Abstract
Non-specific lipid-transfer proteins (nsLTPs) are capable of binding lipid compounds in plant tissues and are coded by the nsLTP genes. Here, we present the analysis of expression of a family of potato (Solanum tuberosum) nsLTP genes that express throughout the developing plant in a highly tissue-specific manner. Three transcript-derived fragments were isolated using an amplified restriction fragment polymorphism-derived technique for RNA fingerprinting that show homology to plant nsLTP genes. These transcript-derived fragments displayed modulated expression profiles related to the development of new tissues, with a peak of transcription around the time of tuberization and just prior to sprout development, at dormancy breakage. In addition, a homologous family of expressed sequence tags was identified whose individual members could be classified according to their tissue specificity. Two subgroups of expressed sequence tags were found to express during tuber life cycle. To study the regulation of potato nsLTP genes, two putative potato nsLTP promoters were isolated and their expression was studied using promoter-marker-gene fusions. The results showed that one of the two promoters directed a highly specific pattern of expression detected in the phloem surrounding the nodes of young plants and in the same tissue of tuber related organs, whereas the second putative promoter showed little tissue or organ specificity. This difference in expression is likely due to a 331-bp insertion present in the tissue-specific promoter.
The potato (Solanum tuberosum) tuber life cycle is a complex multistage process involving stolon formation, tuber initiation, tuber filling, dormancy, and sprouting (Cutter, 1978). Potato tubers constitute underground stems that have undergone a series of morphological changes. Depending on the environmental conditions and the genotype, the potato plant forms stolons at the base of the stem that, after a period of rapid longitudinal growth, swell at their tips initiating tuberization. The onset of tuberization is accompanied by a burst of cell division, and reorientation of the cytoskeleton occurs (Sanz et al., 1996). The morphological differentiation during tuberization is further characterized by a variety of biochemical changes, such as the accumulation of starch and the appearance of new proteins. At tuberization, suppression of growth is imposed on the axillary buds (eyes) of developing tubers leading into dormancy, at the end of which cell division is initiated in the eyes of the tuber, giving rise to sprout development.
Each stage of tuber life cycle is likely to be controlled by a large set of interacting genes throughout the plant. Studies of gene expression using an amplified restriction fragment polymorphism-derived technique for RNA fingerprinting (cDNA-AFLP) during tuber life cycle show that many genes display differential expression and that these can be categorized into groups according to their putative function. The major processes that have been identified during tuber life cycle are related to resource metabolism (starch and protein biosynthesis), stress, and defense, and the regulation of these processes by phytohormones as well as their signal transduction pathways (Bachem et al., 2000). Analysis of the dynamics of gene expression showed that the highest rate of change in gene expression was observed just prior to and during tuber formation and that this activity falls off as tubers grow and eventually enter dormancy. Gene expression dynamics increase again during sprout development (Bachem et al., 2000). A number of process- and tissue-specific transcript-derived fragments (TDFs) have been analyzed in detail. The three TDFs that are the subject of this paper showed high similarity to plant (non-) specific lipid-transfer proteins (nsLTPs).
LTPs in animals and fungi can bind lipids and transfer them between membranes. According to their lipid-binding specificity, they are divided into at least three classes (Wirtz, 1991). Plants LTPs purified until now show similarity to nonspecific (ns) LTPs in animals, as they harbor broad phospholipid substrate specificity in vitro. However, their function in vivo still remains ambiguous. All plant nsLTPs identified so far harbor an N-terminal signal sequence responsible for directing the peptides to the endoplasmic reticulum and, as the K/HDEL retention signal is absent, subsequently to enter the secretory pathway. This has been substantiated by experiments showing that nsLTPs are secreted in vivo (Bernhard et al., 1991; Sterk et al., 1991) and are located in the epidermal cell wall of Arabidopsis (Thoma et al., 1993) or in the epicuticular waxy layer of broccoli (Brassica oleracea; Pyee et al., 1994). Due to the extracellular localization and their ability to bind lipids and fatty acids, nsLTPs have been suggested to have a role in the transfer of lipophylic compounds in the apoplast for epicuticular wax formation (Sterk et al., 1991). According to recent studies, plant nsLTP gene expression is induced as a result of changes in various environmental conditions such as high salt, drought, and temperature extremes, and following biotic stress such as viral, bacterial, and fungal infection (Kader, 1997). Moreover, isolated LTP proteins have been shown to have antimicrobial activity (Cammue et al., 1995). The diverse biochemical activities and expression scenarios make it difficult at this time to draw firm conclusions about the function of nsLTP proteins in the living plant. However, it does seem likely that individual nsLTP genes fulfill different functional roles in different tissues during development.
NsLTP genes have been isolated from a number of di- and monocotyledonous plants (Kader, 1996). Although their nucleotide and deduced protein sequence can diverge considerably, they harbor several common characteristics such as their small size (7–9 kD), a basic pI, and the location of eight conserved Cys residues forming four disulfide bridges. The nsLTP genes are present as small multigene families in all investigated plant species (Kader, 1996) and, as was found in Arabidopsis and tomato (Lycopersicon esculentum), the various members show different expression patterns in individual tissues (Trevino and O' Connell, 1998; Clark and Bohnert, 1999).
The expression of nsLTP genes is developmentally and spatially regulated. Organ and tissue specificity shows a high level of diversity in different species. In general, their expression is detectable in very early stages of plant development (Sterk et al., 1991; Thoma et al., 1994; Vroemen et al., 1996). In mature plants, their expression is associated with young organs rather then the older part of the plant (Fleming et al., 1992; Trevino and O'Connell, 1998). NsLTP genes are expressed mainly in the epidermal tissue of aerial organs (Kader, 1997); however, nonepidermal expression also occurs in other young developing tissues such as in the shoot apex and the floral meristem of tobacco (Nicotiana tabacum; Canevascini et al., 1996), parenchyma cells of the corolla in gerbera (Gerbera hybrida var. Regina; Kotilainen et al., 1994), and in vascular tissues in broccoli (Pyee et al., 1994). Some authors reported expression in epidermal cells of developing root or root initial cells (Canevascini et al., 1996; Sohal et al., 1999), whereas in bean (Phaseolus vulgaris), expression was found to be exclusively to the cortical region of a root-specific nsLTP (Song et al., 1998).
In this paper, we present the isolation of three tuber life cycle-related TDFs with a high sequence similarity to plant nsLTPs. Furthermore, we demonstrate that, using different nsLTP promoters to regulate the expression of a reporter gene, one member of the nsLTP gene family directs expression in the phloem. In addition, via sequence comparison, we have shown that this nsLTP gene is highly homologous to a stolon-specific nsLTP transcript.
