Abstract
UDP-xylose (Xyl) is an important sugar donor for the synthesis of glycoproteins, polysaccharides, various metabolites, and oligosaccharides in animals, plants, fungi, and bacteria. UDP-Xyl also feedback inhibits upstream enzymes (UDP-glucose [Glc] dehydrogenase, UDP-Glc pyrophosphorylase, and UDP-GlcA decarboxylase) and is involved in its own synthesis and the synthesis of UDP-arabinose. In plants, biosynthesis of UDP-Xyl is catalyzed by different membrane-bound and soluble UDP-GlcA decarboxylase (UDP-GlcA-DC) isozymes, all of which convert UDP-GlcA to UDP-Xyl. Because synthesis of UDP-Xyl occurs both in the cytosol and in membranes, it is not known which source of UDP-Xyl the different Golgi-localized xylosyltransferases are utilizing. Here, we describe the identification of several distinct Arabidopsis genes (named AtUXS for UDP-Xyl synthase) that encode functional UDP-GlcA-DC isoforms. The Arabidopsis genome contains five UXS genes and their protein products can be subdivided into three isozyme classes (A–C), one soluble and two distinct putative membrane bound. AtUxs from each class, when expressed in Escherichia coli, generate active UDP-GlcA-DC that converts UDP-GlcA to UDP-Xyl. Members of this gene family have a large conserved C-terminal catalytic domain (approximately 300 amino acids long) and an N-terminal variable domain differing in sequence and size (30–120 amino acids long). Isoforms of class A and B appear to encode putative type II membrane proteins with their catalytic domains facing the lumen (like Golgi-glycosyltransferases) and their N-terminal variable domain facing the cytosol. Uxs class C is likely a cytosolic isoform. The characteristics of the plant Uxs support the hypothesis that unique UDP-GlcA-DCs with distinct subcellular localizations are required for specific xylosylation events.
UDP-Xyl is a nucleotide sugar required for the synthesis of diverse plant cell wall polysaccharides including xyloglucan (Ray, 1980; Hayashi and Matsuda, 1981; White et al., 1993; Baydoun and Brett, 1997), xylan (Rodgers and Bolwell, 1992), and minor plant metabolites (Rose et al., 1996; Martin et al., 1997). UDP-Xyl is also required for the synthesis of glycoproteins (Zeng et al., 1997; Strasser et al., 2000), animal proteoglycans (Gotting et al., 2000; Kuhn et al., 2001), and fungal polysaccharides (Ankel et al., 1967). Biochemical and immunocytochemical studies have provided evidence that the synthesis of these polysaccharides occurs in the lumen of the Golgi apparatus and is catalyzed by various xylosyltransferases that transfer Xyl from UDP-Xyl onto acceptor molecules (Moore et al., 1991; Baydoun and Brett, 1997; Bolwell, 2000).
In plants, the biosynthesis of UDP-Xyl occurs both in the cytosol and in membrane-bound compartments. The synthesis of UDP-Xyl is catalyzed by different membrane-bound and soluble UDP-GlcA decarboxylase (UDP-GlcA-DC) isoforms, all of which convert UDP-GlcA to UDP-Xyl (Neufeld et al., 1958; Feingold et al., 1960; John et al., 1977a, 1977b; Hayashi et al., 1988).
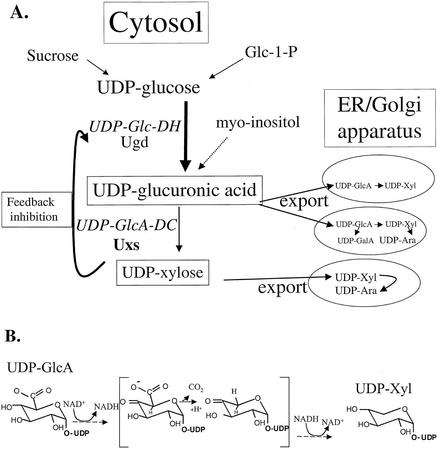
In plants, the major route by which UDP-Xyl is produced is via UDP-Glc (Feingold, 1982; Tenhaken and Thulke, 1996; Gibeaut, 2000; see Fig. 1A). UDP-Glc dehydrogenase (UDP-Glc-DH), a cytosolic enzyme (Stewart and Copeland, 1998), converts UDP-Glc to UDP-GlcA (Seitz et al., 2000). The cytosolic UDP-GlcA-decarboxylase (UDP-GlcA-DC) isozyme converts UDP-GlcA to UDP-Xyl (John et al., 1977a; Kyossev et al., 1995). UDP-Glc-DH, however, is inhibited by UDP-Xyl (Feingold, 1982; Stewart and Copeland, 1998). In addition, UDP-Xyl was also found to regulate in vitro production of UDP-Glc by inhibiting UDP-Glc pyrophosphorylase (Ray and Abdul-Baki, 1968). Such feedback inhibition could regulate the flux of the conversion of UDP-Glc to UDP-GlcA, UDP-Xyl, UDP-Ara, UDP-GalA, and UDP-apiose in plants. These UDP sugars are the source of approximately 40% of the wall polysaccharide mass. UDP-GlcA can also be generated by oxidation of myo-inositol (Loewus and Loewus, 1980), but the relative amount of UDP-GlcA produced in this way is unclear and could be tissue specific (Seitz et al., 2000).
Figure 1.
A, Metabolic routes involved in the synthesis of UDP-Xyl. Synthesis of UDP-Xyl occurs both in the cytosol and in membrane-bound compartments. B, Possible intermediates (in brackets) involved in the enzymatic conversion of UDP-GlcA to UDP-Xyl, based on Feingold (1982).
How the metabolic flux and balance of these diverse UDP sugars is regulated, and how this regulation affects the biosynthesis of plant polysaccharides and glycoproteins, is far from being understood. Furthermore, biochemical studies suggest that the xylosylation of various polymers may take place in different Golgi cisternae (Baydoun and Brett, 1997; Baydoun et al., 2001). Hayashi et al. (1981, 1988) demonstrated that the Golgi consists of membrane-bound UDP-GlcA-DC that readily converts UDP-GlcA to UDP-Xyl upon which a xylosyltransferase incorporates the Xyl residue into xyloglucan polysaccharide. How the different Golgi-localized xylosyltransferases receive their supply of UDP-Xyl is unclear. For example, does a specific Golgi-xylosyltransferase utilize UDP-Xyl made in the cytosol or UDP-Xyl made in the Golgi compartment?
The cellular roles of UDP-Xyl, and the various UDP-GlcA-DC isozymes that are required to generate UDP-Xyl in plants, are complex and many questions related to sugar nucleotide flux (Dalessandro and Northcote, 1977a, 1977b; Amino et al., 1985; Robertson et al., 1995), subcellular compartmentalization, and the sites of UDP-Xyl synthesis remain to be answered. Here, we describe the identification of a functional Arabidopsis gene family encoding soluble and putative membrane-bound isoforms of UDP-GlcA-DC.
