Abstract
A method for the generation of stable activation tag inserts was developed in Arabidopsis using the maize (Zea mays) En-I transposon system. The method employs greenhouse selectable marker genes that are useful to efficiently generate large populations of insertions. A population of about 8,300 independent stable activation tag inserts has been produced. Greenhouse-based screens for mutants in a group of plants containing about 2,900 insertions revealed about 31 dominant mutants, suggesting a dominant mutant frequency of about 1%. From the first batch of about 400 stable insertions screened in the greenhouse, four gain-in-function, dominant activation-tagged, morphological mutants were identified. A novel gain-in-function mutant called thread is described, in which the target gene belongs to the same family as the YUCCA flavin-mono-oxygenase that was identified by T-DNA activation tagging. The high frequency of identified gain-in-function mutants in the population suggests that the En-I system described here is an efficient strategy to saturate plant genomes with activation tag inserts. Because only a small number of primary transformants are required to generate an activation tag population, the En-I system appears to be an attractive alternative to study plant species where the present transformation methods have low efficiencies.
Activation tagging in plants was first proposed as a novel gene isolation method (Walden et al., 1994) and has been effectively applied using T-DNA inserts (Kakimoto, 1996; Kardailsky et al., 1999; Borevitz et al., 2000; Ito and Meyerowitz, 2000; Lee et al., 2000; van der Graaff et al., 2000; Weigel et al., 2000; Huang et al., 2001; Zhao et al., 2001) and the Ac-Ds transposon system (Wilson et al., 1996). These inserts are designed to carry strong activating sequences that can act on genes adjacent to the insertion site and modify their expression. The well-characterized cauliflower mosaic virus (CaMV) 35S enhancer or promoter sequences (Odell et al., 1985) have been used as transcriptional activators in this type of insertion system. Their primary function has been to overexpress the tagged genes to reveal dominant gain-of-function phenotypes. In a number of cases, the CaMV 35S enhancer used in the T-DNA inserts acts by quantitatively increasing the original expression pattern of a gene, rather than by ectopic or constitutive overexpression of genes (Neff et al., 1999; van der Graaff et al., 2000).
The Arabidopsis genome sequence revealed about 25,000 genes that have been predicted by a variety of bioinformatics tools (The Arabidopsis Genome Initiative, 2000). However, the exact biological role for most genes is still unknown, requiring specific biological experiments to uncover their function. Changes in expression of the genes brought about by forward and reverse genetics methods are expected to provide tools to describe their function. Gene knockouts abolishing gene function, using diverse mutagens, are classically employed to reveal the biological role of specific genes. However, recent reverse genetics analysis has shown that most gene knockouts (Bouché and Bouchez, 2001) do not reveal informative phenotypes under “standard” conditions, although some knockouts can reveal phenotypes under specific environmental conditions or when analyzed at different growth stages (Boyes et al., 2001).
A possible explanation for this “phenotype gap” comes from the analysis of the Arabidopsis genome (The Arabidopsis Genome Initiative, 2000). About two-thirds of the genome is duplicated in the form of large chromosomal segments, although during evolution, some of these duplicated genes have diverged to produce new functions. In addition, about 4,000 genes are tandemly repeated as two or more copies. These duplicated and redundant genes would, therefore, not be expected to display a knockout mutant phenotype.
Activation tagging possesses inherent advantages that can overcome some of the limitations of gene disruption, because this technique generates gain-in-function dominant mutations. With a strong constitutive enhancer such as that derived from the CaMV 35S promoter, gene expression can be increased above normal levels (Neff et al., 1999) or even ectopically. In this way, even redundant genes might display an overexpression phenotype if their product is limiting or a change in concentration of gene products creates an imbalance that is manifested as a phenotype. These phenotypes can either directly reveal the gene function or provide a clue to the pathway in which the gene is involved. Moreover, it might be possible to study the function of essential genes that display lethal knockout phenotypes by using activation tagging strategies. The frequency of lethal mutations, essential to the haploid gametophyte and/or the early diploid embryo, is predicted to be around 1 gene every 100 kb (Martienssen, 1998), amounting to about 1,300 genes, some of which might be revealed by gain-in-function phenotypes. For biotechnological applications, the controlled overexpression of genes with specific functions could be easier to employ than gene suppression or knockouts in crops of interest (Huang et al., 2001).
