Abstract
ADP-ribosylation factors (Arf), a family of small GTP-binding proteins, play important roles in intracellular trafficking in animal and yeast cells. Here, we investigated the roles of two Arf homologs, Arf1 and Arf3 of Arabidopsis, in intracellular trafficking in plant cells. We generated dominant negative mutant forms of Arf 1 and Arf3 and examined their effect on trafficking of reporter proteins in protoplasts. Arf1[T31N] inhibited trafficking of H+-ATPase:green fluorescent protein (GFP) and sialyltransferase (ST):GFP to the plasma membrane and the Golgi apparatus. In addition, Arf1[T31N] caused relocalization of the Golgi reporter protein ST:GFP to the endoplasmic reticulum (ER). In protoplasts expressing Arf1[T31N], ST:red fluorescent protein remained in the ER, whereas H+-ATPase:GFP was mistargeted to another organelle. Also, expression of Arf1[T31N] in protoplasts resulted in profound changes in the morphology of the ER. The treatment of protoplasts with brefeldin A had exactly the same effect as Arf1[T31N] on various intracellular trafficking pathways. In contrast, Arf3[T31N] did not affect trafficking of any of these reporter proteins. Inhibition experiments using mutants with various domains swapped between Arf1 and Arf3 revealed that the N-terminal domain is interchangeable for trafficking inhibition. However, in addition to the T31N mutation, motifs in domains II, III, and IV of Arf1 were necessary for inhibition of trafficking of H+-ATPase:GFP. Together, these results strongly suggest that Arf1 plays a role in the intracellular trafficking of cargo proteins in Arabidopsis, and that Arf1 functions through a brefeldin A-sensitive factor.
In eukaryotic cells, a large number of proteins are transported to their final destination after translation by a process called intracellular trafficking. The mechanism of intracellular trafficking has been extensively studied in animal, plant, and yeast cells. Based on numerous studies, the general mechanism of intracellular trafficking is thought to be similar in various organisms (Rothman, 1994; Jahn and Südhof, 1999; Bassham and Raikhel, 2000), implying that the fundamental aspects of intracellular trafficking between the endoplasmic reticulum (ER), Golgi apparatus, vacuole, and plasma membrane are similar in plant cells as well. In fact, many plant proteins have been shown to complement mutations in corresponding proteins in yeast cells (Bassham et al., 1995; Takeuchi et al., 1998). Also, proteins such as clathrin, coatomer subunits of coat protein I (COPI) vesicles, and many small GTP-binding proteins have been identified in plant cells (Memon et al., 1993; Regad et al., 1993; Lebas and Axelos, 1994; Blackbourn and Jackson, 1996; Contreras et al., 2000), although in most cases, the exact biological roles of these proteins have not been directly addressed. However, differences in trafficking also exist between plant and animal cells. It has recently been shown that plant cells have at least two different types of vacuoles: the lytic and storage vacuoles (Paris et al., 1996; Neuhaus and Rogers, 1998; Jauh et al., 1999). The lytic vacuole is thought to be similar to the vacuole of yeast cells or the lysosome of animal cells, whereas an organelle similar to the storage vacuole is absent in yeast and animal cells, implying that transportation of cargo molecules to the storage vacuole is likely unique to plant cells (Matsuoka et al., 1990; Bednarek and Raikhel, 1991; Neuhaus et al., 1991; Chrispeels and Raikhel, 1992; Schroeder et al., 1993; Saalbach et al., 1996; Frigerio et al., 1998). Interestingly, precursor-accumulating vesicles have been shown to transport proteins to the storage vacuole directly from the ER in pumpkin seeds (Hara-Nishimura et al., 1998). In addition, storage proteins such as vicilin have been shown to be sorted to the storage vacuole at the cis-Golgi (Hillmer et al., 2001). Although the presence of these unique pathways in plant cells has been demonstrated, proteins involved in these pathways are largely unknown.
To understand regulation of trafficking in plant cells, we investigated possible roles of ADP-ribosylation factors (Arfs), 20-kD guanine nucleotide-binding proteins that are members of the small ras-like GTPase superfamily. Numerous studies have demonstrated the pivotal role of Arfs on various steps of intracellular trafficking pathway in various organisms (Balch et al., 1992; Dascher and Balch, 1994; Gaynor et al., 1998; Goldberg, 1998; Pacheco-Rodriguez et al., 1998; Spang et al., 1998; Lanoix et al., 1999; Zhao et al., 1999; Jackson and Casanova, 2000). Among various steps of intracellular trafficking, Arf1 has been shown to be specifically involved in retrograde trafficking from the Golgi apparatus to the ER and from the trans-Golgi network to the endosome (Dascher and Balch, 1994; Ooi et al., 1998; Goldberg, 1999; Poon et al., 1999; Roth, 1999; Jackson and Casanova, 2000). During retrograde trafficking, Arf1 has been shown to be involved in the formation of COP1 vesicles (Wieland and Harter, 1999). Also, Arf1 has also been proposed to be involved in the sorting of cargo proteins (Lanoix et al., 1999; Goldberg, 2000). However, it is not known whether Arf plays a role in intracellular trafficking in plant cells. Highly homologous proteins such as Arabidopsis Arf1 and Arf3 have been found in plants (Regad et al., 1993; Memon et al., 1993; Lebas and Axelos, 1994). Also, the fact that brefeldin A (BFA) inhibits intracellular trafficking in plants in much the same way as in animal and yeast cells (Gomez and Chrispeels, 1993; Boevink et al., 1999) suggests that these Arf homologs may also play roles in intracellular trafficking. This notion was further supported by a recent report showing that Arf1 is localized at the Golgi apparatus (Pimpl et al., 2000), as in the case of animal cells (Balch et al., 1992; Dascher and Balch, 1994; Roth, 1999).
In this study, we asked whether Arabidopsis Arf proteins play any role in intracellular trafficking in vivo. To address this question in Arabidopsis, we co-expressed various green fluorescent protein (GFP) reporters and Arf mutants as cargoes and regulators of intracellular trafficking, respectively, and then examined the effect in protoplasts prepared from Arabidopsis leaf tissues.
Here, we present evidence that Arf1 plays a critical role in the trafficking of H+-ATPase and sialyltransferase (ST):GFP to the plasma membrane and the Golgi apparatus, respectively.
RESULTS
In Vivo Functional Assay for Intracellular Trafficking in Protoplasts
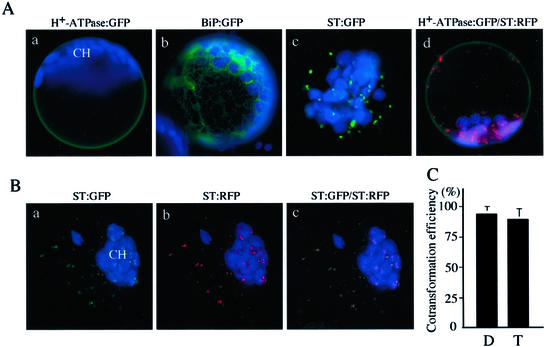
In this study, we developed a new functional assay for intracellular trafficking using protoplasts of Arabidopsis as a model system. The assay consists of two important components, reporter and effector proteins. To monitor intracellular trafficking in the protoplasts using a fluorescent microscope, we used reporter proteins tagged with GFP (Davis and Vierstra, 1998) or red fluorescent protein (RFP; Heikal et al., 2000). We have previously used H+-ATPase:GFP and ST:GFP as a reporter for trafficking to the plasma membrane and the Golgi apparatus, respectively (Kim et al., 2001). Also, we used a GFP fusion protein with chaperon BiP to use as a marker for the ER. As shown in Figure 1A, H+-ATPase:GFP was present at the plasma membrane (Fig. 1A, a and d), and BiP:GFP was present as numerous networks with blots (Fig. 1A, b). ST:GFP gave punctate staining patterns (Fig. 1A, c). Also, we generated similar fusion proteins with RFP—ST:RFP, BiP:RFP, and H+-ATPase:RFP—and compared localization of these RFP fusion proteins with that of corresponding GFP forms. As in the case of ST:GFP, ST:RFP gave punctate staining patterns (Fig. 1A, c and d). When we directly compared localization between ST:GFP and ST:RFP by cotransforming into protoplasts, the green and red punctate stains closely overlapped each other (Fig. 1B, a–c), indicating that both the green and red forms of ST fusion proteins are targeted to the Golgi apparatus. Also, an RFP fusion form of H+-ATPase, H+-ATPase:RFP, was targeted to the plasma membrane as in the case of H+-ATPase:GFP (data not shown). These results suggest that the tetramer formation of RFP (Heikal et al., 2000) per se may not affect localization of these reporter proteins. Another important issue we addressed was efficiency of cotransformation. In this assay, it is essential that both effector and reporter plasmids are expressed in the same protoplasts. Therefore, we first examined the efficiency of cotransformation using two reporter proteins. The efficiency of cotransformation was over 95% when two reporter proteins, BiP:RFP and H+-ATPase:GFP, were used (Fig. 1C). Even with three constructs, the cotransformation efficiency was over 90% (Fig. 1C). These results strongly suggest that the cotransformation approach is a very efficient method of introducing multiple plasmid DNAs into protoplasts.
