Abstract
Abscisic acid (ABA) and stress response from late embryonic growth through early seedling development is regulated by a signaling network that includes the Arabidopsis ABA-insensitive (ABI)5 gene, which encodes a basic leucine zipper transcription factor. We have characterized genetic, developmental, and environmental regulation of ABI5 expression. Although expressed most strongly in seeds, the ABI5 promoter is also active in vegetative and floral tissue. Vegetative expression is strongly induced by ABA, and weakly by stress treatments during a limited developmental window up to approximately 2 d post-stratification, but ABA and some stresses can induce expression in specific tissues at later stages. ABI5 expression is autoregulated in transgenic plants and yeast (Saccharomyces cerevisiae), and stress response appears to involve ABI5-dependent and -independent mechanisms. To determine whether ABI5 is necessary and/or sufficient for ABA or stress response, we assayed the effects of increased ABI5 expression on growth and gene expression. Although overexpression of ABI5 confers hypersensitivity to ABA and sugar, as previously described for ABI4 and ABI3 overexpression lines, it has relatively limited effects on enhancing ABA-responsive gene expression. Comparison of expression of eight ABI5-homologous genes shows overlapping regulation by ABI3, ABI4, and ABI5, suggestive of a combinatorial network involving positive and negative regulatory interactions.
The phytohormone abscisic acid (ABA) regulates many agronomically important aspects of plant growth and development, including seed maturation, dormancy, stress tolerances, and water relations (for review, see Leung and Giraudat, 1998; Rock, 2000; Finkelstein et al., 2002). All of these processes are regulated by additional signals, including other phytohormones, stage-specific regulators, and abiotic stresses. Studies of ABA-deficient mutants have shown that ABA is an essential mediator in triggering some plant responses to abiotic stresses, including drought, salinity, and cold (Rock, 2000; Shinozaki and Yamaguchi-Shinozaki, 2000; Xiong and Zhu, 2001). Dehydration and low temperatures result in elevated levels of ABA, which, in turn, trigger the synthesis of some proteins responsible for drought or freezing tolerance. However, although most of the drought- and salt-induced genes studied to date can be induced by ABA, many aspects of stress response are also mediated by ABA-independent mechanisms (Shinozaki and Yamaguchi-Shinozaki, 2000). These results indicate that ABA participates in only part of a network of stress-signaling mechanisms. Furthermore, even the ABA-dependent portions of this network rely on both independent and partially redundant signaling components, some of which are produced only in specific developmental stages or tissues of the plant.
Components of this signaling network have been identified by a combination of biochemical, cell biological, and forward and reverse genetic approaches. To date, nearly 50 loci in Arabidopsis alone have been demonstrated to function in various aspects of ABA response (for review, see Finkelstein et al., 2002). Their products include transcription factors, protein phosphatases and kinases, RNA binding/processing proteins, GTP-binding proteins, enzymes of phospholipid or phosphoinositide metabolism, and proteins regulating vesicle trafficking or membrane localization of specific proteins. Some of these loci have also been identified independently via diverse genetic screens including defects in response to other phytohormones (ethylene, auxin, or brassinosteroids), abiotic stresses (osmotic, salt, cold, or UV light), sugars (Glc or Suc), or even meristem defects. The repeated identification of a few loci affecting response to multiple signals has led to the suggestions that these genes are points of “cross talk” among signaling pathways or nodes in a regulatory web (for review, see McCourt, 1999; Coruzzi and Zhou, 2001; Gazzarrini and McCourt, 2001; Finkelstein and Gibson, 2002; Finkelstein et al., 2002).
A critical stage in the plant life cycle is the production of viable seeds “programmed” to initiate vegetative growth when conditions are favorable for seedling establishment. ABA contributes to this process by promoting accumulation of storage reserves and acquisition of desiccation tolerance in maturing seeds, and by inhibiting germination and the ensuing loss of desiccation tolerance (for review, see Finkelstein et al., 2002). Among the Arabidopsis genes controlling this aspect of ABA response are those encoding three transcription factors that appear to act combinatorially: ABA-insensitive (ABI)3, ABI4, and ABI5. Although initially described as seed-specific regulators, all three have subsequently been shown to function in various aspects of vegetative growth, including some ABA-regulated gene expression, root branching, plastid differentiation, vegetative meristem quiescence, and floral induction (Rohde et al., 1999, 2000; Söderman et al., 2000; Signora et al., 2001). ABI4, and to a lesser extent ABI5, are also required for the developmental arrest and anthocyanin accumulation induced by exposure to high concentrations of sugars within the first 2 d post-stratification (Arenas-Huertero et al., 2000; Huijser et al., 2000; Laby et al., 2000).
ABI5 differs from ABI3 and ABI4 in that all tested aspects of the null mutant phenotypes are relatively mild. Furthermore, unlike these relatively unique members of the B3- and APETALA2-domain families, it encodes a member of a small subfamily with extensive homology outside the defining domain of its transcription factor class, the basic Leu zippers (bZIPs). BLAST analyses of the Arabidopsis genome have shown that the ABI5-homologous subfamily of Arabidopsis comprises nine genes, each with three conserved charged domains in the amino-half and a bZIP domain near the C terminus (Finkelstein et al., 2002). These have been designated ABA response element binding (AREB), ABA-responsive promoter elements (ABRE) binding factors (ABFs), and Arabidopsis thaliana Dc3 promoter binding factors (AtDPBFs). To date, only ABI5 has been identified by mutation (Finkelstein, 1994; Lopez-Molina and Chua, 2000). However, seven of the other subfamily members have been identified by binding and trans-activation of ABREs and/or homology to sunflower (Helianthus annuus) genes encoding ABRE-binding proteins (Kim et al., 1997; Kim and Thomas, 1998; Choi et al., 2000; Uno et al., 2000; Kim et al., 2002). Furthermore, most have been shown to be ABA- and/or stress-inducible, leading to the suggestion that this subfamily might function redundantly to mediate response to ABA or abiotic stresses. However, it is equally possible that at least some of the different family members might function independently in stage- or stress-specific responses.
In the present work, we have extended our studies of ABI5 regulation to define the stage and tissue specificity of expression under stressed and unstressed conditions. We found that ABI5 is autoregulated and that its promoter is active throughout the plant's life in a variety of specific tissues, but that expression and inducibility by ABA and stress are highest at the transition from mature seeds to seedling growth. In addition, we tested whether overexpressing ABI5 would alter growth responses to ABA or sugar, and we compared ABA-responsive gene expression in transgenic lines overexpressing ABI3, ABI4, or ABI5. These studies show that increased ABI5 expression is sufficient for hypersensitivity to inhibition of growth by ABA or sugar, but affects only a subset of ABA/ABI3/ABI4-regulated genes, including a complex array of positive and negative effects on expression of the ABI5-homologous bZIPs. Our results are consistent with the hypothesis that ABI5 participates in combinatorial regulation of ABA and stress responses involving positive and negative interactions with an array of other regulators that differ among stage-, tissue-, or stress-specific responses.
