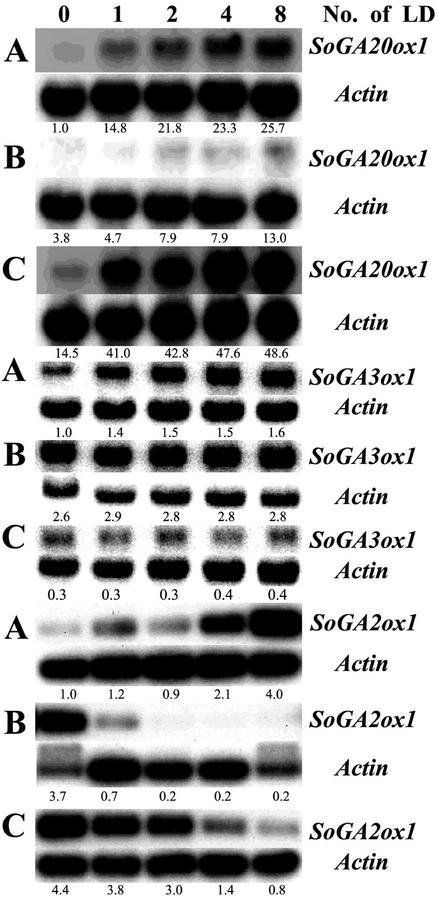
Spinach plants were harvested at different time intervals after transfer from SD to LD conditions to determine the time course of changes in gene expression. The levels of the SoGA20ox1, SoGA3ox1, and SoGA2ox1 transcripts were all up-regulated in petioles in a time-dependent manner (Fig. 5A). This enhancement was much stronger in the case of the SoGA20ox1 gene (26×) than of the SoGA3ox1 gene (1.6×) and of the SoGA2ox1 gene (4×). The levels of the SoGA20ox1 transcripts were also increased time-dependently in young leaves and tips (Fig. 5, B and C). LD conditions caused more rapid accumulation of the SoGA20ox1 transcripts in tips than in petioles and young leaves. However, the SoGA3ox1 mRNA abundance in young leaves and tips was similar or only slightly increased by the LD treatment (Fig. 5, B and C). By contrast, the levels of the SoGA2ox1 transcripts were down-regulated in young leaves and tips (Fig. 5, B and C). The levels of the SoGA2ox1 transcripts were more rapidly decreased in young leaves than in tips. This rapid decrease may compensate for the relatively low increase in the levels of the SoGA20ox1 and SoGA3ox1 transcripts in young leaves to maintain the active GAs. The time course experiment demonstrates the different regulation by the photoperiod of the three GA dioxygenases: (a) expression of SoGA20ox1 changed quickly and strongly in petioles and tips, (b) the SoGA3ox1 transcript level was only slightly changed in petioles, and (c) the SoGA2ox1 transcript level increased in petioles but decreased in young leaves and tips.
Figure 5.
Time course of SoGA20ox1, SoGA3ox1, and SoGA2ox1 expression after spinach plants were transferred from SD to LD conditions. GA20ox1 transcripts increased with increasing duration of LD treatment in petioles (A), young leaves (B), and tips (C). GA3ox1 transcripts increased with increasing duration of LD treatment in petioles (A), but they did not change in young leaves (B), and tips (C) after exposure to increasing numbers of LD. GA2ox1 transcripts increased with increasing duration of LD treatment in petioles (A), but they decreased in young leaves (B), and tips (C). The numbers under the blots indicate the relative amount of transcript after standardization using SoActin as a loading control. The value of each transcript level in petioles in SD was arbitrarily set at 1.0.