Figure 1.
Establishment of an in vivo functional assay for intracellular trafficking in protoplasts. A, Targeting of various reporter proteins. Protoplasts were transformed with various reporter constructs and examined 12 to 48 h after transformation. Transformation efficiency varied from 5% to 40%. Green and red images are GFP and RFP signals, respectively. Autofluorescence of chlorophyll is depicted in blue. CH, Chloroplast. Bar = 20 μm. B, Colocalization of ST:GFP and ST:RFP. Two constructs, ST:GFP and ST:RFP, were introduced into protoplasts and localization of green and red fluorescent signals were examined 24 to 48 h after transformation. Note that the autofluorescence of chlorophyll is depicted in blue. CH, Chloroplast. Bar = 20 μm. C, Estimation of cotransformation efficiency. Equimolar amounts of plasmid DNAs (30 μg in total) were mixed and introduced into protoplasts. To estimate cotransformation efficiency, images of more than 200 transformed protoplasts were taken randomly from samples at 24 h after transformation. To avoid taking images from the same protoplasts more than once, no more than 50 images were captured from each slide. Also, these images were carefully compared to make sure they were not from the same protoplasts. Most of the protoplasts had unique patterns of chloroplasts and RFP or GFP signals, which allowed us to distinguish individual protoplasts. The images were stored in a computer and analyzed for double or triple transformation. Error bars indicate means ± sd (n = 3). D and T indicate protoplasts cotransformed with two plasmids (H+-ATPase:GFP plus BiP:RFP) and three plasmids (H+-ATPase:GFP plus BiP:RFP plus NLS:GFP; Pih et al., 2000), respectively.
Arf1[T31N] May Cause Fusion of the Golgi Membranes to the ER
To understand the biological roles of Arf1 and Arf3, we examined the effect of the GDP-binding dominant-negative mutants Arf1[T31N] and Arf3[T31N] on intracellular trafficking by expressing them transiently in protoplasts derived from leaf tissues of Arabidopsis. The dominant negative mutant form is thought to compete with the wild-type proteins for effector proteins such as guanine nucleotide exchange factor (GEF), thereby inhibiting the activity of the wild-type proteins. In animal cells, Arf1[T28N] causes relocation of Golgi-resident proteins to the ER through the disassembly of the Golgi apparatus (Dascher and Balch, 1994; Peters et al., 1995). We investigated whether Arabidopsis Arf1[T31N] can cause disassembly of the Golgi apparatus as in animal cells. To address this, we examined localization of Golgi-resident proteins in the presence of Arf1[T31N]. As a marker protein for the Golgi resident proteins, we decided to use ST:GFP (Wee et al., 1998; Jin et al., 2001; Kim et al., 2001).
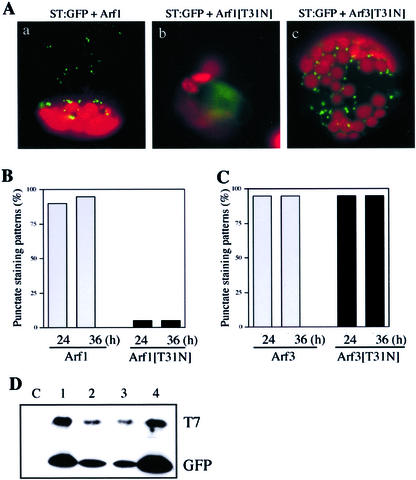
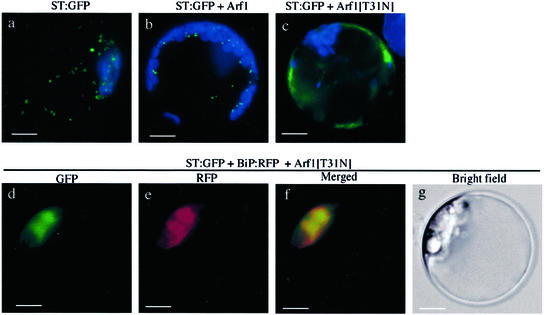
First, we used the transient expression approach by transforming protoplasts with ST:GFP together with Arf1[T31N]. As shown in Figure 2A, green fluorescent signals were present as punctate stains in the protoplasts expressing Arf1 (Fig. 2A, a; Kim et al., 2001), indicating that ST:GFP was localized at the Golgi apparatus. The percentage of transformed protoplasts with punctate stains was more than 95% (Fig. 2B). In contrast, in the presence of Arf1[T31N], diffuse GFP patterns (Fig. 2A, b) were observed in more than 90% of transformed protoplasts (Fig. 2B), indicating that Arf1[T31N] has a profound effect on the localization of ST:GFP or the morphology of the Golgi apparatus. However, Arf3[T31N] did not affect the pattern of ST:GFP (Fig. 2, A and C). To confirm that these Arf proteins were expressed in the protoplasts, we performed protein gel-blot analysis using proteins obtained from the transformed protoplasts. In this case, the wild-type and mutant forms of Arf1 and Arf3 were tagged with the T7 epitope (Zheng et al., 1999). As shown in Figure 2D, the wild-type and mutant forms of Arf1 and Arf3 were all expressed at high levels when detected using a monoclonal T7 antibody. The expression levels of all four Arf proteins appeared to be similar when the level was normalized based on the level of co-expressed GFP. This result strongly suggests that the differential effect of Arf1[T31N] and Arf3[T31N] on the localization of ST:GFP is not due to the difference at the level of these proteins but is likely caused by the functional difference of the two Arf isoforms.
Figure 2.
The effect of Arf1[T31N] on localization of transiently expressed ST:GFP in protoplasts. A, The effect of Arf1[T31N] on localization of ST:GFP. Protoplasts were transformed with various constructs and fluorescent images were obtained 12 to 48 h after transformation. Green and red images are GFP signals and autofluorescence of chlorophyll, respectively. B and C, The percentage of transformed protoplasts with punctate stains in the presence of Arf1 and Arf1[T31N] (B), and Arf3 and Arf3[T31N] (C). Protoplasts were transformed with ST:GFP plus Arf1 (B, Arf1), ST:GFP plus Arf1[T31N] (B, Arf1[T31N]), ST:GFP plus Arf3 (C, Arf3), or ST:GFP plus Arf3[T31N] (C, Arf3[T31N]). Images of more than 200 transformed protoplasts were analyzed as described in Figure 1. Images were analyzed by more than one person who did not know the experimental setting. D, Protein gel-blot analysis of T7-tagged Arf proteins. Total cellular extracts were prepared from protoplasts transformed with T7:Arf constructs. A GFP construct was cotransformed to estimate transformation efficiency. Protein extracts were analyzed by protein gel-blot analysis using a monoclonal anti-T7 antibody (T7) and a polyclonal anti-GFP antibody (GFP). 1, Arf1; 2, Arf1[T31N]; 3, Arf3; and 4, Arf3[T31N]. C, Protein extracts obtained from untransformed protoplasts.