RESULTS
Protein Alignment of AtUXS Gene Family
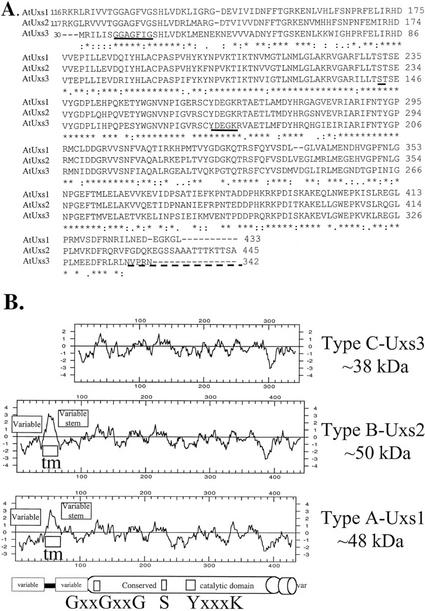
Analysis of Arabidopsis genomic and expressed sequence tag (EST) sequences revealed at least five different GenBank entries (Table I) with amino acid sequence similarity to the UDP-GlcA-DC gene product, Uxs, isolated from C. neoformans (Bar-Peled et al., 2001). The existence in plants of several membrane-bound and soluble UDP-GlcA-DC isozymes is consistent with prior biochemical data (Feingold et al., 1960; John et al., 1977a, 1977b; Hayashi et al., 1988) and suggests that multiple genes for this enzyme are present in plants. We decided to isolate and characterize three isoforms of the UDP-GlcA-DC gene family from Arabidopsis because they contained the conserved functional motifs found in the catalytic domain of Uxs from C. neoformans and also appeared distinct from each other at the N terminus region (see below). Three Arabidopsis cDNA clones designated AtUXS1, AtUXS2, and AtUXS3 were generated using reverse transcription (RT)-PCR. Nucleotide sequence comparison of clone AtUXS1 matched a partial Arabidopsis EST (accession no. be038709) DNA sequence. The gene for AtUXS1 was found to be located on chromosome 3 (accession no. al1232966 or AT3g53520; see Table I). cDNA clone AtUXS2 matched several ESTs as well as an Arabidopsis cDNA clone ATD18MR (Kushnir et al., 1995) with unknown function. The gene for AtUXS2 was also found to be located on chromosome 3 (accession no. al162651 or AT3g62830). cDNA clone AtUXS3 matched several ESTs that share nucleotide identity with a genomic clone on chromosome 5 (accession no. abo16890 or AT5g59290). NCBI BLAST analysis (Altschul et al., 1997) of the three cDNA clones (AtUXS 1–3) isolated from Arabidopsis demonstrated that the encoded proteins share 59%, 61%, and 57% amino acid sequence identity, respectively, to the C. neoformans UDP-GlcA-DC gene product UXS. The amino acid alignment between the conserved regions of the putative Arabidopsis UDP-GlcA-DC isoforms is shown in Figure 2. The Arabidopsis UXS genes and the C. neoformans UXS gene appear to belong to a gene family that shares similarity with dehydratases, dehydrogenases, and epimerases. These genes have several fingerprint sequence motifs, including an N-terminal GxxGxxG sequence that is characteristic of the ADP-binding βαβαβ-fold (Rosmann fold) associated with NAD(P)-binding proteins (Weirenga et al., 1986). The GxxGxxG motif (Fig. 2B) is probably located at amino acid position 126 to 132 in AtUxs1, amino acid position 125 to 131 in AtUxs2, and amino acid position 36 to 42 in AtUxs3. The first step in the dehydratase and epimerase enzyme mechanisms is the abstraction of the 4-hydroxyl proton and hydride transfer from the C4 position of the sugar to NAD+. This mechanism is well studied in UDP-Glc epimerase and 4,6-dehydratase (Baker and Blasco, 1992; Liu et al., 1997). This mechanism requires a catalytic triad whereby Ser and Lys are suggested to activate Tyr to abstract the C4-proton to yield a nucleotide-4-keto sugar intermediate (Fig. 1B). The Uxs gene family contains a characteristic and highly conserved Ser, Tyr, and Lys triad. The Ser residue is probably located at amino acid position 232 in AtUxs1, position 231 in AtUxs2, and position 143 in AtUxs3. The Tyr and Lys residues of the YxxxK motif are probably located at amino acid positions 262 to 266 (AtUxs1), 261 to 265 (AtUxs2), and 176 to 180 (AtUxs3). Although the core catalytic domain of the AtUxs is conserved, variable regions were identified in the Arabidopsis UXS gene family at the N and C terminus (Fig. 2A). Analysis of the three cDNAs using the PSORT program (version 6.4; Hartmann et al., 1989; Nakai and Kanehisa, 1992) indicated that cDNA AtUXS1 and AtUXS2 likely encode putative type II membrane proteins. The N-terminal extension region of protein AtUxs1 and AtUxs2 have low amino acid sequence identity and can be divided into three domains. The first approximately 45-amino acid domain of AtUxs1 and AtUxs2 has a predicted cytoplasmic region (amino acids 1–48 and 1–43, respectively). The cytosolic domain is followed by a 16-amino acid hydrophobic putative membrane-spanning domain (amino acids 49–65 and 44–60, respectively). The putative membrane domain is followed by a “stem domain” (approximately 50 amino acids) that represents a variable amino acid linker region spanning from amino acids 66 to 115 and 61 to 116, respectively. Overall, the N-terminal approximately 120-amino acid-long extension region of AtUxs1 has no sequence similarity to AtUxs2 (or AtUxs4); therefore, we classified them as class A and B putative membrane-bound UDP-GlcA-DC isoforms. The Arabidopsis clone encoding protein AtUxs3 lacks the N-terminal extension region, suggesting that it is a soluble protein. The large (>350 amino acid) C terminus region of AtUxs1 (amino acids 66–433) and AtUxs2 (amino acids 61–445) is proposed to face the lumen of an endomembrane compartment. A variable region at the carboxy terminus was also observed in the different AtUxs (Fig. 2). The last 19 amino acids of AtUxs2 are not found in AtUxs3 and they do not share any homology with the carboxy terminus region of AtUxs1.
Table I.
Chromosomal location and amino acid sequence similarity between Arabidopsis AtUXS and with UXS-like genes in other organisms
| UDP-GlcA-Decarboxylase Genes | Accession no. (Based on The Arabidopsis Information Resource Database or NCBI*a) | Chromosome, Map Position in Arabidopsis | Predicted Molecular Mass of the Protein | Amino Acid Identity/Similarity |
|---|---|---|---|---|
| kD | ||||
| AtUXS1-Arabidopsis | AT3g53520 | 3, ∼80 cM | 48.3 | 68/79 |
| AtUXS2-Arabidopsis | AT3g62830 | 3, ∼100 cM | 49.9 | 100 |
| AtUXS3-Arabidopsis | AT5g59290 | 5, ∼125 cM | 38.5 | 72/83 |
| AtUXS4-Arabidopsis | AT2g47650 | 2, ∼100 cM | 49.9 | 88/92 |
| AtUXS5-Arabidopsis | AT3g46440 | 3, ∼75 cM | 38.3 | 71/83 |
| Uxs-rat | *aak85410 | – | 47 | 63/76 |
| Uxs-C. neoformans | *aak59981 | – | 47.4 | 61/76 |
National Center for Biotechnology Information (NCBI) BLAST analyses of the Arabidopsis genomic database (The Arabidopsis Information Resource) were performed using the AtUXS cDNAs described in this study (AtUXS1, 2, and 3). AtUxs2 was used to compare, by BLAST analysis, the amino acid sequence similarity between the various Uxs. Note: The sequence identity/similarity between AtUxs3 and AtUxs5 is 92/96. The activity of Uxs in Cryptococcus neoformans (Bar-Peled et al., 2001) and in rat (Rattus norvegicus; Moriarity et al., 2002) was confirmed. The activity of AtUxs1, 2, and 3 is confirmed in this publication.
An asterisk indicates NCBI database.
Figure 2.
Amino acid sequence comparison of AtUxs1 to AtUxs2 and AtUxs3. A, ClustalX alignment of the conserved amino acid sequences of AtUxs1, 2, and 3. The N terminus alignment between AtUxs1, 2, and 3 is not shown because of low amino acid sequence identity. Alignment of the AtUxs1 and 2 starts at amino acids 116 and 117, respectively; alignment of AtUxs1 and 3 starts at amino acids 120 and 30, respectively. Identical amino acid residues are indicated by an asterisk, and similar amino acid residues are indicated by a colon. The GxxGxxG and the Ser and YxxxK motifs are underlined. The dashed line demonstrates the unconserved C-terminal region. B, Comparison of the hydropathy plots of AtUxs 1, 2, and 3. Note the variable N-terminal region of AtUxs1 and 2 that spans from amino acid 1 to 48 and 1 to 43, respectively, followed by an approximately 16-amino acid hydrophobic region encoding the putative transmembrane domain (tm), which is followed by a variable “stem domain” in AtUxs1 and 2 that spans from amino acid 66 to 115 and 61 to 116.