RESULTS
Autoregulation of ABI5 Expression
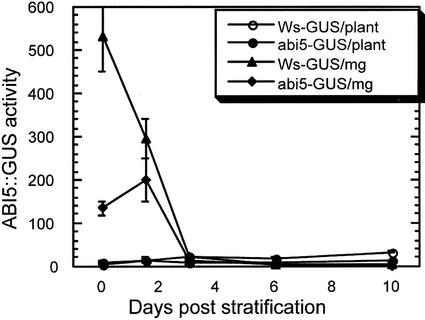
Our initial studies demonstrated that ABI5 transcript accumulation was regulated by ABA and most of the ABI loci (Finkelstein and Lynch, 2000). In the case of abi5 mutants, it was not clear whether the reduced transcript accumulation reflected autoregulation of ABI5 transcription or decreased accumulation of a poorly translated transcript. To distinguish between these possibilities, activity of the ABI5 promoter was assayed in fusions with reporter genes that function in plants or yeast (Saccharomyces cerevisiae). An ABI5::β-glucuronidase (GUS) transgene containing approximately 3 kb of genomic DNA was used to construct a translational fusion including the first 28 codons of ABI5. Transgenic wild-type plants carrying a single copy of the transgene were backcrossed to abi5-1 mutants (which are biochemical nulls) so that ABI5 promoter activity could be compared in isogenic lines, with or without ABI5 function. Comparison of GUS activity in extracts from mature dry seeds showed an approximately 4-fold reduction in the abi5 mutant, demonstrating that ABI5 activity is required for full expression, but that ABI5 is still expressed in the mutant (Fig. 1).
Figure 1.
ABI5- and developmental regulation of ABI5::GUS activity during early seedling growth. GUS activity was measured in dry seeds or germinating seedlings at 1.5, 3, 6, and 10 d post-stratification. Extracts were derived from isogenic ABI5::GUS transgenic lines, differing only at the ABI5 locus. Activity (nanomoles MU produced per hour) is expressed per plant and per milligram of fresh weight. Values shown are the mean ± sd of eight replicate assays.
To determine whether ABI5 expression is sufficient for activation of its promoter, a yeast one-hybrid assay was used to test β-galactosidase activity produced from a series of ABI5 promoter fusions to the reporter gene lacZ. Although the ABI5::GUS translational fusion had led to high-level expression of GUS in plant tissues, a comparable fusion with the lacZ gene was completely inactive in yeast (data not shown), possibly due to the presence of a large intron in the 5′-untranslated region (UTR). Therefore, we constructed transcriptional fusions whose ABI5 promoter fragments terminated upstream of this intron, but included three (ABI5pDra::lacZ) or four (ABI5pHinc::lacZ) predicted ABI5 binding sites (Fig. 2A), based on the DPBF core binding sequence defined by Kim et al. (1997). Expression of ABI5 did not trans-activate the ABI5::lacZ fusions in yeast (data not shown), possibly because the phosphorylation required for activation (Uno et al., 2000) might not occur in yeast. However, cell lines combining the reporter genes with a GAL4 activation domain (GAL4AD)-ABI5 fusion had 3- to 12-fold higher β-galactosidase activity than lines expressing only the GAL4AD protein (Fig. 2B), indicating that the binding domain of ABI5 can target trans-activation of the ABI5 promoter. In contrast, although ectopic expression of ABI3 or ABI4 leads to increased ABI5 expression in transgenic plants (Finkelstein and Lynch, 2000; Söderman et al., 2000), GAL4AD fusions to these transcription factors did not activate the ABI5 promoter.
Figure 2.
ABI5 promoter regulation in yeast. One-hybrid analysis with ABI5 promoter fragments fused to lacZ reporter. A, Promoter fragments used in fusions. Black boxes represent DPBF core recognition sequences. Arrow represents the transcription unit. B, ABI5 promoter-driven β-galactosidase activity produced by GAL4AD fusions to ABI transcription factors (AD-ABIx) or vector control (AD).
Developmental and Environmental Regulation of ABI5 Expression
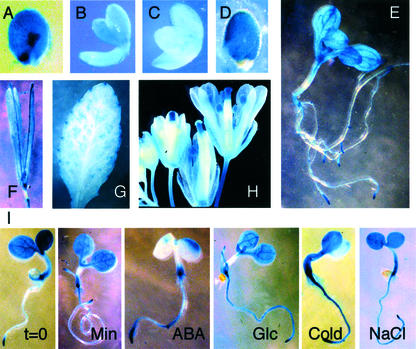
Studies of ABI5 expression to date have shown that transcript levels are highest in mature seeds and flowers, with transcript levels in unstressed vegetative tissues ranging from low to undetectable (Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000). To analyze the tissue specificity of ABI5 promoter activity, ABI5::GUS activity was assayed at various stages through development and in response to assorted stresses. Although previously described as almost undetectable within 2 d after pollination (Lopez-Molina and Chua, 2000), histochemical staining of GUS activity is observed throughout the seed at heart stage (Fig. 3A). Expression continues at low levels throughout the growing embryo (Fig. 3, B and C), reaching a peak at seed maturity (Fig. 3D). ABI5::GUS expression also occurs throughout the silique wall, especially along the suture joining the carpels (Fig. 3F). ABI5::GUS activity per milligram of fresh weight declines rapidly following germination, dropping by approximately 100-fold by 6 d post-stratification due primarily to dilution as seedling fresh weight increases (Fig. 1), possibly accounting for the previous characterization of ABI5 transcript levels as undetectable in 5-d-old seedlings (Lopez-Molina and Chua, 2000). However, total GUS activity per plant approximately doubles over this period, reflecting de novo expression in unstressed vegetative tissue. Vegetative ABI5::GUS expression shows the highest concentrations in root tips, nodes, and major leaf veins of seedlings (Fig. 3E). In older plants, expression is observed in minor veins and at discrete points along the margins of leaves and in flowers, especially in stigmas, maturing anthers, and pedicels (Fig. 3, G and H).
Figure 3.
Localization of ABI5::GUS activity during development or stress response of wild-type plants. A, Seed with heart-stage embryo; B, cotyledon stage embryo; C, mid-maturation stage embryo; D, mature seed; E, 10-d seedling; F, silique; G, leaf from 3-week-old plant; H, flowers; and I, 3-d-old seedlings treated as indicated: t = 0, or 2 d of exposure to minimal medium (Min); ABA, 50 μm ABA; Glc, 250 mm Glc; Cold, 4°C; NaCl, 125 mm NaCl.
