Abstract
Tomato bushy stunt virus and its cell-to-cell movement protein (MP; P22) provide valuable tools to study trafficking of macromolecules through plants. This study shows that wild-type P22 and selected movement-defective P22 amino acid substitution mutants were equivalent for biochemical features commonly associated with MPs (i.e. RNA binding, phosphorylation, and membrane partitioning). This generated the hypothesis that their movement defect was caused by improper interaction between the P22 mutants and one or more host factors. To test this, P22 was used as bait in a yeast (Saccharomyces cerevisiae) two-hybrid screen with a tobacco (Nicotiana tabacum) cDNA library, which identified a new plant homeodomain leucine-zipper protein that reproducibly interacted with P22 but not with various control proteins. These results were confirmed with an independent in vitro binding test. An mRNA for the host protein was detected in plants, and its accumulation was enhanced upon Tomato bushy stunt virus infection of two plant species. The significance of this interaction was further demonstrated by the failure of the homeodomain protein to interact efficiently with two of the well-defined movement-deficient P22 mutants in yeast and in vitro. This is the first report, to our knowledge, that a new plant homeodomain leucine-zipper protein interacts specifically and in a functionally relevant manner with a plant virus MP.
The ability of a plant virus to establish a successful systemic infection requires that it can replicate in the initially infected cell, spread locally by traversing the cell wall through the plasmodesmata, and finally travel to distant parts of the plant via the vascular system. A precise characterization of the virus movement pathway and underlying molecular and biochemical interactions is essential to obtain a comprehensive understanding of the infection process. In addition, such information may expose the existence of general processes that govern transport of endogenous ribonucleoprotein complexes through plants.
The initial transfer of viral material between cells is defined as cell-to-cell movement, which is supported by virus-encoded movement proteins (MPs; Carrington et al., 1996; Gilbertson and Lucas, 1996; Ghoshroy et al., 1997; Nelson and van Bel, 1998; Lazarowitz and Beachy, 1999). MPs of plant viruses with a Tobacco mosaic virus (TMV)-like movement strategy share a number of biochemical characteristics (Carrington et al., 1996; Lazarowitz and Beachy, 1999). Among these is the capacity to bind nucleic acids to form non-virion ribonucleoprotein complexes for transport through plasmodesmata (Citovsky and Zambryski, 1993). MPs associate with microtubules in the cytoskeleton and accumulate in punctate bodies along the cell periphery (Heinlein et al., 1995, 1998; McLean et al., 1995) where they increase the size exclusion limit of plasmodesmata (Ding et al., 1992). Consequently, MPs copurify with the cell wall fraction (Deom et al., 1990). Phosphorylation of MPs by a host kinase (Citovsky et al., 1993) may regulate translation of the viral RNA genome (Karpova et al., 1999) and control the localization of the viral ribonucleoprotein complexes to and through plasmodesmata (Waigmann et al., 2000). These observations collectively reflect the existence of a controlled intracellular trafficking route for ribonucleoprotein complexes that radiates outward through the plasmodesmata.
Despite the common biochemical properties shared among different MPs, their ability to function is determined by the virus-host combination, suggesting a specific interaction with host factors. The TMV P30 and MPs of some other plant viruses bind to a host-encoded pectin methylesterase (Dorokhov et al., 1999; Chen et al., 2000). Different tobacco (Nicotiana tabacum) and/or Arabidopsis proteins, described as a DnaJ protein, a rab acceptor-like protein, and a protein of unknown function, have since been reported to interact with MPs of Tomato spotted wilt virus, Cauliflower mosaic virus, and Turnip crinkle virus (Soellick et al., 2000; Huang et al., 2001; Lin and Heaton, 2001), respectively. In addition, the MP of Tomato spotted wilt virus was recently shown to interact with proteins resembling myosin and kinesin (von Bargen et al., 2001). It has yet to be determined how those unrelated host proteins contribute to virus spread. Various plant proteins including transcription factors also traffic through plasmodesmata (Kragler et al., 2000; Lee et al., 2000; Sessions et al., 2000). Whether this endogenous trafficking is related to virus transport is unknown, but it is tempting to speculate that viruses not only exploit existing transport routes but also commandeer host proteins that have the ability to move through plasmodesmata.
Tomato bushy stunt virus (TBSV; Fig. 1) encodes an MP (P22; Rochon and Johnston, 1991; Dalmay et al., 1993; Scholthof et al., 1995b) that is a typical for many positive-sense RNA viruses (Melcher, 2000). P22 is functional in many plant species, which makes TBSV a good model system for virus movement. A recent mutagenesis study identified three P22 regions enriched for acidic residues that were essential for its movement activity (Fig. 1), but it was not known which biochemical events were affected by the mutations (Chu et al., 1999). The present study shows that wild-type P22 and select P22 amino acid substitution mutants were indistinguishable for several biochemical features commonly associated with viral MPs (e.g. RNA binding, phosphorylation, and membrane targeting). However, two of the P22 mutants were specifically compromised for interaction with a newly identified plant homeodomain Leu-zipper transcription factor. This defined genetic correlation between interaction and function provides supportive evidence for an active involvement of the host factor in directing P22-mediated cell-to-cell transport of TBSV.
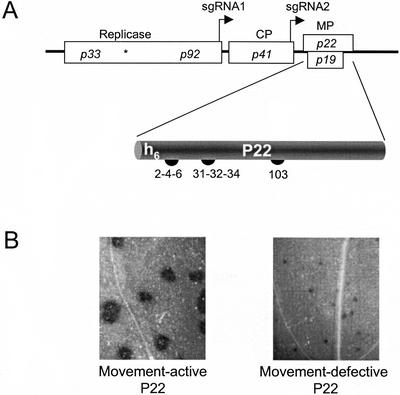
Figure 1.
Schematic representation of the TBSV genome and features of P22 and its movement-defective mutants. A, The TBSV genome. Rectangles represent TBSV open reading frames (ORFs), and the asterisk (*) denotes the p33 amber stop codon to permit read-through translation of the other replicase gene, p92. The transcription initiation sites of two subgenomic RNAs (sgRNA1 and sgRNA2) are indicated by right-angled arrows. CP is the coat protein gene and MP denotes genes for two movement-associated proteins. The p22 ORF product P22 is denoted on the bottom with the position of six N-terminal His residues (h6). The numbers (2-4-6, 31-32-34, and 103) indicate positions of Glu and Asp residues substituted with Ala in the P22 movement-defective mutants used in this study (Chu et al., 1999). B, Comparison of cell-to-cell movement activity of wild-type P22 (left) versus a representative movement-defective P22 mutant (e.g. P22/103; right) using TBSV derivatives expressing the β-glucuronidase reporter gene substituted for the CP gene (Chu et al., 1999).
RESULTS
Biochemical Properties of P22
Purification of Biologically Active P22
Biochemical tests in this study involved the use of His-tagged (his-tag) TBSV P22 fusion proteins (Fig. 1). To ensure that data obtained with those proteins would be biologically relevant, pilot studies were performed to determine whether such manipulations affected the biological activity of P22. Two constructs were generated with his-tag codons at either the 5′ or 3′ end of p22 on the infectious clone of wild-type TBSV. The his-tag insertion at the 3′ end of p22 severely affected the infectivity (data not shown), but transcripts expressing the his-tag at the 5′ end of p22 (NhisP22) established a systemic infection on Nicotiana benthamiana. Symptom development (data not shown) was comparable with that observed upon infection of plants with wild-type virus (Scholthof et al., 1993, 1995b), indicating that the introduction of the his-tag at the N-terminal extremity of P22 did not affect virus movement. Purification of NhisP22 from plants by nickel-nitrilotriacetic acid affinity chromatography required solubilizing the cell wall-associated proteins with 6 m urea (data not shown). Immunoblot analysis of total extracts from infected plants showed that NhisP22 migrated slightly slower than wild-type P22 because of the extra His amino acids, but the overall accumulation was comparable (Fig. 2A). These tests showed that NhisP22 was fully functional for cell-to-cell movement in plants.