AtUxs Gene Isoforms Encode Active UDP-GlA Decarboxylase
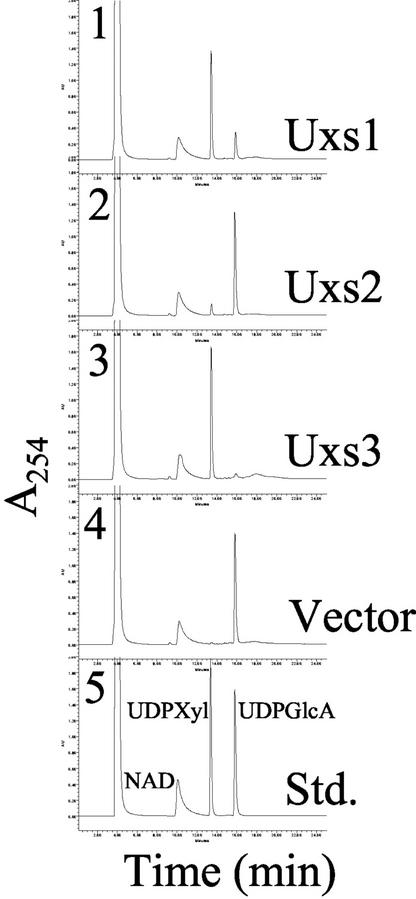
Protein derived from Escherichia coli expressing each gene construct was assayed for the ability to convert UDP-GlcA to UDP-Xyl. Because both proteins AtUxs1 and AtUxs2 have a variable N-terminal extension region each consisting of a putative membrane domain, we express these recombinant proteins in E. coli with or without that hydrophobic domain. The 338-amino acid-long truncated version of AtUxs1 (i.e. AtUxs1Δ1–88; Fig. 3, panel 1) and the full-length soluble AtUxs3 (Fig. 3, panel 3) readily converted substantial amounts of UDP-GlcA into a product that co-eluted like the UDP-Xyl standard on an HPLC column. The 352-amino acid truncated version of AtUxs2 (AtUxs2Δ1–95) converted lower amounts of UDP-GlcA into UDP-Xyl (Fig. 3, panel 2) when compared with AtUxs1 and AtUxs3. For unknown reasons, the expression of AtUxs2 in E. coli resulted in a less active enzyme. E. coli expressing the vector control (Fig. 3, panel 4), full-length AtUxs1 protein, or full-length AtUxs2 protein had no detectable UDP-GlcA-DC activity. The products that comigrated on HPLC-like UDP-Xyl (Fig. 3, panels 1 and 3) were collected and analyzed by 1H-NMR spectroscopy (Table II). The AtUxs1Δ1–88 and AtUxs3 enzymatic products and authentic UDP-Xyl gave comparable one-dimensional proton spectra. The signal corresponding to each proton was assigned using two-dimensional proton-proton correlated spectroscopy and confirmed that the product produced by AtUxs1 and AtUxs3 was UDP-Xyl. The small 3J1H1,H2 coupling constant of 3.5 Hz established that the xylosyl residue had the α-anomeric configuration as expected for UDP-Xyl. Taken together, these data establish that the expressed Arabidopsis genes (AtUXS1 and AtUXS3) encode UDP-GlcA decarboxylases. AtUxs2 gave a product that also comigrated with UDP-Xyl (Fig. 3, panel 2). However, the product was not formed in amounts sufficient for conclusive identification by 1H-NMR spectroscopy. Nevertheless, when taken together, our data suggest that all three genes encode UDP-GlcA-DC; thus, the genes have been named AtUXS1, 2, and 3 (UDP-Xyl Synthase).
Figure 3.
AtUxs 1, 2, and 3 are UDP-GlcA-decarboxylase isozymes. Total soluble protein (10 μg) derived from E. coli expressing, separately: AtUXS1(Δ1–88), 1, Uxs1; AtUXS2(Δ1–95), 2, Uxs2; AtUXS3, 3, Uxs3; or control vector, 4, vector, was incubated for 60 min with 1 mm UDP-GlcA and 1 mm NAD+ for 60 min. The products of the reactions were separated over a Hypersil strong anion exchange (SAX)-HPLC column. 5, Elution positions of standard nucleotides and nucleotide sugars. The chromatography peak that migrated with the same retention time as authentic UDP-Xyl was collected and analyzed by 1H-NMR spectroscopy. Similar results were obtained using total soluble proteins or desalted protein fraction.
Table II.
NMR analysis of the enzymatic reaction products produced by recombinant AtUxs1 and AtUxs3
| Moeity | H1 | H2 | H3 | H4 | H5, 5′ | H6 |
|---|---|---|---|---|---|---|
| Xyl | 5.517 | 3.483 | 3.679 | 3.592 | 3.72 | – |
| 3J1,23.5 | 3J2,38.9 | 3J3,48.9 | – | – | – | |
| 3J1,P7.0 | 3J2,P3.5 | – | – | – | – | |
| Rib | 5.938 | 4.34 | 4.255 | 4.34 | 4.15–4.22 | – |
| Uridine | – | – | – | – | 5.960 | 7.0 |
| – | – | – | – | 5.956 | 7.927 |
UDP-GlcA-DC assays were performed for 60 min with 1 mm UDP-GlcA and the total soluble protein extract derived from E. coli cells expressing recombinant AtUxs1(Δ1–88), AtUxs3, or empty vector control. The reaction products were separated over a Hypersil SAX-HPLC column (Phenomenex, Torrance, CA) and chromatography peaks that migrated with the same retention time as authentic UDP-Xyl were collected and analyzed by 1H-NMR spectroscopy. The chemical shifts (in ppm) of UDP-Xyl enzymatically formed by AtUxs1(Δ1–88), UDP-Xyl enzymatically formed by AtUxs3, and UDP-Xyl standard (std) that was separated on HPLC were obtained in D2O at 25°C and referenced to internal acetone at δ2.218 relative to 3-(trimethylsilyl)-1-propanesulfonic acid. Scalar coupling constants (J, in Hz) for some internal residues are also indicated. Identical chemical shift results were obtained with the standard UDP-Xyl dissolved in D2O. The exact UDP-Xyl chemical shifts were also obtained when HPLC peaks were collected after enzyme assays were carried on with AtUxs3 with or without NAD+.
Biochemical Characterization of AtUxs
Biochemical analyses were performed to characterize the catalytic activity of the Arabidopsis UDP-GlcA-DC expressed in E. coli. This study was restricted to purified AtUxs3 (Fig. 4). AtUxs3 does not require an exogenous supply of metal for activity, nor does EDTA inhibit UDP-Xyl synthesis (Table III). However, the AtUxs activity was affected, to different extents, by the addition of cations. For example, calcium and manganese ions inhibited AtUxs3 by more than 30%. The presence of 5 mm DTT was required during AtUxs3 protein isolation to maintain enzyme activity, and only at high concentrations of DTT (50 mm) was some reduction in enzymatic activity observed. We found the enzyme to be very stable in crude extracts; when stored at −20°C, only a 5% reduction in activity was observed after 1 year. AtUxs (1, 2, or 3) are very specific for UDP-GlcA and none of the nucleotide sugars tested (UDP-Glc, UDP-Gal, UDP-Man, GDP-Glc, or GDP-Man) were used as substrates by the AtUxs (data not shown). The AtUxs also did not convert UDP-GalA (the 4-epimer of UDP-GlcA) into a product, even at high concentrations, suggesting that Uxs display stereospecific recognition of the hydroxyl and hydrogen attached to C4 of GlcA. Further inhibitor studies were carried out with various nucleotides, nucleotide sugars, and co-enzymes (Table IV). UTP, TTP, and TDP strongly inhibit AtUxs3 activity, whereas UMP and TMP had negligible effects on AtUxs3 activity. The activity of partially purified wheat germ UDP-GlcA-DC was reduced by 80% in the presence of 0.3 mm UDP-Xyl (John et al., 1977a), but the Arabidopsis AtUxs3 activity was reduced by only approximately 35% at 2 mm UDP-Xyl (Table IV), suggesting differences between the wheat enzyme and the bacterially expressed Arabidopsis enzymes. Nevertheless, the ability of UDP-Xyl to inhibit UDP-GlcA-DC suggests that end product inhibition is a mechanism whereby the activity of AtUxs could be regulated in vivo.