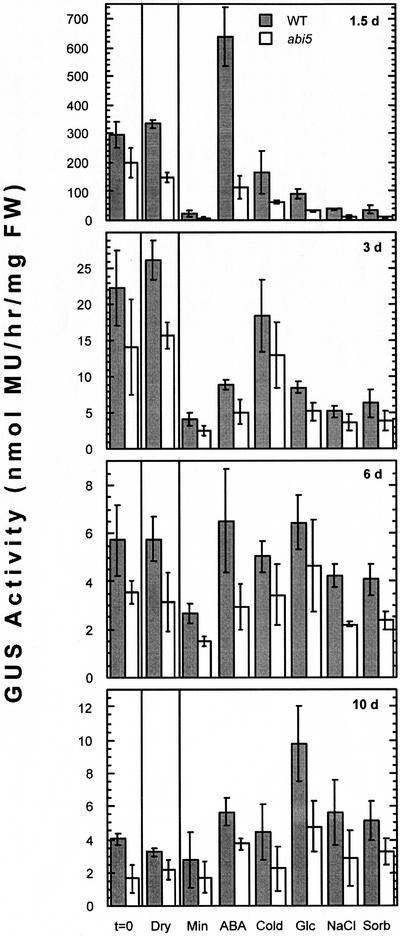
Induction of ABI5 accumulation by ABA was shown to be restricted to a narrow developmental window of approximately 1 to 2 d post-stratification (Lopez-Molina et al., 2001). A similar window has been described for sugar sensitivity of seedling growth (Gibson et al., 2001). Because sugar-induced developmental arrest is slightly dependent on ABI5 function, the window of sugar sensitivity might reflect a limited period of sugar-responsive ABI5 expression. If ABI5 expression is primarily subject to developmental control, other abiotic stress signals (salt, osmotic, drought, or cold) might also become ineffective at approximately the same stage. To test these hypotheses, ABI5::GUS activity was assayed in transgenic lines exposed to stresses at different stages in seedling development. The salt, sugar, and sorbitol stress treatments were designed to be osmotically equivalent and growth inhibiting, but not lethal. All stresses were imposed for 2 d except for drought, which was imposed for 2 h. Exposure to ABA, cold, Glc, or NaCl at 1.5 d post-stratification delayed the decline (cold, Glc, and NaCl treatments) and/or induced de novo ABI5::GUS expression, reflected in increased GUS activity per milligram (ABA treatment; Fig. 4). At later time points, GUS activity per fresh weight is still higher in most of the stressed plants than in the unstressed controls. Although this partly reflects the decreased growth of the stressed plants, comparison of histochemical staining shows that Glc, cold, NaCl, and drought treatment of 3-d seedlings all result in de novo ABI5::GUS expression throughout the root, whereas ABA treatment leads to very strong expression in primary and lateral root tips (Fig. 3I and data not shown). In contrast, seedlings stressed at 6 or 10 d post-stratification showed minimal differences in tissue distribution of ABI5::GUS staining relative to controls (data not shown). However, plants appear to regain some sensitivity to ABA and Glc at 10 d post-stratification (Fig. 4), resulting in increased ABI5-regulated GUS expression overall. Although the applied concentrations of NaCl and sorbitol were sufficient to inhibit growth, they had a relatively minimal impact on total ABI5::GUS activity. Drought treatment also had almost no impact on total ABI5::GUS expression at any time point (Fig. 4). Comparison of ABI5::GUS expression in wild-type and abi5 backgrounds shows that GUS activity is consistently lower in the mutants, suggesting that most of these stress responses are mediated by a combination of ABI5-dependent and -independent mechanisms.
Figure 4.
Stress induction of ABI5 promoter activity in seedlings. GUS activity was measured in seedlings at 1.5, 3, 6, and 10 d post-stratification that were harvested immediately (t = 0), dried on absorbent paper in a closed petri dish for 2 h prior to harvest (Dry), or incubated on control medium (min) or exposed to 50 μm ABA, 125 mm NaCl, 250 mm sorbitol, 250 mm Glc, or 4°C for 2 d prior to harvest. Extracts were derived from isogenic ABI5::GUS transgenic lines, differing only at the ABI5 locus. Activity (nanomoles MU per hour) is expressed per milligram of fresh weight. Values shown are the mean ± sd of duplicate assays on two to four samples per treatment.
Effects of Ectopic ABI5 Expression
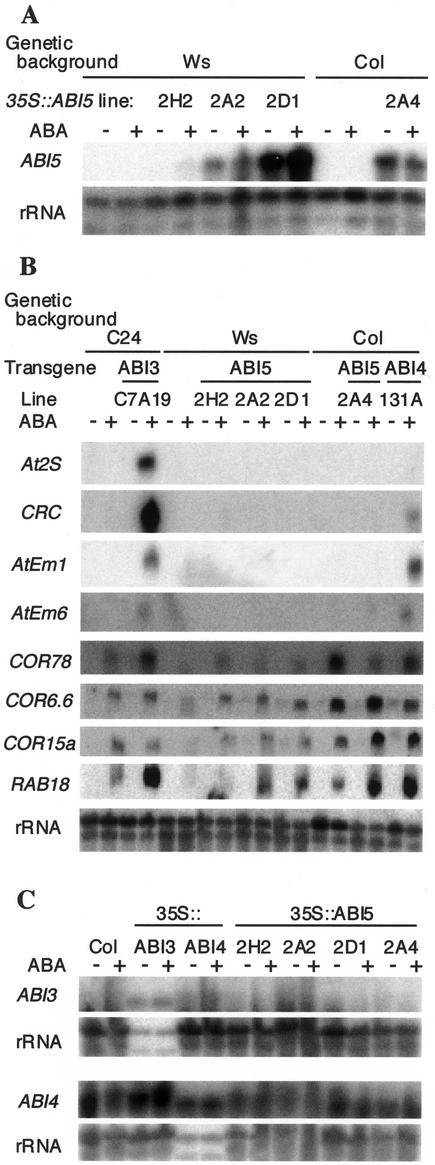
ABI5 function is required for normal regulation of a variety of genes expressed during mid-late embryogenesis and, to a lesser extent, during seedling growth (Finkelstein and Lynch, 2000). Constitutive overexpression of two other transcription factors regulating many of the same genes, ABI3 and ABI4, confers ABA-inducible expression of seed-specific genes in vegetative tissues (Parcy et al., 1994; Söderman et al., 2000). This is accompanied by hyperinduction of ABI5 expression (Söderman et al., 2000), suggesting that ABI5 expression might be an important mediator of these effects on gene expression. To determine whether overexpression of ABI5 is sufficient to confer similar changes in developmental specificity of gene expression, we assayed expression of several marker genes in 2-week-old 35S::ABI5 plants exposed to 50 μm ABA for the last 2 d of growth. Despite having moderate to high levels of ABI5 expression (Fig. 5A), none of the seed-specific marker genes were expressed at detectable levels (Fig. 5B), suggesting that ABI5 must act in combination with the products of regulatory genes normally expressed during seed development. Consistent with this, ABI3 and ABI4 were not overexpressed in these transgenic lines (Fig. 5C). Although ABI5 overexpression did not result in ABA-inducible expression of seed protein genes in vegetative tissues, the 35S::ABI5 lines displayed slightly increased induction of some ABA-inducible transcripts that are normally expressed in seedling tissues (e.g. Cor6.6, Cor15a, and Rab18). Cor78 expression did not show a consistent response to ABI5 overexpression.
Figure 5.
ABA-responsive gene expression in ABI overexpression lines. Plants were grown for 12 d on GM, then transferred to fresh media ± 50 μm ABA for 2 d prior to harvest. RNA gel blots were hybridized to probes corresponding to the indicated genes. A, ABI5 expression in four independent 35S::ABI5 lines and their corresponding wild-type progenitors. Each lane contains 1 μg of total RNA. B, ABA-inducible marker gene expression in 35S::ABI5 lines, representative strong ABI3 and ABI4 overexpression lines, and all corresponding wild types. Each lane contains 5 μg of total RNA. C, ABI3 and ABI4 expression in 35S::ABI5 and wild-type lines. For ABI3 hybridization, 35S::ABI3 lanes contain 0.5 μg and others contain 10 μg of total RNA. For ABI4 hybridization, 35S::ABI4 lanes contain 1 μg and others contain 15 μg of total RNA.