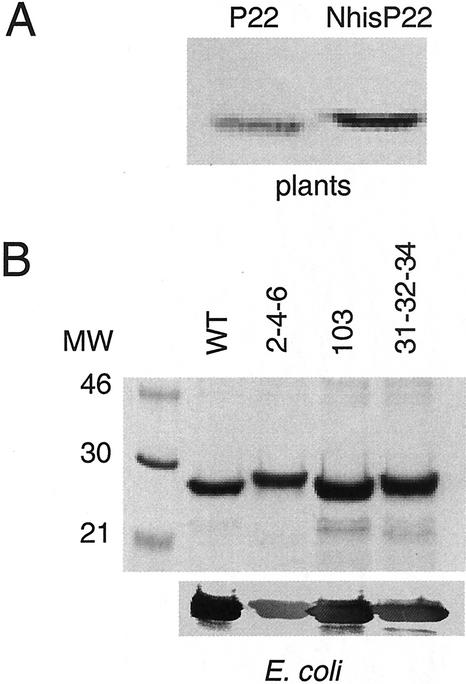
Figure 2.
Expression of NhisP22 in plants and Escherichia coli. A, Immunoblot with P22-specific polyclonal antibodies for detection of P22 in Nicotiana clevelandii plants systemically infected with wild-type TBSV (left) and a mutant expressing NhisP22 (right); the blot is extensively enlarged to visualize the subtle difference in size. B, SDS-PAGE analysis of NhisP22 and the mutant proteins (P22/2-4-6, P22/31-32-34, and P22/103) overexpressed in and purified from E. coli. The 12.5% (w/v) polyacrylamide gel was either stained with Coomassie Brilliant Blue (top) or proteins were transferred to nitrocellulose for detection of P22 with P22-specific polyclonal antibodies (bottom). The molecular mass standards are indicated on the left (in kD). NhisP22 migrates just below the 30-kD marker because of the addition of the his-tag and leader peptide.
To obtain sufficient quantities of NhisP22 (the yield and purity of NhisP22 isolated from plants was insufficient for use in biochemical tests), the protein was overexpressed in Escherichia coli followed by purification. At 37°C, the expression of wild-type P22 in E. coli inhibited the cell growth, but no toxic effect was observed for the mutant constructs expressing P22/2-4-6, P22/31-32-34, and P22/103 (Fig. 1). Upon isopropylthio-β-galactoside induction, P22 accumulated as insoluble inclusion bodies that could only be solubilized and purified with nickel-nitrilotriacetic acid agarose resin in 8 m urea. Likewise, elutions needed to be performed at pH 8 in the presence of 8 m urea with at least 500 mm imidazole. These adaptations resulted in sufficient amounts of proteins that were purified to apparent homogeneity (Fig. 2B, top). Immunoblot assays (Fig. 2B, bottom) showed that the recombinant proteins were specifically recognized by the polyclonal antibodies raised against P22 (Scholthof et al., 1995a, 1995b).
The purified wild-type and mutant P22 proteins were used in comparative analyses regarding the biochemical properties commonly associated with MPs: RNA binding, phosphorylation, and partitioning. It needs to be emphasized at this point that it was not the purpose of these tests to investigate in detail all the parameters and determinants that control the three biochemical activities. Instead, the objective was to compare the behavior of wild-type P22 with that of the selected movement-defective mutants.
RNA Binding
The ability of viral MPs to bind nucleic acids in a cooperative manner has been described for various plant viruses (Citovsky and Zambryski, 1993). Using glutathione-S-transferase (GST)-P22 fusion proteins (Scholthof et al., 1995a), we also obtained cooperative RNA binding in gel retardation assays (data not shown), and RNA binding was similarly evident using a northwestern assay with NhisP22 proteins (Fig. 3A). Because basic amino acids of nucleic acid-binding proteins are generally candidates for interaction with nucleic acids, the change of acidic residues (Asp or Glu) to Ala for the P22 mutants was not predicted to directly affect RNA binding. But whether they imposed an indirect effect could not be ruled out. However, the comparisons showed that binding of 32P-labeled TBSV RNA to the mutant P22/2-4-6, P22/31-32-34, and P22/103 proteins occurred to the same extent as observed for wild type (Fig. 3A). The control proteins, bovine serum albumin (BSA), GST, and a GST-P19 fusion protein did not bind TBSV RNA in these tests (Fig. 3A).
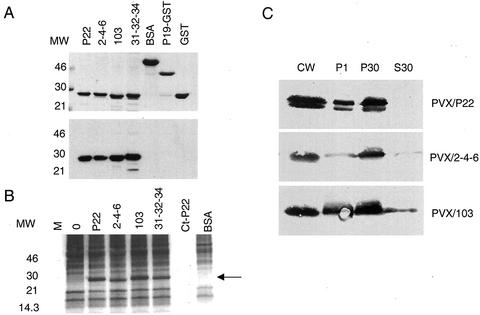
Figure 3.
Biochemical properties of P22 and its movement-defective derivatives. A, Binding of TBSV P22 to 32P-labeled single-stranded TBSV gRNA. Top, Two micrograms of purified NhisP22 proteins and P19-GST and 4 μg of BSA and GST were electrophoresed through a 12.5% (w/v) SDS-polyacrylamide gel and stained with Coomassie Brilliant Blue. The molecular mass standards are indicated on the left. Bottom, After electrophoresis (as in the top) proteins were transferred to a nitrocellulose membrane, and incubated with the full-length 32P-labeled TBSV gRNA followed by exposure to x-ray film. B, In vitro phosphorylation of P22 proteins by a cell wall-associated protein kinase. Eight micrograms of wild-type P22 and movement-defective mutants (P22/2-4-6, P22/31-32-34, and P22/103) was mixed with 25 μL of N. benthamiana cell wall-enriched fraction. M is the lane with molecular mass markers, lane 0 represents a cell wall extract without exogenous P22, and Ct-P22 represents 8 μg of wild-type P22 without the addition of cellular extract. BSA was used as a negative control. Phosphorylated proteins were labeled with [γ-32P]ATP and separated by SDS-PAGE. The dried gel was exposed to an x-ray film, and the position of phosphorylated NhisP22 proteins is marked by the arrow. C, Subcellular localization of P22 in extracts from N. benthamiana 9 d postinoculation with PVX constructs expressing P22 or the movement-defective derivative mutants (PVX/2-4-6 and PVX/103). Leaves were fractionated into cell wall-enriched fractions (CW), organelles and nuclear components (P1), membrane fractions (P30), and a cytosolic fraction (S30). After separation by SDS-PAGE, proteins were transferred to nitrocellulose for immunodetection of P22 with polyclonal P22-specific antiserum. No signal was detected when plants were infected with PVX expressing a nontranslatable p22 gene (pHS160; data not shown; Chu et al., 1999). The significance of the doublet P22 seen for PVX/P22 is not known, but it is not reproducibly associated with functionality for movement.
Phosphorylation
NhisP22 protein was phosphorylated upon incubation with a N. benthamiana cell wall-enriched fraction (Fig. 3B, lane P22). Omission of the plant extract did not result in phosphorylation of P22, confirming that P22 is not capable of autophosphorylation under these circumstances (Fig. 3B, lane Ct-P22). Various unidentified endogenous plant proteins were phosphorylated during the test (Fig. 3B, lane 0), but the extract did not phosphorylate exogenously added BSA. The P22 mutants were also phosphorylated (Fig. 3B), and their movement defect was, therefore, unlikely to be caused by a deficiency in phosphorylation.
Subcellular Localization of P22
Previous fractionation studies showed that P22 expressed from the homologous TBSV genome or the heterologous Potato virus X (PVX) vector (Chapman et al., 1992) partitioned with membrane components (Scholthof et al., 1995a, 1995b). To compare the partitioning of wild-type P22 with that of the mutant proteins, the PVX vector was used to express wild-type and movement-defective P22/2-4-6 and P22/103. The vector RNAs were inoculated onto N. benthamiana followed by subcellular fractionation of extracts (Scholthof et al., 1994) from symptomatic leaves at 9 d postinoculation. P22/31-32-34 was not used in these tests. The results in Figure 3C show that P22 accumulated in fractions enriched for cell wall (CW), organelles and nuclear components (P1), and membranes (P30), but was absent from the soluble fraction (S30). The accumulation of PVX-expressed P22 in the CW fraction was not noticed when P22 was expressed from the homologous TBSV genome (Scholthof et al., 1995a), which suggests that the mode or level of expression may affect the distribution. Nevertheless, the distribution and accumulation of P22/2-4-6 and P22/103 were comparable with what was observed for the wild-type P22 except for a slight presence of P22/103 in the soluble fraction. Therefore, the mutants did not display an obvious aberrance in partitioning.
