Abstract
Four transcripts homologous to K+ transporters of the HAK/KT/KUP family have been characterized from the common ice plant (Mesembryanthemum crystallinum). We report tissue-specific expression of McHAK1 and McHAK4 transcripts abundant in roots, leaves, and stems. McHAK2 was predominantly present in stems and McHAK3 in root tissues. By in situ hybridizations, the McHAKs showed signals in the leaf vascular bundles, mesophyll, and epidermal cells as well as in epidermal bladder cells. In mature roots, transcripts were mainly localized to the vasculature, and in differentiated root tips, the strongest signals were obtained from the epidermis. Expression of McHAK1, McHAK2, and McHAK4 complemented a yeast mutant defective in low- and high-affinity K+ uptake. Growth of the yeast mutant was restored at low-millimolar K+ concentrations and was inhibited by Rb+ and Cs+ but was not affected by Na+. Transcript levels of McHAK1 and McHAK4 increased by K+ starvation and by salt stress of 400 mm NaCl in leaves and roots. Expression of McHAK2 and McHAK3 was stimulated in leaves and was transiently induced in roots in response to high salinity with prestress transcript levels restored in salt-adapted plants. We discuss possible roles for such transporters in ion homeostasis at high salinity.
Multiple mechanisms exist for potassium uptake and transport in higher plants. A biphasic process described by Epstein et al. (1963), requiring high- and low-affinity systems to function at different external K+ concentrations, has long been considered. According to this hypothesis, potassium transporters mediate high-affinity K+ uptake, whereas low-affinity uptake has been relegated to potassium channels (Maathuis and Sanders, 1997). However, new evidence indicates that at least one channel, AKT1, can function in a high-affinity capacity (Spalding et al., 1999). HKT-type transporters were initially considered to mediate high-affinity K+ transport (Schachtman and Schroeder, 1994) and were later shown to function as Na+-K+ symporters in a variety of plant species (e.g. Rubio et al., 1995; Gassmann et al., 1996; Box and Schachtman, 2000; Uozumi et al., 2000; Rus et al., 2001). The Arabidopsis homolog AtHKT1 selectively mediated Na+ but not K+ uptake in Xenopus laevis oocytes, suggesting that HKT-type proteins may not have a significant physiological role for K+ uptake in plant roots (Uozumi et al., 2000). Studies on HKT mutant show that HKT could control sodium uptake in Arabidopsis (Rus et al., 2001). A similar cation selectivity preference for Na+ exists for a rice (Oryza sativa) HKT-type transporters (Horie et al., 2001; D. Golldack, H. Su, F. Quigley, U.R. Kamasani, C. Muñoz-Garay, E. Balders, O.V. Popova, J. Bennett, H.J. Bohnert, and O. Pantoja, unpublished data).
Different candidates for high-affinity K+ uptake became known when another family of K+ transporters, the HAK/KUP transporters, was identified in plants. First isolated from barley (Hordeum vulgare) and Arabidopsis, HAK/KUP transporters clearly show homology to the soil yeast Schwanniomyces occidentalis HAK1 and the bacterium Escherichia coli KUP1 (Santa-Maria et al., 1997; Fu and Luan, 1998; Kim et al., 1998). HAKs from S. occidentalis function as K+/H+ symporters (Haro et al., 1999; Rodriguez-Navarro, 2000), but in plant cells, the transport mechanism has not yet been identified. Plant HAK transporters expressed in yeast mediate both high- and low-affinity K+ uptake. High-affinity K+-uptake activity of both Arabidopsis AtKUP1 and barley HvHAK1 are inhibited by millimolar concentration of Na+ when these transporters are expressed in yeast cells (Santa-Maria et al., 1997; Fu and Luan, 1998). The structure of HAK/KUP transporters has not been solved, but hydrophobicity plots indicate that the protein contains 12 putative transmembrane domains (Kim et al., 1998; Rubio et al., 2000).
Arabidopsis KUP/HAK transporters seem to belong to a large gene family with at least 25 members (including putative open reading frames identified from the GenBank database). There are seven different HAK genes from barley. These genes are expressed in a variety of tissues throughout the plant (Kim et al., 1998). Phylogenetic analysis indicates that they can be categorized into four groups (Rubio et al., 2000), with functional transporters having been characterized in groups I and II. The large redundancy that appeared to exist in the HAK/KUP gene family suggests a primary role in K+ acquisition in plants, probably via expression, distribution, and regulation in various tissues and cell types or under different growth conditions. They might be important for the establishment of cellular ion homeostasis, for instance, when plants are exposed to saline environment.
As part of a study aimed at characterizing the major potassium uptake systems in the halophytic common ice plant (Mesembryanthemum crystallinum) we wished to analyze the role of HAK homologs in this species to gauge their possible involvement in Na+ and K+ homeostasis. Based on the frequency with which the sequences were obtained, we characterized the four most abundant transcripts, McHAK1, McHAK2, McHAK3, and McHAK4, respectively, by functional complementation of a yeast mutant, transcript analysis and in situ hybridizations. We describe cell-specific expression of HAK-type transporters and their modification by salt stress and characterize effects of Na+ on McHAK-mediated K+ transport. We propose differential regulation of McHAK expression in response to salt stress, suggesting the physiological relevance of these transporters for ion homeostasis at high salinity.
RESULTS
Isolation of Four HAK-Type Transporter Homologs from Common Ice Plant
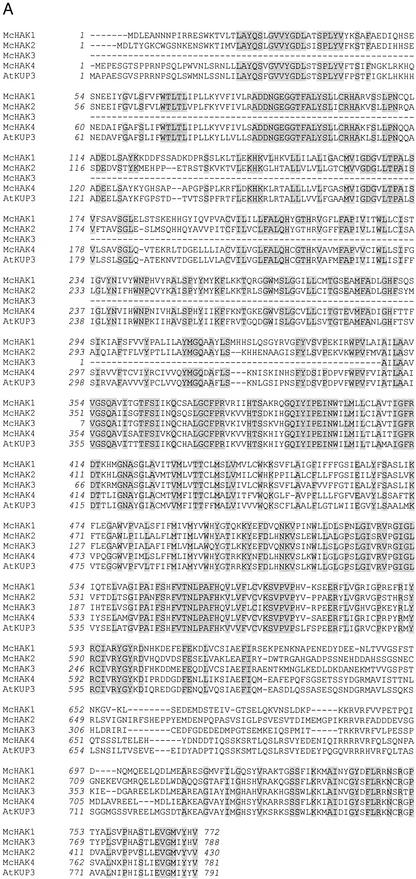
Based on sequence homologies to genes from the AtKT/AtKUP/AtHAK family of putative K+ transporters from Arabidopsis and barley, we used reverse transcriptase (RT)-PCR amplification, cDNA library screening, and 5′- and 3′-RACE amplification to obtain the full-length transcript sequences of McHAK1, McHAK2, and McHAK4 and a partial cDNA-sequence of McHAK3 from common ice plant (Fig. 1). For McHAK1, a 2,919-bp nucleotide sequence was isolated that encodes an open reading frame of 772 amino acids. The open reading frame of McHAK2 includes a protein with 788 amino acids, whereas the McHAK3 sequence isolated was truncated at the 5′ end, and the deduced protein sequence had 430 amino acids. The McHAK4 cDNA sequence was 2,677 bp in length and encodes a protein of 781 amino acids. The predicted proteins of McHAK1, McHAK2, and McHAK4 (Fig. 1B) are characterized by 12 transmembrane domains. Their localization in membranes still awaits careful analysis while computer predictions, which are not strongly supported, place the proteins in the plasma membrane (http://psort.ims.u-tokyo.ac.jp/form.html).