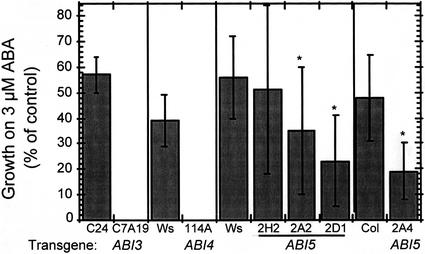
Whereas overexpression of ABI5 had only limited effects on ABA-inducible expression of a small set of ABA-responsive marker genes (Fig. 5B) and growth under osmotic stress (data not shown), it was sufficient to confer hypersensitivity to ABA for inhibition of root growth (Fig. 6). This finding is similar to results described for overexpression of hemagglutinin peptide-tagged ABI5 (Lopez-Molina et al., 2001). The transgenic lines with the strongest ABI5 expression had roughly twice the ABA-induced reduction in growth relative to wild type, whereas overexpression of ABI3 or ABI4 essentially blocks growth under the same conditions, equivalent to at least a 3-fold increase in sensitivity. In addition, overexpression of ABI5 conferred hypersensitivity to sugars for inhibition of seedling growth and induction of anthocyanin accumulation (Fig. 7). The degree of hypersensitivity to ABA and sugar was correlated with the level of ABI5 expression in the transgenic lines. These results suggest that growth control by ABA and sugars may be more tightly correlated with ABI5-regulated expression of genes other than those shown in Figure 5.
Figure 6.
Effects of ABI transcription factor function on ABA hypersensitivity of root growth. After 2 d of growth on hormone-free medium, seedlings were transferred to fresh media containing 0 or 3 μm ABA. New growth was measured after 4 d and is expressed as a percentage of growth on the control medium. Each set represents the average of at least 10 replicates within an individual experiment; sds are expressed as percentages of the control growth. An asterisk indicates 35S::ABI5 lines with significantly increased ABA sensitivity (P = 0.034, 0.0005, or 4 × 10−5 for lines 2A2, 2D1, and 2A4, respectively, based on Student's t test).
Figure 7.
ABI5 overexpression increases seedling sensitivity to sugar. Seedlings of the indicated genotypes were harvested after 9 d of growth on minimal medium, with or without 4% (w/v) Glc. A, Fresh weight per plant. B, Anthocyanin content per fresh weight. Values shown are the mean ± sd of two to four assays per treatment.
The differential response of seed-specific and seedling-inducible genes to ABI5 overexpression (Fig. 5) suggests that developmental specificity may be conferred by other factors regulating these genes. ABI5 is a member of a small subfamily of bZIP proteins, some of which are good candidates for factors contributing to ABA/ABI5-regulated gene expression. For example, some of the ABF/AREBs are correlated with ABA-inducible gene expression in vegetative tissues (Choi et al., 2000; Uno et al., 2000), whereas some AtDPBFs are expressed in seeds and have been shown to form heterodimers with ABI5 (Kim et al., 2002). The overlap in target genes regulated by these transcription factors and the potential for heterodimerization among them has led to the suggestion that these family members might interact and/or have partially redundant functions. Consistent with this, overexpression of ABF3 and ABF4 was recently reported to result in hypersensitivity to ABA, salt, and Glc, as well as producing similar and distinct effects on ABA-regulated gene expression (Kang et al., 2002). Although loss-of-function lines are required to test the specific roles of individual family members, evidence of coordinate expression or regulation could provide further support for the hypothesis of functional redundancy within this family.
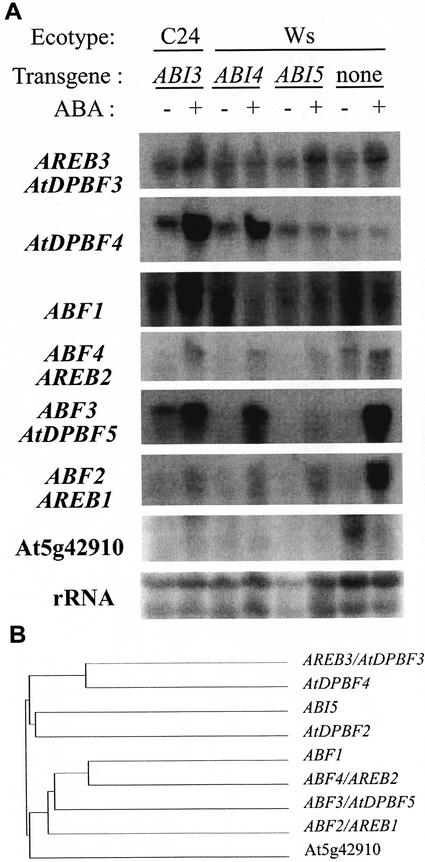
To determine whether any of the ABI5 family members are crossregulated by the ABI transcription factors, transcript accumulation was compared between wild-type and lines strongly overexpressing ABI3, ABI4, or ABI5, treated with or without ABA (Fig. 8). Whereas portions of the encoded proteins are highly conserved at the amino acid level, identity at the nucleotide level is lower and all hybridizations gave distinct patterns, indicating that these results do not reflect cross-hybridization within this gene family. However, some of the probes appeared to hybridize to two transcripts, possibly reflecting alternative splicing. In contrast to ABI5, most of the other ABI5 subfamily members were not overexpressed in any of the transgenic lines. AtDPBF2, the gene with highest homology to ABI5, was not detectably expressed in any of these tissues (data not shown). ABF2/AREB1 even appears underinduced in all of the transgenic lines, whereas ABF3 and/or AtDPBF5 (alternative splicing products of the same gene, differing only by approximately 90 nucleotides of coding sequence and their 3′-UTRs) are strongly repressed only in the 35S::ABI5 line. In contrast, AtDPBF4 is hyperinduced in the ABI3 and ABI4 overexpression lines, but not the ABI5 overexpression line. The observed crossregulation by ABI3 and ABI4, two known regulators of seed gene expression, suggests that AtDPBF4 may also regulate seed gene expression. ABF1 may also be slightly hyperinduced in the 35S::ABI3 line, but this could also reflect a subtle difference between the C24 and Wassilewskija (Ws) ecotypes. The only family member identified solely as a predicted gene (At5g42910) is repressed by ABA in wild type, and even its basal expression appears to be reduced in the 35S::ABI lines. These results indicate that the crossregulation is relatively specific and may reflect functional relationships among genes involved in coordinate regulation.
Figure 8.
ABI5-homologous bZIP expression in ABI overexpression lines. A, RNA gel-blot analyses of three strong 35S::ABI lines (isolates C7A19, 114A, and 2D1 for ABI3, ABI4, and ABI5 overexpression, respectively) hybridized to cDNAs or PCR fragments corresponding to the indicated genes. All reported names are shown for genes isolated independently in multiple labs. Each lane contains 7 μg of total RNA. B, Tree depicting homology relationships among Arabidopsis ABI5-homologous family members. The corresponding AGI or GenBank accession numbers for the named genes, followed by the bZIP designations proposed by Jakoby et al. (2002), are given in parentheses: AtDPBF2 (At3g44460; AtbZIP67); AtDPBF3/AREB3 (At3g56850; AtbZIP66); AtDPBF4 (At2g41070; AtbZIP12); AtDPBF5/ABF3 (At4g34000; AtbZIP37); ABF1 (At1g49720; AtbZIP35); ABF2/AREB1 (AF093545; AtbZIP36); and ABF4/AREB2 (At3g19290; AtbZIP38). Although ABF2 and AREB1 have been independently cloned as cDNAs, the corresponding genomic sequence has not yet been identified.