It is very likely that particular regions on the P22 protein that were not targeted on the mutants used in this study are essential for one or more of the biochemical features tested above. However, the search for such regions was not the objective of this study. Here, we specifically focused on the three available movement-defective P22 mutants to compare their biochemical properties with wild-type P22.
Interaction of P22 with a Novel Protein Encoded by an N. tabacum cDNA
The biochemical studies did not support the hypothesis that the tested P22 movement mutants were dysfunctional for common biochemical properties (i.e. RNA binding, phosphorylation, or subcellular localization). By this process of elimination, the possibility arose that one or more of the movement-defective P22 mutants were compromised for interaction with one or more host proteins. Because P22 belongs to the TMV MP-like class of MPs (Melcher, 2000), we first tested whether P22 interacted with pectin methylesterase that was shown to interact with TMV MP (Chen et al., 2000). However, in our yeast (Saccharomyces cerevisiae) two-hybrid analysis or with an in vitro overlay test, no interaction occurred between pectin methylesterase and TBSV P22 (data not shown). Therefore, it was hypothesized that the P22 mutants were defective for interaction with one or more unknown host proteins.
To identify potential P22-interactive host factors, a yeast two-hybrid screen was employed using P22 as bait to identify binding proteins encoded by prey plasmids representing a N. tabacum (cv Turk) cDNA library. The cDNA library and the bait plasmid pSTT91/p22 were cotransformed into the yeast strain L40, and approximately 2 × 106 transformants were obtained. An interaction was considered positive upon His prototrophic growth and expression of the β-galactosidase gene. Prey plasmids were rescued from 12 His+/β-gal+ yeast colonies, and the specificity of the interaction was verified by transformation into yeast alone, in combination with pSTT91 expressing the structural nuclear protein lamin (a common control; Bartel et al., 1993), or in combination with the original P22 bait plasmid pSTT91/p22. Six of these clones were found to self-activate the transcription of one or both reporter genes, two showed a non-specific interaction with lamin, whereas others lost their ability to give a positive interaction with P22 upon serial passages. However, during these tests one host factor cDNA (p3.5) consistently and reproducibly gave a positive result (Fig. 4).
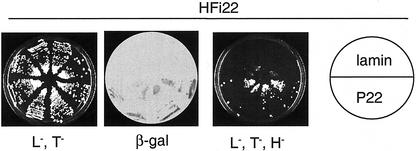
Figure 4.
Specific interaction of P22 with the HFi22 protein encoded by a N. tabacum cDNA. Yeast L40 cells were cotransformed with either the lamin bait construct (pSTT91/lamin) and HFi22 prey plasmids (sections on top one-half of plates) or the P22 bait plus HFi22 plasmids (bottom one-half of plates). Colonies were grown in the absence of Leu and Trp (L−,T−; left) as indicators that both bait and prey plasmids were present in the yeast cells, and a β-galactosidase assay (β-gal) was performed to test for protein interaction (middle). No β-galactosidase reaction was observed with colonies transformed only with the P22 bait or HFi22 prey plasmids (not shown). The plate on the right shows colonies grown in absence of Leu, Trp, and His (L−,T−,H−). Compared with the L−,T− plate, the colony density on this representative L−,T−,H− plate is lower but the size of individual colonies is comparable.
The p3.5 cDNA plasmid encoding the host factor interacting with P22 (HFi22) was further characterized. The HFi22 cDNA (p3.5) was rescued from yeast and was used to transform E. coli for large-scale single-colony plasmid purification by CsCl-ethidium bromide (EtBr) density centrifugation followed by retransformation with various bait proteins into yeast (Fig. 4; Table I). No interaction occurred between HFi22 and a bait containing p22 in the opposite orientation (22p), confirming that the positive result obtained with the HFi22/P22 combination was attributable to protein interactions rather than to nonspecific DNA effects. The specificity of the HFi22-P22 interaction was further evident from the absence of interactions between HFi22 and lamin, TBSV CP, P33, or the p6.6 MP (P6.6) of Panicum mosaic virus, a distantly related member within the Tombusviridae (Turina et al., 1998). TBSV P19, the movement-associated suppressor of gene silencing (Scholthof et al., 1995c; Chu et al., 2000; Qiu et al., 2002), also interacted with HFi22, but this was less pronounced than observed for the HFi22/P22 combination (Table I).
Table I.
Interaction between HFi22 and various proteins upon cotransformation of yeast with corresponding plasmids
| HFi22 Plusa | –b | P22 | P19 | P33 | CP | 22p | P6.6 | Lamin |
|---|---|---|---|---|---|---|---|---|
| Colony growth in L−, T− medium | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| Interactionc | – | +++ | + | – | – | – | – | – |
The HFi22-expressing plasmid (p3.5) was cotransformed with bait plasmids expressing either TBSV P22, P19, P33 or CP, and the 22p plasmid contains p22 in the reverse orientation. Two other control bait plasmids expressed P6.6 of Panicum mosaic virus or lamin.
In this column, only the HFi22-expressing prey plasmid was present, and these colonies were prototrophic only for leucine, whereas for the other columns colonies were also prototrophic for tryptophan because both prey and bait plasmids were present.
The results represent the combined data of growth on histidine-depleted medium and the β-gal assay on colonies grown in parallel in presence of histidine. The +++ indicates rapid growth in absence of histidine and that a blue color appeared 1.5 h after the start of the β-gal reaction. A + indicates a slower growth rate under histidine selection and that the blue color appeared after prolonged incubation (>10 h). A minus (–) indicates no growth in absence of histidine and no blue color developed upon an extended β-gal reaction.
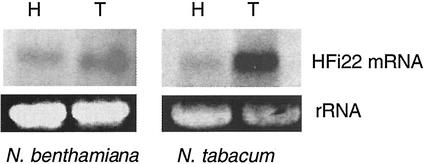
Induction of HFi22 mRNA Accumulation after TBSV Infection
Northern analyses with total RNA from several hosts for TBSV revealed a single mRNA (estimated between 1 and 2 kb) that hybridized to an HFi22-specific probe (Fig. 5). HFi22 mRNA levels were abundant in N. tabacum, a host which supports local infections with TBSV (and served as the donor for the cDNA library). The mRNA was also readily detectable in N. benthamiana, which supports a systemic infection with TBSV. These results were reproducibly verified with reverse transcriptase-PCR assays (data not shown). The HFi22 mRNA was also detectable, albeit at low levels, in pepper (Capsicum annuum) and tomato (Lycopersicon esculentum; data not shown), which under conditions used in our laboratory (e.g. 24°C, daytime temperature) support systemic or local infections, respectively.
Figure 5.
Induction of HFi22 mRNA accumulation upon infection of plants with TBSV. Total RNA was collected from healthy (H) N. benthamiana and N. tabacum or from plants 9 d after inoculation with TBSV (T). RNA was electrophoresed, and loading was verified based on an EtBr-staining signal obtained for the 28S ribosomal RNA (rRNA). RNA was transferred to a membrane and hybridized to an HFi22 cDNA specific probe.
To monitor the effect of TBSV infection on HFi22 mRNA accumulation, total RNA was collected from leaves inoculated 9 d previously with TBSV. The hybridization results showed that the accumulation of HFi22 mRNA was enhanced upon infection compared with its accumulation in healthy plants (Fig. 5). This increase was noticeable in N. benthamiana but was especially prominent in N. tabacum (Fig. 5).