Figure 1.
Characterization of the McHAK transcripts from common ice plant. A, Alignment of the predicted amino acid sequences of McHAK1 (AF367864), McHAK2 (AF367865), McHAK3 (AF367866), McHAK4, and AtKUP3 (AAF19432). B, Hydropathy plot of McHAK1 and McHAK2 (Kyte and Doolittle, 1982). Putative transmembrane domains are indicated (I–XII). C, Phylogenetic tree of members of the HAK subfamilies (ClustalX, only full-length cDNAs included).(Figure continues on facing page.)
Although the deduced amino acid sequences of McHAKs show low homology to the HAK1 from S. occidentalis (e.g. 33% identity between McHAK1 and SoHAK1), they do share high homology with HAK-type proteins from other plants. Their homology to Arabidopsis HAK/KUP transporters varies from 68.3% to 83.7% in similarity and from 58.7% to 78.7% in identity. McHAK2 shows a particularly high homology to AtKT2 (78.8% identity), which is also illustrated by a phylogenetic tree for all of the full-length (except McHAK3) plant HAK/KUP proteins, including 17 Arabidopsis HAKs (Fig. 1C). According to Rubio et al. (2000) all plant HAK/KUP transporters fall into four groups. In Figure 1C, HAK-transporters of groups I to III are identified. All four McHAKs belong to group II; each aligned to an Arabidopsis ortholog. The existence of closely related orthologs among different species suggests that these genes are highly conserved structurally and may be functionally related as well. McHAK4 seems to belong to group II too, but in a different subgroup from the other three McHAKs (the highest identity in BlastX searches are AAF14830 and AtKUP3).
Southern-type hybridizations with genomic DNA were performed to study the complexity of McHAK genes in the genome of common ice plant. Using the full-length coding sequence of McHAK1 as a probe, a complex pattern of bands (not shown) was obtained, whereas single bands hybridized to a probe corresponding to 3′-untranslated regions (UTRs; not shown). Copy number reconstitution indicated a signal strength equivalent to at least four different genes of the McHAK1 subfamily in the genome of common ice plant, which very likely is an under-representation but indicates that the probes used are not cross-hybridizing indiscriminately.
Tissue and Cell Specificity of McHAK Expression
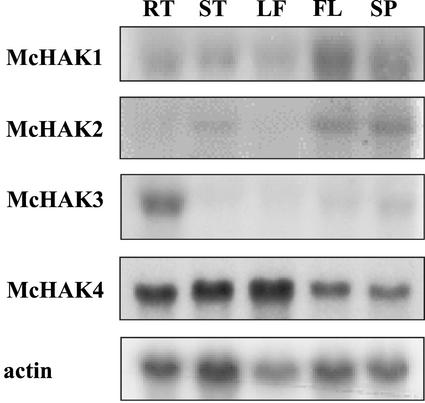
By RNA-blot hybridizations the tissue specificity of expression of the four McHAK homologs from common ice plant was probed (Fig. 2). The 3′-UTR fragments of McHAK1, 2, and 3 and an expressed sequence tag (EST) sequence of McHAK4 (350 bp in length) were used as probes. McHAK1 was expressed primarily in roots and weakly in the stem and leaf, yet the signals in unstressed tissues were never as strong as those in flower and seedpod, which only exist in salinity-stressed plants. McHAK2 was expressed in the stem at low abundance under nonstress conditions, and similar to McHAK1, expression in reproductive tissues was apparent. McHAK3 is root specific and seemed to be one of the major HAK transporters in roots under normal growth conditions. McHAK4 is highly expressed in all tissues investigated.
Figure 2.
Tissue-specific expression of McHAK genes. 3′-UTR regions of McHAK1, McHAK2, and McHAK3 and the EST sequence AI547371 of McHAK4 were hybridized to total RNA extracted from unstressed plants (except flower and seed pot). Tissues were specified as root (RT), stem (ST), leaf (LF), flower (FL), and seed pot (SP). Actin was used as a loading control. McHAK1, 2.8 kb (lower band, 2.7 kb); McHAK2, 2.9 kb; McHAK3, 2.8 kb (lower band, 2.7 kb); and McHAK4, 2.9 kb.
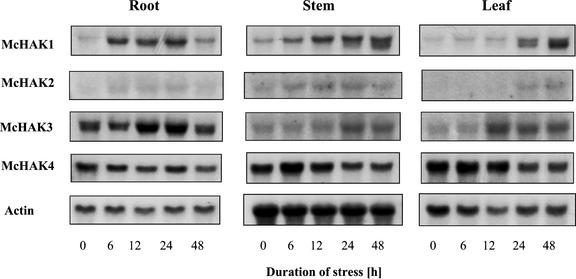
Expression of McHAK genes under saline conditions was investigated by RNA-blot analyses in root, stem, and leaf tissues. Five- to six-week-old plants of common ice plant were treated with 400 mm NaCl for the time periods indicated (Fig. 3). Expression of McHAK1 was up-regulated transiently in root tissue. The McHAK1 transcript level increased significantly after 6 h of salt stress, and remained high until 24 h of stress, after which the signal decreased to a lower level. In stem and leaf, the signals kept increasing for up to 48 h. The expression patterns of McHAK2 and McHAK3 were similar to that of McHAK1, except for a much lower abundance for McHAK2 and the strongest signal detected in root for McHAK3. The double bands observed with McHAK1 and McHAK3 are most likely not attributable to cross-hybridization because the 3′-UTRs that were used show only 72% sequence identity. They might indicate alternative splicing or mark the expression of unknown HAK isoforms, which could imply that additional HAK isoforms are salinity stress-responsive. The close relationship of the two genes is also reflected by a high bootstrap value, 100, at the branch separating them in the phylogenetic tree (Fig. 1C). McHAK4 showed constitutive expression under saline conditions in all tissues after the salt application.