Next, we investigated the localization of the green fluorescent signals of ST:GFP in the presence of Arf1[T31N]. To address this question, we performed colocalization of ST:GFP with BiP:RFP, the ER marker protein. In contrast to the control protoplasts (Fig. 3a), the diffuse green fluorescent signal of ST:GFP closely overlapped the red fluorescent signal of BiP:RFP in the presence of Arf1[T31N] (Fig. 3d), indicating that ST:GFP was localized at the ER. In addition, the morphology of the ER network was profoundly changed in the presence of Arf1[T31N], implying that Arf1[T31N] may also affect the morphology of the ER (see below and Fig. 10). However, this transient expression approach cannot clearly address the question of relocation of ST:GFP from the Golgi apparatus to the ER because trafficking of ST:GFP from the ER to the Golgi apparatus may be blocked at the ER by co-expressed Arf1[T31N]. Therefore, we carried out the relocation experiment using protoplasts obtained from transgenic plants expressing ST:GFP constitutively. As shown in Figure 4, ST:GFP in the transgenic plants was found as punctate stains (Fig. 4a), as would be expected for Golgi localization. Also, expression of Arf1 did not change the pattern of ST:GFP (Fig. 4b). In contrast, when the protoplasts from the transgenic plants were transformed with Arf1[T31N], green fluorescent signals were present as diffuse patterns (Fig. 4c) similar to those seen with transiently expressed ST:GFP.
Figure 3.
Colocalization of ST:GFP and BiP:RFP in the presence of Arf1[T31N]. Protoplasts were transformed with ST:GFP, BiP:RFP, and Arf1[T31N] and localization of reporter proteins was examined. Green and red images are GFP and RFP signals, respectively. The blue signals in d indicate autofluorescence of chlorophyll. Bar = 20 μm.
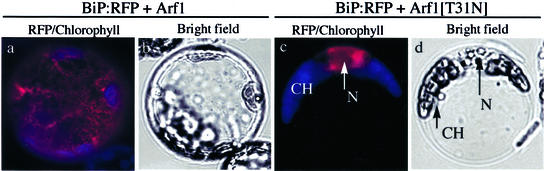
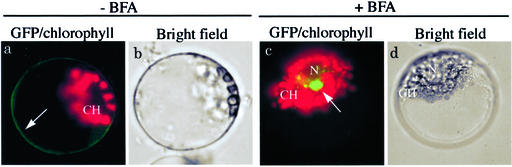
Figure 10.
The effect of Arf1[T31N] and BFA on morphology of the ER. Protoplasts were transformed with either Arf1 plus BiP:RFP or Arf1[T31N] plus BiP:RFP and examined between 12 and 36 h after transformation. a and c are representative fluorescent images of protoplasts, and b and d are bright-field images of a and c, respectively. Red images are RFP signals of BiP:RFP. Autofluorescence of chlorophyll is depicted in blue. N and CH indicate the nucleus and chloroplast, respectively.
Figure 4.
The effect of Arf1[T31N] on the localization of constitutively expressed ST:GFP. Protoplasts obtained from transgenic plants constitutively expressing ST:GFP were transformed with the constructs indicated. The effect of these Arf1 proteins on the localization of ST:GFP was examined 24 h after transformation. Green and red images are GFP and RFP signals, respectively. The blue signals in a, b, and c indicate autofluorescence of chlorophyll. Bar = 20 μm.
Next, we transformed protoplasts prepared from transgenic plants with Arf1[T31N] and BiP:RFP to examine the localization of ST:GFP in the presence of Arf1[T31N]. The green fluorescent signals of ST:GFP closely overlapped the red fluorescent signals of BiP:RFP (Fig. 4, d–f) in the presence of Arf1[T31N]. These results strongly suggest that the Golgi apparatus may be disassembled in the presence of Arf1[T31N], which results in fusion of the Golgi membranes together with Golgi resident proteins such as ST:GFP to the ER in Arabidopsis protoplasts, as in animal cells (Dascher and Balch, 1994; Peters et al., 1995). However, we cannot rule out the possibility that ST:GFP that had been localized at the Golgi apparatus may be turned over during incubation time with Arf1[T31N] and that ST:GFP that was localized at the ER in the presence of Arf1[T31N] may result from de novo synthesis. To address this question, we examined the half-life of ST:GFP in the protoplasts in the presence of cycloheximide, an inhibitor of protein synthesis. As shown in Figure 5, in the presence of cycloheximide, protein synthesis was nearly completely inhibited (Fig. 5, lane C). At this condition, the amount of ST:GFP was nearly the same for up 36 h when the protein level was normalized based on the loading control, indicating that degradation of ST:GFP is relatively minor for 36 h if there is any. Thus, these results indirectly support the notion that ST:GFP localized at the Golgi apparatus is likely relocated to the ER in the presence of Arf1[T31N], instead of being turned over, during 24 h of incubation time.
Figure 5.
Stability of ST:GFP in protoplasts in the presence of cycloheximide. Protoplasts were transformed with ST:GFP and incubated for expression of ST:GFP for 12 h. At 12 h after transformation, cycloheximide (100 μg mL−1) was added and further incubated for the indicated periods of time. The level of ST:GFP was examined from protein extracts prepared from protoplasts by western-blot analysis using a monoclonal anti-GFP antibody. As a control (lane C), cycloheximide was added to protoplasts right after transformation. En indicates an endogenous protein that was detected by the monoclonal anti-GFP antibody and used as a loading control for the western-blot analysis.
Arf1[T31N], But Not Arf3[T31N], Inhibits Trafficking of H+-ATPase:GFP to the Plasma Membrane
We next examined the effect of Arf1[T31N] on trafficking of H+-ATPase:GFP to the plasma membrane. As shown in Figure 6A, H+-ATPase:GFP was correctly targeted to the plasma membrane in the presence of Arf1 (Fig. 6A, b). However, when Arf1[T31N] was co-expressed with the reporter protein, the green fluorescent signal of H+-ATPase:GFP was present as punctate stains or aggregates in the protoplasts (Fig. 6A, c1–c4, arrowheads) and not at the plasma membrane. The targeting efficiency of H+-ATPase:GFP to the plasma membrane was more than 95% in control protoplasts (Fig. 6B), whereas it dropped to less than 5% in the presence of Arf1[T31N] (Fig. 6B), indicating that co-expression of Arf1[T31N] resulted in very efficient inhibition. In contrast, Arf3[T31N] did not affect trafficking of H+-ATPase:GFP to the plasma membrane (Fig. 6, C and D). Together, these results suggest that Arf1, but not Arf3, plays a role in the trafficking of H+-ATPase:GFP to the plasma membrane.
Figure 6.