In Vitro Binding of HFi22 to P22
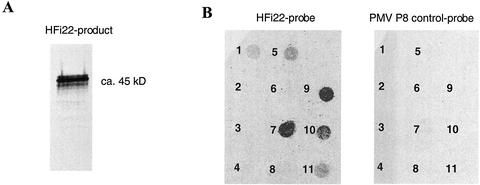
To obtain HFi22 protein, a PCR product was generated containing a 5′ T7 promoter sequence followed by an optimal translational start context (Scholthof et al., 1999a) to generate in vitro HFi22 mRNA transcripts. In vitro translation of these transcripts resulted in the synthesis of a product that, upon mixing with NhisP22, could be precipitated with P22-specific antiserum to yield a major protein band that migrated approximately as a 45-kD protein (Fig. 6A). To verify the specificity of the in vitro P22-HFi22 interaction, the radiolabeled HFi22 product was used to overlay a membrane on which 1 μg of immobilized native purified proteins was spotted (Fig. 6B). These tests showed that HFi22 did not bind to various control proteins (Fig. 6B, details in legend). In contrast to the yeast two-hybrid results, HFi22 did not bind to P19. The interaction observed between HFi22 and the mix of proteins constituting the molecular size markers is probably attributable to the interaction with lysozyme (Fig. 5B, spots 1 and 5).
Figure 6.
Specific interaction of HFi22 with P22 in vitro. A, In vitro-translation product of HFi22 transcripts labeled with [35S]Met and mixed with NhisP22 was coimmunoprecipitated with P22-specific antibodies followed by SDS-PAGE and exposure of the dried gel to x-ray film. B, Overlay of [35S]Met-labeled HFi22-translation product onto nitrocellulose with 1-μg spots of the following proteins: 1, Rainbow molecular size markers (mix of myosin, phosphorylase b, bovine serum albumin, ovalbumin, carbonic anhydrase, trypsin inhibitor, and lysozyme; Amersham, Piscataway, NJ); 2, alkaline phosphatase; 3, ribonuclease; 4, bovine serum albumin; 5, lysozyme; 6, GST; 7, P22; 8, GST-P19; 9, P22/2-4-6; 10, P22/103; and 11, P22/31-32-34. The concentration of proteins present on the membrane was verified before and after loading (see text). No binding occurred with a control translation mix of 35S-labeled P8 of Panicum mosaic virus (Turina et al., 2000; right) or when an untranslatable HFi22 transcript was used to program the in vitro translation (data not shown).
The in vitro binding experiments reproducibly showed that the HFi22 protein had a very strong affinity for P22 (Fig. 6B, spot 7). The specificity was further confirmed because translation mixes programmed with transcripts from an untranslatable HFi22 transcript (data not shown) or containing radiolabeled p8 protein (P8) of Panicum mosaic virus (a common control in the laboratory), did not give a positive signal (Fig. 6). Therefore, these independent tests confirmed the results of the yeast two-hybrid system that the interaction between HFi22 and P22 was specific.
Compromised Interactions between HFi22 and Movement-Defective P22 Mutants
The in vitro binding assays showed that the affinity of HFi22 for P22/2-4-6 (Fig. 6B, spot 9) was comparable with its binding to wild-type P22 (Fig. 6B, spot 7). This suggested that the movement defect of P22/2-4-6 was not related to its binding to HFi22. In contrast, HFi22 had a reduced affinity for P22/103 and P22/31-32-34 (Fig. 6B, spots 10 and 11 compared with spot 7). Because these movement-defective proteins were fully functional for MP-associated biochemical properties (Fig. 3), these findings supported the hypothesis that their compromised binding to HFi22 was responsible for the movement-defect.
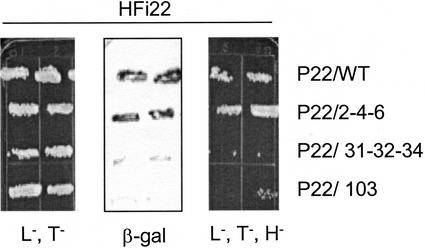
To verify the in vitro results, the genes encoding P22/2-4-6, P22/31-32-34, and P22/103 were inserted into the bait vector for cotransformation of yeast with the HFi22 cDNA. Again, as for the in vitro test (Fig. 6) the interaction between HFi22 and P22/2-4-6 was similar to that observed for wild-type P22 (Fig. 7). However, yeast colonies cotransformed with HFi22 and P22/31-32-34 or P22/103 grew very poorly on triple dropout medium (His−, Leu−, Trp−) and the β-galactosidase activity was very low (Fig. 7), showing that these mutant P22 proteins were compromised for interaction with HFi22.
Figure 7.
Compromised interaction between HFi22 and two movement-defective P22 mutant proteins in yeast. Yeast cells were cotransformed with HFi22 prey plasmid and different P22 bait plasmids as indicated. Duplicate colonies were grown in the absence of Trp and Leu (L−,T−; left), followed by a β-galactosidase assay (middle), or colonies were grown in the absence of Trp, Leu, and His (L−,T−,H−; right).
The in vivo yeast two-hybrid assay and the in vitro overlay independently demonstrated a clear genetic correlation between the involvement of P22 amino acids at positions 31-32-34 and 103 in cell-to-cell movement and their contribution to the interaction with HFi22.
HFi22 Is a Homeodomain Leu-Zipper Protein
Nucleotide sequence analyses showed that the HFi22 cDNA was 1,040 bp in length and contained a 3′-poly(A) stretch, and one uninterrupted ORF was present in-frame with the activation domain of the pGAD prey plasmid. The precise size of the HFi22 mRNA has not yet been determined, but nucleotide sequence comparison of HFi22 with an homologous protein (Tang et al., 2001) suggests that the HFi22 ORF only lacks the coding sequence for the extreme four N-terminal residues. The HFi22 ORF was predicted to encode a protein of 308 amino acids (Fig. 8), which showed homology to many plant-specific homeodomain Leu-zipper proteins. The most extensive similarity (52% identity) was found with a 275-amino acid homeodomain Leu-zipper protein from soybean (Glycine max; Tang et al., 2001). The homology is particularly high (approximately 92% identity) at the N-terminal 85-amino acid portion of the protein. Within this highly conserved region, the homeobox signature is located between amino acids 49 and 72. Three potential Leu zipper motifs L(x)6L(x)6L(x)6L are present at positions 76 to 97, 83 to 104, or 90 to 111. A putative Tyr phosphorylation site is located between amino acids 74 and 80, and a very highly charged region is located between amino acids 151 and 170.
Figure 8.
Amino acid sequence of the HFi22 protein (GenBank accession no. AY 101610). The homeobox signature is underlined. The Tyr phosphorylation site is denoted by the top line, and the three important amino acids are in italics. The six Leu residues of the three possible Leu zippers are noted in bold. Based on sequence comparisons, three yet unknown amino acids are present between the G at position 1 and the upstream Met (see text).
DISCUSSION
TBSV P22 Biochemical Properties
During the past century, many biological and biochemical concepts have been discovered through research on plant viruses (Creager et al., 1999; Scholthof et al., 1999b). Likewise, contemporary studies on virus movement improve our understanding of transport of macromolecules through plants. TBSV provides a particularly good model system to study virus movement because its cell-to-cell movement is controlled by a genetically well-characterized protein (P22), which is functional in numerous plants of various families (Martelli et al., 1988; Russo et al., 1994; Scholthof et al., 1995b).
The movement-defective P22 proteins used in this study were as stable as wild-type P22 in planta, and they were also equally active for the elicitation of wild-type defense responses in Nicotiana edwardsonii and Nicotiana glutinosa (Chu et al., 1999). These features suggested that their overall three dimensional structure was not compromised. Therefore, it was likely that the targeted charged amino acids (Asp and Glu), predicted to be exposed on the protein surface, were required for specific interaction with viral proteins or RNA, or with host factors. In a separate study, we obtained evidence that P22 does not self-interact, neither did it interact with other TBSV proteins in a yeast two-hybrid system, or with NhisP19 pull-down experiments from infected plants (J.-W. Park, B. Desvoyes, and H.B. Scholthof, unpublished data). This led us to investigate interactions of P22 with viral RNA and host factors and to compare such biochemical properties of purified P22 with that of the movement-defective P22 mutants.