Figure 4.
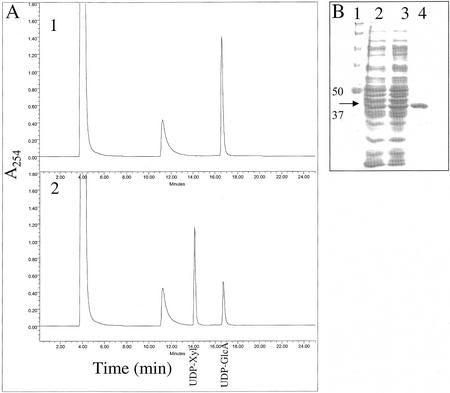
Purification of active UDP-GlcA-DC (AtUxs3). A, Purified AtUxs3 was incubated with 1 mm UDP-GlcA and 1 mm NAD+ for 0 (1) or 15 (2) min. The products of the reactions were separated over a Phenomenex SAX-HPLC column. The retention times of std UDP-Xyl and UDP-GlcA are indicated. B, SDS-PAGE of AtUxs3 during purification. Total soluble E. coli protein expressing AtUXS3 (lane 2) was desalted (lane 3) and purified over an Ni column (lane 4). Lane 1, Marker proteins with the indicated molecular masses. The molecular mass of recombinant AtUxs3 (42.6 kD; arrow) is larger than the native AtUxs3 (38.5 kD) because of the N-terminal His-6 amino acid tag fusion that facilitates purification.
Table III.
Effect of additives on AtUxs3 activity
| Additive and Concentration in Assay | Relative Uxs Activitya |
|---|---|
| % | |
| Water | 100 |
| 5 mm EDTA | 97 |
| 10 mm MgCl2 | 88 |
| 10 mm MnCl2 | 65 |
| 10 mm CaCl2 | 70 |
| 10 mm MgSO4 | 87 |
| 50 mm NH4Cl | 89 |
| 50 mm CHO2NH3 | 93 |
| 50 mm NaCl | 89 |
| 50 mm Dithiothreitol (DTT) | 86 |
| 5% (w/v) Suc | 101 |
| 5% (v/v) Glycerol | 108 |
Protein was separately mixed with each additive for 30 min on ice. 1 mm UDP-GlcA was added and the reactions were incubated for 20 min at 30oC.
The data are the average relative amount of UDP-Xyl produced compared with the control (water) from two experiments and the values varied by no more then 10%. 100% corresponds to 42 nmol UDP-Xyl produced.
Table IV.
Effect of nucleotide and nucleotide sugars on AtUxs3 activity
| Additive and Concentration in the Assay | Relative Uxs Activitya |
|---|---|
| % | |
| Water | 100 |
| 2 mm UDP-Glc | 93 |
| 2 mm UDP-Gal | 94 |
| 2 mm GDP-Man | 101 |
| 2 mm GDP-Glc | 102 |
| 2 mm UDP-Xyl | 65 |
| 2 mm NAD+ | 110 |
| 2 mm NADH | 104 |
| 2 mm NADPH | 100 |
| 2 mm NADP+ | 100 |
| 1 mm UMP | 85 |
| 1 mm UDP | 17 |
| 1 mm UTP | 14 |
| 1 mm TMP | 93 |
| 1 mm TDP | 26 |
| 1 mm TTP | 33 |
| 2 mm GMP | 94 |
| 2 mm GDP | 92 |
| 2 mm GTP | 89 |
| 2 mm CMP | 98 |
| 2 mm CDP | 95 |
Protein was separately mixed with each additive for 30 min on ice. One millimolar UDP-GlcA was added and the reactions were incubated for 20 min at 30oC. Data are the average relative amounts of UDP-Xyl produced compared with the control (no additives). One hundred percent corresponds to 40 nmol UDP-Xyl produced.
Each value is the mean of duplicate reactions and the values varied by no more than 10%.
None of the AtUxs studied (1–3) require exogenous NAD+ to convert UDP-GlcA to UDP-Xyl, suggesting that NAD+ is strongly associated with AtUxs as has been observed with other plant isoforms (Feingold et al., 1960; John et al., 1977a; Feingold, 1982; Hayashi et al., 1988). Inhibition studies were carried out with various co-enzymes (Table IV) in an effort to compete with the enzyme-associated-NAD+. NADH, NADP+, or NADPH, at 2 mm concentration, did not inhibit Arabidopsis AtUxs3 activity. However, NADH completely inactivates the Uxs derived from C. neoformans (Bar-Peled et al., 2001). Taken together, these results suggest that the Arabidopsis Uxs proteins contain tightly bound NAD+.
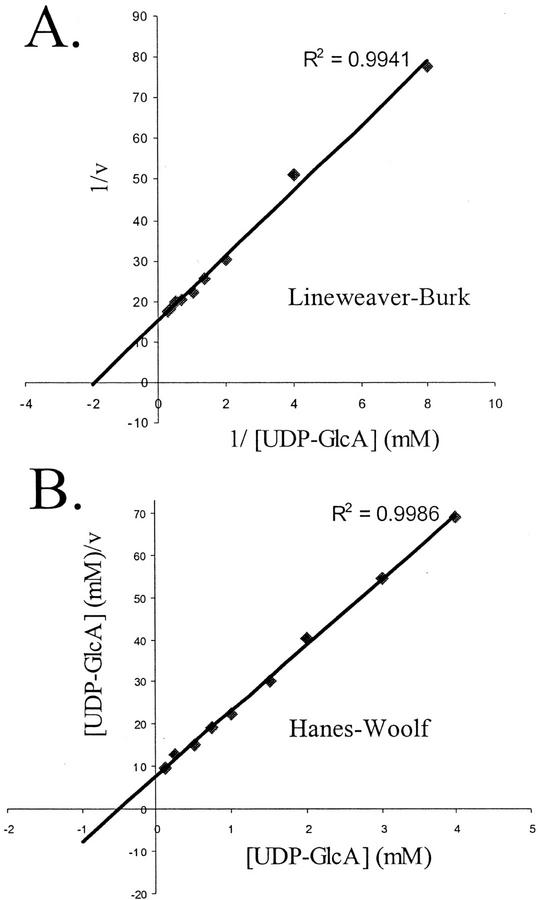
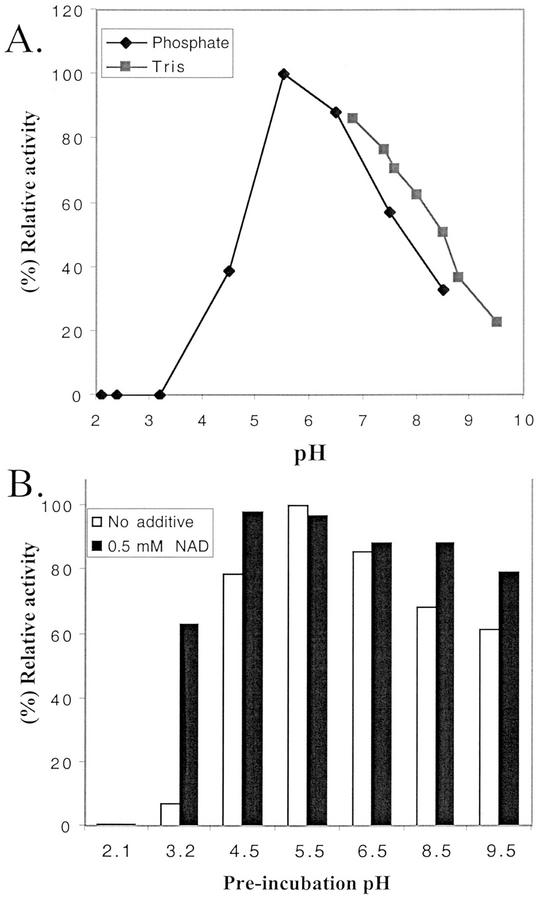
Initial kinetic studies indicate that the amount of UDP-Xyl formed from UDP-GlcA by AtUxs is linear for up to 20 min. Further kinetic studies performed at 15 min indicated that AtUxs3 has an apparent Km of 0.51 mm based on Lineweaver-Burk and Hanes-Woolf plots (Fig. 5, A and B) at a UDP-GlcA concentration ranging between 0.125 and 4 mm. Partially purified AtUxs1 has an apparent Km of 0.19 mm (data not shown). Similar Km values (0.53 and 0.18 mm) were obtained for the two isozymes partially purified from wheat germ (John et al., 1977a). The Golgi membrane-associated UDP-GlcA-DC from soybean (Glycine max) has an apparent Km of 0.24 mm, whereas the soluble isozymes have a higher Km value of 0.7 mm (Hayashi et al., 1988), suggesting that the membrane-bound isozymes have a higher affinity for UDP-GlcA. AtUxs3 had a broad pH range and activity was observed between pH 4.5 and 9.5, with maximum activity at pH 5.5 (Fig. 6A). Approximately 30% of AtUxs3 activity was obtained at pH 9.5 and complete inactivation occurred at pH values below 3.2 (Fig. 6A). Interestingly, pre-incubation of the enzyme with NAD+ at pH 3.2 for a period of 30 min retained 60% of the UDP-GlcA DC activity (Fig. 6B) when compared with controls. The enzyme also demonstrated activity over a broad temperature range. AtUxs3 was active between 22°C and 42°C, with maximum activity at 30°C (data not shown). No activity was observed at temperatures higher than 55°C.