Effect of Light and Darkness on Gene Expression
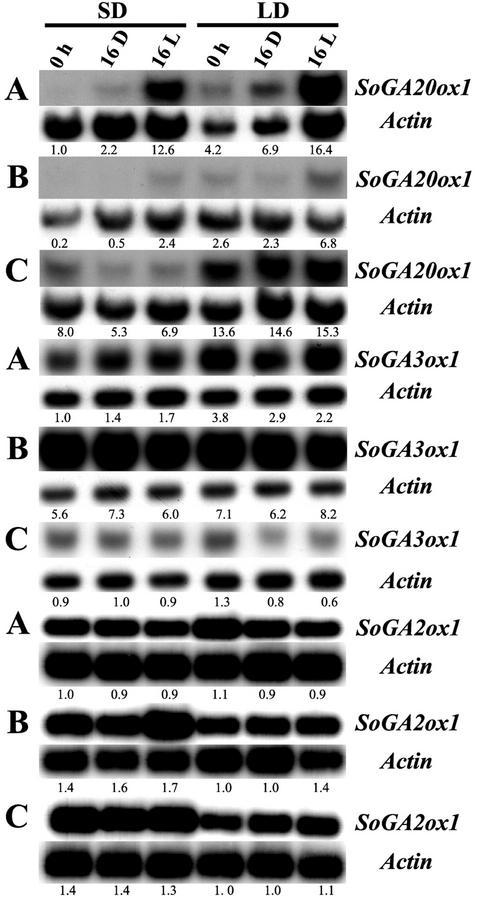
To investigate the effect of light and darkness on gene expression, spinach plants grown in SD and LD conditions were harvested at the end of the 8-h high-intensity light period (0 h), at the end of 16 h of darkness (16 D), or at the end of 16 h of low-intensity light (16 L). The levels of the SoGA20ox1, SoGA3ox1, and SoGA2ox1 transcripts were determined in petioles, young leaves, and tips (Fig. 6). Compared with the levels in each group at the end of the 8-h high-intensity light period (0 h), additional light (16 L) greatly increased the levels of the SoGA20ox1 transcripts in petioles of plants that were initially in SD conditions (13× compared with SD at 0 h) and also in petioles in LD conditions (4× compared with LD at 0 h; Fig. 6A). The levels of the SoGA20ox1 transcripts in tips were not changed by additional light (Fig. 6C). A long dark treatment (16 D) did not affect the levels of the SoGA20ox1 transcripts in petioles, young leaves, and tips in either SD or LD conditions. This means that the levels of SoGA20ox1 transcript are regulated much more by light than by darkness. Exposure to weak light or dark treatment had little effect on the levels of the SoGA3ox1 and SoGA2ox1 transcripts (Fig. 6).
Figure 6.
Effects of light and darkness on the transcript level of SoGA20ox1, SoGA3ox1, and SoGA2ox1 in petioles (A), young leaves (B), and tips (C). Spinach plants were harvested at the end of the 8-h high-intensity light period (0 h) and after transfer from SD or LD conditions to 16 h of darkness (16 D) or to 16 h of weak incandescent light (16 L). The numbers under the blots indicate the relative amount of each transcript after standardization using SoActin as a loading control. The value of each transcript level in petioles at 0 h in SD was arbitrarily set at 1.0.
Expression in Other Organs
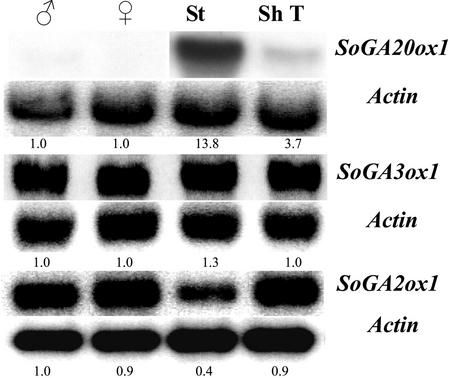
To determine other organ-specific expression patterns of the GA dioxygenases, male and female flowers, stems, and shoot tips were harvested from spinach plants that had been exposed to LD conditions for more than 3 weeks (Fig. 7). The shoot tips included the upper 1 cm; stems were the next 1 cm of the elongated stem. The SoGA20ox1 mRNA was more abundant in stems than in shoot tips, and little transcript was detected in flowers. By contrast, SoGA3ox1 and SoGA2ox1 were highly expressed in male and female flowers, stems, and shoot tips.
Figure 7.
Expression of SoGA20ox1, SoGA3ox1, and SoGA2ox1 in male flowers (♂), female flowers (♀), stems (St), and shoot tips (ShT). Spinach plants were harvested after more than 3 weeks in LD conditions. In this case, the shoot tips included the upper 1 cm of the shoots; stems were the next 1 cm of the elongated stems. The numbers under the blots indicate the relative amount of each transcript after standardization using SoActin as a loading control. The value of the transcript level of each gene in male flowers was arbitrarily set at 1.0.