Insertion of a his-tag coding region at the C terminus of P22 severely compromised viral replication presumably by disrupting an essential cis-acting RNA element (Ray and White, 1999; Park et al., 2000). However, an N-terminal his-tag P22 (NhisP22) was active for movement, which encouraged a strategy to use E. coli-mediated expression and subsequent purification of NhisP22 proteins. The use of these proteins in biochemical tests showed that the RNA-binding properties of P22 and the P22 movement-defective mutants were comparable, which suggested that they form native ribonucleoprotein complexes with the genomic RNA. Phosphorylation of MPs plays an important regulatory role for movement (Lee and Lucas, 2001), and our results showed that P22 was phosphorylated by a protein kinase associated with N. benthamiana and N. tabacum cell walls. Although acidic amino acids (Glu and Asp) affected in the TBSV P22 mutants used in this study were unlikely substrates for phosphorylation, they could provoke subtle protein conformational changes to prevent kinase access. However, our tests showed that the movement-defect of the mutant P22 proteins was not caused by a deficiency in phosphorylation. Likewise, the P22/2-4-6 and P22/103 movement-defective proteins appeared to properly associate with membranes, including those in plasmodesmata embedded within the cell wall. This agrees with preliminary results from confocal microscopy studies with GFP-P22 fusion proteins, which suggest that the mutant P22/103 behaves similarly to wild-type P22 regarding the membrane association (T. Rubio, H.B. Scholthof, and A.O. Jackson, unpublished data). This is reminiscent of TMV P30 movement-deficient mutants that associate with plasmodesmata (Boyko et al., 2000; Mas and Beachy, 2000). These observations suggest that subcellular localization of MPs is not strictly linked to the movement function but that other factors are involved.
Biologically Significant Interaction of P22 with a New Homeodomain Protein
Biochemical characterization of P22 and its mutants led to the hypothesis that some P22 mutants failed to properly associate with one or more host proteins. To test this, a yeast two-hybrid screen was performed with a N. tabacum cDNA library. This yielded a cDNA clone encoding a protein (HFi22) that specifically interacted with P22. Using several variations on western overlay assays with crude plant extracts (Dorokhov et al., 1999; Chen et al., 2000), we have noticed that P22 binds to an approximately 45-kD N. tabacum cell wall-associated protein (data not shown). Although this protein is similar in size to the approximately 45-kD in vitro-translated HFi22 product, it remains to be determined whether these are the same proteins.
The binding of HFi22 to P22 was verified upon overlay of membranes with in vitro-translated HFi22. Thus, the specific and reproducible binding of HFi22 to P22 in two independent assays (yeast two-hybrid and overlay) affirms that this reflects a specific protein-protein interaction. Furthermore, HFi22 mRNA accumulated in hosts for TBSV, most notably in N. tabacum, and the expression of HFi22 mRNA was substantially enhanced upon TBSV infection of N. tabacum, and to a lesser extent in N. benthamiana. These observations provide evidence that HFi22 is a biologically significant participant in TBSV infections. We previously established that P22 does not induce any noticeable symptoms in N. tabacum (Scholthof et al., 1995a). Therefore, the P22-HFi22 interaction is unlikely involved in activation of a typical gene-for-gene resistance response. Instead, it is likely that the responsiveness of HFi22 expression to TBSV infection is a reflection of its participation in the infection process.
Cotransformation of yeast with plasmids expressing HFi22 and the defective P22 mutants P22/31-32-34 or P22/103 resulted in reduced growth on His-depleted medium and a lower β-galactosidase activity compared with that observed for wild-type P22, indicating that the interaction was ineffective. Essentially the same results were obtained during in vitro binding tests that also showed a weaker binding of HFi22 to P22/103 and most notably to P22/31-32-34. In contrast, the N-terminal mutant P22/2-4-6 behaved like wild-type P22 regarding its binding to HFi22, which like P22/103 or P22/31-32-34 also had acidic amino acids changed (Chu et al., 1999). These results signify that the essential amino acids at positions 2-4-6 are required for an as-yet-unknown reason—perhaps they interact with a host receptor? However, the most important evidence from this observation is that not every charged cluster on the P22 protein is required for interaction with HFi22. Instead, this is specifically determined by the two charged regions represented by positions 31-32-34 and 103. Considerations that these amino acids are not involved in MP-associated biochemical interactive events discussed above and that these defined amino acids that interact with HFi22 are also crucial for the movement function (Chu et al., 1999) provide genetic arguments that the interaction is biologically relevant.
Virus Movement and Plant Homeodomain Proteins
The predicted HFi22 protein sequence showed a high degree of similarity with a class of plant-specific homeodomain Leu-zipper proteins (Chan et al., 1998). This represents an entirely new class of proteins for interaction with a virus MP. Other plant proteins reported to interact with MPs (Dorokhov et al., 1999; Chen et al., 2000; Soellick et al., 2000; Huang et al., 2001; Lin and Heaton, 2001) have no similarity with homeodomain Leu-zipper proteins. However, the homeodomain transcription factor KNOTTED 1 traffics between cells through the plasmodesmata and is able to specifically transport its mRNA (Lucas et al., 1995; Lee et al., 2000). Other transcription factors and regulatory proteins and their mRNAs also move between cells (Hake, 2001; Jackson, 2001). Examples are DEF in snapdragon (Antirrhinum majus) and LEAFY and APETALA1 in Arabidopsis that have also been shown to move from cell to cell (Perbal et al., 1996; Lee et al., 2000; Sessions et al., 2000) or even long distance (Kim et al., 2001).
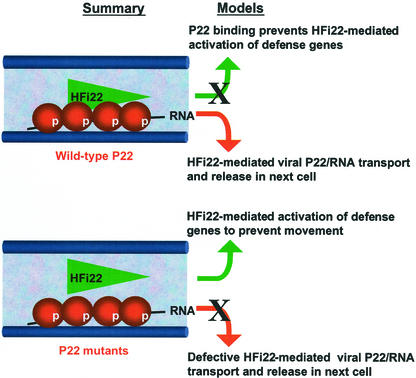
Our observations, in combination with the above cited literature, invite the hypothesis that binding of HFi22 to P22 enables directional transport of P22/RNA complexes through plasmodesmata for cell-to-cell movement (Fig. 9). A direct interpretation of our results is that the wild-type P22/RNA complex enlists HFi22 in order to be transported through plasmodesmata (Fig. 9). Failure of the P22 mutants to use HFi22 for this purpose may account for the entrapment of mutant P22/RNA complexes inside plasmodesmata. In a second, mutually inclusive scenario, it is possible that binding of wild-type P22 to HFi22 prevents (i.e. suppresses) the transcription factor from activating expression of one or more broad-acting defense genes, whereas the failure of the P22 mutants to bind to HFi22 results in a defense that prevents movement (Fig. 9). This could be related to a recent suggestion that TMV P30 and other virus MPs interact with a transcriptional coactivator protein to influence host gene expression (Matsushita et al., 2001).
Figure 9.
A summary of findings on P22 properties and its interaction with HFi22 and a model how HFi22 might participate in P22-mediated movement. Summary, The wild-type and mutant P22 (beads) residues are phosphorylated (p), form a complex with viral RNA, and are partitioned to membranes, including those membranes (thick bars) that traverse plasmodesmata in cell walls. Wild-type P22 interacts with HFi22 (triangle), but the mutant P22 proteins fail to establish this interaction. Model, Two mutually inclusive mechanisms are indicated. In the first, cell-to-cell movement occurs because P22 prevents HFi22-mediated activation of defense genes (X on top), whereas in the second, HFi22 enables and directs channeling of the wild-type P22/RNA complex through plasmodesmata, but this is defective for the P22 mutants (X on bottom).