Figure 5.
Kinetic studies of AtUxs3. UDP-GlcA-DC assays were performed with increasing amounts of UDP-GlcA (0.125–4 mm). Duplicate assays were conducted for 15 min at 30°C. Lineweaver-Burk and Hanes-Woolf plots were obtained with an R2 > 0.96.
Figure 6.
Effect of pH on the activity of AtUxs3. A, Assays were carried out for 20 min at 30°C with 1 mm UDP-GlcA in 0.1 m sodium-phosphate or Tris-HCl at the indicated pH values. B, Protein was pre-incubated with or without 0.5 mm NAD+ at the indicated pHs for 30 min on ice. The pH was then adjusted to neutrality, and 1 mm UDP-GlcA and 1 mm NAD+ were added. Duplicate reactions were incubated for 20 min at 30°C. The data are the mean and varied by no more than 5%. The relative amount of UDP-Xyl produced is plotted; 100% equals 41 nmol of UDP-Xyl produced.
Expression of AtUxs1, 2, and 3 in Arabidopsis
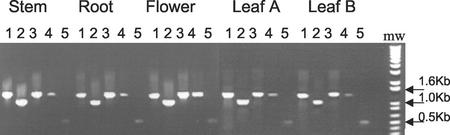
The expression of gene specific AtUxs transcripts was determined by RT-PCR. Transcripts of AtUXS1, 2, and 3 were observed in all tissue examined (Fig. 7). Transcript encoding the putative membrane-bound AtUxs2 (class B) was expressed at similar levels as the transcript encoding the soluble AtUxs3 isoforms (Fig. 7, lanes 2 and 3). Transcript of the gene encoding class A putative membrane-bound isoform AtUxs1 (Fig. 7, lane 4) was lower in most tissues compared with class B (AtUXS2) and AtUXS3. However, it appears that in flowers, the AtUXS1 expression level is higher compared with the other tissues.
Figure 7.
Expression of AtUXS1, 2, and 3 in Arabidopsis. Total RNA isolated from flowers, stems, roots, and rosette leaves of fully mature plants (Leaf A, 6-week-old plants), or rosette leaves of 3-week-old plants (Leaf B) was used to amplify by RT-PCR AtUXS-specific cDNA. The following gene-specific transcripts were amplified by RT-PCR: 4, 1,317-bp AtUXS1; 3, 1,341-bp AtUXS2; and 2, 1,046-bp AtUXS3. As internal RT-PCR controls, the amplification of the CAB79762 gene whose cDNA sequence is expressed in all EST databases examined resulted in a predicted 1,302-bp DNA fragment (5), and a portion of the actin gene was RT-PCR amplified using degenerated primers (McKinney et al., 1995) to yield a 495-bp fragment (1). The data are representative of at least three independent RT-PCR reactions.
DISCUSSION
The UXS gene family members we have cloned are not unique to Arabidopsis. Members of this gene family are found (based on EST databases) in monocots and dicots and appear to be expressed throughout all stages of plant growth and development. The UXS gene family is not restricted to higher plants because it was also identified in a cDNA library from a green alga, Chlamydomonas reinhardtii. In addition, UXS homologs were identified in human (Homo sapiens), rat (Rattus norvegicus), Drosophila melanogaster, and bacteria genomic databases, indicating that Uxs proteins are evolutionarily conserved. Two more UXS-like sequences (AtUXS4 and AtUXS5) were identified in the Arabidopsis genome (Table I) and cDNA library, but they were not analyzed because they each share high amino acid sequence identity with AtUxs2 and AtUxs3, respectively (AtUXS4, accession no. at2g47650, which shares 92% protein amino acid sequence similarity to AtUxs2; and AtUXS5, accession no. at3g46440, which shares 95% amino acid similarity to AtUxs3). AtUXS1, 2, and 5 are located on chromosome III of Arabidopsis, AtUXS 3 is on chromosome V, and AtUXS4 is located on chromosome II.
The first step in the conversion of UDP-GlcA to UDP-Xyl, the oxidation to UDP-4-keto-GlcA, is thought to require NAD+ (Feingold, 1982). Studies of UDP-GlcA decarboxylase activity from Cryptococcus laurentii indicated an absolute requirement for exogenous NAD+ (Ankel and Feingold, 1966). However, the isozymes from plants do not require an exogenous supply of NAD+ (Ankel and Feingold, 1965; John et al., 1977a; Feingold, 1982; Hayashi et al., 1988) and this study indicates that the Arabidopsis AtUxs3, as well as AtUxs1 and AtUxs2, do not require an exogenous NAD+. Thus, our data are consistent with the notion that NAD+ is tightly bound to the enzymes (Ankel and Feingold, 1965; John et al., 1977). Similar to the enzyme from wheat germ, NADH has no discernible affect on Arabidopsis Uxs3 (Table IV), Uxs1, or Uxs2 (data not shown). However, it should be noticed that the Uxs from C. neoformans is completely inactivated by NADH (Bar-Peled et al., 2001). These dissimilarities between the effect of NADH on the plant AtUxs and the C. neoformans enzymes may result from differences in their affinities for the co-enzyme. This is made more likely by the demonstration that UDP-4-Glc epimerase from human and E. coli have different affinities for NAD+ (Thoden et al., 2000). The bacterial UDP-4-Glc epimerase was shown, by x-ray crystallography, to form 19 hydrogen bonds with NAD+, whereas only 11 bonds are proposed to occur between NAD+ and the human UDP-4-Glc epimerase (Thoden et al., 2000).