DISCUSSION
We isolated a full-length cDNA clone encoding GA 3-oxidase and two full-length cDNA clones encoding GA 2-oxidase from spinach. Heterologous expression in E. coli showed that the product of the SoGA3ox1 gene is able to catalyze 3β-hydroxylation of GA20 to GA1, whereas the products of the SoGA2ox genes catalyze 2β-hydroxylation of GA20 and GA1, both C19-GAs with a γ-lactone in the A ring of the GA structure (Fig. 1). In addition, recombinant SoGA2ox1 converts GA53 to GA97, both C20-GAs (Fig. 1). The recombinant proteins of GA 2-oxidases from other species specifically 2β-hydroxylate C19-GAs (Thomas et al., 1999; Elliott et al., 2001). So far, few exceptions have been reported to this rule: GA15, a C20-GA, was 2β-hydroxylated by PcGA2ox1 and AtGA2ox2 (Thomas et al., 1999); and GA44, also a C20-GA (Fig. 1), was 2β-hydroxylated to GA98 by OsGA2ox1 (Sakamoto et al., 2001). Both of these GAs have a δ-lactone in the A ring. When this lactone ring was opened, GA15 was no longer metabolized by recombinant PcGA2ox1, but it was (although less efficiently) by AtGA2ox2 (Thomas et al., 1999). Spinach produces large amounts of 2β-hydroxy-GA53 (=GA97), and smaller amounts of GA98 (= 2β-hydroxy-GA44), GA99 (= 2β-hydroxy-GA19; Mander et al., 1996), and GA110 (= 2β-hydroxy-GA12; Owen et al., 1998). Substrate specificity of the 2-oxidases for the possible precursors of these 2β-hydroxy-C20-GAs remains to be determined.
Expression of SoGA3ox1 was very little affected by the photoperiod (Figs. 4 and 5), so that it is possible that the conversion of GA20 to GA1 in LD is limited by the amount of GA20 substrate available and not by the enzyme level. Although expression of SoGA2ox1 was down-regulated by LD, this may be only a minor factor in the maintenance of a high GA1 level, because SoGA2ox1 transcripts are much less abundant than those of SoGA2ox2, and expression of the latter is not under photoperiodic control (Fig. 4). In a different system, de-etiolation of pea seedlings, GA 3-oxidase was down-regulated by light, whereas GA 2-oxidase was up-regulated. This resulted in decreased GA1, increased GA8 levels, and a reduction in growth rate (Reid et al., 2002).
The increase in growth rate of petioles of spinach in LD occurs during the first 16 h of weak supplementary light (Zeevaart, 1971). This may be mainly because of the increase in SoGA20ox1 transcripts during the additional 16 h of light rather than the small increase in SoGA3ox1 and decrease in SoGA2ox1 transcripts (Fig. 6A). The results in Figure 6 show that moving spinach plants from LD to 16 h darkness did not reduce the transcript level of any of the GA dioxygenases studied. On the other hand, Gilmour et al. (1986) reported that enzyme activities for the conversions GA53 to GA44 and GA19 to GA20, both representing GA 20-oxidase activity, declined quickly in plants in darkness. It is possible, therefore, that regulation of GA 20-oxidase by photoperiod is not only at the transcriptional level, but that there may be posttranscriptional control as well. Thus, transcript levels may not necessarily be indicative of enzyme activity. Further work on photoperiodic control of GA biosynthesis in spinach should therefore attempt to compare the protein level and enzymatic activity of GA 20-oxidase.
The low expression of the SoGA20ox1 gene in spinach flowers (Fig. 7) may mean that it has no role in flower development, unlike the LeGA20ox1 and LeGA20ox2 genes, which are highly expressed in tomato (Lycopersicon esculentum) during flower bud and early fruit development (Rebers et al., 1999). Thus, another GA 20-oxidase gene may be involved in flower development in spinach.
Up-regulation of GA biosynthesis by LD starts at the beginning of the pathway with increased ent-kaurene biosynthesis (Zeevaart and Gage, 1993), and in the later stages, it is mainly attributable to the much increased expression of SoGA20ox1. It is surprising that expression of SoGA3ox1, encoding the product that catalyzes the pivotal step of 3β-hydroxylation, shows little fluctuation in response to the photoperiod (Fig. 4). Although expression of SoGA2ox1 is down-regulated in tips and young leaves of plants in LD, the overall effect of LD on GA metabolism is that more GA29 (Metzger and Zeevaart, 1980, 1982) and GA8 (Wu et al., 1996) are produced in plants in LD than in SD. Thus, it appears that in SD, the steps catalyzed by GA 20-oxidase are enzyme limited, whereas GA 3-oxidase and GA 2-oxidase are limited by the amount of substrate available. In LD, a new balance is established between GA1 synthesis and deactivation, increasing the GA1 level to such an extent that the threshold value required for stem elongation is exceeded (Zeevaart et al., 1993).