CONCLUSION
Although a number of plant proteins have recently been identified that interact with virus MPs (Dorokhov et al., 1999; Chen et al., 2000; Soellick et al., 2000; Huang et al., 2001; Lin and Heaton, 2001; von Bargen et al., 2001), how these participate in movement is not known for any of those because their biological role remains to be determined. This is also the challenge ahead for the HFi22-P22 interaction. But in comparison, our results with TBSV P22 have provided one of the most precise correlations between MP function and its ability to interact with a host homeodomain protein not described previously, to our knowledge. We are optimistic that future efforts to either overexpress or inactivate the HFi22 gene in planta should resolve its explicit role in the TBSV infection process to further our understanding of virus transport and movement of macromolecules in plants.
MATERIALS AND METHODS
Expression of His-Tag P22 in Planta
A sequence encoding six His residues was introduced at the 5′ end of p22 by PCR using the forward primer gcccccagttc atg gct cat cac cat cac cat cac gat act gaa tac gaa caa; this primer contains the PflmI site (underlined) present at the 5′ end of p22, the start codon (italics), codons encoding the six His residues (bold), and p22 5′ nucleotides. The reverse primer is complementary to the 3′ end of pTBSV-100 (Scholthof and Jackson, 1997). The 950-bp PCR product was digested with PflmI and SalI and was used to replace the homologous PflmI-SalI fragment in pTBSV-100. Transcripts of the resulting construct, pT22Nhis, were generated in vitro using T7 RNA polymerase and inoculated onto Nicotiana benthamiana as described previously (Scholthof et al., 1993).
Purification of P22 Expressed in Escherichia coli
Plasmids pTBSV-100, pT22/2-4-6, pT22/31-32-34, and pT22/103 (Chu et al., 1999) were used as a source for TBSV wild-type and mutant p22 genes. The p22 genes were released by digestion with PflmI (nucleotide 3,848) filled-in with DNA Polymerase I Klenow fragment (Klenow treatment), followed by digestion with SalI (nucleotide 4,499). The fragments were gel-purified and cloned into the expression vector pRSETC (Invitrogen, Carlsbad, CA) that had been digested with BamHI (Klenow treated) and XhoI. The 5′ junction of each constructs was sequenced to verify the preservation of the reading frame. The resulting construct was introduced into E. coli BL21 (DE3, pLysS); 200-mL cultures were incubated at 30°C for pRSETCp22T (expressing wild-type P22) and at 37°C for the mutant constructs until the A600 reached 0.7 unit. Protein expression was induced by adding isopropylthio-β-galactoside (final concentration, 1 mm) and cultures were incubated for an additional 3 h at 30°C, as above. The bacterial pellets were collected by low speed centrifugation (5 min, 5,000g), resuspended in 10 mL of TN buffer (20 mm Tris-HCl, pH 8, and 100 mm NaCl) containing 5 mm imidazole, and sonicated on ice by three pulses of 30 s each. The P22 inclusion bodies were recovered by centrifugation and resuspended in 8 mL of the TNU buffer (TN containing 8 m urea) and 5 mm imidazole. The bacterial lysates were clarified and incubated in batch with 600 μL of nickel-nitrilotriacetic acid agarose beads (Qiagen USA, Valencia, CA) for 1 h at 25°C. The beads were washed four times with 5 mL of TNU buffer containing 35 mm imidazole, and the recombinant P22 was eluted with 500 mm imidazole in TNU buffer (two times 500 μL). The his-tag P22 was dialyzed twice against 1 L of 20 mm Tris, pH 7.4, and 5% (v/v) glycerol, aliquoted, and kept at −70°C. Protein concentrations were determined using a bicinchoninic acid assay (Pierce, Rockford, IL).
Northwestern Assay, Subcellular Fractionation, Phosphorylation, and RNA Hybridization
RNA-binding assays were performed essentially as described previously (Desvoyes and Scholthof, 2000). For fractionation studies, capped transcripts of the PVX constructs were inoculated onto plants (Scholthof et al., 1993, 1995a; Chu et al., 1999), followed by subcellular fractionation of infected tissue following a routine procedure (Scholthof et al., 1994). Preparations of the cell wall-enriched fraction and phosphorylation assays were also adapted from previous protocols (Citovsky et al., 1993). RNA isolation, denaturing gel electrophoresis, and hybridizations with randomly primed gel-isolated DNA were all performed according to standard procedures (Sambrook et al., 1989; Scholthof et al., 1993, 1995c).
Yeast (Saccharomyces cerevisiae) Two-Hybrid Assay
The yeast two-hybrid screens were performed with the yeast strain L40 (Hollenberg et al., 1995), a bait plasmid pSTT91 (a pBTM116 derivative), and the prey plasmid pGAD424 (CLONTECH Laboratories, Palo Alto, CA) containing a cDNA library of tobacco (Nicotiana tabacum) fused with the activation domain of GAL4. The library contains over 2 × 106 different cDNAs with an average size of 1.3 kb cloned as EcoRI-XhoI fragments into the EcoRI-SalI sites of pGAD424. All of these materials were a generous gift from V. Citovsky (State University of New York, Stony Brook). The PflmI (Klenow treated) to SalI TBSV cDNA fragment containing p22 was cloned into the pSTT91 plasmid that was digested with BamHI (Klenow treated) and SalI to yield pSTT91/p22. Yeast L40 cells were transformed with both plasmids using a standard transformation protocol (Hollenberg et al., 1995), and the transformants were plated onto a synthetic dropout medium lacking Leu, Trp, and His and incubated at 30°C for 4 to 8 d. Visible colonies were tested for their β-galactosidase activity (Hollenberg et al., 1995), and positives were selected by passaging on Leu-Trp dropout medium. Plasmid DNA was prepared from these positive clones and transformed into E. coli and used for large scale CsCl-EtBr gradient DNA isolation (Sambrook et al., 1989). The purified prey plasmids were used for cotransformations and DNA sequence analyses. Details on cloning of control bait constructs used in Table I will be described elsewhere (H.B. Scholthof, unpublished data), but in essence, they all involved direct or PCR-mediated in-frame cloning of ORFs into pSTT91.
In Vitro Overlay
The N. tabacum HFi22 protein was obtained by in vitro transcription/translation (TNT T7, Promega, Madison, WI). For this purpose, a PCR product was generated to serve as a template for in vitro transcription. PCR was performed with HFi22 cDNA template and a forward primer T7–3.5wt2 (5′-cc gga att cct aat acg act cac tat aga gaa caa gac caa acc atg gaa atg ggc acg aga tta-3′, 100 pmol) and reverse primer GADSeqRev (5′-atg gtg cac gat gca cag-3′, 56 pmol) in presence of 2 mm MgSO4, 250 μm each of dNTP, and 2 units of Vent DNA Polymerase (New England BioLabs, Beverly, MA). The T7–3.5wt2 primer contains a 5′ EcoRI site (underlined), a T7 promoter (bold), an optimal 5′ leader and translational start-site context (italics; Scholthof et al., 1999a) with two in-frame start codons (bold italics), and the 3′ sequences represent the 5′ end of the HFi22 cDNA. Two microliters of the PCR product was used as template for the in vitro transcription/translation reaction in presence of [35S]Met (Scholthof et al., 1999a) according to the manufacturer's instructions (Promega, Madison, WI). The HFi22 in vitro-translated product was analyzed by SDS-PAGE.
One microgram of the pure native proteins, as quantified with Bradford reagent (Sigma-Aldrich, St. Louis) was blotted onto a nitrocellulose membrane through a “slot-blot” manifold (Bio-Rad, Richmond, CA), and presence of proteins was verified with Ponceau S staining of membranes. Blots were blocked for 2 h in 140 mm NaCl, 10 mm Tris-HCl, pH 7.4, 2 mm EDTA, 1% (w/v) BSA, 0.1% (v/v) Tween 20, and 2 mm dithiothreitol (Chen et al., 2000) at room temperature. Then [35S]Met labeled in vitro-translation mixes were added to the blots for overnight incubation at 4°C. Subsequently the blots were washed four times (5 min each) in Tween-Tris-buffered saline (Tris-buffered saline plus 0.05% [v/v] Tween 20) and exposed overnight on x-ray film.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
ACKNOWLEDGMENTS
We thank Vitaly Citovsky for his various contributions and assistance. We thank Karen-Beth G. Scholthof, Wenping Qiu, Jeff Batten, and Rustem Omarov for valuable suggestions during the experiments and preparation of the manuscript. We appreciate the contribution of the GUS-assay results by Meihua Chu.