RESULTS
RNA Fingerprinting
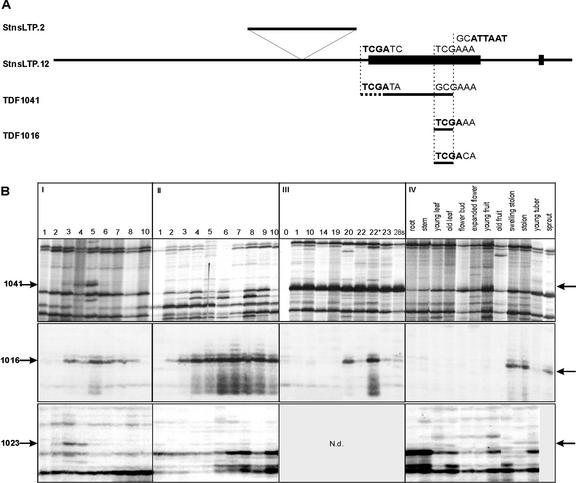
A synchronous in vitro tuberization, dormancy, and sprouting system has been developed to study gene expression throughout the potato tuber life cycle (Hendriks et al., 1991; Bachem et al., 1998). In the course of this study, around 300 TDFs were isolated corresponding to genes with a modulated expression pattern and related to key events during development (Bachem et al., 2000). Among these, three TDFs (TDF1016, TDF1023, and TDF1041) showed sequence similarity to plant nsLTPs. A comparison between the three TDFs revealed that their nucleotide sequence is largely identical. TDF1016 and TDF1023 have the same length (58 bp) and overlap with the 3′ region of TDF1041, which is 266 bp long, due to the absence of the restriction recognition site forming the termini of the TDFs 1016 and 1023. The sequence differences and the relative positions of the three TDFs compared with the genomic clones (described below) are illustrated in Figure 1A.
Figure 1.
Expression pattern and relative location of three potato TDFs corresponding to potato nsLTP genes: TDFs 1016, 1023, and 1041. A, The location of the three TDFs is shown relative to the StnsLTP.2/12 clones. The sequences indicated above the TDFs show the nucleotide sequence variations in and around the TaqI site (bold), giving rise to the different amplification products in the cDNA-AFLP fingerprint in comparison with the sequences found in the genomic clones. The AseI site (bold), present in all cases, is shown above the genomic clones. The dotted line in TDF1041 denotes a region of reduced sequence homology to the genomic sequences. In the genomic clone StnsLTP.2, an insertion of 331 bp is positioned above the common sequence. The exons of the StnsLTP genes are indicated by black blocks. B, I, (8% [w/v] Suc) Ten separate time points of tuber development (days) based on templates isolated from in vitro explants grown under tuber inducing conditions. The arrows indicate the location of the respective TDF; II, (8% [w/v] Suc + gibberellin [GA]) The same developmental period as in I, however, GA was added to the growth medium, suppressing tuber formation; III, Dormancy and sprouting, and the numbers refer to time points in weeks after harvest. The difference between the samples taken at weeks 22 and 22* is that the latter was harvested from a batch that had started sprouting for 1 to 2 weeks, whereas sample 22 had no sprouts at all. IV, RNA fingerprints generated from different potato plant tissues as indicated.
The expression pattern visualized with cDNA-AFLP showed an increase in gene expression linked to new organ development such as tuber and sprout (Fig. 1B, I and III, respectively). Using a growth medium that inhibits tuber formation (Fig. 1B, II) the nsLTP TDFs showed altered expression patterns in comparison with the tuberization medium and compared with each other (Fig. 1B). On tuberization medium, all three TDFs showed increased expression from d 3. TDF1016 shows expression throughout the 10 d, whereas TDF1023 and TDF1041 show a sharp declining expression from d 5 (Fig. 1B, I). In tuberization inhibition medium, the expression related to TDF1016 is induced a day earlier (d 2) and it continues to increase over the 10 d. In contrast, TDFs 1023 and 1041 show very low levels of transcription on the tuberization inhibition medium (Fig. 1B, II). During the early part of dormancy, TDFs 1016 and 1041 appear to be transcribed weakly and their transcription increases shortly before sprouting (Fig. 1B, III, week 20). When tested in different tissues, all the nsLTP TDFs showed a highly organ-specific expression profile that was detectable mainly in the stolon and sprout samples (Fig. 1B, IV).
Expression Studies
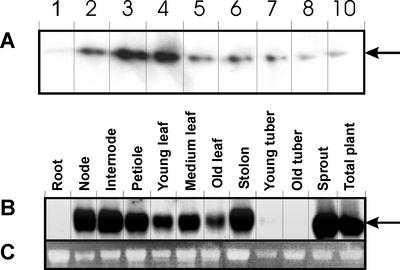
To examine the overall expression pattern of potato nsLTP genes and to confirm the results of the cDNA-AFLP experiments, northern-blot analysis was carried out. Total RNA was isolated from in vitro-grown tubers harvested in daily intervals (1–10 d, as used for the cDNA-AFLP method). A 312-bp-long fragment was used as a probe from the coding region of the genomic clone StnsLTP.2 described below. Figure 2A shows that potato nsLTP genes are expressed weakly on d 1, show the highest expression on d 3 through 4, and their expression declines thereafter.
Figure 2.
Northern analysis of StnsLTP gene expression during tuber development and in different potato tissues. A, Total RNA was isolated from the same 10 developmental stages of tuber development as shown in Figure 1. B, Expression of StnsLTP in different potato tissues. The 312-bp fragment of the StnsLTP.2 genomic clone representing the coding region (amplified as described in “Materials and Methods”) was used as a probe for hybridization in A and B. The arrow indicates the transcript(s) of potato StnsLTP genes. C, Transcript level of 26S RNA of samples used in the northern analysis of the tissues.
To be able to detect the tissue specificity of nsLTP genes in potato, a similar analysis was carried out for different tissues representing RNA isolated from root, stem, leaf, tuber, stolon, and sprout. As shown in Figure 2B, no expression can be detected in roots, whereas high expression is visible in the nodes, internodes, petioles, and leaf tissues. Tubers, stolons, and sprouts show nsLTP gene expression at different levels. A low amount of transcript was detectable in young and old tubers, whereas the nsLTP mRNAs were more abundant in sprout and stolon tissues.
Characterization of Two StnsLTP Genes and the Analysis of Homologous Expressed Sequence Tag (EST) Sequences
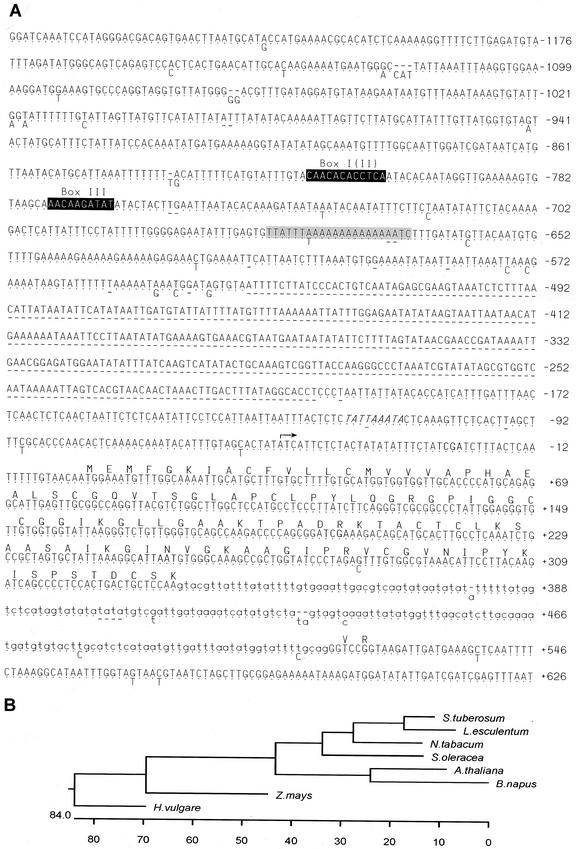
Using the TDF1041 as a probe to screen the genomic potato library, two clones (StnsLTP.2 and StnsLTP.12) were isolated and the nucleotide sequence was determined (Fig. 3A). According to the sequence data, both clones contain an open reading frame with a high degree of similarity to plant nsLTP genes. Figure 3B shows a phylogenetic tree of the potato StnsLTP.2-derived amino acid sequence compared with sequences from other species. The level of similarity ranges from 80% in other Solanaceous plants, decreasing to 44% in the Brassicacea, and to 42% in the monocot barley (Hordeum vulgare).