DISCUSSION
Comparison of ABI5 transcript accumulation in mature mutant and wild-type seeds has demonstrated that normal accumulation depends on wild-type function of ABI1, ABI2, ABI3, and ABI5, as well as normal ABA biosynthesis (Finkelstein and Lynch, 2000). In addition, ABI5 expression can be induced by ABA in vegetative tissue, and this response is greatly amplified in lines expressing ABI3 or ABI4 under control of the cauliflower mosaic virus 35S promoter (Finkelstein and Lynch, 2000; Söderman et al., 2000). These results collectively indicated that ABA and all of the ABI loci could regulate ABI5 expression, but did not address whether the regulation was direct or indirect. We have examined regulation of ABI5 promoter activity using fusions to reporter genes and found that this promoter is active in more tissues than those of ABI3 or ABI4 (Parcy et al., 1994; Rohde et al., 1999; Söderman et al., 2000), and that ABI5 is necessary for full ABI5 expression in plants. Furthermore, a GAL4AD-ABI5 fusion was sufficient to activate ABI5::lacZ expression in yeast in the absence of any other plant transcription factors. Although a nonhybrid ABI5 did not regulate its promoter in yeast, analyses of GAL4BD fusions to various domains of ABI5 have shown that full-length ABI5 does not function as a transcriptional activator in yeast (Nakamura et al., 2001), possibly because it requires modification such as phosphorylation and/or a conformational change to become active. Taken together, these results imply direct autoregulation of ABI5 expression, but also demonstrate ABI5-independent regulation in plants. In contrast, ABI3 and ABI4 were not sufficient to trans-activate the ABI5 promoter in yeast, suggesting that their effects in plants are indirect, multifactorial, or require some form of posttranslational modification that does not occur in yeast. These results suggest that ABI5 expression is activated by ABI3 and/or ABI4 or other stage/tissue-specific regulators, and can then contribute to maintaining its expression by autoregulation.
ABI5 is expressed in a variety of distinct cell types through development, showing some overlap with ABF3 and ABF4 expression in vegetative and floral tissues (Kang et al., 2002), and it is likely that its physiological role varies among tissues and stages. The highest ABI5 expression is observed in mature dry seeds, where it has been shown to regulate some late embryogenesis abundant genes (Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000; Delseny et al., 2001), at least some of which are correlated with desiccation tolerance (Swire-Clark and Marcotte, 1999). A similar role might be played in anthers, where maturing pollen must also acquire desiccation tolerance. In seedlings, expression is highest in root tips (primary and lateral), nodes, and major veins; when effective, ABA or stress treatments accentuate these expression patterns and may slightly expand the distribution of ABI5 expression to include tissues that might otherwise be growing rapidly, e.g. those present throughout new leaves. ABI5 has been proposed to act as a mediator of ABA repression of growth at the onset of seedling development (Lopez-Molina et al., 2001), possibly via effects on cell-cycle machinery (Wang et al., 1998), and might continue this role in selected tissues well after commitment to seedling growth. Specific effects of ABA on promoting initiation of lateral roots, but inhibiting their elongation, have been implicated in response to drought (Vartanian et al., 1994). More recently, ABA, ABI5, and ABI4 have been shown to mediate the inhibitory effect of NO3− on lateral root development, possibly via effects on sensing the C/N balance regulating lateral root growth (Signora et al., 2001). Localized expression of ABI5 might allow it to function as a sensor, becoming stabilized and active by ABA-dependent changes in phosphorylation state under stress conditions. In addition, ABI5 expression might be high under unstressed conditions in tissues that have low rates of cell division, such as maturing vascular tissue.
As previously described for ABI5 protein accumulation (Lopez-Molina et al., 2001), ABI5 promoter activity was highly inducible by ABA when applied within 2 d post-stratification, but was only weakly ABA responsive at later time points. Cold and Glc slightly induced ABI5::GUS expression at most time points, especially in roots, indicating that these stress responses were not limited to the same developmental window as ABA response. In contrast, the NaCl, sorbitol, and drought treatments had little or no effect on ABI5::GUS activity per milligram of fresh weight, suggesting that the reported increase in ABI5 protein accumulation in response to salt or drought might be a largely posttranscriptional effect, possibly mediated by phosphorylation-induced changes in protein stability (Lopez-Molina et al., 2001).
Previous studies of the ABI transcription factors have shown that overexpression of ABI3 or ABI4 resulted in hypersensitivity to ABA inhibition of root growth and ABA-induced vegetative expression of genes normally expressed primarily in seeds, including ABI5 (Parcy et al., 1994; Finkelstein and Lynch, 2000; Söderman et al., 2000). Observation of additional crossregulation among these ABI loci led to the suggestion that these transcription factors act combinatorially to regulate subsets of ABA-inducible and/or seed-specific genes. Subsequent studies demonstrated direct physical interactions between ABI5 and ABI3 or their rice (Oryza sativa) homologs in yeast (Hobo et al., 1999; Nakamura et al., 2001), as well as synergistic interactions between ABI5 and VP1 or their rice homologs in mediating ABA-activated gene expression in transient expression assays in plant protoplasts (Hobo et al., 1999; Gampala et al., 2002). ABI5 overexpression has been shown to resemble ABI3 overexpression in that it confers hypersensitivity to ABA inhibition of root growth and promotes slightly more efficient water retention (Parcy et al., 1994; Parcy and Giraudat, 1997; Lopez-Molina et al., 2001). Furthermore, overexpression of ABI3 or ABI5 confers increased sensitivity to Glc inhibition of seedling growth (Finkelstein et al., 2002) even though abi3 and abi5 mutants have not been recovered in sugar-sensing screens. Similar results have recently been described for overexpression of ABF3 and ABF4 (Kang et al., 2002). In combination with the observation that abi5 mutants are mildly sugar resistant (Arenas-Huertero et al., 2000; Huijser et al., 2000; Laby et al., 2000), these results suggest that ABI5 can mediate sugar response, but that its activity is not normally a limiting factor, possibly because other functionally redundant proteins can partially compensate for loss of ABI5 function. Further evidence for a role in sugar response is provided by the observation that ABI5 expression is induced by Glc. However, even the transgenic lines with the strongest ABI5 expression were not as hypersensitive to ABA or sugar as the lines that strongly overexpress ABI3 or ABI4 (Fig. 6; Finkelstein et al., 2002). Furthermore, we have found that ABI5 overexpression does not increase vegetative expression of ABI3, ABI4, or the seed-specific genes regulated by the combinatorial activity of these transcription factors. The specific genes regulated by ABI5 in any given tissue might be determined by combinatorial interactions between ABI5 and ABI3 or other appropriate and available transcription factors.