Footnotes
This work was supported by the Texas Agricultural Experiment Station (grant no. TEX08387), by the U.S. Department of Agriculture/CSREES-National Research Initiative Competitive Grants Program (grant no. 99–35303–8022), by the Texas Higher Education Coordinating Board Advanced Technology Program (grant no. 000517–0070–1999), and by the S.R. Noble Foundation, Inc.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.004754.
LITERATURE CITED
- Bartel P, Chien C, Sternglanz R, Fields S. Elimination of false positives that arise in using the two-hybrid system. Biotechniques. 1993;14:920–924. [PubMed] [Google Scholar]
- Boyko V, van der Laak J, Ferralli J, Suslova E, Kwon M-O, Heinlein M. Cellular targets of functional and dysfunctional mutants of tobacco mosaic virus. J Virol. 2000;74:11339–11346. doi: 10.1128/jvi.74.23.11339-11346.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington JC, Kasschau KD, Mahajan SK, Schaad MC. Cell-to-cell and long distance transport of viruses in plants. Plant Cell. 1996;8:1669–1681. doi: 10.1105/tpc.8.10.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan RL, Gago GM, Palena CM, Gonzalez DH. Homeoboxes in plant development. Biochim Biophys Acta. 1998;1442:1–19. doi: 10.1016/s0167-4781(98)00119-5. [DOI] [PubMed] [Google Scholar]
- Chapman S, Kavanagh T, Baulcombe D. Potato virus X as a vector for gene expression in plants. Plant J. 1992;2:549–557. doi: 10.1046/j.1365-313x.1992.t01-24-00999.x. [DOI] [PubMed] [Google Scholar]
- Chen M-H, Sheng J, Hind G, Handa AK, Citovsky V. Interaction between the tobacco mosaic virus movement protein and host cell pectin methylesterases is required for viral cell-to-cell movement. EMBO J. 2000;19:913–920. doi: 10.1093/emboj/19.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu M, Desvoyes B, Turina M, Noad R, Scholthof HB. Genetic dissection of tomato bushy stunt virus p19-protein-mediated host-dependent symptom induction and systemic invasion. Virology. 2000;266:79–87. doi: 10.1006/viro.1999.0071. [DOI] [PubMed] [Google Scholar]
- Chu M, Park J-W, Scholthof HB. Separate regions on the tomato bushy stunt virus p22 protein mediate cell-to-cell movement versus elicitation of effective resistance responses. Mol Plant-Microbe Interact. 1999;12:285–292. [Google Scholar]
- Citovsky V, McLean BG, Zupan JR, Zambryski P. Phosphorylation of tobacco mosaic virus cell-to-cell movement protein by a developmentally regulated plant cell wall-associated protein kinase. Genes Dev. 1993;7:904–910. doi: 10.1101/gad.7.5.904. [DOI] [PubMed] [Google Scholar]
- Citovsky V, Zambryski P. Transport of nucleic acids through membrane channels: snaking through small holes. Annu Rev Microbiol. 1993;47:167–197. doi: 10.1146/annurev.mi.47.100193.001123. [DOI] [PubMed] [Google Scholar]
- Creager ANH, Scholthof K-BG, Citovsky V, Scholthof HB. Tobacco mosaic virus: pioneering research for a century. Plant Cell. 1999;11:301–308. doi: 10.1105/tpc.11.3.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay T, Rubino L, Burgyan J, Kollar A, Russo M. Functional analysis of cymbidium ringspot virus genome. Virology. 1993;194:697–704. doi: 10.1006/viro.1993.1310. [DOI] [PubMed] [Google Scholar]
- Deom CM, Schubert KR, Wolf S, Holt CA, Lucas WJ, Beachy RN. Molecular characterization and biological function of the movement protein of tobacco mosaic virus in transgenic plants. Proc Natl Acad Sci USA. 1990;87:3284–3288. doi: 10.1073/pnas.87.9.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvoyes B, Scholthof K-BG. RNA:protein interactions associated with satellites of panicum mosaic virus. FEBS Lett. 2000;485:25–28. doi: 10.1016/s0014-5793(00)02177-3. [DOI] [PubMed] [Google Scholar]
- Ding B, Haudenshield JS, Hull RJ, Wolf S, Beachy RN, Lucas WJ. Secondary plasmodesmata are specific sites of localization of the tobacco mosaic virus movement protein in transgenic tobacco plants. Plant Cell. 1992;4:915–928. doi: 10.1105/tpc.4.8.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorokhov YL, Makinen K, Frolova OY, Merits A, Saarinen J, Kalkkinen N, Atabekov JG, Saarma M. A novel function for a ubiquitous plant enzyme pectin methylesterase: the host-cell receptor for the tobacco mosaic virus movement protein. FEBS Lett. 1999;461:223–228. doi: 10.1016/s0014-5793(99)01447-7. [DOI] [PubMed] [Google Scholar]
- Ghoshroy S, Lartey R, Sheng J, Citovsky V. Transport of proteins and nucleic acids through plasmodesmata. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:27–49. doi: 10.1146/annurev.arplant.48.1.27. [DOI] [PubMed] [Google Scholar]
- Gilbertson RL, Lucas WJ. How do viruses traffic on the “vascular highway.”. Trends Plant Sci. 1996;1:260–268. [Google Scholar]
- Hake S. Transcription factors on the move. Trends Genet. 2001;17:2–3. doi: 10.1016/s0168-9525(00)02163-6. [DOI] [PubMed] [Google Scholar]
- Heinlein M, Epel BL, Padgett HS, Beachy RN. Interaction of tobamovirus movement protein with the plant cytoskeleton. Science. 1995;270:1983–1985. doi: 10.1126/science.270.5244.1983. [DOI] [PubMed] [Google Scholar]
- Heinlein M, Padgett HS, Gens JS, Pickard BG, Casper SJ, Epel BL, Beachy RN. Changing patterns of localization of the tobacco mosaic virus movement protein and replicase to the endoplasmic reticulum and microtubules during infection. Plant Cell. 1998;10:1107–1120. doi: 10.1105/tpc.10.7.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenberg SM, Sternglanz R, Cheng PF, Weintraub H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Andrianov VM, Han Y, Howell SH. Identification of Arabidopsis proteins that interact with the cauliflower mosaic virus (CaMV) movement protein. Plant Mol Biol. 2001;47:663–675. doi: 10.1023/a:1012491913431. [DOI] [PubMed] [Google Scholar]
- Jackson D. The long and the short of it: signalling development through plasmodesmata. Plant Cell. 2001;13:2569–2570. doi: 10.1105/tpc.13.12.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpova OV, Rodionova NP, Ivanov KI, Kozlovsky SV, Dorokhov YL, Atabekov JG. Phosphorylation of tobacco mosaic virus movement protein abolishes its translation repressing ability. Virology. 1999;161:20–24. doi: 10.1006/viro.1999.9842. [DOI] [PubMed] [Google Scholar]
- Kim M, Canio W, Kessler S, Sinha HN. Developmental changes due to long-distance movement of a homeobox fusion transcript in tomato. Science. 2001;293:287–289. doi: 10.1126/science.1059805. [DOI] [PubMed] [Google Scholar]
- Kragler F, Monzer J, Xoconostle-Cazares B, Lucas WJ. Peptide antagonists of the plasmodesmal macromolecular trafficking pathway. EMBO J. 2000;19:2856–2868. doi: 10.1093/emboj/19.12.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarowitz SG, Beachy RN. Viral movement proteins as probes for intracellular and intercellular trafficking in plants. Plant Cell. 1999;11:535–548. doi: 10.1105/tpc.11.4.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-Y, Lucas WJ. Phosphorylation of viral movement proteins: regulation of cell-to-cell trafficking. Trends Microbiol. 2001;9:5–8. doi: 10.1016/s0966-842x(00)01901-6. [DOI] [PubMed] [Google Scholar]