The effect of Arf1 and Arf3 mutants on trafficking of H+-ATPase:GFP to the plasma membrane. A, The effect of Arf1 mutants on trafficking of H+-ATPase:GFP. Protoplasts were transformed with the constructs indicated, and localization of H+-ATPase:GFP was examined at 12 to 36 h after transformation. Arrows indicate green fluorescent signals at the plasma membrane and arrowheads indicate punctate stains in the protoplasts. CH, Chloroplasts. Bars = 20 μm. B and C, Targeting efficiency of H+-ATPase:GFP in the presence of various forms of Arfs. To determine a targeting efficiency estimate, images of more than 200 transformed protoplasts at each time point were analyzed based on the GFP patterns of H+-ATPase:GFP as described in Figure 2. The patterns shown in A, a, were considered to indicate that H+-ATPase:GFP was targeted to the plasma membrane. The patterns shown in A, c-1 to c-4, were considered to indicate that the protein was not targeted to the membrane. D, The effect of Arf3 mutants on the trafficking of H+-ATPase:GFP. Protoplasts were transformed with H+-ATPase:GFP alone (a), Arf3 plus H+-ATPase:GFP (b), and Arf3[T31N] plus H+-ATPase:GFP (c), and localization of reporter protein was examined between 12 and 36 h after transformation. The green and red images indicate GFP signals and autofluorescence of chlorophyll, respectively. Arrows indicate green fluorescent signals at the plasma membrane in the protoplasts. CH, Chloroplasts. Bars = 20 μm.
In animal cells, BFA and the dominant negative mutant of Arf1 have been shown to have similar effects on intracellular trafficking (Sata et al., 1998; Morinaga et al., 1999). BFA has also been shown to inhibit intracellular trafficking in plant cells (Gomez and Chrispeels, 1993; Boevink et al., 1999). Therefore, we investigated the effect of BFA on trafficking of H+-ATPase:GFP to the plasma membrane in the protoplast. As shown in Figure 7, a green fluorescent signal was present as punctate stains around the nucleus in the presence of BFA (Fig. 7c) as observed in the presence of Arf1[T31N] (Fig. 6A, c1–c4), suggesting that, as in animal cells, Arf1 may play a role in the trafficking of H+-ATPase:GFP to the plasma membrane through a BFA-sensitive factor (Sata et al., 1998; Morinaga et al., 1999).
Figure 7.
The effect of BFA on the targeting of H+-ATPase:GFP. Targeting of H+-ATPase:GFP was examined in protoplasts in the presence (+BFA) and absence (−BFA) of 30 μg mL−1 BFA. BFA was added right after transformation. Green and red images are GFP and chlorophyll, respectively. N and CH indicate nucleus and chloroplast, respectively. Arrows indicate H+-ATPase:GFP.
To further understand the role of Arf1 in trafficking in plant cells, we attempted to determine the localization of H+-ATPase:GFP in the presence of Arf1[T31N]. To address this question, H+-ATPase:GFP and Arf1[T31N] were transformed into protoplasts together with BiP:RFP. As shown in Figure 8A, the green fluorescent signals were present as a few punctate stains that did not overlap the red fluorescent signals of BiP:RFP (Fig. 8A, d–f), indicating that H+-ATPase:GFP is not localized at the ER. Interestingly, it appeared that the behavior of H+-ATPase:GFP was different from that of ST:GFP in the presence of Arf1[T31N]. To confirm this notion, protoplasts were cotransformed with H+-ATPase:GFP, ST:RFP, and Arf1[T31N], and localization of these reporter proteins was examined. As shown in Figure 8B, H+-ATPases:GFP did not overlap ST:RFP in protoplasts transformed with H+-ATPase:GFP, ST:RFP, and Arf1[T31N]. Together, these results suggest that H+-ATPase:GFP was not present at the ER but at some other organelle.
Figure 8.
Localization of H+-ATPase:GFP in the presence of Arf1[T31N]. A, Localization of H+-ATPase. Protoplasts were transformed with three sets of constructs, either H+-ATPase:GFP, BiP:RFP, and Arf1 (a–c) or H+-ATPase:GFP, BiP:RFP, and Arf1[T31N] (d–f). Localization of green fluorescent signals was examined 12 to 36 h after transformation. Arrows and arrowheads indicate H+-ATPase:GFP and BiP:RFP, respectively. B, Protoplasts were transformed with H+-ATPase:GFP, ST:RFP, and Arf1 or H+-ATPase:GFP, ST:RFP, and Arf1[T31N], and localization of reporter proteins was examined.
Motifs in the Middle and C-Terminal Regions Are Necessary for Inhibition of Trafficking by Arf1
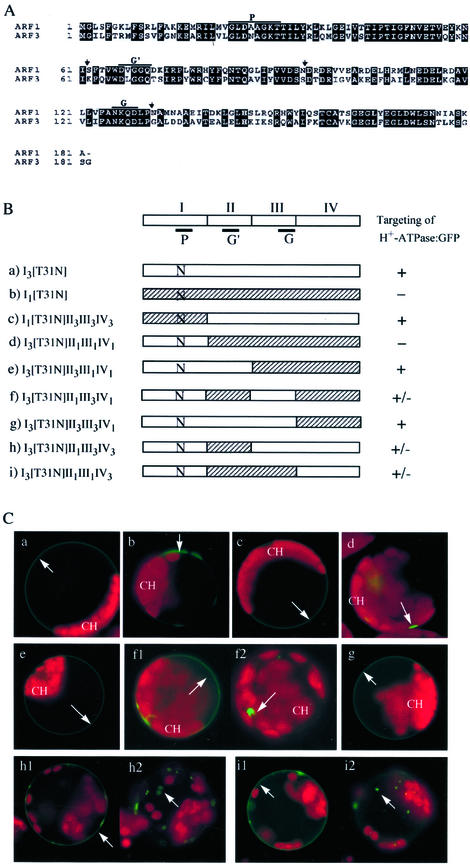
Arf has been shown to be regulated by various effector molecules such as GEFs and GTPase-activating proteins (Kahn et al., 1994; Goldberg, 1998, 1999). These proteins are known to interact with specific domains of Arf1, such as switches 1 and 2 (Goldberg, 1998, 1999). To define which domains of Arf1 are necessary for its role in intracellular trafficking, we made Arf1 and Arf3 mutants with swapped domains. The Arf molecules were divided into four domains as shown in Figure 9, A and B. Domain I (amino acid residues 1–61) is the N-terminal region and includes the P-loop motif (P) of the GTP-binding domain. Domains II (amino acid residues 62–95) and III (amino acid residues 96–130) include the second (G′) and third (G) GTP-binding motifs, respectively. Finally, domain IV (amino acid residues 131–181 for Arf1 and 131–182 for Arf3) is the C-terminal region downstream of the third GTP-binding motif. These domains were exchanged to make hybrid mutants. In addition to domain exchanges, all these constructs had the dominant-negative mutation [T31N] at the P-loop.
Figure 9.
Trafficking of H+-ATPase:GFP in the presence of various hybrid mutants between Arf1[T31N] and Arf3[T31N]. A, Sequence alignment of Arf1 and Arf3 isoforms of Arabidopsis. P, G′, and G indicate the three GTP-binding motifs. Vertical arrows indicate the boundary of the domains. B, Schematic presentation of various hybrid constructs between Arf1[T31N] and Arf3[T31N]. The first (I), second (II), third (III), and fourth (IV) domains are regions containing amino acid residues from 1 to 61, 62 to 95, 96 to 130, and 131 to 181, respectively. The subscripts 1 and 3 in the names of the constructs indicate domains derived from Arf1 and Arf3, respectively. Ns in the diagrams indicate T31 N mutations. P, G′, and G indicate the three GTP-binding motifs. C, H+-ATPase:GFP was transformed into protoplasts together with various hybrid constructs. The trafficking of H+-ATPase:GFP was examined 20 to 30 h after transformation. a through i represent protoplasts transformed with constructs a through i, respectively, in B. Green and red fluorescent signals are GFP and chlorophyll, respectively. At least three independent transformation experiments were carried out for each condition. Arrows indicate the green fluorescent signals of H+-ATPase:GFP. CH, Chloroplast.