Previous literature on UDP-GlcA decarboxylase activities has indicated that the subcellular localization of the enzymes from different sources varies. Although some UDP-GlcA-DC isoforms are cytosolic (i.e. soluble; Feingold et al., 1960; Ankel and Feingold, 1965; John et al., 1977a; Hayashi et al., 1988), other isoforms are membrane bound (Feingold, 1982; Hayashi et al., 1988). The Arabidopsis UDP-GlcA-DC AtUxs3 and AtUxs5 appear to be soluble, whereas the AtUxs1, AtUxs2, and AtUxs4 proteins have hydrophobic N-terminal extensions that suggest a type II transmembrane protein topology. In plants, Hayashi et al. (1988) provided evidence for the localization of a UDP-Glc-DC activity in Golgi membranes. Thus, it is likely that the Arabidopsis AtUxs1, AtUxs2, and AtUxs4 may function in the Golgi apparatus. In animal systems, UDP-GlcA was found to be transported into Golgi vesicles using the SQV7 gene product (Berninsone et al., 2001), and UDP-GlcA-DC activity was also found to face the Golgi lumen (Kearns et al., 1993). That the orthologs of the Arabidopsis UXS gene found in human, rat, and D. melanogaster databases appear to encode membrane proteins also argues for Golgi localization of some of the AtUxs. The fact that the three (AtUXS 1–3) genes were expressed in all tissue examined (Fig. 7) prompts us to speculate that the cellular localization of each AtUxs isozyme may indicate a specific metabolic function for each unique UDP-GlcA-DC. Further research aimed at localizing each AtUxs and understanding the functional role of each isozyme is under way.
MATERIALS AND METHODS
Cloning and RT-PCR Analysis of Arabidopsis UDP-GlcA-DC Genes
Arabidopsis genomic and expressed tag cDNA databases were BLAST searched using the amino acid sequence of a UDP-GlcA-DC gene (USX, GenBank accession no. AF385328) from Cryptococcus neoformans (Bar-Peled et al., 2001). Several ESTs and genomic sequences were identified and used to design primers to obtain the corresponding Arabidopsis genes by RT-PCR. In brief, total RNA was isolated (Chomczynski, 1993) from flowers, rosette leaves, or stems obtained from 3- or 6-week-old Arabidopsis ecotype Columbia plants. RNA was also isolated from roots obtained from 4-week-old Arabidopsis plants grown in liquid media as described by Bar-Peled and Raikhel (1997). RNA from each tissue was reverse transcribed using 1 mm oligo(dT) primer and 200 units of SuperScript II-reverse transcriptase (Life Technologies/Gibco-BRL, Cleveland). After incubation with 2 units of RNase H (Life Technologies/Gibco-BRL), the resulting reverse transcriptase products were used as a template for PCR using high-fidelity Platinum Taq DNA polymerase (Life Technologies/Gibco-BRL) and gene-specific primers. For amplification of AtUXS1 (clone no. 42), the sense primer 40-1S 5′ agt-act-atg-aag-cag-ctt-cac-aag-caa-atg-agc-tc and the antisense primer 40-2AS 5′ gcg-gcc-gct-tag-aga-cct-tta-cct-tcg-tct-tcg were used. For amplification of AtUXS2 (clone no. 71), the sense primer 70-1#S 5′ cat-atg-gcg-agc-gag-ctg-atc-aat-cgg-cg and the antisense primer 70-2AS 5′ gcg-gcc-gct-caa-gct-gaa-gtt-gtc-ttg-gtg-gtg-g were used. For amplification of AtUXS3 (clone no. 101), the primer set used was 101-1S sense primer 5′ aga-att-ccc-atg-gca-gct-aca-agt-gag-aaa-cag, and the antisense primer used was 101-2AS 5′ gcg-gcc-gct-tag-ttt-ctt-ggg-acg-tta-agc-ctt-ag. RT-PCR reaction products were cloned into pCR2.1-TOPO plasmid (Invitrogen, Carlsbad, CA), sequenced, and the nucleotide sequence was submitted to GenBank (accession no. AF387787, AtUxs1; accession no. AF387788, AtUxs2; and accession no. AF387789, AtUxs3). The DNA coding region of each gene was further subcloned from plasmid pCR2.1 into a pET Escherichia coli expression vector (Novagen, Madison, WI), resulting in clones pMBP (nos. 42, 71, and 101). Truncated versions of Uxs1 and Uxs2, that lack the transmembrane domain, were generated as follows. Clone 43 (AtUXS1Δ1–88 amino acids) and clone 71b (AtUXS2Δ1–71 amino acids) were generated by digesting clone 42 with SalI-XhoI, or clone 71 with BamHI-NotI, respectively. Each DNA was inserted in-frame into the pET vector (Novagen) digested with the same enzymes.
The protein sequence for UDP-GlcA-DC from Arabidopsis has been deposited in the GenBank database (GenBank accession nos.: AtUxs1, AAK70880; AtUxs2, AAK70881; and AtUxs3, AAK70882).
For AtUXS expression studies in Arabidopsis (Columbia), samples of total RNA (3 μg; from flowers of 6-week-old plants, fully expanded rosette leaves of 6-week-old plants, stems of 6-week-old plants, roots of 4-week-old plants, or rosette leaves of 3-week-old plants) were reverse transcribed into cDNA in 20-μL reactions with 200 units of SuperScript II reverse transcriptase (Invitrogen) and primed with 1 mm oligo(dT) in the manufacturer's recommendation buffer. One-twentieth of each of the reverse-transcribed products was used as template for PCR reactions using 0.5 units of Taq DNA polymerase (Boehringer Mannheim/Roche, Basel), manufacturer's buffer, 1.5 mm MgCl2, and 1 mm gene-specific sense and antisense primers: AtUxs1, 40-1S 5′ agt-act-atg-aag-cag-ctt-cac-aag-caa-atg-agc-tc and 40-2AS 5′ gcg-gcc-gct-tag-aga-cct-tta-cct-tcg-tct-tcg; AtUxs2, 70-1#S 5′ cat-atg-gcg-agc-gag-ctg-atc-aat-cgg-cg and 70-2AS 5′ gcg-gcc-gct-caa-gct-gaa-gtt-gtc-ttg-gtg-gtg-g; and AtUxs3, 101-1S 5′ aga-att-ccc-atg-gca-gct-aca-agt-gag-aaa-cag and 101-2AS 5′ gcg-gcc-gct-tag-ttt-ctt-ggg-acg-tta-agc-ctt-ag. As internal RT-PCR controls, we used actin primers ACT119S and ACT284A (McKinney et al., 1995; a gift from R. Meagher, University of Georgia, Athens), and primers encoding highly expressed gene product cab79762 based on EST databases (183-cab79762 BspHI/S 1 5′ATC-ATG-atg-cct-tca-ata-gaa-gat-gag-ctg-ttt-c and 183-cab79762 BamHI/AS2 5′ GGA-TCC-tta-atg-tac-aag-ctt-ggc-ttt-agt-att-g). One-fifth of each sample and DNA Mr marker were resolved on a 1% (w/v) agarose gel and stained with ethidium bromide. All RNA samples were determined to be free of genomic DNA contamination.