MATERIALS AND METHODS
Plant Material and Growing Conditions
Spinach (Spinacia oleracea L. cv Savoy Hybrid 612) plants were grown in a SD growth chamber for approximately 6 or 7 weeks as described previously (Zeevaart and Gage, 1993). SD conditions consisted of 8 h of light from fluorescent tubes and incandescent bulbs of approximately 300 μmol m−2 s−1 at 23°C, followed by 16 h darkness at 20°C. For LD conditions, the 8-h main light period was followed by 16 h of weak light from incandescent bulbs of 10 μmol m−2 s−1 at 20°C.
RT-PCR
RT-PCR was used for the cloning of GA 3-oxidase and GA 2-oxidases. First-strand cDNA from total RNA was synthesized by SuperScript II RNase H-minus Reverse Transcriptase (Invitrogen, Carlsbad, CA), using oligo(dT) primers at 42°C for 1 h. The cDNA product was then used in PCR reactions containing Taq polymerase and several pairs of degenerate primers (Table I). For GA 3-oxidase, degenerate primers were synthesized based on conserved regions of GA 3-oxidases from Arabidopsis (Chiang et al., 1995; Yamaguchi et al., 1998) and pea (Lester et al., 1997). For GA 2-oxidase, the degenerate primers were synthesized based on conserved regions of GA 2-oxidases from runner bean and Arabidopsis (Thomas et al., 1999). These primers were used in all combinations in PCR reactions with first-strand cDNA as a template. The PCR amplifications were performed with a DNA Thermal Cycler (RTC-200, MJ Research, Waltham, MA) in a 50-μL reaction containing 1× PCR buffer (Invitrogen), 1.5 mm MgCl2, 200 μm dNTP, 2.5 pmol of each primer, 1 unit of Taq DNA polymerase (Invitrogen). The reaction mixtures were heated to 94°C for 3 min and then subjected to 30 cycles of 94°C for 1 min, 48°C for 1 min, and 72°C for 45 s. A final extension was performed at 72°C for 10 min. A 5-μL volume of the PCR reaction products was analyzed by 1.2% (w/v) agarose gel electrophoresis, purified with a Wizard PCR purification system (Promega, Madison, WI), and then cloned into the pGEM Teasy vector (Promega). Analysis of the DNA sequences was carried out using the DNASTAR program (DNASTAR, Inc., Madison, WI). Multiple sequence alignments were performed using the Clustal W 1.8 Multiple Sequence Alignment program and printed using BOXSHADE 3.21 (http://www.ch.embnet.org/software/Box_form.html).
RACE and Cloning
5′- and 3′-RACE of spinach GA 3-oxidase cDNA and GA 2-oxidase cDNA were performed using the SMART RACE cDNA Amplification Kit from BD Biosciences Clontech. Poly(A+) RNA was isolated from spinach using the PolyATtract mRNA Isolation System IV (Promega) according to the manufacturer's instructions. After RT with primers supplied by BD Biosciences Clontech, the first-strand cDNA was used directly in 5′- and 3′-RACE PCR reactions. Primary PCR amplification reactions were achieved using a high-fidelity enzyme (Pfu Turbo polymerase; Stratagene, La Jolla, CA) and gene-specific primers JZ282 and JZ298 for GA 2-oxidase1, JZ375 and JZ574 for GA 2-oxidase2, and JZ445 and JZ444 for GA 3-oxidase1 to generate the 5′- and 3′-cDNA fragments, respectively. The PCR reaction consisted of the first denaturation for 3 min at 94°C, a series of 30 cycles (1 min at 94°C, 1 min at 54°C or 55°C, and 1 min at 72°C) with a final extension for 5 min at 72°C using a thermal cycler (RTC-200, MJ Research). A 5-μL aliquot of the RT-PCR and RACE reaction solution was analyzed by 1.2% (w/v) agarose gel electrophoresis. PCR products were purified and cloned into the pCR-Script Cam SK(+) cloning vector from Stratagene. These constructs were sequenced.