The effects of these mutants on trafficking of H+-ATPase:GFP were examined in protoplasts after transformation. As shown in Figure 9C, the mutant I3[T31N]II1III1IV1, consisting of domain I of Arf3 [T31N] and domains II, III, and IV of Arf1, completely inhibited trafficking of H+-ATPase:GFP to the plasma membrane (Fig. 9C, d). This indicates that the first N-terminal domain of Arf3 can functionally replace that of Arf1. However, the opposite mutant I1[T31N]II3III3IV3, consisting of domain I of Arf1[T31N] and domains II, III, and IV of Arf3 did not inhibit trafficking (Fig. 9C, c), which suggests that the T31 N mutation and additional motif(s) in the downstream region of Arf1 are necessary for inhibition of trafficking. When constructs I3[T31N]II3III1IV1 (Fig. 9C, e) and I3[T31N]II3III3IV1 (Fig. 9C, g) were introduced into protoplasts, they did not inhibit trafficking, indicating that domain II of Arf1 is critical for inhibition. Next, we examined the effect of the domain III on the inhibition using I3[T31N]II1III3IV1 (Fig. 9C, f1 and f2). When I3[T31N]II1III3IV1 was introduced together with H+-ATPase:GFP, green fluorescent signals of H+-ATPase:GFP were present at the plasma membrane (Fig. 9C, f1) and in the cytosol as punctate stains (Fig. 9C, f2), indicating that the trafficking of H+-ATPase:GFP was partially inhibited. These results suggest that domain III is necessary for complete inhibition. Finally, we examined the effect of domain IV on the trafficking using mutants I3[T31N]II1III3IV3 (Fig. 9C, h1 and h2) and I3[T31N]II1III1IV3 (Fig. 9C, i1 and i2). When these mutants were expressed in protoplasts, green fluorescent signals were present at the plasma membrane (Fig. 9C, h1 and i1) and in the cytosol as punctate stains (Fig. 9C, h2 and i2), indicating a partial inhibition by these mutants. These results indicate that domain IV is also necessary for complete inhibition. Therefore, it is likely that protein-binding motifs in domains II, III, and IV may interact with Arf1-specific effector proteins.
Arf1[T31N] Affects Morphology of the ER
Throughout this study, we noticed that red fluorescent signals of BiP:RFP were present as diffuse patterns around the nucleus in protoplasts when Arf1[T31N] was co-expressed. One possibility is that the morphology of the ER might have been changed due to expression of Arf1[T31N]. To address this question, we compared the morphology of the ER in the presence of Arf1 and Arf1[T31N] in protoplasts using BiP:GFP as a reporter for the ER. As described previously, the red fluorescent signals of BiP:GFP were present as numerous networks with blobs throughout the protoplast in the presence of co-expressed Arf1 (Fig. 10). In contrast, protoplasts expressing Arf1[T31N] gave diffuse patterns of BiP:GFP, clearly indicating that this pattern was caused by Arf1[T31N]. Boevink et al. (1999) has reported a similar observation; namely, the ER network is transformed into large lamellae or sheets in the presence of BFA in tobacco (Nicotiana tabacum) cells. In yeast, mutations at Arf1 also caused exaggeration of the ER membrane (Yahara et al., 2001). Thus, one possible explanation is that in the presence of Arf1[T31N], fusion of the Golgi membranes to the ER and inhibition of anterograde trafficking may cause expansion of the ER networks, which results in profound changes in the ER morphology as shown here.
DISCUSSION
In animal and yeast cells, involvement of Arf in retrograde trafficking from the Golgi apparatus to the ER has been extensively characterized (Balch et al., 1992; Haun et al., 1993; Kahn et al., 1994; Moss and Vaughan, 1998; Spang et al., 1998; Lanoix et al., 1999; Poon et al., 1999; Zhao et al., 1999). Use of the dominant negative mutant of Arf1 and BFA, an inhibitor of Arf1, has been instrumental in elucidating the role of Arf1. In the presence of the dominant negative mutant or BFA, the Golgi structure is disassembled and fused to the ER, thereby resulting in relocation of the Golgi-resident proteins to the ER and eventually inhibiting anterograde trafficking. In this study, we used similar approaches using a dominant negative mutant of Arf1, Arf1[T31N], and examined the effect of Arf1[T31N] on trafficking of ST:GFP and H+-ATPase:GFP to their final destinations. In the presence of Arf1[T31N], ST:GFP was present at the ER as a diffuse pattern. In addition, Arf1[T31N] caused relocation of ST:GFP from the Golgi apparatus to the ER, suggesting that Arf1[T31N] may cause disassembly of the Golgi apparatus in plant cells as in animal cells. The behavior of ST:GFP is very similar to the known effect of the dominant negative mutant of Arf1 in animal and yeast cells. However, interestingly, the inhibition pattern of H+-ATPase:GFP by Arf1[T31N] was quite different from that of ST:GFP. In the presence of Arf1[T31N], H+-ATPase:GFP was not targeted to the plasma membrane, indicating that Arf1[T31N] inhibits trafficking of H+-ATPase:GFP to the plasma membrane. However, in contrast to ST:GFP, it was not present in the ER but instead present at an unidentified organelle as punctate stains.
It has been shown that certain cargo molecules can take alternative pathways and be transported to other organelles when the normal trafficking pathway is inhibited. In Bright-Yellow 2 cells, sporamin is secreted into media when trafficking of sporamin to the central vacuole was inhibited by wortmannin (Matsuoka et al., 1995). Also, in yeast cells with mutations in both AP3-μ and VPS45 genes, vacuolar alkaline phosphatase is transported via an alternate intracellular route and present as punctate stains in the cell instead of being transported to the vacuolar membrane (Stepp et al., 1997). At the moment, the route for trafficking of H+-ATPase:GFP to the plasma membrane is not known in plant cells. One possible route for trafficking of H+-ATPase:GFP is from the ER to the plasma membrane through the Golgi apparatus, although we were not able to see colocalization of H+-ATPase:GFP with the Golgi marker, ST:RFP (data not shown). This could be because H+-ATPase:GFP is present at the Golgi apparatus only for short time and does not accumulate there in high enough level to be detected. Thus, it is possible that H+-ATPase:GFP may be mistargeted to other organelles in the presence of Arf1[T31N] because of blocking of the normal trafficking pathway through the Golgi apparatus. It has recently been shown that BFA treatment resulted in accumulation of plasma membrane H+-ATPase at the BFA-induced intracellular compartment in Arabidopsis root cells (Geldner et al., 2001). Thus, it is possible that in the presence of Arf1[T31N] H+-ATPase:GFP accumulates at a compartment similar to the BFA-induced intracellular compartment. In addition to the inhibition of normal intracellular trafficking of ST:GFP and H+-ATPase:GFP, Arf1[T31N] caused profound changes in ER morphology, which is likely due to fusion of the Golgi membranes to the ER and inhibition of anterograde trafficking.
BFA treatments had the same effect on trafficking of these reporters as Arf1[T31N], indicating that Arf1[T31N] may play a role in intracellular trafficking through a BFA-sensitive factor. BFA has been shown to inhibit Arf1 function by stabilizing the transitory complexes of Sec7 domain-Arf1-GDP (Sata et al., 1998; Morinaga et al., 1999; Robineau et al., 2000). Thus, it is likely that similar GEF proteins may also be present in plant cells. One possible role of Arf1 in intracellular trafficking is that Arf1 may also be involved in the formation of COPI vesicles at the Golgi apparatus as proposed in animal cells (Lanoix et al., 1999; Roth, 1999; Wieland and Harter, 1999). In fact, Arf1 has recently been shown to be localized at the Golgi apparatus in plant cells (Pimpl et al., 2000). Also, the presence of coatomer subunits of COPI vesicles in plant cells has been reported (Contreras et al., 2000).