Some of the likely candidates for transcription factors that interact with ABI5 are other members of the ABI5-homologous subfamily of bZIP proteins. However, despite the fact that most of these proteins were initially identified on the basis of binding to ABREs, and some can confer increased ABA-inducible reporter gene expression in protoplasts, they show widely varying degrees of induction by ABA and the ABI transcription factors (Choi et al., 2000; Uno et al., 2000; Finkelstein et al., 2002). In fact, different ABI5-homologous family members show either positive or negative response to overexpression of any given ABI transcription factor, which may reflect similar or antagonistic interactions, respectively. The structural similarities of this subfamily are not paralleled in their expression patterns. Given that the ABI transcription factors are not expressed constitutively throughout development, the ABI5-homologous genes that they regulate are also likely to vary in their developmental specificity. Consistent with this, the reported ABA inducibility of individual family members varies substantially, possibly reflecting differential responses of 3- to 5-week-old plants exposed to ABA hydroponically (Choi et al., 2000; Uno et al., 2000) or 2-week-old plants grown on solid medium (Fig. 7). It is noteworthy that the two family members displaying the strongest ABA inducibility in 2-week-old wild-type plants, ABF2/AREB1 and ABF3/AtDPBF5, differ greatly in their regulation by the ABI genes. These results collectively argue against simple functional redundancy within the ABI5-homologous family. However, they do not exclude the possibility that some family members will interact, synergistically or antagonistically, to regulate specific target genes. Hetero- and homodimerization among sunflower homologs of this family have already been documented (Kim and Thomas, 1998), and there is ample precedent for positive and negative interactions among dimerizing bZIP proteins (for review, see Hurst, 1995).
In summary, we have demonstrated that ABI5 expression extends throughout the life of the plant in a variety of specific tissues and in response to a variety of stresses, rather than being limited to the stages associated with its previously characterized role in mediating seed maturation and ABA response in very young seedlings. Despite the breadth of its expression pattern, loss of ABI5 function has relatively subtle phenotypic effects, suggesting that it plays a minor or a functionally redundant role in many tissues. Comparison of ABI3, ABI4, and ABI5 overexpression lines has revealed similar and distinct effects of these transcription factors, consistent with complex regulatory interactions among them and other factors such as the other members of the ABF/AREB/AtDPBF family. The fact that ABI5 overexpression is sufficient for many aspects of ABA- and sugar-hypersensitive growth, but only a subset of ABA-inducible genes, should help provide us with a way to identify the ABA-regulated genes that are most tightly correlated with growth effects.
MATERIALS AND METHODS
Plant Material
The abi5-1 mutant line was isolated from the Ws background, as described by Finkelstein (1994). The 35S::ABI3 line (C7A19 isolate) was constructed in the C24 background, as described by Parcy et al. (1994). The 35S::ABI4 lines (114A and 131A isolates) were constructed in the Ws and Columbia backgrounds, respectively, as described by Söderman et al. (2000). ABI5::GUS and 35S::ABI5 transgenic lines were constructed in the Columbia and Ws backgrounds by vacuum infiltration (Bechtold et al., 1993) with Agrobacterium tumefaciens carrying the plasmids described below. An ABI5::GUS transgenic line in the Ws background was crossed to the abi5 mutant, and ABA- and kanamycin-resistant F2 individuals were selected and screened for homozygosity of the transgene and the abi5 mutation in the F3 generation.
For RNA isolation from 2-week-old plants, seeds were surface sterilized in 5% (v/v) hypochlorite and 0.02% (v/v) Triton X-100, and then rinsed two to four times with sterile water before being plated on GM (Valvekens et al., 1988) composed of 0.5× Murashige-Skoog nutrients (Murashige and Skoog, 1962) and 1% (w/v) Suc solidified with 0.55% (w/v) agar. The dishes were incubated 3 d at 4°C to break any residual dormancy, and were then transferred to 22°C in continuous light (50–70 μE m−2 sec−1). After 12 d, plants were transferred to fresh GM with 1% (w/v) Suc, 0.7% (w/v) agar, and 0 or 50 μm ABA (mixed isomers; Sigma, St. Louis) for an additional 2 d before harvest. All tissues for RNA isolation were weighed, flash frozen in liquid nitrogen, and stored at −70°C until extraction.
For histochemical staining of GUS activity in developing seeds, plants were grown in soil in continuous light at 22°C. To test for induction of ABI5 promoter activity by specific stresses, seedlings were grown aseptically on Murashige-Skoog medium with 1% (w/v) Suc and 0.55% (w/v) agar. After 1.5, 3, 6, or 10 d at 22°C in continuous light, seedlings were harvested immediately or transferred to minimal medium (Haughn and Somerville, 1986) with 0.7% (w/v) agar and 50 μm ABA, 0.125 m NaCl, 0.250 m sorbitol, 0.250 m Glc, or no supplement for an additional 2 d before harvest. Cold stress was imposed by incubation for 2 d in a dimly illuminated cold room at 4°C. Drought stress was imposed by letting the seedlings dry for 2 h on 3MM paper (Whatman, Beverly, MA) in a closed petri dish. GUS activity was assayed histochemically and fluorometrically in replicate samples.
For Glc sensitivity assays, seeds were grown aseptically on minimal medium (Haughn and Somerville, 1986) containing 0.7% (w/v) agar with or without 4% (w/v) Glc. The dishes were incubated 3 d at 4°C, and were then transferred to 22°C in continuous light. Plants were harvested and weighed after 9 d, and then stored at −70°C until used for extraction of anthocyanins.
Transgene Construction
An ABI5 overexpression transgene was constructed by fusing an ABI5 cDNA into pGA643 (An et al., 1988) with 23 bp of 5′-UTR between the cauliflower mosaic virus 35S promoter and the initiating codon of ABI5. An ABI5::GUS transgene was constructed in pBI101 as a translational fusion containing approximately 3 kb of upstream genomic sequence and the first 28 codons of ABI5. Although there was substantial quantitative variability among the transgenic lines, the qualitative expression patterns were very similar. The data presented displays results with one representative transgene in wild-type and abi5 backgrounds. Binary plasmids carrying the transgenes were introduced into A. tumefaciens line GV3101 by direct transformation, followed by selection for growth on appropriate antibiotics (kanamycin for pBI101 derivatives, and tetracycline and kanamycin for pGA643 derivatives).
One-Hybrid Assays of Transcription Activation Function
Two overlapping restriction fragments from the promoter region of ABI5 were fused upstream of the lacZ gene in the plasmid pLGΔ178, a derivative of the plasmid pLGΔ312 (Guarente and Mason, 1983). These fragments were a 1,028-bp HincII fragment that ended 109 bp upstream of the transcription start site of ABI5 and an 819-bp DraI fragment that ended 33 bp downstream of the transcription start site of ABI5. Each construct was used to cotransform the yeast cell line PJ69–4A where the second plasmid was pGAD (a plasmid containing the activation domain of GAL4), or its derivatives encoding ABI3, ABI4, or ABI5 fused in frame with the activation domain of GAL4. All transformations were performed using the EZ transformation kit (Bio 101, Vista, CA) following the manufacturer's instructions. The activation domain and pLGΔ178 plasmids were maintained by growing the yeast on media lacking Leu and uracil.
Quantitative assays of GAL4AD-driven β-galactosidase gene expression were performed as described at http://www.fhrc.org/labs/gottschling/yeast/Bgal.html. All data presented are the averages ± sd of assays on at least three independent transformants.