Figure 3.
Northern-type hybridization of the expression of McHAK during NaCl stress. Six-week-old plants were stressed with 400 mm NaCl for the time indicated. Total RNA from different tissues were probed with either the 3′-UTR region of McHAK1, McHAK2, and McHAK3, respectively, or the EST fragment of McHAK4 (AI547371). Apparent Mrs are as in Figure 2.
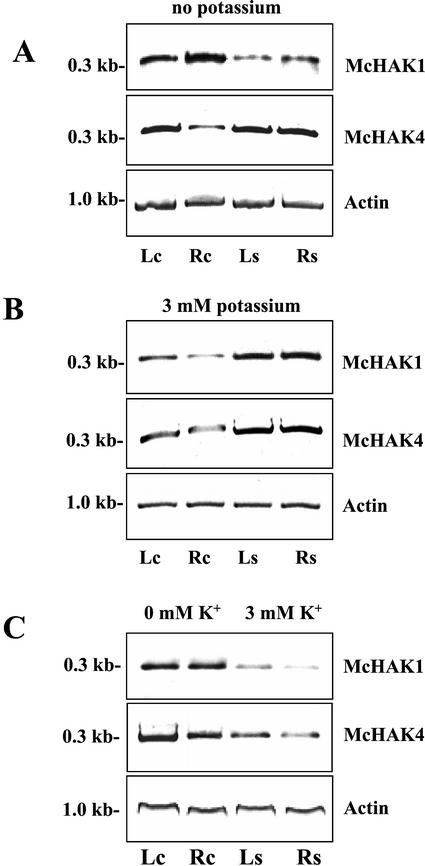
To analyze the effects of K+ nutrition and Na+ application on the expression of McHAK, we quantitated the transcript abundance of McHAK1 and McHAK4 in leaf and root tissue of 5-week-old common ice plant by RT-PCR amplification with gene-specific oligonucleotide primers for the coding region of the genes (Fig. 4). The plants were adapted to nutrient solutions containing either 3 mm K+ or solutions without K+ added that contained 10 to 16 μm K+ due to impurities from other salts (in the following this solution is referred to as 0 mm K+). Expression of McHAK1 and McHAK4 could be detected in leaves and roots of the common ice plant at 0 and at 3 mm K+. To test the effect of salinity stress on the expression of McHAK1 and McHAK4, 5-week-old common ice plants were exposed to 400 mm NaCl for 72 h in nutrition solution containing either 0 or 3 mm K+. The transcript levels of McHAK1 and McHAK4 increased in leaf and root tissue in plants adapted to 0 and 3 mm K+ in response to salinity stress (Fig. 4, A and B). Direct comparison of McHAK1 and McHAK4 expression in plants adapted to either 0 or 3 mm K+ showed higher transcript levels for both genes in plants adapted to low K+ medium and decreased at 3 mm external K+ (Fig. 4C).
Figure 4.
Effects of NaCl treatment and K+ starvation on the expression of McHAK1 and McHAK4 in common ice plant. Transcript amounts were quantified by RT-PCR using template cDNA obtained from leaves (Lc) and roots (Rc) from nonstressed control plants and from leaves (Ls) and roots (Rs) from plants treated with 400 mm NaCl for 72 h. Plants were adapted to nutrition solution without K+ added (A) and to nutrition solution containing 3 mm K+ (B). C, Comparison of transcript amounts from plants adapted to 0 mm K+ and 3 mm K+. For PCR, gene-specific primers as outlined in “Materials and Methods” were used. Transcripts were amplified in the linear range of amplification with 26 cycles for both transcripts (A), 28 cycles for McHAK1 and 27 cycles for McHAK4 (B), and 27 cycles for both transcripts (C). Actin was amplified as a loading control.
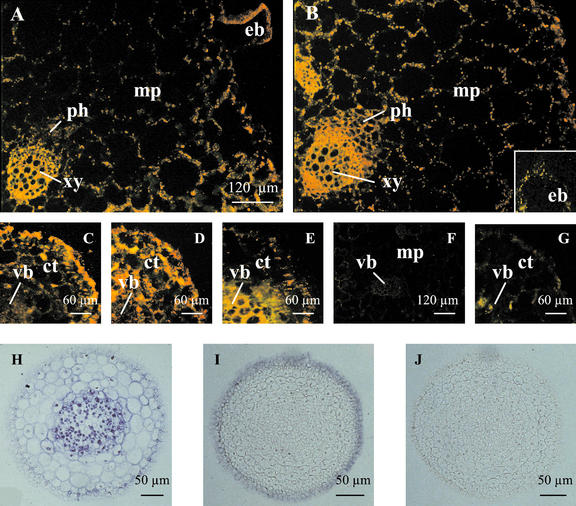
In situ hybridizations were performed for McHAK1, McHAK3, and McHAK4 with identical results in at least two separate repeat experiments. McHAK2 was not included due to its low-transcript abundance and the sensitivity of the experimental setup. Cell specificity of expression was studied for McHAK4 in common ice plant adapted to 0 mm K+ (Fig. 5). In leaf sections of nonstressed plants, McHAK4 was expressed in mesophyll and epidermal cells with similar signal strength. Higher transcript abundance was observed in xylem parenchyma and epidermal bladder cells that are a morphological characteristic of common ice plant. In salt-stressed leaves, signal strength increased in mesophyll and epidermal cells, and expression was exceptionally stimulated in phloem cells. Signal strength conversely decreased in the epidermal bladder cells.
Figure 5.
Cell specificity of McHAK expression in common ice plant. A through G, In situ hybridization of McHAK4 to tissue cross sections from common ice plant adapted to nutrition solution without K+ added. A, Leaf, control; antisense. B, Leaf, 72 h 400 mm NaCl, antisense. C, Root tip, control; antisense. D, Root tip, 72 h, 400 mm NaCl; antisense. E, Root, 72 h, 400 mm NaCl; antisense. F, Leaf, control; sense. G, Root, 72 h, 400 mm NaCl; sense. H and J, In situ hybridization of McHAK1 and McHAK3 to tissue cross sections from common ice plant adapted to nutrition solution with 3 mm K+ added. H, Root tip, 12 h, 400 mm NaCl; McHAK1, antisense. I, Root tip, 12 h, 400 mm NaCl; McHAK3, antisense. J, Root tip, 12 h, 400 mm NaCl; McHAK3, sense. mp, Mesophyll; ph, phloem; xy, xylem; eb, epidermal bladder cell; ct, cortex; vb, vascular bundle.