To further investigate the role that Arf1 plays in intracellular trafficking, we carried out inhibition studies using various mutants with swapped domains between Arf1 and Arf3. These studies revealed that the N-terminal regions of Arfs were interchangeable. However, interestingly, I3[T31N]II3III1IV1 and I3[T31N]II3III3IV1 did not inhibit trafficking of H+-ATPase:GFP to the plasma membrane at all. These results demonstrated the importance of domain II for the function of Arf1 in intracellular trafficking. One of the protein interaction domains of Arf1, named the switch 2 (amino acid residues from 70–80), is located in the region designated as domain II in this study and has been shown to be involved in interaction with effector proteins, such as GEFs and GTPase-activating proteins (Moss and Vaughan, 1998; Goldberg, 1999). However, the fact that mutants I3[T31N]II1III3IV1, I3[T31N]II1III3IV3, and I3[T31N]II1III1IV3 gave only partial inhibition of trafficking of the H+-ATPase:GFP to the plasma membrane suggests that motifs in domains III and IV of Arf1 are also necessary for the function of Arf1. Domain III of Arf1 defined in this study contains the third GTP-binding motif. However, it is possible that domain III may contain additional motifs involved in interaction with effector protein for the function of Arf1. Domain IV contains the C-terminal Arf consensus motif, TCAT (Kahn et al., 1994). Thus, the inhibition experiments using various domain-swapped mutants clearly demonstrated the functional difference between Arf1 and Arf3. The difference probably resulted from the difference in the interaction of Arf isoforms with effector proteins. The motif in each domain may be involved in interaction with isoform-specific effector proteins. However, it is also possible that the motif in domain II of Arf isoforms may play a major role in interaction with an Arf isoform-specific effector protein, and motifs in domains III and IV may increase binding affinity of the Arf isoform-specific effector bound to the motif of domain II. Thus, depletion of the Arf1-specific effector protein by the dominant negative mutant of Arf1 may result in inhibition of intracellular trafficking.
In conclusion, although Arf1 and Arf3 isoforms of Arabidopsis are highly homologous to each other, only Arf1 but not Arf3 was shown to be involved in the intracellular trafficking pathways we examined. Similarly, in yeast and animal cells, multiple isoforms of Arfs have been found, but only certain isoforms have been shown to play roles in intracellular trafficking (Jackson and Casanova, 2000). However, we cannot rule out the possibility that Arf3 may also be involved in other steps or other routes of intracellular trafficking that we did not address in this study. In animal cells, Arf6 has been shown to be involved in endocytosis and membrane recycling at the plasma membrane (D'Souza-Schorey et al., 1998). Another possibility is that Arf3 may be involved in other biological process, as in the case of Arf6 in actin polymerization in animal cells (Al-Awar et al., 2000). Further studies are necessary to define the biological role of the Arf3 isoform in Arabidopsis.
MATERIALS AND METHODS
Growth of Plants
Arabidopsis (ecotype Columbia) was grown on soil at 20°C to 25°C in a greenhouse with a 16/8-h light/dark cycle. Leaf tissues were harvested from the plants and immediately used for protoplast isolation.
Generation of Various Constructs
Arf1 (accession no. M95166) and Arf3 (accession no. X77385) cDNAs were PCR amplified from a lambda cDNA library using gene-specific primers (Arf1-5, TTGGACGACCATCGGCGTTAAG; Arf1-3; TCCATCTATGCCTTGCTTGCGA; Arf3-5, CCAATCGAAGAAGAAGATGGGA; and Arf3-3, CCTTATGGGGAATATTAAGTGA). The nucleotide sequences of both cDNAs were confirmed through DNA sequencing. Point mutants of Arf1 and Arf 3 were generated by PCR. The primers used were: Arf1[T31N], GGTAAGAACACTATCCTCTACAAGC and GATAGTGTTCTTACCAGCAGCATCG; Arf3[T31N], GGAAAAAACACAATCCTATATCGGC and GATTGTGTTTTTTCCAGCATTGTCG. To generate mutants with swapped domains, common restriction sites were introduced into the two Arf isoforms by PCR using primers that did not change the amino acid sequence. The primer sequences used were: Arf1F62HindIII, CATAAGCTTCACCGTGTGGGA; Arf1R62HindIII, GCGAAGCTTATGTTCTTGTATTCAAC; Arf1F95XhoI, GACTCGAGTGATCGTGACCGTGTTG; Arf1R95XI, GCCTCGAGTCCACAACAAAGATAAG; Arf3R62HIII, GCGAAGCTTATATTGTTGTATTGCAC; Arf3F62HIII, CGCAAGCTTTCAGGTCTGGGATTT; Arf3F130BgII, ATAGATCTTCCCGGTGCACT, and Arf3R130BHI, GCGGATCCTGCTTGTTTGCAA. The sequence of all of the PCR products was confirmed through DNA sequencing. These fragments were then fused using the common restriction sites after restriction digestion. Finally, these constructs were introduced into an expression vector that has the cauliflower mosaic virus 35S promoter and the nopaline synthetase terminator. To add the T7 epitope tag to the N termini of wild types and dominant negative mutants of Arf1 and Arf3, BamHI and XhoI fragments encoding Arf1, Arf1[T31N], Arf3, and Arf3[T31N] were ligated into the BamHI and XhoI sites of pET21a (+) (Novagen, Madison, WI). XbaI and XhoI fragments encoding the T7-Arf clones were subsequently isolated and placed downstream of the cauliflower mosaic virus 35S promoter.
Generation of Transgenic Plants
A binary vector, pST:GFP, was constructed by replacing the β-glucuronidase coding region of pBI121 with ST:GFP and transformed into Agrobacterium tumefaciens by electroporation. Arabidopsis was then transformed according to the vacuum-infiltration method (Clough and Bent, 1998). Transgenic plants were selected on Murashige-Skoog plates containing 50 mg L−1 kanamycin.
Transient Expression of Arf Constructs and in Vivo Targeting of Reporter Proteins
For expression in protoplasts, all of the chimeric GFP fusion constructs were placed under the control of the 35S promoter in a pUC vector. Plasmids were purified using Qiagen (Valencia, CA) columns according to the manufacturer's protocol. The plasmids were introduced into Arabidopsis protoplasts that had been prepared from leaf tissues by polyethylene glycol-mediated transformation (Jin et al., 2001; Kim et al., 2001; Lee et al., 2001). Expression of the fusion constructs was monitored at various time points after transformation and images were captured with a cooled CCD camera and a Zeiss (Jena, Germany) Axioplan fluorescence microscope. The filter sets used were XF116 (exciter, 474AF20; dichroic, 500DRLP; and emitter, 510AF23), XF33/E (exciter, 535DF35; dichroic, 570DRLP; and emitter, 605DF50), and XF137 (exciter, 540AF30; dichroic, 570DRLP; and emitter, 585ALP; Omega, Inc., Brattleboro, VT) for GFP, RFP, and auto-fluorescence of chlorophyll, respectively. Data were then processed using Adobe Photoshop software (Adobe Systems, Mountain View, CA) and presented in pseudo-color format.
ACKNOWLEDGMENT
We thank Dr. John Rogers (University of Washington, Pullman) for the suggestions and helpful discussion for the work.
Footnotes
This work was supported by a grant from National Creative Research Initiatives from the Ministry of Science and Technology.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.003624.