Measurement of GUS Activity
Soluble extracts of seeds were assayed fluorometrically for GUS activity using 4-methylumbelliferyl glucuronide (Rose Scientific Ltd., Edmonton, AB, Canada) as substrate as described in (Jefferson et al., 1987).
GUS activity in intact plants was detected histochemically by infiltration with 5-bromo-4-chloro-3-indolyl-β-glucuronic acid as described in (Jefferson et al., 1987). Plant material was cut and incubated in GUS staining solution containing 50 mm sodium phosphate, pH 7.0, 0.1% (v/v) Triton X-100, K3/K4 FeCN 0.5 mm, and 1 mm 5-bromo-4-chloro-3-indolyl-β-glucuronic acid at 37°C overnight. Tissues were cleared of chlorophyll in ethanol. Photographs of whole-mounted tissues were taken using a stereomicroscope.
RNA Gel-Blot Analysis
RNA was isolated from vegetative tissue by hot phenol extraction as described previously (Finkelstein, 1993). RNA concentrations were estimated based on absorbance at 260 and 280 nm.
Total RNA (5–20 μg lane−1) was size fractionated on MOPS-formaldehyde gels (Sambrook et al., 1989), and was then transferred to Magna Nylon membranes (Osmonics, Westborough, MA) using 20× SSPE as blotting buffer. RNA was bound to the filters by UV-crosslinking (120 mJ cm−2 at 254 nm). Uniformity of loading and transfer was assayed qualitatively by methylene blue staining of the filters (Herrin and Schmidt, 1988). Transcripts from At2S3, CRC, AtEm1, ABI3, and the RAB18 homolog were detected by hybridization to cDNA clones as described by Söderman et al. (2000), labeled by random priming to a specific activity of 108 cpm μg−1. The COR6.6, COR15a, and COR78 transcripts were detected by hybridization to cDNA clones as described by Hajela et al. (1990). The AtEm6 mRNA was detected by hybridization to a genomic clone encompassing the entire transcribed region and 0.8-kb 5′-flanking sequences. The ABI5 probe was a PCR-amplified genomic fragment excluding most of the conserved bZIP domain. The ABI4 probe was an EcoRI fragment from a cDNA clone encompassing all but the first two and final codons of the coding sequence. Hybridization probes for the AtDPBF transcripts were full-length cDNA clones as described by S. Kim, J. Ma, P. Perret, Z. Li, and T. Thomas (unpublished data); the remaining ABF/AREB family members were detected by hybridization to exon-specific PCR fragments corresponding to the following segments of the coding sequences (ABF1, nucleotides 286–941; ABF2, nucleotides 324–1,050; ABF4, nucleotides 2–1,016; and At5g42910, nucleotides 60–881). Hybridization conditions were 50% (v/v) formamide, 5× SSPE, 5× Denhardt's, 0.1% (w/v) SDS, and 200 μg mL−1 DNA at 43°C or 7% (w/v) SDS, 0.5 m sodium-phosphate, pH 7.2, 1 mm EDTA, and 1% (w/v) bovine serum albumin at 65°C for 16 to 24 h (Church and Gilbert, 1984) in a Hyb-Aid rotisserie oven. Filters were washed twice at 60°C in 2× SSC and 0.1% (w/v) SDS, and once at 60°C in 0.2× SSC and 0.1% (w/v) SDS for 30 to 60 min.
Root Growth Assays
For root growth assays, seeds were surface sterilized before being plated on GM containing 0.7% (w/v) agar. Petri plates were incubated for 3 d at 4°C and were then transferred to 22°C in continuous light. After 2 d, germinated seedlings were transferred to GM supplemented with 0 or 3 μm ABA. Plates were incubated vertically, with seedlings placed with their root tips pointing up such that new root growth would occur along the surface of the plates in the opposite direction from the original growth. New root growth was measured after 4 d, and then average growth was calculated for each genotype and treatment and was expressed as a percentage of the growth on hormone-free media.
Measure of Anthocyanin Content
Anthocyanins were extracted as described by Chia et al. (2000) by homogenization in methanol/HCl (99:1), using between 20 and 150 μL solvent mg−1 fresh weight. Extracts were cleared by centrifugation, and then absorbances were measured at 530 and 657 nm. Relative anthocyanin concentrations were calculated and expressed relative to tissue fresh weight according to the formula: [OD530 − (0.25 × OD657)][extract volume]/fresh weight.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
ACKNOWLEDGMENTS
We thank Drs. Jerome Giraudat and Francois Parcy for the ABI3 overexpression line, Dr. Michel Delseny for the storage protein and lea cDNAs, Dr. Michael Thomashow for the COR cDNAs, and Dr. Terry Thomas for sharing results and cDNA clones for the AtDPBFs prior to publication. We thank Ana Citrin, Christian Snowden, and Aimee Sunseri for technical assistance with one-hybrid assays and transgenic plant analyses.
Footnotes
This work was supported by the National Science Foundation (grant no. IBN99–82779 to R.R.F.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.005793.
LITERATURE CITED
- An G, Ebert P, Mitra A, Ha S. Binary vectors. In: Gelvin S, Schilperoort R, editors. Plant Molecular Biology Manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1988. , A3 pp 1–19. [Google Scholar]
- Arenas-Huertero F, Arroyo A, Zhou L, Sheen J, Leon P. Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes Dev. 2000;14:2085–2096. [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris Life Sci. 1993;316:1194–1199. [Google Scholar]
- Chia DW, Yoder TJ, Reiter W-D, Gibson SI. Fumaric acid: an overlooked form of fixed carbon in Arabidopsis and other plant species. Planta. 2000;211:743–751. doi: 10.1007/s004250000345. [DOI] [PubMed] [Google Scholar]
- Choi H, Hong J, Ha J, Kang J, Kim S. ABFs, a family of ABA-responsive element binding factors. J Biol Chem. 2000;275:1723–1730. doi: 10.1074/jbc.275.3.1723. [DOI] [PubMed] [Google Scholar]
- Church G, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coruzzi GM, Zhou L. Carbon and nitrogen sensing and signaling in plants: emerging “matrix effects.”. Curr Opin Plant Biol. 2001;4:247–253. doi: 10.1016/s1369-5266(00)00168-0. [DOI] [PubMed] [Google Scholar]
- Delseny M, Bies-Etheve N, Carles C, Hull G, Vicient C, Raynal M, Grellet F, Aspart L. Late embryogenesis abundant (LEA) protein regulation during Arabidopsis seed maturation. J Plant Physiol. 2001;158:419–427. [Google Scholar]
- Finkelstein R. Abscisic acid-insensitive mutations provide evidence for stage-specific signal pathways regulating expression of an Arabidopsis late embryogenesis-abundant gene. Mol Gen Genet. 1993;238:401–408. doi: 10.1007/BF00291999. [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SSL, Rock CD. Abscisic acid signaling in seeds and seedlings. Plant Cell. 2002;14:S15–S45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R, Gibson SI. ABA and sugar interactions regulating development: “cross-talk” or “voices in a crowd”? Curr Opin Plant Biol. 2002;5:26–32. doi: 10.1016/s1369-5266(01)00225-4. [DOI] [PubMed] [Google Scholar]