In tissue sections from root tips of nonstressed control plants, McHAK4 expression was concentrated in epidermal cells, and weaker signals were found in cells of the outer cortex layers and the vascular tissue. In root tips from salt-stressed plants, cells of the cortex showed up-regulation of the expression of McHAK4 to similar signal strength as it was found in the epidermal cells. In older roots, transcription of McHAK4 occurred in the vascular tissue, in cells of the outer cortex, and in epidermal cells. In comparison with the salt-stressed roots (Fig. 5E), the signals in cortex cells of nonstressed plants were slightly reduced (not shown).
For McHAK1 and McHAK2, we used root tissue of plants adapted to 3 mm K+ that were stressed by 400 mm NaCl for 12 h (Fig. 5, H–J). In young root, McHAK1 mRNA was detected mainly in stelar cells (Fig. 5H). As the root matured, signals also appeared in epidermal cells (data not shown). McHAK3 mRNA was mainly detected in epidermal cells of both young root and developing stele structure (Fig. 5I). The expression of McHAK1 and McHAK3 in epidermal cells suggested their roles in potassium uptake from the soil and their possible involvement in cation loading into vascular tissues.
Na+ Accumulation in Common Ice Plant Depends on the External K+ Concentration
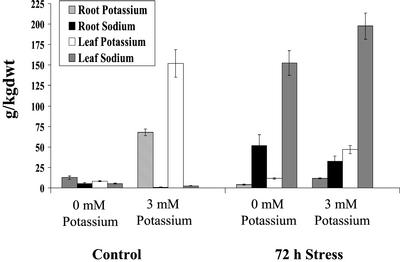
To determine the dependence of Na+ accumulation on the external K+ concentration, Na+ and K+ accumulation were measured by inductively coupled plasma atomic emission spectrometer (ICP-AES; Fig. 6). Five-week-old common ice plant plants were adapted to 0 and 3 mm K+, respectively, and either grown as control plants or stressed with 400 mm NaCl for 72 h. In comparison with plants grown at 3 mm K+, the K+ content was reduced in leaves and roots of plants grown at 0 mm K+. Salt-stressed plants that were grown at 3 mm K+ accumulated Na+ very efficiently in leaves, whereas the root Na+ content was about 15% of the leaf content. Under salt stress the K+ content decreased dramatically in common ice plant and was about 30% in leaves and about 15% in roots of nonstressed plant material. In leaves of salt-stressed plants adapted to 0 mm K+, the Na+ accumulation was reduced to about 70% in comparison with salt-stressed common ice plant at 3 mm K+. However, the root Na+ concentration was increased in comparison with plants adapted to 3 mm K+.
Figure 6.
Accumulation of K+ and Na+ in common ice plant plants grown hydroponically in nutrition solution without K+ added or in nutrition solution containing 3 mm K+. Plants were grown as nonstressed control plants or treated with 400 mm NaCl for 72 h (n = 6).
McHAK1, McHAK2, and McHAK4 Complement Yeast Mutants Deficient in K+ Uptake
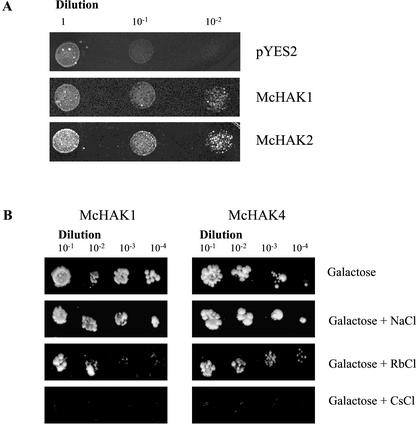
To test whether McHAK-type transporters mediate K+ uptake, functional yeast complementation was used (Fig. 7). The full-length coding sequences of McHAK1, McHAK2, and McHAK4 were cloned in the yeast expression vector pYES under control of the inducible GAL-promoter and transformed into the strain CY162 of Saccharomyces cerevisiae that has deletions in the TRK1 and TRK2 potassium transporter genes (Ko and Gaber, 1991). The mutant yeast strain requires concentrations of 100 mm K+ for growth and is unable to grow at low K+ concentrations. Positive transformants of CY162 containing the McHAK1, McHAK2, or McHAK4 constructs were able to grow at K+ concentrations of 3 and 7 mm, respectively, when the promoter was induced. Effects of other monovalent cations on the growth of the yeast strain CY162 was tested for McHAK1 and McHAK4 expressing cells. Addition of 150 mm NaCl did not affect the growth of the yeast expressing McHAK1 and McHAK4, whereas the growth of the transformants was inhibited by the addition of 150 mm RbCl and 15 mm CsCl.
Figure 7.
Complementation of a yeast TRK1/TRK2 mutant with McHAK1, McHAK2, and McHAK4. The yeast strain CY162 (Ko and Gaber, 1991) was transformed with either the empty vector pYES2 or with McHAK1, McHAK2, and McHAK4, respectively, cloned into the yeast expression vector pYES2. Positive transformants were grown on Gal-SC containing 3 mm K+ (A) or 7 mm K+ (B), or Gal-SC supplemented with 150 mm RbCl, 150 mm NaCl, or 15 mm CsCl.
DISCUSSION
McHAK Transcripts Are Expressed in Specific Tissues and Cells
K+ transporters of the HAK/KT/KUP family have been characterized from Arabidopsis and barley and were shown to function as a high- and low-affinity K+ uptake system, respectively. Here, we isolated four transcripts from common ice plant that share about 70% homology with HAK-type transporters from Arabidopsis and barley on the amino acid level, whereas the homology to the bacterial K+ transporter KUP from E. coli and to the fungal HAK1 is about 30%. For the common ice plant HAK-type transporter homologs, 12 transmembrane domains can be identified as they were also predicted for other plant HAKs. According to calculations with PSORT (http://psort.ims.u-tokyo.ac.jp/form.html), the McHAK proteins are localized to the plasma membrane. HAK-type transporters are members of multigene families in Arabidopsis, barley, and common ice plant, and the different genes show tissue-specific expression patterns. The barley HvHAK1 was only detected in roots (Santa-Maria et al., 1997), Arabidopsis AtKT1 and AtKT2 were found in roots and leaves (Quintero and Blatt, 1997), whereas Arabidopsis AtKUP1 was predominantly expressed in roots (Fu and Luan, 1998). Kim et al. (1998) detected four AtKUP-transporters in roots, stems, leaves, and flowers of Arabidopsis.