LITERATURE CITED
- Al-Awar O, Radhakrishna H, Powell NN, Donaldson JG. Separation of membrane trafficking and actin remodeling functions of ARF6 with an effector domain mutant. Mol Cell Biol. 2000;20:5998–6007. doi: 10.1128/mcb.20.16.5998-6007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch WE, Kahn RA, Schwaninger R. ADP-ribosylation factor is required for vesicular trafficking between the endoplasmic reticulum and the cis-Golgi compartment. J Biol Chem. 1992;267:13053–13061. [PubMed] [Google Scholar]
- Bassham DC, Gal S, da Silva Conceicao A, Raikhel NV. An Arabidopsis syntaxin homologue isolated by functional complementation of a yeast pep12 mutant. Proc Natl Acad Sci USA. 1995;92:7262–7266. doi: 10.1073/pnas.92.16.7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham DC, Raikhel NV. Unique features of the plant vacuolar sorting machinery. Curr Opin Cell Biol. 2000;12:491–495. doi: 10.1016/s0955-0674(00)00121-6. [DOI] [PubMed] [Google Scholar]
- Bednarek SY, Raikhel NV. The barley lectin carboxyl-terminal propeptide is a vacuolar protein sorting determinant in plants. Plant Cell. 1991;3:1195–1206. doi: 10.1105/tpc.3.11.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackbourn HD, Jackson AP. Plant clathrin heavy chain: sequence analysis and restricted localisation in growing pollen tubes. J Cell Sci. 1996;109:777–786. doi: 10.1242/jcs.109.4.777. [DOI] [PubMed] [Google Scholar]
- Boevink P, Martin B, Oparka K, Santa Cruz S, Hawes C. Transport of virally expressed green fluorescent protein through the secretory pathway in tobacco leaves is inhibited by cold shock and brefeldin A. Planta. 1999;208:392–400. [Google Scholar]
- Chrispeels MJ, Raikhel NV. Short peptide domains target proteins to plant vacuoles. Cell. 1992;68:613–616. doi: 10.1016/0092-8674(92)90134-x. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Contreras I, Ortiz-Zapater E, Castilho LM, Aniento F. Characterization of COPI coat proteins in plant cells. Biochem Biophys Res Commun. 2000;273:176–182. doi: 10.1006/bbrc.2000.2918. [DOI] [PubMed] [Google Scholar]
- Dascher C, Balch WE. Dominant inhibitory mutants of ARF1 block endoplasmic reticulum to Golgi transport and trigger disassembly of the Golgi apparatus. J Biol Chem. 1994;269:1437–1448. [PubMed] [Google Scholar]
- Davis SJ, Vierstra RD. Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol Biol. 1998;36:521–528. doi: 10.1023/a:1005991617182. [DOI] [PubMed] [Google Scholar]
- D'Souza-Schorey C, van Donselaar E, Hsu VW, Yang C, Stahl PD, Peters PJ. ARF6 targets recycling vesicles to the plasma membrane: insights from an ultrastructural investigation. J Cell Biol. 1998;140:603–616. doi: 10.1083/jcb.140.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigerio L, de Virgilio M, Prada A, Faoro F, Vitale A. Sorting of phaseolin to the vacuole is saturable and requires a short C-terminal peptide. Plant Cell. 1998;10:1031–1042. doi: 10.1105/tpc.10.6.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor EC, Chen CY, Emr SD, Graham TR. ARF is required for maintenance of yeast Golgi and endosome structure and function. Mol Biol Cell. 1998;9:653–670. doi: 10.1091/mbc.9.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N, Friml J, Stierhof YD, Jurgens G, Palme K. Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature. 2001;413:425–428. doi: 10.1038/35096571. [DOI] [PubMed] [Google Scholar]
- Goldberg J. Structural basis for activation of ARF GTPase: mechanisms of guanine nucleotide exchange and GTP-myristoyl switching. Cell. 1998;95:237–248. doi: 10.1016/s0092-8674(00)81754-7. [DOI] [PubMed] [Google Scholar]
- Goldberg J. Structural and functional analysis of the ARF1-ARFGAP complex reveals a role for coatomer in GTP hydrolysis. Cell. 1999;96:893–902. doi: 10.1016/s0092-8674(00)80598-x. [DOI] [PubMed] [Google Scholar]
- Goldberg J. Decoding of sorting signals by coatomer through a GTPase switch in the COPI coat complex. Cell. 2000;100:671–679. doi: 10.1016/s0092-8674(00)80703-5. [DOI] [PubMed] [Google Scholar]
- Gomez L, Chrispeels MJ. Tonoplast and soluble vacuolar proteins are targeted by different mechanisms. Plant Cell. 1993;5:1113–1124. doi: 10.1105/tpc.5.9.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara-Nishimura I, Shimada T, Hatano K, Takeuchi Y, Nishimura M. Transport of storage proteins to protein storage vacuoles is mediated by large precursor-accumulating vesicles. Plant Cell. 1998;10:825–836. doi: 10.1105/tpc.10.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haun RS, Tsai SC, Adamik R, Moss J, Vaughan M. Effect of myristoylation on GTP-dependent binding of ADP-ribosylation factor to Golgi. J Biol Chem. 1993;268:7064–7068. [PubMed] [Google Scholar]
- Heikal AA, Hess ST, Baird GS, Tsien RY, Webb WW. Molecular spectroscopy and dynamics of intrinsically fluorescent proteins: coral red (dsRed) and yellow (Citrine) Proc Natl Acad Sci USA. 2000;97:11996–12001. doi: 10.1073/pnas.97.22.11996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillmer S, Movafeghi A, Robinson DG, Hinz G. Vacuolar storage proteins are sorted in the cis-cisternae of the pea cotyledon Golgi apparatus. J Cell Biol. 2001;152:41–50. doi: 10.1083/jcb.152.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CL, Casanova JE. Turning on ARF: the sec7 family of guanine-nucleotide exchange factors. Trends Cell Biol. 2000;10:60–67. doi: 10.1016/s0962-8924(99)01699-2. [DOI] [PubMed] [Google Scholar]
- Jahn R, Südhof TC. Membrane fusion and exocytosis. Annu Rev Biochem. 1999;68:863–911. doi: 10.1146/annurev.biochem.68.1.863. [DOI] [PubMed] [Google Scholar]
- Jauh GY, Phillips TE, Rogers JC. Tonoplast intrinsic protein isoforms as markers for vacuolar functions. Plant Cell. 1999;11:1867–1882. doi: 10.1105/tpc.11.10.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin JB, Kim YA, Kim SJ, Lee SH, Kim DH, Cheong GW, Hwang I. A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell. 2001;13:1511–1526. doi: 10.1105/TPC.000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn RA, Clark J, Rulka C, Stearns T, Zhang C, Randazzo PA, Terui T, Cavenagh M. Mutational analysis of Saccharomyces cerevisiae ARF1. J Biol Chem. 1994;270:143–150. doi: 10.1074/jbc.270.1.143. [DOI] [PubMed] [Google Scholar]
- Kim DH, Eu YJ, Yoo CM, Kim YW, Pih KT, Jin JB, Kim SJ, Stenmark H, Hwang I. Trafficking of phosphatidylinositol 3-phosphate from the trans-Golgi network to the lumen of the central vacuole in plant cells. Plant Cell. 2001;13:287–301. doi: 10.1105/tpc.13.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanoix J, Ouwendijk J, Lin CC, Stark A, Love HD, Ostermann J, Nilsson T. GTP hydrolysis by arf-1 mediates sorting and concentration of Golgi resident enzymes into functional COP I vesicles. EMBO J. 1999;18:4935–4948. doi: 10.1093/emboj/18.18.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebas M, Axelos M. A cDNA encoding a new GTP-binding protein of the ADP-ribosylation factor family from Arabidopsis. Plant Physiol. 1994;106:809–810. doi: 10.1104/pp.106.2.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Kim DH, Kim YW, Hwang I. Identification of a signal that distinguishes between the chloroplast outer envelope membrane and the endomembrane system in vivo. Plant Cell. 2001;13:2175–2190. doi: 10.1105/tpc.010232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka K, Bassham DC, Raikhel NV, Nakamura K. Different sensitivity to wortmannin of two vacuolar sorting signals indicates the presence of distinct sorting machineries in tobacco cells. J Cell Biol. 1995;130:1307–1318. doi: 10.1083/jcb.130.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka K, Matsumoto S, Hattori T, Machida Y, Nakamura K. Vacuolar targeting and post-translational processing of the precursor to the sweet potato tuberous root storage protein in heterologous plant cells. J Biol Chem. 1990;265:19750–19757. [PubMed] [Google Scholar]