Figure 3.
DNA sequence and analysis of the two StnsLTP genes and their associated 5′-regulatory region. A, The sequence of the StnsLTP.2 clone is shown in full, with the dots below representing identical bases in the StnsLTP.12 clone. Where differences occur between the sequences, the appropriate base is shown below and deletions are shown as dashes. Similarities to consensus boxes in the nsLTP promoters are shown by inverse text, and the relevant name, referred to in the text, is shown above. Another conserved promoter box is shaded, and the suggested TATA-box is shown italicized. The arrow at base number −50 shows the putative transcriptional start site. The potential coding region of the StnsLTP gene is shown as an amino acid sequence above the nucleotide sequence and the intron is indicated in lowercase. All numbering is given relative to the site of initiation of translation. The restriction sites giving rise to the three TDFs are shown in boldface. B, Phylogenetic tree of derived amino acid sequences of various plant nsLTPs compared with the potato sequence (tomato, X56040; tobacco, X62395; S. oleracea, M58635; Arabidopsis, M80567; oilseed rape [Brassica napus], AJ245873; maize [Zea mays], P19656; and barley [Hordeum vulgare], AF109195).
In the coding and upstream regions, the two clones differ primarily in the length of the putative promoter region, as the StnsLTP.2 clone is 331 bp longer due to an insertion (Figs. 1A and 3A). The additional minor changes between the genomic clones and the restriction sites giving rise to the three TDFs are shown in Figure 3A. Although TDFs 1016 and 1023 are almost identical to the sequence of the genomic clones, the nucleotide sequence homology of TDF1041 begins at nucleotide 35 (Figs. 1A and 3), giving rise to a shorter open reading frame beginning at the third Met (position 43) in the StnsLTP clones (Fig. 3A).
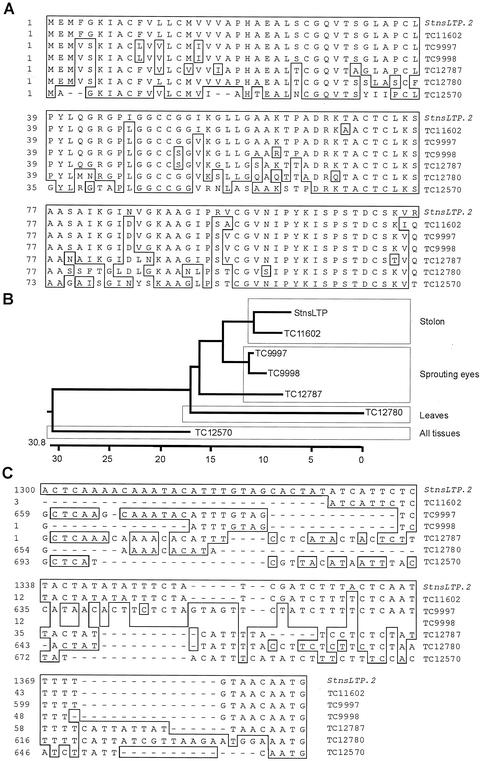
To be able to further characterize the genomic clones, a nucleotide sequence comparison with the potato database (Potato Gene Index, StGI, The Institute for Genomic Research, Rockville, MD) was carried out. Six highly homologous tentative consensuses (TCs) composed of potato ESTs were found. A comparison of the conceptual translations of the coding region from StnsLTP.2 and all TCs revealed that the TC11602 has the highest similarity to the StnsLTP.2 (Fig. 4A). Interestingly, TC11602 is composed of ESTs derived mainly from stolon RNA. The next three most similar TCs (TC9997, TC9998, and TC12787) could be grouped as originating from sprouting eyes of tubers. Closely related to the latter groups is TC12780 composed of leaf ESTs. The least related to the stolon group is TC12570, containing ESTs from all potato organs tested (leaves, Phytophthora infestans-challenged leaves, stolon, and sprouting eye). The relationship between the deduced StnsLTP.2 protein and the above mentioned TCs is shown in a phylogenetic tree (Fig. 4B). This analysis indicates that different classes of nsLTP genes exist in potato, and that these express more or less specifically throughout the plant.
Figure 4.
Analysis of relationships between different potato nsLTP sequences. A, Derived amino acid sequences of available potato EST compositions (TCs) and the genomic clone StnsLTP.2. Boxed amino acids represent those with 100% similarity to StnsLTP.2. B, Phylogenetic tree of the sequences shown in A. The boxed names are groups of sequences that have been shown to express primarily in the indicated tissues. C, Nucleotide sequence alignment of the same TCs and the genomic clone StnsLTP.2 in the 5′-untranslated region. The boxed residues show 100% homology to the StnsLTP.2 gene.
Characterization of Potential Regulatory Elements and Structure of the StnsLTP Genes
The 5′-untranslated regions of the StnsLTP.2 and StnsLTP.12 genomic sequences were compared with the TC sequences described above. TC11602 is composed of four EST sequences, three that are derived from swelling stolons and that begin at the same nucleotide in the 5′ region on the gene. The sequence of the StnsLTP genomic clones differed only in one nucleotide of the 5′-untranslated region of TC11602. The 5′ terminus of TC11602 maps at position −51 from the putative translational start site (+1) in the genomic clones. The sequence context around the adenine residue at position −51 (CTATATCAT) closely resembles the sequence (CTCATCAA), which has been identified as a consensus for transcriptional start sites in plant genes (Joshi, 1987). However, the other TCs with high levels of similarities in the coding region diverged significantly in their 5′-untranslated regions (Fig. 4C).
A putative TATA-box (−120 to −112 bp) was identified in the 5′-noncoding region of the potato genomic clones (Fig. 3A). A comparison with the eukaryotic promoter database (GenBank) revealed that certain regions of the putative StnsLTP promoter have homology to one or two particular promoter regions of a number of plant genes. It is interesting that one region (Fig. 3A, shaded box) of the putative StnsLTP promoters showed homology to six other plant promoters, including the promoters of a soybean (Glycine max) heat shock gene, several chalcone synthase genes, a potato proteinase inhibitor, and a tomato 1-aminocyclopropane-1-carboxylic acid synthase. In addition to this, known consensus sequences to nsLTP boxes were found (Thoma et al., 1994); a particularly good match to Box I is located between positions −816 and −805 and to Box III between positions −775 and −766. A potential Box II could be identified overlapping with Box I (Fig. 3A).
The homology between the genomic clones and TC11602 breaks down at position 336 (CAA/gta; Fig. 3A) and returns 183 bp later at position 516 (cag/GGT). The additional homology of six nucleotide residues covers a coding region of two additional amino acids, which is followed by two consecutive stop codons in the StnsLTP genes. This 2-exon structural organization resembles that of other nsLTP genes (Figs. 3A and 1A; Kader, 1996). Based on this data, the StnsLTP.2 and 12 genes encode a polypeptide of 114 amino acids (11.4 kD) in which two conservative amino acid changes and five nonconservative substitutions occur, in comparison with the translated product encoded by TC11602. Overall, 36.0% of the predicted residues in the protein are hydrophobic. According to the hydropathy plot, hydrophobic amino acids are concentrated in the amino terminus, indicating the presence of a signal sequence in analogy to other plant nsLTPs. Of the 10 Cys residues, eight are highly conserved in the mature protein of all plant nsLTP that are thought to participate in the formation of disulfide bonds (Kader, 1996).