Northern-Blot Analysis
Leaf blades, petioles, young leaves, tips (Zeevaart and Gage, 1993), and other organs were harvested, frozen immediately in liquid N2, and stored at −80°C. Northern blots were prepared by electrophoresis of 30 μg of total RNA in the presence of formaldehyde (Sambrook et al., 1989), and RNA was transferred to nitrocellulose. Full-length cDNAs were labeled with [32P]dCTP by the Random Primers DNA Labeling System (Invitrogen) and the radioactive probes were used to probe northern blots. Hybridization was carried out at 42°C using a 50% (v/v) formamide system (Sambrook et al., 1989). Membranes were washed twice for 10 min in 2× SSC at room temperature and then twice for 10 min in 0.2× SSC with 0.1% (w/v) SDS at 65°C (high stringency). At low stringency, the membrane was washed once for 10 min in 2× SSC at room temperature and then washed again for 10 min in 2× SSC at 65°C.
The relative amounts of mRNA were determined with a PhosphorImager (Molecular Dynamics, Sunnyvale, CA). Each blot was first probed with one of the various cDNAs, and then stripped and reprobed with SoActin. The ratio of the GA dioxygenase mRNA to Actin mRNA was used to compare the mRNA levels in various organs and treatments. Each experiment was repeated at least three times with similar results.
Quantitative RT-PCR
The first-strand cDNA synthesis reactions were carried out using 5 μg of each total RNA sample, random hexamer primer, and avian RNase H-minus reverse Transcriptase. A pair of primers (JZ385 and JZ386) was used to amplify a 401-bp fragment of GA 2-oxidase1. JZ389 and JZ390 primers (Table I) were used to amplify a 324-bp fragment of the SoActin gene as a control. Both of these fragments can be distinguished from any contaminant of genomic DNA by size. The RT-PCR products were cloned and sequenced to verify the sequences of the fragments of the SoGA2ox1 and SoActin genes. The cDNA solutions (2.5 μL) were used as a template in a standard 50-μL PCR reaction. Expression of SoGA2ox1 and SoActin was compared at 29 cycles where both PCR reactions were progressing exponentially. Ten microliters of PCR reaction products was separated by electrophoresis, blotted, and hybridized.
Expression of Recombinant GA 3-Oxidase and GA 2-Oxidase Proteins
Coding regions of 2-oxidases were produced by PCR with the following primer set designed from the RACE product: JZ300 and JZ301 for mature GA 2-oxidase1 (PspAI and XhoI sites inserted to facilitate cloning) and JZ582 and JZ619 (BamHI and XhoI sites inserted to facilitate cloning) for mature GA 2-oxidase2. For GA 3-oxidase, JZ509 and JZ510 (PspAI and SalI sites inserted to facilitate cloning) were used to get the coding region. The resulting PCR fragments were cloned into a pCR-Script Cam SK(+) cloning vector and then digested with PspA1/XhoI, BamHI/XhoI, or PspA1/SalI, respectively, and subcloned into the corresponding restriction sites of the pGEX-5X-2 vector (Amersham Biosciences, Piscataway, NJ). These full-length cDNA clones (pGEXGA3ox1, pGEXGA2ox1, or pGEXGA2ox2) were transformed into Escherichia coli, strain BL21pLysS. Fifty milliliters of freshly cultured cells was added to 1 L of Luria-Bertani medium with 100 mg L−1 ampicillin and incubated at 37°C with vigorous shaking. When the optical density at 600 nm reached 0.6, isopropyl-β-d-thiogalactopyranoside was added to give a final concentration of 3 mm, and the culture was incubated for another 2 h. The cells were harvested and suspended in lysis buffer (100 mm Tris-HCl, pH 7.5, and 10 mg mL−1 lysozyme), and incubated at room temperature for 10 min. Crude cell extracts were obtained by using a Sonifier Cell Disruptor 200 (Branson Ultrasonics, Danbury, CT) carried out for 10 cycles (10 × 10 s) on ice. The lysates were submerged in liquid N2 for 2 min and then thawed in an ice bath for 15 min (Johnson and Hecht, 1994). The lysates were centrifuged at 13,000 rpm for 30 min, and the supernatant was stored at −80°C until used for enzyme assays.