In common ice plant, McHAK transcripts show gene-specific expression patterns at different abundance levels. McHAK4 was the most abundant transcript and was found in all tissues and especially in nonflowering plants. McHAK3, showed moderate abundance in roots whereas the other two genes were present at low-abundance in unstressed tissues, McHAK1 is all tissues, McHAK2 in the stems. For McHAK1, McHAK3, and McHAK4, respectively, we studied their cell-specific expression pattern, which had not previously been reported for any plant. In differentiated roots close to the meristematic root tip, we found the highest transcript levels in the root epidermis. In mature roots and leaves, the strongest hybridization signals were detected in the vascular tissue. These data indicate that McHAKs seem to have a role in mediating root K+ uptake and that they could be involved in plant long-distance K+ transport through loading and/or unloading in the vasculature. Also, the strong signals found in parenchyma cells of the vasculature may suggest importance for K+ influx and maintenance of osmotic pressure in developing cells, thus regulating cell elongation. In an Arabidopsis mutant carrying a T-DNA insertion in the HAK-homolog TRH1, both Rb+ uptake and root hair growth were inhibited, indicating that e.g. the TRH1-mediated K+ uptake is specifically required for elongation of root hair cells (Rigas et al., 2001). In common ice plant, additional signals from in situ hybridizations were found in root cortex cells and in leaf mesophyll. The location of HAK-transporter transcripts in most cell types of common ice plant indicates involvement of these K+ transport proteins in symplastic K+ transport and distribution within the plant and, thus, supports and extends the hypothesis by Fu and Luan (1998) that HAK homologs are a major transport system for K+ in plants.
McHAK Expression Is Regulated in Response to K+ Starvation and High Salinity
Functions of common ice plant McHAK proteins as K+ transporters were shown by yeast complementation. In the yeast strain CY162 that carries deletions in the K+ uptake systems TRK1/TRK2, the ability to grow on millimolar K+ concentrations was restored by expression of McHAK1, McHAK2, and McHAK4. Interestingly, the mutant CY162 cells expressing McHAK1 and McHAK4 grew poorly in the presence of Rb+ and Cs+, but Na+ showed less effect. These data suggested that common ice plant HAK-type proteins mediate the transport of K+, Rb+, and Cs+ but not that of Na+. Cs+ transport with high efficiency has also been reported for the KUP-transporter from E. coli (Bossemeyer et al., 1989). K+ uptake in CY162 expressing Arabidopsis AtKUP1 was inhibited by Cs+ (Fu and Luan, 1998). As shown by Kim et al. (1998), AtKUP1 complemented the E. coli mutant TK2463 that requires 25 mm K+ for growth and growth was completely inhibited by 10 mm Cs+. Although the mediation of Cs+ transport by common ice plant McHAKs is similar to the ion selectivity of HAK homologs from other organisms, the Na+ discrimination that we observed for McHAKs has not been reported for HAK-type transporters from other plants. In CY162 expressing Arabidopsis AtKt2, Rb+ uptake was not affected by Na+ up to 10 mm but was inhibited at higher concentrations (Quintero and Blatt, 1997), whereas for common ice plant HAK transporters, Na+ concentrations of 150 mm did not affect the growth of the complemented yeast mutant. The different discrimination of Na+ observed for Arabidopsis and for common ice plant could be attributable to differences in the primary amino acid sequence of the transporters that might influence selectivity. Arabidopsis AtHAK5, for example, did not complement the trk1/trk2 yeast strain, but a mutant sequence, Athak5-1, with the substitution L776H, which changed Vmax, was able to restitute K+ uptake in yeast (Rubio et al., 2000). Similarly, alterations of the sequences could affect selectivity.
In plant roots, discrimination between K+ and Rb+ has not been observed, and accordingly, both ions compete for import through K+ transport systems (Rubio et al., 2000). Kinetic analyses revealed that K+ and Cs+ also compete for influx into plant roots (White and Broadley, 2000). Two other plant K+ transport systems, inward-rectifying K+ channels and HKT-type transporters, show different ion selectivity compared with the HAK-type transporters. Inward-rectifying K+ channels show high, but not exclusive, selectivity for K+ over all other alkali cations (Schachtman et al., 1992; Maathuis and Sanders, 1997; Amtmann and Sanders, 1999). Wheat (Triticum aestivum) HKT1 functions as a K+/Na+ symporter. In contrast, the Arabidopsis homolog AtHKT1 mediates Na+ influx when expressed in X. laevis oocytes (Rubio et al., 1995; Uozumi et al., 2000), and AtHKT1 represents a route for sodium influx at least in some plants (Rus et al., 2001). Such a conclusion has also been drawn for the rice HKT1-homolog, OsHKT, which appears to represent an alkali ion transporter (D. Golldack, H. Su, F. Quigley, U.R. Kamasani, C. Muñoz-Garay, E. Balders, O.V. Popova, J. Bennett, H.J. Bohnert, and O. Pantoja, unpublished data). In Arabidopsis, wheat, and barley, application of Na+ inhibited, or at least did not affect, K+ uptake, whereas a stimulation of import would be expected for substantial HKT-mediated K+ transport, indicating that HKT-type transporters might not have a significant physiological role in K+ uptake into plant roots (Maathuis et al., 1996; Box and Schachtman, 2000). In accordance, we consider HAK-type transporters to be strong candidates for substantial K+ uptake and distribution in plants.
McHAK transporters did not mediate Na+ uptake when heterologously expressed in yeast, but the transcript levels of all four McHAK were enhanced transiently or permanently in response to salinity stress. To understand the physiological role of McHAK up-regulation for the salt adaptation of common ice plant, we compared the ion contents in control plants and in salt-stressed plants grown at low millimolar and low micromolar K+ concentrations, respectively. At both external K+ concentrations, treatment with 400 mm NaCl for 72 h resulted in a dramatic decrease of the root and leaf K+ content, demonstrating that salinity stress resulted in K+ starvation of the plants. Similarly, the K+ starvation that was induced by transferring common ice plant plants from low millimolar to low micromolar K+ in the medium resulted in transcriptional stimulation of all McHAKs. The most parsimonious explanation is that the increased expression of McHAK in salt-stressed common ice plant is not a result of increased Na+ uptake by the plants under high salinity, which is substantial during the first few days of salt stress, but that it is due to salt-induced K+ starvation. In addition, these data indicate that McHAK is highly selective for K+ over Na+ and is likely to transport specifically K+ while excluding Na+ in common ice plant.