Transgenics
To analyze the tissue-specific nature and the developmental regulation of the StnsLTP genes, the two 5′-untranslated regions (1.2 and 0.9 kb long) were amplified by PCR and cloned into the pMP2490 vector as transcriptional fusions with the β-glucuronidase (gus)A::intr/green fluorescent protein (gfp) genes. The constructs were introduced into potato cultivar Karnico via Agrobacterium tumefaciens cocultivation (Visser, 1991). For the StnsLTP.2 and StnsLTP.12 constructs, 21 and 22 kanamycin-resistant lines were regenerated, respectively. These lines were further propagated and grown in the greenhouse. To analyze gusA gene expression under the control of the putative StnsLTP promoters, only tetraploid lines with normal phenotype were examined further. Thus, for StnsLTP.2 and StnsLTP.12, 19 and 16 independent lines were analyzed, respectively. Two independent transgenic lines (StnsLTP.2-19 and -16) with higher activity than other lines and the control were chosen for further detailed analysis. Because these two showed the same expression pattern, the data obtained from the line StnsLTP.2-19 is presented below. Southern analysis of genomic DNA from these lines showed that line StnsLTP.2-19 carries around three to four copies of the transgene, whereas StnsLTP.2-16 had one to two copies (data not shown). Among the 16 transgenic lines carrying the StnsLTP.12 promoter-gusA::intr/gfp fusion, none of the transformants showed a similarly high level of expression compared with the StnsLTP.2-19 line, although all 16 lines did harbor one or more copies of the transgene (data not shown). Several lines did show a faint blue coloration after staining in all analyzed tissues, which could, however, not be localized to any particular tissue or developmental stage. This result indicates that the 331-bp insertion present in StnsLTP.2 and absent in StnsLTP.12 controls the tissue specificity in the corresponding gene.
Detailed Histological Analysis of GUS Activity in Transgenic Lines
In the transgenic line harboring the StnsLTP.2 promoter-gusA::intr/gfp fusion, GUS activity was detectable throughout the growth of the stem. In the first internodal/nodal region of the plants grown from tubers, GUS activity is visible in the whole ring of the vascular bundles (Fig. 5A). In mature plants (1–3 months), GUS activity is restricted to the three major bicollateral vascular bundles in the internodal region (Fig. 5B). The intensity of staining depends on the age and location of the examined section. The activity was most prominent in the vascular ring of the young nodes, close to the apex, and the intensity of blue staining decreased, becoming more restricted to main vascular bundles in the lower parts of the plant. In plant tissues entering senescence where the nodes have necrotic leaves or abscission wounds and somewhat lignified stems, no expression whatsoever was visible (data not shown).
Figure 5.
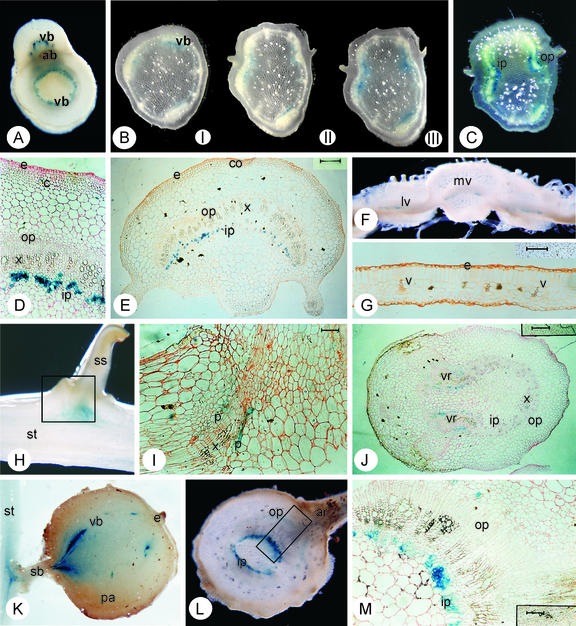
Spatial expression of the GUS gene expressed under the control of the StnsLTP.2 promoter. A, Hand-cut transverse section through the third node (from the base) of a 2-week-old plant showing the vascular ring (vr) of the stem and the emerging petiole showing blue staining in the inner phloem. The axillary bud (ab) is seen adjacent to the petiole. B, Serial hand-cut transverse sections between the fifth and fourth node (from the apical node) of a 2.5-month-old plant. Section I is taken from just above the fifth node and sections II and III show sections taken at 3-mm consecutive intervals toward the node. Section III shows the appearance of the petiolar vascular traces. In all sections, the inner phloem shows blue staining. C, Hand-cut transverse sections of a 3.5-month-old plant just below the fourth node (from the apex). The staining of inner and outer phloem is visible (For A through C, sections of around 0.5 mm were taken and the diameter of the stems was 4 to 6 mm). D, Eight-micrometer transverse section through the fourth node (from the apex) from a 1.5-month-old plant. The epidermis (e), cortex (c), outer phloem (op), xylem (x), and inner phloem (ip) are indicated (Bar = 125 μm). E, Twenty-micrometer transverse section of a petiole from the fourth node from a 1.5-month-old plant showing blue staining in the inner phloem. The epidermis (e), collenchyma (co), outer phloem (ip), xylem (x), and outer phloem (op) are indicated (Bar = 400 μm). F, Hand-cut transverse section of a leaf (1 mm) showing the mid-rib with the mid-vein (mv) and adjacent lamina with lateral veins (lv). Blue staining is visible in the inner phloem of the vascular tissues. G, Twenty-micrometer section of the same leaf lamina as F showing the epidermis (e) and the peripheral leaf veins (v; Bar = 125 μm). H, Longitudinal hand-cut section of a stolon (st) to secondary stolon (ss) junction. The rectangle indicates the position of section I. I, Twenty-micrometer section of a stolon junction. Staining is visible in the emerging vascular ring of the secondary stolon. Phloem (p) and xylem (x) cell layers are shown (Bar = 125 μm). J, Twenty-micrometer transverse section of a stolon with emerging secondary stolons. Staining is visible in the vascular ring (vr). Inner phloem (ip), outer phloem (op), and xylem (x) are also indicated (Bar = 400 μm). K, Longitudinal hand-cut section of a tuber attached to the stolon. The staining runs through the vascular bundles (vb). The stolon (st), stolon branch (sb), the terminal eye (e), and the parenchyma (pa) are indicated. The tuber was about 10 mm in diameter. L, Hand-cut transverse section of a (tuber) sprout with emerging adventitious root (ar). The stained inner phloem (ip) and the outer phloem (op) are indicated. The rectangle shows the location of the section shown in M. The sprout was around 5 mm in diameter. M, Twenty-micrometer transverse section of a sprout showing the internal and external phloem (ip and op, respectively) where the staining is located (Bar = 125 μm).
The intensity of GUS activity can be observed to change over serial node-to-node section. The lowest GUS activity is detected just above the node from which the activity increases toward the next node upwards, particularly in the vascular bundles (leaf traces) that lead through the petiole to the leaves (Fig. 5, B and C). After the branch point of the node, the vascular bundles in the stem harbor only a trace of the activity before the branch point, whereas the activity of the vascular bundles in the petiole is comparable with its counterparts in the nodes. This activity continues into the main vein and in the primary and secondary veins in the leaves (Fig. 5F), but with decreasing activity toward the leaf peak and the lamina edges. No activity can be seen in the minor veins (Fig. 5G). In a similar manner, coloration is not detectable in the dormant axillary bud (Fig. 5A). However, when development of a side shoot begins from an axillary bud, GUS activity reappears in the vascular tissues (data not shown).