- Memon AR, Clark GB, Thompson GA., Jr Identification of an ARF type low molecular mass GTP-binding protein in pea (Pisum sativum) Biochem Biophys Res Commun. 1993;193:809–813. doi: 10.1006/bbrc.1993.1697. [DOI] [PubMed] [Google Scholar]
- Morinaga N, Adamik R, Moss J, Vaughan M. Brefeldin A inhibited activity of the sec7 domain of p200, a mammalian guanine nucleotide-exchange protein for ADP-ribosylation factors. J Biol Chem. 1999;274:17417–17423. doi: 10.1074/jbc.274.25.17417. [DOI] [PubMed] [Google Scholar]
- Moss J, Vaughan M. Molecules in the ARF orbit. J Biol Chem. 1998;273:21431–21434. doi: 10.1074/jbc.273.34.21431. [DOI] [PubMed] [Google Scholar]
- Neuhaus JM, Rogers JC. Sorting of proteins to vacuoles in plant cells. Plant Mol Biol. 1998;38:127–144. [PubMed] [Google Scholar]
- Neuhaus JM, Sticher L, Meins F, Jr, Boller T. A short C-terminal sequence is necessary and sufficient for the targeting of chitinases to the plant vacuole. Proc Natl Acad Sci USA. 1991;88:10362–10366. doi: 10.1073/pnas.88.22.10362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi CE, Dell'Angelica EC, Bonifacino JS. ADP-ribosylation factor 1 (ARF1) regulates recruitment of the AP-3 adaptor complex to membranes. J Cell Biol. 1998;142:391–402. doi: 10.1083/jcb.142.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco-Rodriguez G, Meacci E, Vitale N, Moss J, Vaughan M. Guanine nucleotide exchange on ADP-ribosylation factors catalyzed by cytohesin-1 and its Sec7 domain. J Biol Chem. 1998;273:26543–26548. doi: 10.1074/jbc.273.41.26543. [DOI] [PubMed] [Google Scholar]
- Paris N, Stanley CM, Jones RL, Rogers JC. Plant cells contain two functionally distinct vacuolar compartments. Cell. 1996;85:563–572. doi: 10.1016/s0092-8674(00)81256-8. [DOI] [PubMed] [Google Scholar]
- Peters PJ, Hsu VW, Ooi CE, Finazzi D, Teal SB, Oorschot V, Donaldson JG, Klausner RD. Overexpression of wild-type and mutant ARF1 and ARF6: distinct perturbations of nonoverlapping membrane compartments. J Cell Biol. 1995;128:1003–1017. doi: 10.1083/jcb.128.6.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pih KT, Yi MJ, Liang YS, Shin BJ, Cho MJ, Hwang I, Son D. Molecular cloning and targeting of a fibrillarin homolog from Arabidopsis. Plant Physiol. 2000;123:51–58. doi: 10.1104/pp.123.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimpl P, Movafeghi A, Coughlan S, Denecke J, Hillmer S, Robinson DG. In situ localization and in vitro induction of plant COP1-coated vesicles. Plant Cell. 2000;12:2219–2235. doi: 10.1105/tpc.12.11.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon PP, Cassel D, Spang A, Rotman M, Pick E, Singer RA, Johnston GC. Retrograde transport from the yeast Golgi is mediated by two ARF GAP proteins with overlapping function. EMBO J. 1999;18:555–564. doi: 10.1093/emboj/18.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regad F, Bardet C, Tremousaygue D, Moisan A, Lescure B, Axelos M. cDNA cloning and expression of an Arabidopsis GTP-binding protein of the ARF family. FEBS Lett. 1993;316:133–136. doi: 10.1016/0014-5793(93)81201-a. [DOI] [PubMed] [Google Scholar]
- Robineau S, Chabre M, Antonny B. Binding site of brefeldin A at the interface between the small G protein ADP-ribosylation factor 1 (ARF1) and the nucleotide-exchange factor Sec7 domain. Proc Natl Acad Sci USA. 2000;97:9913–9918. doi: 10.1073/pnas.170290597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth MG. Snapshots of ARF1: implications for mechanisms of activation and inactivation. Cell. 1999;97:149–152. doi: 10.1016/s0092-8674(00)80723-0. [DOI] [PubMed] [Google Scholar]
- Rothman JE. Mechanisms of intracellular protein import. Nature. 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- Saalbach G, Rosso M, Schumann U. The vacuolar targeting signal of the 2S albumin from Brazil nut resides at the C terminus and involves the C-terminal propeptide as an essential element. Plant Physiol. 1996;112:975–985. doi: 10.1104/pp.112.3.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sata M, Donaldson JG, Moss J, Vaughan M. Brefeldin A-inhibited guanine nucleotide-exchange activity of Sec7 domain from yeast Sec7 with yeast and mammalian ADP ribosylation factors. Proc Natl Acad Sci USA. 1998;95:4204–4208. doi: 10.1073/pnas.95.8.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder MR, Borkhsenious ON, Matsuoka K, Nakamura K, Raikhel NV. Colocalization of barley lectin and sporamin in vacuoles of transgenic tobacco plants. Plant Physiol. 1993;101:451–458. doi: 10.1104/pp.101.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Matsuoka K, Hamamoto S, Schekman R, Orci L. Coatomer, Arf1p, and nucleotide are required to bud coat protein complex I-coated vesicles from large synthetic liposomes. Proc Natl Acad Sci USA. 1998;95:11199–11204. doi: 10.1073/pnas.95.19.11199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepp JD, Huang K, Lemmon SK. The yeast adaptor protein complex, AP-3, is essential for the efficient delivery of alkaline phosphatase by the alternate pathway to the vacuole. J Cell Biol. 1997;139:1761–1774. doi: 10.1083/jcb.139.7.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi M, Tada M, Saito C, Yashiroda H, Nakano A. Isolation of a tobacco cDNA encoding Sar1 GTPase and analysis of its dominant mutations in vesicular traffic using a yeast complementation system. Plant Cell Physiol. 1998;39:590–599. doi: 10.1093/oxfordjournals.pcp.a029409. [DOI] [PubMed] [Google Scholar]
- Wee EG-T, Sherrier DJ, Prime TA, Dupree P. Targeting of active sialyltransferase to the plant Golgi apparatus. Plant Cell. 1998;10:1759–1768. doi: 10.1105/tpc.10.10.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland F, Harter C. Mechanisms of vesicle formation: insights from the COP system. Curr Opin Cell Biol. 1999;11:440–446. doi: 10.1016/s0955-0674(99)80063-5. [DOI] [PubMed] [Google Scholar]
- Yahara N, Ueda T, Sato K, Nakano A. Multiple roles of Arf1 GTPase in the yeast exocytic and endocytic pathways. Mol Biol Cell. 2001;12:221–238. doi: 10.1091/mbc.12.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Helms JB, Brunner J, Wieland FT. GTP-dependent binding of ADP-ribosylation factor to coatomer in close proximity to the binding site for dilysine retrieval motifs and p23. J Biol Chem. 1999;274:14198–14203. doi: 10.1074/jbc.274.20.14198. [DOI] [PubMed] [Google Scholar]
- Zheng H, von Mollard GF, Kovaleva V, Stevens TH, Raikhel NV. The plant vesicle-associated SNARE AtVTI1a likely mediates vesicle transport from the trans-Golgi network to the prevacuolar compartment. Mol Biol Cell. 1999;10:2251–2264. doi: 10.1091/mbc.10.7.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]