The molecular mechanisms for Na+ uptake by plants have not been identified, but nonspecific uptake of this ion via K+ transport systems has been suggested (Schachtman and Liu, 1999; Blumwald et al., 2000). It seems that proteins encoded by the Hkt-type genes, with their ion specificity depending on the species, could provide one major port of entry (Rus et al., 2001). In addition, evidence for entry through cyclic nucleotide-gated channels and (similar to the wheat) LCT1-type channels is mounting (Schuurink et al., 1998; Amtmann et al., 2001; Maeser et al., 2001). For common ice plant, it is clear that the strongly expressed transcript and highly abundant protein of the root-specific, inward-rectifying K+ channel MKT1 is transcriptionally inhibited, and protein amounts decrease after salt stress (Su et al., 2001). Thus, inward-rectifying K+ channels are unlikely candidates for Na+ uptake in salt-stressed common ice plant. We now demonstrate that common ice plant McHAK show high discrimination of K+ over Na+, which could make them specific mediators for K+ uptake at high salinity. According to these results we suggest that HAK-type transporters represent the major K+ uptake system and that they are responsible for intercellular distribution in salt-stressed common ice plant thus selectively maintaining K+ uptake under high-salinity conditions. We consider the McHAK genes in common ice plant to be major contributors to potassium homeostasis under high-salinity conditions both by facilitating uptake and transport through the vasculature. By their up-regulation, they seem to alleviate potassium starvation. It is suggested that this may be also a major function of HAK proteins in other organisms when uptake through potassium channels is not provided or possible when other alkali cations are present in high concentration.
MATERIALS AND METHODS
Plant Material
Common ice plant (Mesembryanthemum crystallinum) plants were grown in a growth chamber with 10 h of light (300 μE m−2 s−1, 23°C) and 14 h of dark (18°C) with 50% relative humidity. Common ice plant seeds were germinated in vermiculite soaked with one-half-strength Hoagland nutrition solution (Ostrem et al., 1987). Three weeks after germination, seedlings were transferred to hydroponic culture with one-half-strength Hoagland nutrition solution, and aeration of the hydroponic tanks was started 1 week after transferring the plants. Seedlings for the construction of cDNA libraries were harvested at the age of 2 weeks. For salt-stress treatments, the nutrient solution was supplemented with 400 mm NaCl for the time periods indicated. Unstressed control plants were grown in parallel and harvested at the same time. All plants were harvested 5 h after the start of illumination except in the case of plants stressed for various time periods. For experiments with leaf material, the second youngest leaf pair of the plants was used.
Isolation and Characterization of Transcripts
A 1.7-kb PCR product of McHAK1 was amplified with degenerated primers (5′-AAYGAYAAYGGNGARGGNGG-3′ and 5′-TATCCRTANCGNGCNACRCA-3′) from common ice plant cDNA. A 940-bp fragment of McHAK1 was used to screen cDNA libraries of common ice plant seedlings, mature leaves, and roots. A full-length cDNA McHAK1 and two partial cDNAs McHAK2 and McHAK3 were isolated. A full-length clone of McHAK2 was obtained by 5′-RACE amplification (Invitrogen, Carlsbad, CA).
Also, for McHAK1 and McHAK4, partial cDNA sequences homologous to the Arabidopsis KUP-type transporters were identified in the EST database with the GenBank accession numbers AA933556 and AI547371. The 5′ ends of both common ice plant sequences and the 3′ end of the sequence AA933556 were obtained using 5′- and 3′-RACE amplification (SMART RACE cDNA amplification, CLONTECH, Palo Alto, CA). The 5′- and 3′-RACE PCR products were cloned in the vector PCR-TOPO-2.1 (Invitrogen) and sequenced. Full-length sequences were obtained by PCR with Advantage2 polymerase mix (CLONTECH) from SMART RACE cDNA. The full-length cDNAs were cloned in PCR-TOPO-2.1 (Invitrogen) and sequenced.
Nucleic Acid Isolation and Hybridization, RT-PCR, and Measurement of Na+
For Southern-type analysis, genomic DNA was extracted by SDS and KOAc extraction followed by isopropanol and ethanol precipitation and subsequent LiCl precipitation to remove RNA. Ten grams of seedling leaf tissues was ground in liquid nitrogen, mixed gently with 30 mL of extraction buffer (100 mm Tris-HCl pH 8.0, 500 mm NaCl, 50 mm EDTA, and 0.07% [v/v] mercaptoethanol). Then 2 mL of 20% (w/v) SDS was added, and the mix was incubated at 65°C for 10 min, after which 10 mL of 5 m KOAc pH 7.5 was added. The extract was incubated on ice for 20 min followed by centrifugation at 2,500g for 15 min. The supernatant was then precipitated by 0.6 volume of isopropanol and 70% (v/v) ethanol, respectively, at −20°C. The pellet was dissolved in 10 mL of Tris-EDTA buffer, followed by the addition of 10 mL of 4 m LiCl and incubation at 4°C for 4 h. The supernatant was extracted gently with phenol/chloroform and then extracted with chloroform twice. Finally, the genomic DNA was precipitated by 0.1 volume of 3 m NaOAc (pH 5.2) and 2 volumes of ethanol, and the pellet was dissolved in appropriate volumes of Tris-EDTA buffer. Ten micrograms of DNA was digested with different restriction enzymes, separated in 0.7% (w/v) agarose gels in 0.5× Tris-borate/EDTA buffer, hybridized according to the membrane instruction manual (Stratagene, La Jolla, CA), and probed with 32P-labeled DNA fragments (ICN Biomedicals, Irvine, CA).