The GUS activity of the stem vascular bundles is also detectable in the underground counterpart of the stem: the stolon, tuber, and sprout. In addition to the staining of the vascular bundles throughout the stolon, strong blue staining is also visible at the stolon branches (Fig. 5, H–J) and the stolon-tuber junctions (Fig. 5K). Freshly harvested and stored tubers independent of their age show GUS activity in their vascular bundles. In dormant tubers, the vascular bundles that lead to the eyes (axillary buds) also show a stronger blue coloration compared with pith cells. In line with the expression patterns in the stem side shoots, the newly developed sprouts of germinating tubers show high GUS activity in their vascular system (Fig. 5L).
Solanaceous plants have a bicollateral vascular system in which one part of the phloem occurs on the outside (external or abaxial) and another on the inside (internal or adaxial) of the xylem. To distinguish in which tissue of the vascular bundle the gusA gene is active, plastic-embedded material was used to gain 8- and/or 20-μm sections. As Figure 5 shows, in young internodes/nodes (Fig. 5D), petioles (Fig. 5E), and sprouts (Fig. 5M), primarily the internal phloem shows GUS activity, whereas at later stages, weak activity is detectable in the outer phloem as well (Fig. 5, C and M). No activity was detected in root or flower tissues, and no background GUS activity was detected in any tissues of the untransformed potato plants treated with 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (data not shown).
In Situ Hybridization
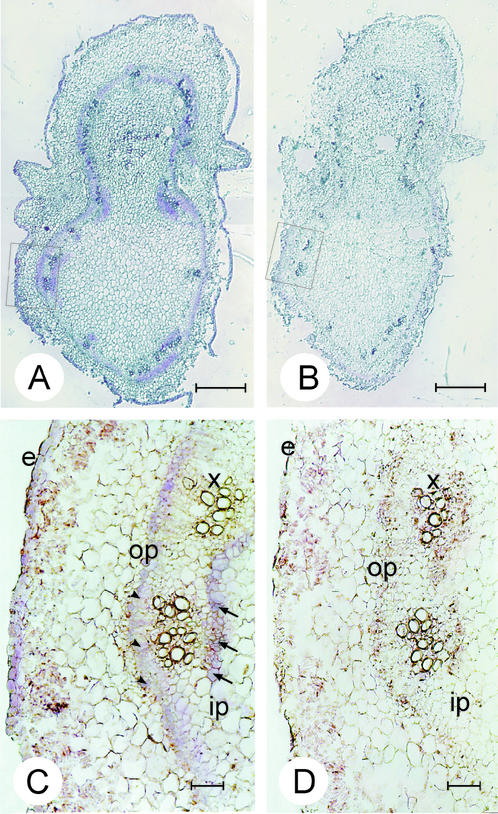
To visualize StnsLTP gene transcription in the vascular bundles, in situ hybridization was carried out to detect StnsLTP transcripts in this tissue. As mentioned above, expression studies indicated that nodes with petioles showed well-defined GUS activity, thus 12-μm consecutive cross sections were taken from the fourth node of 1.5-month-old plants. The sections were hybridized with a sense and antisense nsLTP.2 RNA as described in “Materials and Methods.” Figure 6A shows the entire cross section of the node, with vascular ring showing hybridization to the antisense RNA. A hybridization signal is also detectable in the epidermis above the background. When hybridization is carried out with the sense RNA, no signal is detected in any tissue (Fig. 6B). At higher magnification, hybridization can be seen in cells of the inner phloem (Fig. 6C, ip) and outer phloem (Fig. 6C, op). Signal is also detectable in the epidermal cells (Fig. 6C, e). No other tissue shows hybridization signal above background when compared with the sense control (Fig. 6D).
Figure 6.
Spatial expression of potato nsLTP genes in stem tissues of potato visualized by in situ hybridization. Consecutive 12-μm transverse sections of the fourth node were hybridized with the antisense (A and C) and sense (B and D) in vitro-synthesized StnsLTP.2 RNA. In sections A and B, the bar = 1,000 μm and in sections C and D, the bar = 100 μm. The arrowheads point out hybridization signal in the outer phloem cells, and the arrows indicate signal in the inner phloem cells. e, Epidermis; op, outer phloem; ip, inner phloem; x, xylem.
DISCUSSION
In this paper, we describe the isolation and characterization of a number of potato nsLTP genes with particular regard to their expression. We show that a wide variety of nsLTP genes are expressed in potato plants and that some of these have a high degree of tissue specificity and express at key stages of potato tuber life cycle. The expression studies carried out on the two potato nsLTP promoters demonstrate that the highly homologous nsLTP genes are regulated in a strikingly different manner and that, in one case, the expression appears to be primarily in the inner phloem adjacent to stem and stolon nodes.
There has been much speculation about the in vivo function of LTPs in plants (Kader, 1996). From the in vitro activity of binding cutin monomers and from the high levels of expression in the epidermis, it was suggested that plant LTPs may be primarily responsible for the production of the epicuticular wax layer (Sterk et al., 1991), although direct evidence for this function is not available. Furthermore, the expression in root tissues—devoid of cuticle—and the lack of response to wounding appears to contradict this as their only function (Sohal et al., 1999). Our data localizing StnsLTP.2 promoter-driven gene expression to the phloem also makes the direct participation of the nsLTP.2 gene product in epicuticular wax production unlikely. The antimicrobial action of nsLTPs has also been demonstrated in vitro and in vivo (Cammue et al., 1995). Also, fungal (Garcia-Olmedo et al., 1995) and viral infection (Sohal et al., 1999) induces the expression of nsLTP genes. Such an antipathogenic role could also be envisaged in potato, particularly as it is known that potato produces a range of different prophylactic defense proteins in tubers. Likewise, plants are known to have large amounts of functionally similar proteins in their phloem sap (Yoo et al., 2000). Potato plants expressing antisense StnsLTP.2 RNA under control of their own promoter did not help resolve the question of the function of plant nsLTP proteins in vivo because no observable phenotype different from untransformed plants was found under the conditions tested (data not shown). These results indicate that other StnsLTP proteins can probably complement the function of the StnsLTP.2 protein.
However, the expression of potato nsLTP genes indicates an apparently important role in new organ development, for example, during potato tuber life cycle. The expression profile of the three highly related TDFs displayed by cDNA-AFLP shows a short induction in the stolon just prior to tuber formation and again an increased expression at sprout development at the end of the dormancy period. The expression profile revealed by the northern analysis can be viewed as the sum of all potato nsLTP gene expression as partly displayed by cDNA-AFLP for temporal and spatial expression patterns. High GUS enzyme activity is detected when the gusA::int gene is under the control of the StnsLTP.2 promoter during stolon and sprout formation and in the branch points of stem nodes. The latter spatial expression of the StnsLTP.2 gene is further corroborated by the in situ hybridization data. A relatively high histochemical staining is also observed in the tubers, which is limited, however, to the vascular bundles surrounding the eyes. This spatial restriction in expression helps to explain the low signals found in northern analysis of whole tubers.