Enzyme Assays and Product Identification
The assays for GA 3-oxidase activity and GA 2-oxidase activities were performed with approximately 30,000 dpm of 14C-labeled GAs. [2H2]GAs were used only for product identification. The reaction mixture (100 μL) contained 100 mm Tris buffer (pH 7.5), 0.5 mm FeSO4, 5 mm 2-oxoglutarate, 5 mm ascorbate, and 80 μL of bacterial extract. Cofactors were added again after 1 h. The mixture was incubated at 30°C for up to 6 h with gentle shaking. For identification of products, the assays were scaled up to 500 μL. The products were isolated by reverse phase HPLC with an on-line FLO-ONE detector (Radioanalytic, Tampa, FL; Zeevaart and Gage, 1993). For identification of the products by gas chromatography (GC)-mass spectrometry (Zeevaart et al., 1993), the GC was equipped with a DB-5MS capillary column (30-m × 0.32-mm × 0.25-μm film, J&W Scientific, Folsom, CA), which was operated in splitless mode. The oven temperature was kept at 100°C for 1 min after sample injection and then programmed from 100°C to 230°C at 40°C min−1, from 230 to 280°C at 8°C min−1, and finally to 300°C at 20°C min−1. Specific activities of substrates and products were determined by GC-selected ion monitoring of the molecular ion clusters (Zeevaart and Gage, 1993).
Distribution of Materials
Upon written request, all novel materials described in this publication will be made available in limited quantities and in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
ACKNOWLEDGMENTS
We thank Bev Chamberlin (Michigan State University Mass Spectrometry Facility) for her assistance with GC-mass spectrometry. We are grateful to Dr. Xiangqian Ma (Michigan State University) for her helpful comments on the manuscript.
Footnotes
This work was supported by the U.S. Department of Agriculture (grant no. 97–35304–4692) and by the U.S. Department of Energy (grant no. DE–FG02–91ER20021).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.008581.
LITERATURE CITED
- Chiang H-H, Hwang I, Goodman HM. Isolation of the Arabidopsis GA4locus. Plant Cell. 1995;7:195–201. doi: 10.1105/tpc.7.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowling RJ, Kamiya Y, Seto H, Harberd NP. Gibberellin dose-response regulation of GA4gene transcript levels in Arabidopsis. Plant Physiol. 1998;117:1195–1203. doi: 10.1104/pp.117.4.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott RC, Ross JJ, Smith JJ, Lester DR, Reid JB. Feed-forward regulation of gibberellin deactivation in pea. J Plant Growth Regul. 2001;20:87–94. [Google Scholar]
- Gilmour SJ, Zeevaart JAD, Schwenen L, Graebe JE. Gibberellin metabolism in cell-free extracts from spinach leaves in relation to photoperiod. Plant Physiol. 1986;82:190–195. doi: 10.1104/pp.82.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P, Phillips AL. Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci. 2000;5:523–530. doi: 10.1016/s1360-1385(00)01790-8. [DOI] [PubMed] [Google Scholar]
- Itoh H, Tanaka-Ueguchi M, Kawaide H, Chen X, Kamiya Y, Matsuoka M. The gene encoding tobacco gibberellin 3-oxidase is expressed at the site of GA action during stem elongation and flower organ development. Plant J. 1999;20:15–24. doi: 10.1046/j.1365-313x.1999.00568.x. [DOI] [PubMed] [Google Scholar]
- Itoh H, Tanaka-Ueguchi M, Sentoku N, Kitano H, Matsuoka M, Kobayashi M. Cloning and functional analysis of two gibberellin 3β-oxidase genes that are differently expressed during the growth of rice. Proc Natl Acad Sci USA. 2001;98:8909–8914. doi: 10.1073/pnas.141239398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BH, Hecht MH. Recombinant proteins can be isolated from Escherichia colicells by repeated cycles of freezing and thawing. BioTechnology. 1994;12:1357–1360. doi: 10.1038/nbt1294-1357. [DOI] [PubMed] [Google Scholar]
- Lester DR, Ross JJ, Davies PJ, Reid JB. Mendel's stem length gene (Le) encodes a gibberellin 3-oxidase. Plant Cell. 1997;9:1435–1443. doi: 10.1105/tpc.9.8.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester DR, Ross JJ, Smith JJ, Elliott RC, Reid JB. Gibberellin 2-oxidation and the SLN gene of Pisum sativum. Plant J. 1999;19:65–73. doi: 10.1046/j.1365-313x.1999.00501.x. [DOI] [PubMed] [Google Scholar]