Protein Expression, Purification, and Enzyme Assays
E. coli strain BL21(DE3) pLysS (Novagen), carrying the various pMBP vectors or control pET vector alone, was induced with 1 mm isopropyl-β-d-thiogalactoside for approximately 3 h at approximately 25°C. Cells were collected, washed with cold water, resuspended in lysis buffer (50 mm Tris-HCl [pH 8] containing 20% [v/v] glycerol, 1 mm EDTA, 5 mm DTT, and 0.5 mm phenylmethylsulfonyl fluoride), and ruptured in a French Press. The suspension was centrifuged (20,000g for approximately 30 min, 4°C), and the supernatant fraction was collected, fractionated on Sephadex G-25 column as described by Bar-Peled et al. (1991), and stored at −20°C. AtUxs3 was furthered purified over a nickel column as described by Bar-Peled et al. (2001). The standard 50-μL UDP-GlcA-DC (Uxs) assays contained 0.1 m Tris HCl (pH 7.4; or 0.1 m sodium-phosphate [pH 5.5]), 1 mm UDP-GlcA, and up to 10 μg of protein. Where indicated, 1 mm NAD+ was added to the std assay. Assays were performed at 30°C for 20 min (unless otherwise specified) and stopped by the addition of 50 μL of phenol:chloroform (1:1 [v/v]). The mixture was vortexed and centrifuged at 16,000g for 5 min at room temperature. The aqueous phase was retained, and the lower phase re-extracted with 80 μL of water. The two aqueous phases were combined and analyzed by HPLC using SAX-Hypersil or SAX-Phenosphere ion-exchange columns (250 × 4.6 mm). Nucleotide sugars were separated at 1.5 mL min−1 using a potassium-phosphate gradient (Bar-Peled et al., 1991) or at 1 mL min−1 using an ammonium-formate gradient (2–600 mm in 25 min; Bar-Peled et al., 2001). Nucleotides and nucleotide sugars were detected by UV A254 (PDA, Waters, Milford, MA), and the elution times of the assay products were compared with authentic nucleotide sugar standards (Sigma, St. Louis).
1H-NMR Spectroscopic Analysis of the Products Formed by AtUxs1, 2, and 3
UV-absorbing peaks eluting from the SAX column in ammonium formate were collected. Each sample was lyophilized, dissolved in water, relyophilized twice, and exchanged twice with 99.96% D2O. Proton NMR spectroscopy was performed at 25°C on an Inova spectrometer (Varian, Palo Alto, CA) operating at 500 and 600 MHz (Bar-Peled et al., 2001).
ACKNOWLEDGMENTS
The authors thank Dr. J. Glushka (Complex Carbohydrate Research Center, Athens, GA) for performing NMR spectroscopic analyses, Dr. R. Meagher (University of Georgia) for discussions and the generous gift of the actin primers, and Drs. D. Mohnen and M. O'Neill (University of Georgia) for constructive comments on the manuscript.
Footnotes
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.009654.
LITERATURE CITED
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amino SI, Takeuchi Y, Kamamine A. Changes in enzyme activities involved in the formation and interconversion of UDP-sugars during cell cycle in a synchronous culture of Catharanthus roseus. Physiol Plant. 1985;64:111–117. [Google Scholar]
- Ankel H, Ankel E, Feingold DS, Schutzbach JS. Formation of UDP-d-xylose in algae. Biochim Biophys Acta. 1967;136:172–175. doi: 10.1016/0304-4165(67)90339-x. [DOI] [PubMed] [Google Scholar]
- Ankel H, Feingold DS. Biosynthesis of uridine diphosphate d-xylose: I. Uridine diphosphate glucuronate carboxy-lyase of wheat germ. Biochemistry. 1965;4:2468–2475. doi: 10.1021/bi00865a024. [DOI] [PubMed] [Google Scholar]
- Ankel H, Feingold DS. Biosynthesis of uridine diphosphate d-xylose: II. Uridine diphosphate d-glucuronate carboxy-lyase of Cryptococcus laurentii. Biochemistry. 1966;5:182–189. doi: 10.1021/bi00865a024. [DOI] [PubMed] [Google Scholar]
- Baker ME, Blasco R. Expansion of the mammalian 3 beta-hydroxysteroid dehydrogenase/plant dihydroflavonol reductase superfamily to include a bacterial cholesterol dehydrogenase, a bacterial UDP-galactose-4-epimerase, and open reading frames in vaccinia virus and fish lymphocystis disease virus. FEBS Lett. 1992;301:89–93. doi: 10.1016/0014-5793(92)80216-4. [DOI] [PubMed] [Google Scholar]
- Bar-Peled M, Griffith CL, Doering TL. Functional cloning and characterization of a UDP-glucuronic acid decarboxylase: The pathogenic fungus Cryptococcus neoformanselucidates UDP-xylose synthesis. Proc Natl Acad Sci USA. 2001;98:12003–12008. doi: 10.1073/pnas.211229198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled M, Lewinsohn E, Fluhr R, Gressel J. UDP-rhamnose:flavanone-7-O-glucoside-2-O-rhamnosyltransferase, purification and characterization of an enzyme catalyzing the production of bitter compounds in citrus. J Biol Chem. 1991;266:20953–20959. [PubMed] [Google Scholar]
- Bar-Peled M, Raikhel NV. Characterization of AtSEC12 and AtSAR1. Proteins likely involved in endoplasmic reticulum and Golgi transport. Plant Physiol. 1997;114:315–324. doi: 10.1104/pp.114.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baydoun E, Brett CT. Distribution of xylosyltransferases and glucuronosyltransferases within the Golgi apparatus in etiolated pea (Pisum sativumL.) epicotyls. J Exp Bot. 1997;311:1209–1214. [Google Scholar]
- Baydoun EAH, Abdel-Massih RM, Dani D, Rizk SE, Brett CT. Galactosyl- and fucosyltransferases in etiolated pea cipcotyls: product identification and sub-cellular localization. J Plant Physiol. 2001;158:145–150. [Google Scholar]
- Berninsone P, Hwang HY, Zemtseva I, Horvitz HR, Hirschberg CB. SQV-7, a protein involved in Caenorhabditis elegansepithelial invagination and early embryogenesis, transports UDP-glucuronic acid, UDP-N-acetylgalactosamine, and UDP-galactose. Proc Natl Acad Sci USA. 2001;98:3738–3743. doi: 10.1073/pnas.061593098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolwell PG. Biosynthesis of plant cell wall polysaccharides. Trends Glycosci Glycotechnol. 2000;12:143–160. [Google Scholar]
- Chomczynski P. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques. 1993;15:536–537. [PubMed] [Google Scholar]
- Dalessandro G, Northcote DH. Possible control sites of polysaccharides synthesis during cell growth and wall expansion of pea seedlings (Pisum sativumL.) Planta. 1977a;134:39–44. doi: 10.1007/BF00390092. [DOI] [PubMed] [Google Scholar]
- Dalessandro G, Northcote DH. Changes in enzymic activities of nucleoside diphosphate sugar interconversions during differentiation of cambium to xylem in sycamore and poplar. Biochem J. 1977b;162:267–279. doi: 10.1042/bj1620267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feingold DS. Aldo (and keto) hexoses and uronic acids. In: Loewus FA, Tanner W, editors. Plant Carbohydrates I, Intracellular Carbohydrates (Encyclopedia of Plant Physiology New Series) 13A. Heidelberg: Springer-Verlag; 1982. pp. 3–76. [Google Scholar]
- Feingold DS, Neufeld EF, Hassid WZ. The 4-epimerization and decarboxylation of uridine diphosphate d-glucuronic acid by extracts from Phaseolus aureusseedlings. J Biol Chem. 1960;235:910–913. [PubMed] [Google Scholar]
- Gibeaut DM. Nucleotide sugars and glycosyltransferases for synthesis of cell wall matrix polysaccharides. Plant Physiol Biochem. 2000;38:69–80. [Google Scholar]
- Gotting C, Kuhn J, Zhan R, Brinkmann T, Kleesiek K. Molecular cloning and expression of human UDP-d-xylose:proteoglycan core protein β-d-xylosyltransferase and its first isoform XT-II. J Mol Biol. 2000;304:517–528. doi: 10.1006/jmbi.2000.4261. [DOI] [PubMed] [Google Scholar]