Of particular biological interest is the fact that TDFs 1041 and 1023 do not show induction in the absence of tuber formation (on medium with GA), indicating some process specificity in expression for tuberization. In contrast, TDF1016 shows a marked increase in expression when on tuber inhibition medium containing GA. This expression pattern is similar to the one described for the Stgan gene that was shown to affect potato stem growth, including stolon and sprout length (Bachem et al., 2001). It remains unclear whether the induction of gene expression corresponding to TDF1016 is a direct affect of GA or a secondary affect arising from the change in developmental program. However, it is becoming evident that GA is an important hormone for the regulation of tuberization and tuber life cycle in general (Carrera et al., 1999; Bachem et al., 2001).
A wider analysis of potato nsLTP gene expression, including data from the recently established potato EST databases, also indicates a role of highly related nsLTP genes in the early part of tuber life cycle as well as toward the end of dormancy and during sprouting by the exclusivity and/or abundance in cDNA banks from a particular organs. Other less related nsLTP ESTs from the database showed a different spatial expression profile, appearing with greater abundance in nontuber-related tissues such as leaves. The phylogenetic grouping of nsLTP sequences correlates closely with the spatial expression pattern and indicates that the StnsLTP clones described here are of the “stolon type.” It cannot be excluded that some of the TCs show a grouping due to the differences in potato varieties used for the RNA production (var. Bintje and Kennebec); however, because several of the TCs contain ESTs from both varieties, it makes this possibility less likely.
Due to the small size, particularly of TDFs 1016 and 1023, it remains difficult to ascribe the genomic clones directly to one of the isolated TDFs. However, the sequence identity of TDF1016 to both genomic clones suggests that this TDF is likely to correspond to the StsnLTP.2 gene and to TC11602, based on the derived expression profile. As the first 45 nucleotides of TDF1041 are radically different from the equivalent StnsLTP sequences, the gene corresponding to this TDF is likely to be of a different type. Because the first 20 to 25 amino acid residues of nsLTPs generally code for transit peptides (Kotilainen et al., 1994), the mature TDF1041-type protein may have a different cellular location.
The sequence analysis of both StnsLTP promoters showed similarities to sequences necessary for transcription initiation and to conserved boxes typical for plant nsLTPs genes. We conclude that the spatial and temporal gene regulation conferred by the StnsLTP.2 promoter is likely to be due to the 331-bp insertion, absent in the StnsLTP.12 promoter and lacking this expression specificity. It is interesting to note that the sequence unique to StnsLTP.2 carries no known cis-acting elements for plant nsLTP genes or for general promoter functions. Further work will be necessary to determine the exact nature of this putative regulatory element and other factors that interact with the sequence to give rise to the observed tissue specificity of expression.
MATERIALS AND METHODS
Isolation of nsLTP Genomic Clones
The StnsLTP genomic clones were isolated by screening a genomic library constructed from potato (Solanum tuberosum var. Desiree) kindly provided by Prof. Uwe Sonnewald (Institut fur Pflanzengenetik und Kulturpflanzenforschung, Gathersleben, Germany). This library was constructed by ligating Sau3A partially digested total DNA into the BamHI site of the pBK-cytomegalovirus phagemid of the λ-ZAP Express vector system. Using the probe TDF1041, two positive clones (StnsLTP.2 and StnsLTP.12) were isolated and as the restriction enzyme digestion patterns were different, both clones were sequenced on both strands using vector- and gene-specific primers.
In Vitro Tuberization Plant Material for the RNA Fingerprinting Technique
In vitro-grown axillary bud products of nodal cuttings from potato (cv Bintje) were grown on tuber induction medium (8% [w/v] Suc) and on a medium inhibiting tuber induction and growth (8% [w/v] Suc + 5 μm GA4+7), and were then harvested daily as described earlier (Bachem et al., 1996; Xu et al., 1998). The microtubers used for production of the dormancy template were taken from the same synchronized tuberization system. Tuber samples were collected during dormancy on a weekly basis. Total RNA, poly(A)+ RNA isolations, and template preparation were carried out as described by Bachem et al. (1996, 1998) using AseI and TaqI restriction enzymes. To display the three TDFs described here, primers with GC-selective extensions on the AseI end were used for all three cases and AA (TDF1016), CA (TDF1023), and TA (TDF1041) extensions were used on the TaqI end.
Plant Material, RNA Isolation, and Northern Analysis
Potato plants grown from seed tubers and under standard greenhouse conditions (16 h of light at 20°C and 8 h of dark at 18°C) were harvested for RNA isolation. Leaves were harvested from plants at the 11- to 12-leaf stage (1.5-month-old plants), whereas for other tissues, 3-month-old plants were used. For the “young leaf” sample, emerging sink leaves were collected, which were not wider then 7 to 8 mm, whereas for the “medium leaf” samples, sink leaves were a maximum of 1.5 cm in width. For the “old leaf” samples, fully expanded source leaves were used. For the internode samples, only stem tissues were used, whereas the nodes with the axillary buds were analyzed separately. The petioles were 5 to 6 cm long from which the top emerging leaves were cut off. For the root samples, actively growing roots were taken.
For plant material involved in tuberization, the following samples were collected: “stolons” with no swellings or only just emerging swellings; “young tuber,” tubers smaller than 5 mm in diameter; and “old tubers,” stored tubers larger than 10 mm in diameter. Etiolated sprouts were harvested at 4 to 6 cm long where terminal leaves were removed. The sprouts were collected from stored tubers that had been potted into moist compost and allowed to sprout in the dark.
Total RNA was isolated as described previously (Bachem et al., 1998). Equal amounts of total RNA (40 μg) were separated on 1.5% (w/v) agarose-formaldehyde gels and blotted onto Hybond N nylon filters (Amersham, Buckinghamshire, UK). Filters were hybridized at 65°C and 2× SSC with the 312-bp long amplified fragment of the StnsLTP.2 genomic clone using oligonucleotides 5′-[C−21TTTACTCAATTT−8]-3′ and 5′-[G291CCACAAACTCTAG278]-3′ for the amplification (base numbering shown in subscript refers to numbering in Fig. 3A).
DNA Constructs
For construction of the StnsLTP promoter-reporter gene transcriptional fusions, the pMP2490 plasmid (kindly provided by Prof. Herman P. Spaink, Leiden, The Netherlands) was used. This vector is a promoterless derivative of the pMP2482 construct (Quaedvlieg et al., 1998), which contains the gusA::intr and the gfp reporter genes as a translational fusion.
Using the synthetic oligonucleotides 5′-[GGATCCA−1244TAGGGACGACAG−1232]-3′ and 5′-[GGATCCA−8AAATTGAGTAAAGATCGA−21]-3′ partially complementary to the upstream and downstream regions of the putative promoter in the StnsLTP.2 clone, a 1,269-bp fragment was produced (base numbering shown in subscript refers to numbering in Fig. 3A). Using the same primers on the StnsLTP.12 clone as a template, a 931-bp-long DNA fragment could be amplified. Both amplification products were initially cloned into the pGEMT-Easy vector (Promega, Leiden, The Netherlands). Using the introduced BamHI site into the synthetic oligonucleotides at the termini of the amplified region, the inserts were cut out of the pGEMT-Easy vector and were recloned into pMP2490. The integrity of the amplified promoter region and junction between the promoter and the reporter gene were confirmed by sequence determination.