- Lee J-Y, Yoo B-C, Lucas WJ. Parallels between nuclear-pore and plasmodesmal trafficking of information molecules. Planta. 2000;210:177–187. doi: 10.1007/PL00008124. [DOI] [PubMed] [Google Scholar]
- Lin B, Heaton LA. An Arabidopsis thaliana protein interacts with a movement protein of turnip crinkle virus in yeast cells and in vitro. J Gen Virol. 2001;82:1245–1251. doi: 10.1099/0022-1317-82-5-1245. [DOI] [PubMed] [Google Scholar]
- Lucas WJ, Bouche-Pillon S, Jackson DP, Nguyen L, Baker L, Ding B, Hake S. Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science. 1995;270:1980–1985. doi: 10.1126/science.270.5244.1980. [DOI] [PubMed] [Google Scholar]
- Martelli GP, Gallitelli D, Russo M. Tombusviruses. In: Koenig R, editor. The Plant Viruses. New York: Plenum Publishing; 1988. pp. 13–72. [Google Scholar]
- Mas P, Beachy RN. Role of microtubules in the intracellular distribution of tobacco mosaic virus movement protein. Proc Natl Acad Sci USA. 2000;97:12345–12349. doi: 10.1073/pnas.97.22.12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita Y, Deguchi M, Youda M, Nishiguchi M, Nyunoya H. The tomato mosaic tobamovirus movement protein interacts with a putative transcriptional coactivator KELP. Mol Cells. 2001;12:57–66. [PubMed] [Google Scholar]
- McLean BG, Zupan J, Zambryski PC. Tobacco mosaic virus movement protein associates with the cytoskeleton in tobacco cells. Plant Cell. 1995;7:2101–2114. doi: 10.1105/tpc.7.12.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher U. The “30K” superfamily of viral movement proteins. J Gen Virol. 2000;81:257–266. doi: 10.1099/0022-1317-81-1-257. [DOI] [PubMed] [Google Scholar]
- Nelson RS, van Bel AJE. The mystery of virus trafficking into, through and out of vascular tissue. Cell Biol Physiol. 1998;59:476–533. [Google Scholar]
- Park J-W, Desvoyes B, Scholthof HB. Tomato bushy stunt virus RNA accumulation is affected by interdependent cis-acting RNA elements. Phytopathology. 2000;90:S59. doi: 10.1128/JVI.76.24.12747-12757.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perbal MC, Haughn G, Saedler H, Schwarz-Sommer Z. Non-cell-autonomous function of the Antirrhinum floral homeotic proteins DEFICIENS and GLOBOSA is exerted by their polar cell-to-cell trafficking. Development. 1996;122:3433–3441. doi: 10.1242/dev.122.11.3433. [DOI] [PubMed] [Google Scholar]
- Qiu WP, Park J-W, Scholthof HB. Tombusvirus P19-mediated suppression of virus induced gene silencing is controlled by genetic and dosage features that influence pathogenicity. Mol Plant-Microbe Interact. 2002;15:269–280. doi: 10.1094/MPMI.2002.15.3.269. [DOI] [PubMed] [Google Scholar]
- Ray D, White KA. Enhancer-like properties of an RNA element that modulates tombusvirus RNA accumulation. Virology. 1999;256:162–171. doi: 10.1006/viro.1999.9630. [DOI] [PubMed] [Google Scholar]
- Rochon DM, Johnston JC. Infectious transcripts from cloned cucumber necrosis virus cDNA: evidence for a bifunctional subgenomic mRNA. Virology. 1991;181:656–665. doi: 10.1016/0042-6822(91)90899-m. [DOI] [PubMed] [Google Scholar]
- Russo M, Burgyan J, Martelli GP. Molecular biology of Tombusviridae. Adv Virus Res. 1994;44:381–428. doi: 10.1016/s0065-3527(08)60334-6. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Scholthof HB, Desvoyes B, Kuecker J, Whitehead E. The biological activity of two tombusvirus proteins translated from nested genes is influenced by dosage control via context-dependent leaky scanning. Mol Plant-Microbe Interact. 1999a;12:670–679. [Google Scholar]
- Scholthof HB, Jackson AO. The enigma of pX: a host dependent cis-acting element with variable effects on tombusvirus RNA accumulation. Virology. 1997;237:56–65. doi: 10.1006/viro.1997.8754. [DOI] [PubMed] [Google Scholar]
- Scholthof HB, Morris TJ, Jackson AO. The capsid protein gene of tomato bushy stunt virus is dispensable for systemic movement and can be replaced for localized expression of foreign genes. Mol Plant-Microbe Interact. 1993;6:309–322. [Google Scholar]
- Scholthof HB, Scholthof K-BG, Jackson AO. Identification of tomato bushy stunt virus host-specific symptom determinants by expression of individual genes from a potato virus X vector. Plant Cell. 1995a;7:1157–1172. doi: 10.1105/tpc.7.8.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholthof HB, Scholthof K-BG, Kikkert M, Jackson AO. Tomato bushy stunt virus spread is regulated by two nested genes that function in cell-to-cell movement and host-dependent systemic invasion. Virology. 1995b;213:425–438. doi: 10.1006/viro.1995.0015. [DOI] [PubMed] [Google Scholar]
- Scholthof K-BG, Hillman BI, Modrell B, Heaton LA, Jackson AO. Characterization and detection of sc4: a sixth gene encoded by sonchus yellow net virus. Virology. 1994;204:279–288. doi: 10.1006/viro.1994.1532. [DOI] [PubMed] [Google Scholar]
- Scholthof K-BG, Scholthof HB, Jackson AO. The tomato bushy stunt virus replicase proteins are coordinately expressed and membrane associated. Virology. 1995c;208:365–369. doi: 10.1006/viro.1995.1162. [DOI] [PubMed] [Google Scholar]
- Scholthof K-BG, Shaw JG, Zaitlin M. Tobacco Mosaic Virus: One Hundred Years of Contributions to Virology. St. Paul: American Phytopathological Society; 1999b. [Google Scholar]
- Sessions A, Yanofsky MF, Weigel D. Cell-cell signalling and movement by the floral transcription factors LEAFY and APETALA1. Science. 2000;289:779–781. doi: 10.1126/science.289.5480.779. [DOI] [PubMed] [Google Scholar]
- Soellick T-R, Uhrig JF, Buchner GL, Kellmann J-W, Schreier PH. The movement protein NSm of tomato spotted wilt tospovirus (TSWV): RNA binding, interaction with the TSWV N protein, and identification of interacting plant proteins. Proc Natl Acad Sci USA. 2000;97:2373–2378. doi: 10.1073/pnas.030548397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Sadka A, Morishige DT, Mullet JE. Homeodomain leucine zipper proteins bind to the phosphate response domain of the soybean VspB tripartite promoter. Plant Physiol. 2001;125:797–809. doi: 10.1104/pp.125.2.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turina M, Desvoyes B, Scholthof K-BG. A gene cluster encoded by panicum mosaic virus is associated with virus movement. Virology. 2000;266:120–128. doi: 10.1006/viro.1999.0069. [DOI] [PubMed] [Google Scholar]
- Turina M, Maruoka M, Monis J, Jackson AO, Scholthof K-BG. Nucleotide sequence and infectivity of a full-length cDNA clone of panicum mosaic virus. Virology. 1998;241:141–155. doi: 10.1006/viro.1997.8939. [DOI] [PubMed] [Google Scholar]
- von Bargen S, Salchert K, Paape M, Piechulla B, Kellmann J-W. Interactions between the tomato spotted wilt virus movement protein and plant proteins showing homologies to myosin, kinesin, and DnaJ-like chaperons. Plant Physiol Biochem. 2001;39:1083–1093. [Google Scholar]
- Waigmann E, Chen M-H, Bachmaier R, Ghoshroy S, Citovsky V. Regulation of plasmodesmal transport by phosphorylation of tobacco mosaic virus cell-to-cell movement protein. EMBO J. 2000;19:4875–4884. doi: 10.1093/emboj/19.18.4875. [DOI] [PMC free article] [PubMed] [Google Scholar]