- Hartmann E, Rapoport TA, Lodish HF. Predicting the orientation of eukaryotic membrane-spanning proteins. Proc Natl Acad Sci USA. 1989;86:5786–5790. doi: 10.1073/pnas.86.15.5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Koyama T, Matsuda K. Formation of UDP-xylose and xyloglucan in soybean Golgi membranes. Plant Physiol. 1988;87:341–345. doi: 10.1104/pp.87.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Matsuda K. Biosynthesis of xyloglucan in suspension-cultured soybean cells. Occurrence and some properties of xyloglucan 4-β-d-glucosyltransferase and 6-α-d-xylosyltransferase. J Biol Chem. 1981;256:11117–11122. [PubMed] [Google Scholar]
- John KV, Schutzbach JS, Ankel H. Separation and allosteric properties of two forms of UDP-glucuronate carboxy-lyase. J Biol Chem. 1977a;252:8013–8017. [PubMed] [Google Scholar]
- John KV, Schwartz NB, Ankel H. UDP-glucuronate carboxy-lyase in cultured chondrocytes. J Biol Chem. 1977b;252:6707–6710. [PubMed] [Google Scholar]
- Kearns AE, Vertel BM, Schwartz NB. Topography of glycosylation and UDP-xylose production. J Biol Chem. 1993;268:11097–11104. [PubMed] [Google Scholar]
- Kuhn J, Gotting C, Schnolzer M, Kempf T, Brinkman T, Kleesiek K. First isolation of human UDP-d-xylose: proteoglycan core protein β-d-xylosyltransferase secreted from cultured JAR choriocarcinoma cells. J Biol Chem. 2001;276:4940–4947. doi: 10.1074/jbc.M005111200. [DOI] [PubMed] [Google Scholar]
- Kushnir S, Babiychuk E, Kampfenkel K, Belles-Boix E, van Montagu M, Inzé D. Characterization of Arabidopsis thalianacDNAs that render yeasts tolerant toward the thiol-oxidizing drug diamide. Proc Natl Acad Sci USA. 1995;92:10580–10584. doi: 10.1073/pnas.92.23.10580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyossev ZN, Drake RR, Kyosseva SV, Elbein AD. Synthesis of a new photoaffinity probe, 5-azido-[32P]UDP xylose, by UDP glucuronate carboxylase from wheat germ. Eur J Biochem. 1995;228:109–112. [PubMed] [Google Scholar]
- Liu Y, Thoden JB, Kim J, Berger E, Gulick AM, Ruzicak FJ, Holden HM, Frey PA. Mechanistic roles of tyrosine 149 and serine 124 in UDP-galactose 4-epimerase from Escherichia coli. Biochemistry. 1997;36:10675–10684. doi: 10.1021/bi970430a. [DOI] [PubMed] [Google Scholar]
- Loewus FA, Loewus MW. Myo-inositol: biosynthesis and metabolism. In: Stumpf PK, Conn EE, editors. The Biochemistry of Plants. Vol. 3. New York: Academic Press; 1980. pp. 43–76. [Google Scholar]
- Martin RC, Mok MC, Mok DW. Protein processing and auxin response in transgenic tobacco harboring a putative cDNA of zeatin O-xylosyltransferase from Phaseolus vulgaris. Plant J. 1997;12:305–312. doi: 10.1046/j.1365-313x.1997.12020305.x. [DOI] [PubMed] [Google Scholar]
- McKinney EC, Ali N, Traut A, Feldmann KA, Belostotsky DA, McDowell JM, Meagher RB. Sequence-based identification of T-DNA insertion mutations in Arabidopsis: actin mutants act2-1 and act4-1. Plant J. 1995;8:613–622. doi: 10.1046/j.1365-313x.1995.8040613.x. [DOI] [PubMed] [Google Scholar]
- Moore PJ, Swords KM, Lynch MA, Staehelin LA. Spatial organization of the assembly pathways of glycoproteins and complex polysaccharides in the Golgi apparatus of plants. J Cell Biol. 1991;112:589–602. doi: 10.1083/jcb.112.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarity JL, Hurt KJ, Resnick AC, Storm PB, Laroy W, Schnaar RL, Snyder SH. Nucleotide, protein UDP-glucuronate decarboxylase, a key enzyme in proteoglycan synthesis: cloning, characterization, and localization. J Biol Chem. 2002;277:16968–16975. doi: 10.1074/jbc.M109316200. [DOI] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld EF, Feingold DS, Hassid WZ. Enzymatic conversion of uridine diphosphate d-glucuronic acid to uridine diphosphate galacturonic acid, uridine diphosphate xylose, and uridine diphosphate arabinose. J Am Chem Soc. 1958;80:4430. [Google Scholar]
- Ray P. Cooperative action of the β-glucan transferases and the UDP-xylose xylosyl transferases of Golgi membranes in the synthesis of xyloglucan-like polysaccharides. Biochim Biophys Acta. 1980;629:431–444. doi: 10.1016/0304-4165(80)90149-x. [DOI] [PubMed] [Google Scholar]
- Ray PM, Abdul-Baki AA. Regulation of cell wall synthesis in response to auxin. In: Wightman F, Setterfield G, editors. Biochemistry and Physiology of Plant Growth Substances. Ottawa: Runge Press; 1968. pp. 647–658. [Google Scholar]
- Robertson D, McCormack BA, Bolwell GP. Cell wall polysaccharide biosynthesis and related metabolism in elicitor-stressed cells of French bean (Phaseolus vulgarisL.) Biochem J. 1995;306:745–750. doi: 10.1042/bj3060745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers MW, Bolwell GP. Partial purification of Golgi-bound arabinosyltransferase and two isoforms of xylosyltransferase from French bean (Phaseolus vulgarisL.) Biochem J. 1992;288:817–822. doi: 10.1042/bj2880817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose A, Glassgen WE, Hopp W, Seitz HU. Purification and characterization of glycosyltransferases involved in anthocyanin biosynthesis in cell-suspension cultures of Daucus carotaL. Planta. 1996;198:397–403. doi: 10.1007/BF00620056. [DOI] [PubMed] [Google Scholar]
- Seitz B, Klos C, Wurm M, Tenhaken R. Matrix polysaccharide precursors in Arabidopsiscell walls are synthesized by alternate pathways with organ-specific expression patterns. Plant J. 2000;21:537–546. doi: 10.1046/j.1365-313x.2000.00696.x. [DOI] [PubMed] [Google Scholar]
- Stewart DC, Copeland L. Uridine 5′-diphosphate-glucose dehydrogenase from soybean nodules. Plant Physiol. 1998;116:349–355. [Google Scholar]
- Strasser R, Mucha J, Mach L, Altmann F, Wilson IB, Glossl J, Steinkellner H. Molecular cloning and functional expression of β1,2-xylosyltransferase cDNA from Arabidopsis thaliana. FEBS Lett. 2000;472:105–108. doi: 10.1016/s0014-5793(00)01443-5. [DOI] [PubMed] [Google Scholar]
- Tenhaken R, Thulke O. Cloning of an enzyme that synthesizes a key nucleotide-sugar precursor of hemicellulose biosynthesis from soybean: UDP-glucose dehydrogenase. Plant Physiol. 1996;112:1127–1134. doi: 10.1104/pp.112.3.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoden JB, Wohlers TM, Fridovich-Keil JL, Holden HM. Crystallographic evidence for Tyr 157 functioning as the active site base in human UDP-galactose 4-epimerase. Biochemistry. 2000;39:5691–5701. doi: 10.1021/bi000215l. [DOI] [PubMed] [Google Scholar]
- Weirenga RK, Terpstra P, Hol WGJ. Prediction of the occurrence of the ADP-binding beta alpha beta-fold in proteins, using an amino acid sequence fingerprint. J Mol Biol. 1986;187:101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]
- White AR, Xin Y, Pezeshk V. Xyloglucan glucosyltransferase in Golgi membranes from Pisum sativum(pea) Biochem J. 1993;264:231–238. doi: 10.1042/bj2940231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Bannon G, Thomas VH, Rice K, Drake R, Elbein A. Purification and specificity of β1,2-xylosyltransferase, an enzyme that contributes to the allergenicity of some plant proteins. J Biol Chem. 1997;272:31340–31347. doi: 10.1074/jbc.272.50.31340. [DOI] [PubMed] [Google Scholar]