- Mander LN, Owen DJ, Croker SJ, Gaskin P, Hedden P, Lewis MJ, Talon M, Gage DA, Zeevaart JAD, Brenner ML et al. Identification of three C20-gibberellins: GA97 (2β-hydroxy-GA53), GA98 (2β-hydroxy-GA44) and GA99 (2β-hydroxy-GA19) Phytochemistry. 1996;43:23–28. doi: 10.1016/0031-9422(96)00251-8. [DOI] [PubMed] [Google Scholar]
- Metzger JD, Zeevaart JAD. Effect of photoperiod on the levels of endogenous gibberellins in spinach as measured by combined gas chromatography-selected ion current monitoring. Plant Physiol. 1980;66:844–846. doi: 10.1104/pp.66.5.844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger JD, Zeevaart JAD. Photoperiodic control of gibberellin metabolism in spinach. Plant Physiol. 1982;69:287–291. doi: 10.1104/pp.69.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen DJ, Mander LN, Storey JMD, Huntley RP, Gaskin P, Lenton JR, Gage DA, Zeevaart JAD. Synthesis and confirmation of structure for a new gibberellin, 2β-hydroxy-GA12 (GA110), from spinach and oil palm. Phytochemistry. 1998;47:331–337. doi: 10.1016/s0031-9422(97)00577-3. [DOI] [PubMed] [Google Scholar]
- Rebers M, Kaneta T, Kawaide H, Yamaguchi S, Yang Y-Y, Imai R, Sekimoto H, Kamiya Y. Regulation of gibberellin biosynthesis genes during flower and early fruit development of tomato. Plant J. 1999;17:241–250. doi: 10.1046/j.1365-313x.1999.00366.x. [DOI] [PubMed] [Google Scholar]
- Reid JB, Botwright NA, Smith JJ, O'Neill DP, Kerckhoffs LHJ. Control of gibberellin levels and gene expression during de-etiolation in pea. Plant Physiol. 2002;128:734–741. doi: 10.1104/pp.010607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Kobayashi M, Itoh H, Tagiri A, Kayano T, Tanaka H, Iwahori S, Matsuoka M. Expression of a gibberellin 2-oxidase gene around the shoot apex is related to phase transition in rice. Plant Physiol. 2001;125:1508–1516. doi: 10.1104/pp.125.3.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Talon M, Zeevaart JAD. Gibberellins and stem growth as related to photoperiod in Silene armeriaL. Plant Physiol. 1990;92:1094–1100. doi: 10.1104/pp.92.4.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talon M, Zeevaart JAD, Gage DA. Identification of gibberellins in spinach and effects of light and darkness on their levels. Plant Physiol. 1991;97:1521–1526. doi: 10.1104/pp.97.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SG, Philips AL, Hedden P. Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc Natl Acad Sci USA. 1999;96:4698–4703. doi: 10.1073/pnas.96.8.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Gage DA, Zeevaart JAD. Molecular cloning and photoperiod-regulated expression of gibberellin 20-oxidase from the long-day plant spinach. Plant Physiol. 1996;110:547–554. doi: 10.1104/pp.110.2.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Kamiya Y. Gibberellin biosynthesis: its regulation by endogenous and environmental signals. Plant Cell Physiol. 2000;41:251–257. doi: 10.1093/pcp/41.3.251. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Smith MW, Brown RGS, Kamiya Y, Sun T. Phytochrome regulation and differential expression of gibberellin 3β-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell. 1998;10:2115–2126. doi: 10.1105/tpc.10.12.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart JAD. Effects of photoperiod on growth rate and endogenous gibberellins in the long-day rosette plant spinach. Plant Physiol. 1971;47:821–827. doi: 10.1104/pp.47.6.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart JAD, Gage DA. ent-Kaurene biosynthesis is enhanced by long photoperiods in the long-day plants Spinacia oleracea L. and Agrostemma githagoL. Plant Physiol. 1993;101:25–29. doi: 10.1104/pp.101.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart JAD, Gage DA, Talon M. Gibberellin A1is required for stem elongation in spinach. Proc Natl Acad Sci USA. 1993;90:7401–7405. doi: 10.1073/pnas.90.15.7401. [DOI] [PMC free article] [PubMed] [Google Scholar]