Total RNA from leaves and roots of common ice plant was isolated by SDS and phenol extraction, in the presence of aurin tricarboxylic acid (Su et al., 2001). Northern analyses were performed according to standard procedures with 20 μg of total RNA per lane (Sambrook et al., 1989). The detection probes corresponding to the 3′-noncoding fragments of McHAK1, McHAK2, and McHAK3 and the EST fragment of McHAK4, respectively, were prepared by PCR and labeled with 32P-labeled DNA fragments (ICN Biomedicals). Filters were washed with 0.1× SSC at 42°C for 30 min. Signal detection was performed with autoradiography.
cDNA for RT-PCR amplification was synthesized from each 3 μg of total RNA with SuperScript RT II (Invitrogen) in 20-μL reactions. After synthesis, the cDNA was diluted 1:10 and 10-μL aliquots of cDNA were used as template for PCR amplifications in 50-μL standard reactions. The following sequence specific forward and reverse primers were used for PCR amplifications: 5′-GAAGAGCTGATGGATCTA-3′, 5′-CATGCCAACTT CAATGAG-3′, 5′-GTATGGCACTACGGTACA-3′, and 5′-CCATATCGTGCGATGCAT-3′.
For PCR, the following cycle parameters were used: 94°C for 1.5 min in the first cycle, followed by 1 min at 94°C, 1 min at 55°C, and 2 min at 72°C with cycle numbers as indicated, and a final extension at 72°C for 10 min. The PCR products were separated in 1.7% (w/v) agarose gels and stained with ethidium bromide. Photographic images were obtained with a gel documentation system (INTAS, Göttingen, Germany). The fragments were cloned in PCR-TOPO-II (Invitrogen) and sequenced.
Analyses of K+ and Na+ concentrations were performed with an ICP-AES (Jobin-Yvon JY 70, Instruments S.A., Logjumea, France) according to Brune et al. (1995).
In Situ Hybridization
Sections of root and leaf tissue were fixed with formaldehyde-acetic acid, dehydrated, and embedded with Paraplast Plus (Fisher Scientific, Pittsburgh). Sections of 10 μm were mounted on poly-l-Lys-coated microscopic slides. PCR-TOPO-II plasmid harboring a partial cDNA sequence from the coding region of McHAK4 as described above and pBS-KS harboring 3′-untranslated fragments of McHAK1 and McHAK3 were linearized by restriction digestion and sense and antisense RNA transcripts were synthesized by SP6 and T7 RNA polymerase with digoxigenin-UTP (Roche Diagnostics, Mannheim, Germany) as a label. In situ hybridizations were performed as described before (Golldack and Dietz, 2001). Signal detection was performed with antidigoxigenin alkaline phosphatase-conjugated Fab fragments, naphthol-AS-phosphate, Fast Red TR, 5-bromo-4-chloro-3-indolyl-phosphate, and nitroblue tetrazolium, respectively, as substrates (Roche Diagnostics). Microscopic images were obtained with an Axioskop fluorescence microscope (Zeiss, Oberkochen, Germany) and a cooled CCD-Camera coupled to an Axioskop fluorescence microscope (Zeiss), respectively, and processed through Axiovision (Zeiss) and Adobe Photoshop (Adobe Systems, Mountain View, CA).
Yeast Complementation
The coding sequences of McHAK1, McHAK2, and McHAK4 were cloned in pYES2 (Invitrogen) under control of the inducible Gal1 promoter. The constructs were transformed into the Saccharomyces cerevisiae strain CY162 (Ko and Gaber, 1991) by the lithium acetate method according to Gietz et al. (1992). Transformants were selected on Glc-containing SC-agar plates without uracil supplemented with 100 mm K+. Positive transformants of McHAK1 and McHAK2 were transferred to yeast nitrogen base liquid medium with 20 mm of K+, 2% (w/v) Gal, and 2% (w/v) Suc, to induce expression of HAK genes. After 48 h, the liquid cultures were diluted and plated onto LS (Ramos et al., 1985) agar plates supplemented with 3 mm of K+, 2% (w/v) Gal, and 2% (w/v) Suc. As an alternative, positive McHAK1 and McHAK4 yeast transformants were directly transferred on Gal-containing SC-agar plates without uracil and with 7 mm K+. Also, growth of the positive transformants was studied on the same medium but supplemented with 150 mm NaCl, 150 mm RbCl, and 15 mm CsCl, respectively.
Supplementary Material
ACKNOWLEDGMENT
We are grateful to Elfriede Reisberg (University of Würzburg) for performing the ICP-AES measurements.
Footnotes
This work was supported by the Arizona Agricultural Experiment Station and by the Deutsche Forschungsgemeinschaft, Bonn (to D.G.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.001149.
LITERATURE CITED
- Amtmann A, Fischer M, Marsh EL, Stefanovic A, Sanders D, Schachtman DP. The wheat cDNA LCT1 generates hypersensitivity to sodium in a salt-sensitive yeast strain. Plant Physiol. 2001;126:1061–1071. doi: 10.1104/pp.126.3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amtmann A, Sanders D. Mechanisms of Na+uptake by plant cells. Adv Bot Res. 1999;29:75–112. [Google Scholar]
- Blumwald E, Aharon GS, Apse MP. Sodium transport in plant cells. Biochim Biophys Acta. 2000;1465:140–151. doi: 10.1016/s0005-2736(00)00135-8. [DOI] [PubMed] [Google Scholar]
- Bossemeyer D, Schlosser A, Bakker EP. Specific cesium transport via the Escherichia coli Kup (TrkD) K+-uptake system. J Bacteriol. 1989;171:2219–2221. doi: 10.1128/jb.171.4.2219-2221.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box S, Schachtman DP. The effect of low concentrations of sodium on potassium uptake and growth of wheat. Aust J Plant Physiol. 2000;27:175–182. [Google Scholar]
- Brune A, Urbach W, Dietz KJ. Differential toxicity of heavy-metals is partly related to a loss of preferential extraplasmic compartmentation: a comparison of Cd-stress, Mo-stress, Ni-stress and Zn-stress. New Phytol. 1995;129:403–409. [Google Scholar]
- Epstein E, Rains DW, Elzam OE. Resolution of dual mechanisms of potassium absorption by barley roots. Proc Natl Acad Sci USA. 1963;49:684–692. doi: 10.1073/pnas.49.5.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H-H, Luan S. AtKUP1: a dual-affinity K+ transporter from Arabidopsis. Plant Cell. 1998;10:63–73. doi: 10.1105/tpc.10.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann W, Rubio F, Schroeder JI. Alkali ion selectivity of the wheat root high-affinity potassium transporter HKT1. Plant J. 1996;10:869–882. doi: 10.1046/j.1365-313x.1996.10050869.x. [DOI] [PubMed] [Google Scholar]