Generation and Analysis of the Transgenic Lines
Internodal stem sections from in vitro-grown plants of potato cv Karnico (tetraploid) were used for transformation by Agrobacterium tumefaciens cocultivation (Visser, 1991). Each transgenic line was controlled for ploidy level via flow cytometric analysis (Plant Cytometry Services, Scheindel, The Netherlands). For histochemical analysis of the tetraploid transgenic lines, two types of plant material were used. For initial stages of tuber life cycle, sections were made from plants originating from in vitro-grown material, whereas for the analysis of dormant tubers, stored greenhouse material was used. To analyze expression in sprouts, young internodes, and nodes, plant material originating from seed tubers was used. To study expression patterns in other tissues, tissue culture and greenhouse plant material were used.
All transgenic lines were histochemically screened for GUS activity. The first, fifth, 10th, 20th, and second to last internode-node region (nodes counted from the apex) were analyzed. Also, young and old leaf, root, and young tuber samples were tested. In the case of positive lines, every node, internode, as well as stolon, dormant tuber, sprout, and flower sample was examined. Hand-cut sections of 0.5- to 1-mm thickness were infiltrated under low pressure, first with phosphate buffer (50 mm Na2HPO4 and 50 mm KH2PO4, pH 7.0), and then stained with phosphate buffer containing 1 mg mL−1 5-bromo-4-chloro-3-indolyl-β-d-glucuronide, 10 mm EDTA, 5 mm K4Fe(CN)6, and 5 mm K3Fe(CN)6, both for at least 30 min. The plant material was subsequently stained for 4 to 16 h at 37°C in the dark. After staining, tissues were screened for the appearance of blue color. Following this, the tissues were rinsed in 70% (v/v) ethanol and photographed, or were fixed in phosphate buffer containing 0.25% (v/v) glutaraldehyde and 4% (w/v) paraformaldehyde overnight at 4°C with rotation. After washing and dehydration, the sections were embedded in Historesin (Cambridge Instruments, Heidelberg) using a 5:1, 3:1, 1:1, 1:3, 1:5, and 0:1 96% (v/v) ethanol:historesin series at 4°C for at least 1 h in each solution. The additional steps were carried out according to the manufacturer's recommendations. Sections of 8 and 20 μm were taken using a rotary microtome (model 2055; Leica-Jung, Rijswijk, The Netherlands). Additional staining was carried out in ruthenium red. No quantitative assay to measure GUS activity was undertaken, as the activity was localized in a very limited number of cells.
Similar assays were carried out to test GFP expression. However, no significant activity was found in any potato tissues tested due to high levels of autofluorescence. Only the control construct (35S-cauliflower mosaic virus promoter-driven gusA::intr-gfp) showed GFP-related fluorescence above background level (data not shown).
In Situ Hybridization
RNA in situ hybridizations were performed as described by Cox and Goldberg (1988) except for the labeling of the RNA probes. This was carried out according to manufacturer recommendations with dioxigenin-11-rUTP using a nucleic acid kit (Roche Molecular Biochemicals, Summerville, NJ). Fourth and fifth nodes from 1.5-month-old potato plants were fixed in 4% (w/v) paraformaldehyde, 0.25% (v/v) glutaraldehyde, and 100 mm NaCl for 90 min under vacuum at room temperature. The samples were dehydrated and embedded in paraffin (Para Clean; Klinipath, Duiven, The Netherlands). Transversal sections of 12-μm thickness were cut and transferred to glass slides. Paraffin was removed with xylene and the tissues were step-wise rehydrated. After proteinase-K treatment, the sections were hybridized with 150 to 300 ng of probe. To synthesize RNA probes (sense and antisense), PCR was performed using clone STnsLTP.2 as template with the primers 5′-[A7TGTTTGGCAAAATTGC23]-3′ and 5′-[G333GAGCAGTCAGTGGAGG317]-3′. The PCR product was then cloned in both orientations in pGEMT-Easy vector. Sense and antisense probes have been synthesized with primer T7 after digestion with enzyme SalI. In all in situ experiments, the sense and antisense 265 rRNA probes were used as negative and positive controls, respectively.
Hybridization was done in the dark for 20 h at 42°C, after which the tissues were washed with 4× SSC. Excess probe was removed by treatment of the tissues with RNase-A. After incubation of the sections in blocking solution (100 mm Tris-HCl, pH 7.5, 2% [w/v] bovine serum albumin [fraction V], and 1% [v/v] Triton X-100) for 30 min, the sections were treated with antidigoxygenin antigen binding fragment alkaline phosphate conjugate for 1 h at room temperature. The sections were washed three times in 100 mm Tris-HCl (pH 7.5) and were subsequently incubated in the dark at room temperature in reaction buffer (100 mm Tris-HCl, pH 9.5, 100 mm NaCl, and 5 mm MgCl2) containing 0.45 mg mL−1 nitroblue tetrazolium chloride and 0.175 mg mL−1 5-bromo-4-chloro-3-indolyl phosphate. A positive signal appeared after 20 h as purple staining. The reaction was stopped by washing the sections with 10 mm Tris-HCl (pH 8.0) and 1 mm EDTA for 5 min at room temperature, followed by two washes in distilled water. The slides were finally mounted in glycergel (DAKO Glycergel; Mounting Medium, Uithoorn, The Netherlands).
Image and Data Processing
The images presented were taken using cameras (Agfa Ultra, 180°/50ASA; Zeiss, Jena, Germany) mounted on a microscope (Axiophot; Zeiss). Photographic and autoradiographic images were scanned and processed for printing using the CorelDraw 9.0 program (Corel, Ottawa, Ontario, Canada).
Nucleotide and amino acid sequence comparisons were carried out with the Megalign program (DNA-STAR, London) for multiple alignment (gap penalty, 10; gap length penalty, 10 for amino acid sequences and gap penalty, 20; gap length penalty, 2 for nucleic acid sequence).
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
ACKNOWLEDGMENTS
We thank Dr. Niek Appeldoorn for fruitful discussions and critical reading of the manuscript. We also thank Marjan Bergervoet and Dirkjan Huigen for their excellent technical assistance with plant transformation as well as care and analysis of the transgenic plants.
Footnotes
This research was supported in part by the Technology Foundation Stichting Technische Wetenschappen (grant no. WBI 4923).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.004762.