- Finkelstein R, Lynch T. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell. 2000;12:599–609. doi: 10.1105/tpc.12.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR. Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J. 1994;5:765–771. [Google Scholar]
- Gampala SSL, Finkelstein RR, Sun SM, Rock CD. ABA INSENSITIVE-5 interacts with ABA signaling effectors in rice protoplasts. J Biol Chem. 2002;277:1689–1694. doi: 10.1074/jbc.M109980200. [DOI] [PubMed] [Google Scholar]
- Gazzarrini S, McCourt P. Genetic interactions between ABA, ethylene and sugar signaling pathways. Curr Opin Plant Biol. 2001;4:387–391. doi: 10.1016/s1369-5266(00)00190-4. [DOI] [PubMed] [Google Scholar]
- Gibson SI, Laby RJ, Kim D. The sugar-insensitive1 (sis1) mutant of Arabidopsis is allelic to ctr1. Biochem Biophys Res Commun. 2001;280:196–203. doi: 10.1006/bbrc.2000.4062. [DOI] [PubMed] [Google Scholar]
- Guarente L, Mason T. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell. 1983;32:1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- Hajela R, Horvath D, Gilmour S, Thomashow M. Molecular cloning and expression of cor (cold-regulated) genes in Arabidopsis thaliana. Plant Physiol. 1990;93:1246–1252. doi: 10.1104/pp.93.3.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughn G, Somerville C. Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol Gen Genet. 1986;204:430–434. [Google Scholar]
- Herrin D, Schmidt G. Rapid, reversible staining of Northern blots prior to hybridization. BioTechniques. 1988;6:196–200. [PubMed] [Google Scholar]
- Hobo T, Kowyama Y, Hattori T. A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc Natl Acad Sci USA. 1999;96:15348–15353. doi: 10.1073/pnas.96.26.15348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijser C, Kortstee A, Pego J, Weisbeek P, Wisman E, Smeekens S. The Arabidopsis SUCROSE UNCOUPLED-6 gene is identical to ABSCISIC ACID INSENSITIVE-4: involvement of abscisic acid in sugar responses. Plant J. 2000;23:577–585. doi: 10.1046/j.1365-313x.2000.00822.x. [DOI] [PubMed] [Google Scholar]
- Hurst H. bZIP proteins. Protein Profile. 1995;2:105–168. [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002;7:106–111. doi: 10.1016/s1360-1385(01)02223-3. [DOI] [PubMed] [Google Scholar]
- Jefferson R, Kavanagh T, Bevan M. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Choi H, Im M, Kim SY. Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell. 2002;14:343–357. doi: 10.1105/tpc.010362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Chung H-J, Thomas TL. Isolation of a novel class of bZIP transcription factors that interact with ABA-responsive and embryo-specification elements in the Dc3 promoter using a modified yeast one-hybrid system. Plant J. 1997;11:1237–1251. doi: 10.1046/j.1365-313x.1997.11061237.x. [DOI] [PubMed] [Google Scholar]
- Kim SY, Ma J, Li Z, Thomas TL (2002) Arabidopsis ABI5 subfamily members have distinct DNA-binding and transcriptional activities. Plant Physiol (in press) [DOI] [PMC free article] [PubMed]
- Kim SY, Thomas TL. A family of novel basic leucine zipper proteins binds to seed-specification elements in the carrot Dc3 gene promoter. J Plant Physiol. 1998;152:607–613. [Google Scholar]
- Laby R, Kincaid M, Kim D, Gibson S. The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J. 2000;23:587–596. doi: 10.1046/j.1365-313x.2000.00833.x. [DOI] [PubMed] [Google Scholar]
- Leung J, Giraudat J. Abscisic acid signal transduction. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:199–222. doi: 10.1146/annurev.arplant.49.1.199. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Chua N-H. A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol. 2000;41:541–547. doi: 10.1093/pcp/41.5.541. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua N-H. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci USA. 2001;98:4782–4787. doi: 10.1073/pnas.081594298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCourt P. Genetic analysis of hormone signaling. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:219–243. doi: 10.1146/annurev.arplant.50.1.219. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Nakamura S, Lynch T, Finkelstein R. Physical interactions between ABA response loci of Arabidopsis. Plant J. 2001;26:627–635. doi: 10.1046/j.1365-313x.2001.01069.x. [DOI] [PubMed] [Google Scholar]
- Parcy F, Giraudat J. Interactions between the ABI1 and the ectopically expressed ABI3 genes in controlling abscisic acid responses in Arabidopsis vegetative tissues. Plant J. 1997;11:693–702. doi: 10.1046/j.1365-313x.1997.11040693.x. [DOI] [PubMed] [Google Scholar]
- Parcy F, Valon C, Raynal M, Gaubier-Comella P, Delseny M, Giraudat J. Regulation of gene expression programs during Arabidopsis seed development: roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell. 1994;6:1567–1582. doi: 10.1105/tpc.6.11.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock C. Pathways to abscisic acid-regulated gene expression. New Phytol. 2000;148:357–396. doi: 10.1046/j.1469-8137.2000.00769.x. [DOI] [PubMed] [Google Scholar]
- Rohde A, Kurup S, Holdsworth M. ABI3 emerges from the seed. Trends Plant Sci. 2000;5:418–419. doi: 10.1016/s1360-1385(00)01736-2. [DOI] [PubMed] [Google Scholar]
- Rohde A, Van Montagu M, Boerjan W. The ABSCISIC ACID-INSENSITIVE 3 (ABI3) gene is expressed during vegetative quiescence processes in Arabidopsis. Plant Cell Environ. 1999;22:261–270. [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol. 2000;3:217–223. [PubMed] [Google Scholar]
- Signora L, Smet I, Foyer C, Zhang H. ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. Plant J. 2001;28:655–662. doi: 10.1046/j.1365-313x.2001.01185.x. [DOI] [PubMed] [Google Scholar]
- Söderman E, Brocard I, Lynch T, Finkelstein R. Regulation and function of the Arabidopsis ABA-insensitive4 (ABI4) gene in seed and ABA response signaling networks. Plant Physiol. 2000;124:1752–1765. doi: 10.1104/pp.124.4.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swire-Clark GA, Marcotte WR. The wheat LEA protein Em functions as an osmoprotective molecule in Saccharomyces cerevisiae. Plant Mol Biol. 1999;39:117–128. doi: 10.1023/a:1006106906345. [DOI] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA. 2000;97:11632–11637. doi: 10.1073/pnas.190309197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvekens D, Van Montagu M, Van Lijsebettens M. Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartanian N, Marcotte L, Giraudat J. Drought rhizogenesis in Arabidopsis thaliana. Plant Physiol. 1994;104:761–767. doi: 10.1104/pp.104.2.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Qi Q, Schorr P, Cutler AJ, Crosby W, Fowke LC. ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J. 1998;15:501–510. doi: 10.1046/j.1365-313x.1998.00231.x. [DOI] [PubMed] [Google Scholar]
- Xiong L, Zhu J-K. Abiotic stress signal transduction in plants: Molecular and genetic perspectives. Physiol Plant. 2001;112:152–166. doi: 10.1034/j.1399-3054.2001.1120202.x. [DOI] [PubMed] [Google Scholar]