- Gietz D, St Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golldack D, Dietz KJ. Salt-induced expression of the vacuolar H+-ATPase in the common ice plant is developmentally controlled and tissue specific. Plant Physiol. 2001;125:1643–1654. doi: 10.1104/pp.125.4.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haro R, Sainz L, Rubio F, Rodriguez-Navarro A. Cloning of two genes encoding potassium transporters in Neurospora crassa and expression of the corresponding cDNAs in Saccharomyces cerevisiae. Mol Microbiol. 1999;31:511–520. doi: 10.1046/j.1365-2958.1999.01192.x. [DOI] [PubMed] [Google Scholar]
- Horie T, Yoshida K, Nakayama H, Yamada K, Oiki S, Shinmyo A. Two types of HKT transporters with different properties of Na+ and K+ transport in Oryza sativa. Plant J. 2001;27:129–138. doi: 10.1046/j.1365-313x.2001.01077.x. [DOI] [PubMed] [Google Scholar]
- Kim EJ, Kwak JM, Uozumi N, Schroeder JI. AtKUP1: an Arabidopsisgene encoding high-affinity potassium transport activity. Plant Cell. 1998;10:51–62. doi: 10.1105/tpc.10.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko CH, Gaber RF. TRK1 and TRK2 encode structurally related K+ transporters in Sacchararomyces cerevisiae. Mol Cell Biol. 1991;8:4266–4273. doi: 10.1128/mcb.11.8.4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Maathuis FJM, Sanders D. Regulation of K+ absorption in plant root cells by external K+: interplay of different K+transporters. J Exp Bot. 1997;48:451–458. doi: 10.1093/jxb/48.Special_Issue.451. [DOI] [PubMed] [Google Scholar]
- Maathuis FJM, Verlin D, Smith FA, Sanders D, Fernandez JA, Walker NA. The physiological relevance of Na+-coupled K+-transport. Plant Physiol. 1996;112:1609–1616. doi: 10.1104/pp.112.4.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeser P, Thomine S, Schroeder JI, Ward JM, Hirschi K, Sze H, Talke IN, Amtmann A, Maathuis FJ, Sanders D et al. Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 2001;126:1646–1667. doi: 10.1104/pp.126.4.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrem JA, Olson SW, Schmitt JM, Bohnert HJ. Salt stress increases the level of translatable mRNA for phosphoenolpyruvate carboxylase in Mesembryanthemumregenerates. Dev Biol. 1987;167:239–251. doi: 10.1104/pp.84.4.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero FJ, Blatt MR. A new family of K+-transporters from Arabidopsisthat are conserved across phyla. FEBS Lett. 1997;415:206–211. doi: 10.1016/s0014-5793(97)01125-3. [DOI] [PubMed] [Google Scholar]
- Ramos J, Contreras P, Rodriguez-Navarro A. A potassium transport mutant of Saccharomyces cerevisiae. Arch Microbiol. 1985;143:88–93. [Google Scholar]
- Rigas S, Debrosses G, Haralampidis K, Vicente-Agullo F, Feldmann K, Grabov A, Dolan L, Hatzopoulos P. Trh1 encodes a potassium transporter required for tip growth in Arabidopsisroot hairs. Plant Cell. 2001;13:139–151. doi: 10.1105/tpc.13.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Navarro A. Potassium transport in fungi and plants. Biochim Biophys Acta. 2000;1469:1–30. doi: 10.1016/s0304-4157(99)00013-1. [DOI] [PubMed] [Google Scholar]
- Rubio F, Gassmann W, Schroeder JI. Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science. 1995;270:1660–1663. doi: 10.1126/science.270.5242.1660. [DOI] [PubMed] [Google Scholar]
- Rubio F, Santa-Maria GE, Rodriguez-Navarro A. Cloning of Arabidopsisand barley cDNAs encoding HAK potassium transporters in root and shoot cells. Physiol Plant. 2000;109:34–43. [Google Scholar]
- Rus A, Yokoi S, Sharkuu A, Reddy M, Lee B-H, Matsumoto TK, Koiwa H, Zhi J-K, Bressan RA, Hasegawa PM. AtHKT1 is a salt tolerance determinant that controls Na+entry into plant roots. Proc Natl Acad Sci USA. 2001;98:14150–14155. doi: 10.1073/pnas.241501798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Santa-Maria GE, Rubio F, Dubcovsky J, Rodgriguez-Navarro A. The HAK1gene of barley is a member of a large gene family and encodes a high-affinity potassium transporter. Plant Cell. 1997;9:2281–2289. doi: 10.1105/tpc.9.12.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtman D, Liu W. Molecular pieces to the puzzle of the interaction between potassium and sodium uptake in plants. Trends Plant Sci. 1999;4:281–287. doi: 10.1016/s1360-1385(99)01428-4. [DOI] [PubMed] [Google Scholar]
- Schachtman DP, Schroeder JI. Structure and transport mechanism of a high-affinity potassium uptake transporter from higher plants. Nature. 1994;370:655–658. doi: 10.1038/370655a0. [DOI] [PubMed] [Google Scholar]
- Schachtman DP, Schroeder JI, Lucas WJ, Anderson JA, Gaber RF. Expression of an inward-rectifying potassium channel by the ArabidopsisKAT1 cDNA. Science. 1992;258:1654–1658. doi: 10.1126/science.8966547. [DOI] [PubMed] [Google Scholar]
- Schuurink RC, Shartzer SF, Fath A, Jones RL. Characterization of a calmodulin-binding transporter from the plasma membrane of barley aleurone. Proc Natl Acad Sci USA. 1998;95:1944–1949. doi: 10.1073/pnas.95.4.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding EP, Hirsch RE, Lewis DR, Qi Z, Sussman MR, Lewis BD. Potassium uptake supporting plant growth in the absence of AKT1 channel activity: inhibition by ammonium and stimulation by sodium. J Gen Physiol. 1999;113:909–918. doi: 10.1085/jgp.113.6.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H, Golldack D, Katsuhara M, Zhao C, Bohnert HJ. Expression and stress-dependent induction of potassium channel transcripts in the common ice plant. Plant Physiol. 2001;125:604–614. doi: 10.1104/pp.125.2.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uozumi N, Kim EJ, Rubio F, Yamaguchi T, Muto S, Tsuboi A, Bakker EP, Nakamura T, Schroeder JI. The Arabidopsis HKT1 gene homolog mediates inward Na+ currents in Xenopus laevis oocytes and Na+ uptake in Saccharomyces cerevisiae. Plant Physiol. 2000;122:1249–1259. doi: 10.1104/pp.122.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White PJ, Broadley MR. Mechanisms of cesium uptake by plants. New Phytol. 2000;147:241–256. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.