LITERATURE CITED
- Bachem CWB, Horvath BM, Trindade L, Claassens M, Davelaar E, Jordi W, Visser RGF. A potato tuber expressed mRNA with homology to steroid dehydrogenases affects gibberellin levels and plant development. Plant J. 2001;25:595–604. doi: 10.1046/j.1365-313x.2001.00993.x. [DOI] [PubMed] [Google Scholar]
- Bachem CWB, Oomen RJFJ, Visser RGF. Transcript imaging with cDNA-AFLP: a step-by-step protocol. Plant Mol Biol Rep. 1998;16:157–173. [Google Scholar]
- Bachem CWB, van der Hoeven RS, de Bruijn SM, Vreugdenhil D, Zabeau M, Visser R. Visualisation of differential gene expression using a novel method of RNA finger-printing based on AFLP: analysis of gene expression during potato tuber development. Plant J. 1996;9:745–753. doi: 10.1046/j.1365-313x.1996.9050745.x. [DOI] [PubMed] [Google Scholar]
- Bachem CWB, van der Hoeven RS, Lucker J, Oomen RJFJ, Casarini E, Jacobsen E, Visser RGF. Functional genomic analysis of potato tuber life-cycle. Potato Res. 2000;43:297–312. [Google Scholar]
- Bernhard WR, Thoma RS, Botella J, Somerville CR. Isolation of a cDNA clone for spinach lipid transfer protein and evidence that the protein is synthesized by the secretory pathway. Plant Physiol. 1991;95:164–170. doi: 10.1104/pp.95.1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammue BP, Thevissen K, Hendriks M, Eggermont K, Goderis IJ, Proost P, Van Damme J, Osborn RW, Guerbette F, Kader JC. A potent antimicrobial protein from onion seeds showing sequence homology to plant lipid transfer proteins. Plant Physiol. 1995;109:445–455. doi: 10.1104/pp.109.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canevascini S, Caderas D, Mandel T, Fleming AJ, Dupuis I, Kuhlemeier C. Tissue-specific expression and promoter analysis of the tobacco Itp1 gene. Plant Physiol. 1996;112:513–524. doi: 10.1104/pp.112.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera E, Jackson SD, Prat S. Feedback control and diurnal regulation of gibberellin 20-oxidase transcript levels in potato. Plant Physiol. 1999;119:765–774. doi: 10.1104/pp.119.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AM, Bohnert HJ. Cell-specific expression of genes of the lipid transfer protein family from Arabidopsis thaliana. Plant Cell Physiol. 1999;40:69–76. doi: 10.1093/oxfordjournals.pcp.a029476. [DOI] [PubMed] [Google Scholar]
- Cox KH, Goldberg RB. Analysis of plant gene expression. In: Shaw CH, editor. Analysis of Plant Gene Expression. Oxford: IRL Press; 1988. pp. 1–34. [Google Scholar]
- Cutter EG. Structure and Development of the Potato Plant. London: Chapman and Hall; 1978. [Google Scholar]
- Fleming AJ, Mandel T, Hofmann S, Sterk P, de Vries SC, Kuhlemeier C. Expression pattern of a tobacco lipid transfer protein gene within the shoot apex. Plant J. 1992;2:855–862. [PubMed] [Google Scholar]
- Garcia-Olmedo F, Molina A, Segura A, Moreno M. The defensive role of nonspecific lipid-transfer proteins in plants. Trends Microbiol. 1995;3:72–74. doi: 10.1016/s0966-842x(00)88879-4. [DOI] [PubMed] [Google Scholar]
- Hendriks T, Vreugdenhil D, Stiekema WJ. Patatin and four serine proteinase inhibitor genes are differentially expressed during potato tuber development. Plant Mol Biol. 1991;17:385–394. doi: 10.1007/BF00040633. [DOI] [PubMed] [Google Scholar]
- Joshi CP. An inspection of the domain between putative TATA box and translation start site in 79 plant genes. Nucleic Acids Res. 1987;15:6643–6653. doi: 10.1093/nar/15.16.6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kader JC. Lipid-transfer proteins in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:627–654. doi: 10.1146/annurev.arplant.47.1.627. [DOI] [PubMed] [Google Scholar]
- Kader JC. Lipid transfer proteins: a puzzling family of plant proteins. Trends Plant Sci. 1997;2:66–70. [Google Scholar]
- Kotilainen M, Helariutta Y, Elomaa P, Paulin L, Teeri TH. A corolla- and carpel-abundant, non-specific lipid transfer protein gene is expressed in the epidermis and parenchyma of Gerbera hybrida var. Regina (Compositae) Plant Mol Biol. 1994;26:971–978. doi: 10.1007/BF00028863. [DOI] [PubMed] [Google Scholar]
- Pyee J, Yu H, Kolattukudy PE. Identification of a lipid transfer protein as the major protein in the surface wax of broccoli (Brassica oleracea) leaves. Arch Biochem Biophys. 1994;311:460–468. doi: 10.1006/abbi.1994.1263. [DOI] [PubMed] [Google Scholar]
- Quaedvlieg NE, Schlaman HR, Admiraal PC, Wijting SE, Stougaard J, Spaink HP. Fusions between green fluorescent protein and β-glucuronidase as sensitive and vital bifunctional reporters in plants. Plant Mol Biol. 1998;38:861–873. doi: 10.1023/a:1006182623165. [DOI] [PubMed] [Google Scholar]
- Sanz MJ, Mingo Castel A, van Lammeren AAM, Vreugdenhil D. Changes in the microtubular cytoskeleton precede in vitro tuber formation in potato. Protoplasma. 1996;191:46–54. [Google Scholar]
- Sohal AK, Pallas JA, Jenkins GI. The promoter of a Brassica napus lipid transfer protein gene is active in a range of tissues and stimulated by light and viral infection in transgenic Arabidopsis. Plant Mol Biol. 1999;41:75–87. doi: 10.1023/a:1006232700835. [DOI] [PubMed] [Google Scholar]
- Song JY, Choi DW, Lee JS, Kwon YM, Kim SG. Cortical tissue-specific accumulation of the root-specific ns-LTP transcripts in the bean (Phaseolus vulgaris) seedlings. Plant Mol Biol. 1998;38:735–742. doi: 10.1023/a:1006008117795. [DOI] [PubMed] [Google Scholar]
- Sterk P, Booij H, Schellekens GA, Van Kammen A, De Vries SC. Cell-specific expression of the carrot EP2 lipid transfer protein gene. Plant Cell. 1991;3:907–921. doi: 10.1105/tpc.3.9.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma S, Hecht U, Kippers A, Botella J, De Vries S, Somerville C. Tissue-specific expression of a gene encoding a cell wall-localized lipid transfer protein from Arabidopsis. Plant Physiol. 1994;105:35–45. doi: 10.1104/pp.105.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma S, Kaneko Y, Somerville C. A non-specific lipid transfer protein from Arabidopsis is a cell wall protein. Plant J. 1993;3:427–436. doi: 10.1046/j.1365-313x.1993.t01-25-00999.x. [DOI] [PubMed] [Google Scholar]
- Trevino MB, O'Connell MA. Three drought-responsive members of the nonspecific lipid-transfer protein gene family in Lycopersicon pennellii show different developmental patterns of expression. Plant Physiol. 1998;116:1461–1468. doi: 10.1104/pp.116.4.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser R. Regeneration and transformation of potato by Agrobacterium tumefaciens. In: Lindsey K, editor. Plant Tissue Culture Manual: Fundamentals and Applications. B5. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1991. pp. 1–9. [Google Scholar]
- Vroemen CW, Langeveld S, Mayer U, Ripper G, Jurgens G, van Kammen A, de Vries SC. Pattern formation in the Arabidopsis embryo revealed by position-specific lipid transfer protein gene expression. Plant Cell. 1996;8:783–791. doi: 10.1105/tpc.8.5.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz KW. Phospholipid transfer proteins. Annu Rev Biochem. 1991;60:73–99. doi: 10.1146/annurev.bi.60.070191.000445. [DOI] [PubMed] [Google Scholar]
- Xu X, van Lammeren AAM, Vermeer E, Vreugdenhil D. The role of gibberellin, abscisic acid, and sucrose in the regulation of potato tuber formation in vitro. Plant Physiol. 1998;117:575–584. doi: 10.1104/pp.117.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo BC, Xiang Y, Campbell LR, Hull RJ, Xoconostle-Cazares B, Monze RJ, Lee JY, Ullman DE, Lucas WJ. Characterization of Cucurbita maxima phloem serpin-1 (CmPS-1): a developmentally regulated elastase inhibitor. J Biol Chem. 2000;275:35122–35128. doi: 10.1074/jbc.M006060200. [DOI] [PubMed] [Google Scholar]