Most activation tag collections have been generated using the Agrobacterium tumefaciens T-DNA as a tag (Ito and Meyerowitz, 2000; van der Graaff et al., 2000; Weigel et al., 2000; Huang et al., 2001). Detailed analysis of a number of mutants identified using T-DNA activation tagging has recently contributed to a better understanding of this gene activation system and also resulted in the identification of novel genes. One such example is the discovery of the role of members of the family of flavin mono-oxygenases in auxin biosynthesis (Zhao et al., 2001), which had not been characterized previously. In Madagascar periwinkle (Catharantus roseus), a positive selection system was used to isolate the ORCA3 gene, which codes for a jasmonate-responsive transcriptional regulator of primary and secondary metabolism (Van der Fits and Memelink, 2000). Activation tagging has also been employed as a novel approach to isolate suppressor mutants of known mutant phenotypes (Neff et al., 1999). These examples demonstrate the versatility of the uses of this tagging system and encourage further studies and developments.
One of the major disadvantages of T-DNA-based activation systems is the creation of complex integration patterns and chromosomal rearrangements near the insertion site (Gheysen et al., 1990; De Neve et al., 1997; Nacry et al., 1998). This could affect either the activity or the interaction of the T-DNA integrated enhancer sequences with the adjacent plant DNA. The use of single stable transposon insertions as the activation tags could overcome this problem and would also allow targeted tagging of genes near the original insertion site. This could give more information about specific members of linked gene family clusters.
The En-I (Spm-dSpm) system of maize (Zea mays) is an efficient tool for heterologous transposon tagging in Arabidopsis (Aarts et al., 1995; Wisman et al., 1998; Speulman et al., 1999; Tissier et al., 1999). Two features of the En-I system make it particularly suitable for the selection of stable transposants: the high frequency of independent transpositions (Aarts et al., 1995) and the ability to transpose to unlinked locations (Speulman et al., 1999), rendering selections for segregated transpositions efficient.
A significant development in transposon technology has been the use of greenhouse positive and negative selectable markers to select for stable transpositions (Tissier et al., 1999), which we have also employed in the research reported here. This system makes use of the selectable markers BAR (De Block et al., 1987; Thompson et al., 1987) conferring resistance to the herbicide Basta and SU1 (O'Keefe et al., 1994) that converts the pro-herbicide R7402 (DuPont, Wilmington, DE) into the herbicide sulfonylurea inhibiting or reducing the growth of plants that contain it. The selection for stable transposition can be performed in soil in the greenhouse, avoiding the need for media and seed sterilization. Both markers are carried on a single construct that also bears the nonautonomous element and a source of transposase (an En/Spm element that lacks the terminal-inverted repeats and is, thus, immobile). The selectable BAR gene is placed inside the nonautonomous I/dSpm element and the negatively selectable marker SU1 in the adjacent T-DNA allowing selection against the transposase source. The application of both Basta and R7402 to the progeny seedlings of transformants containing both elements leads to selection of plants bearing stable germinal insertions. Here, we have applied the double selection system to develop and generate I/dSpm inserts containing a multiple-enhancer element for large-scale activation tagging.
RESULTS
Development of an En-I Transposon System
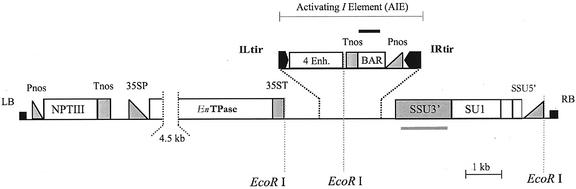
The En-I transposon system from maize was used to construct vectors suitable for developing collections of stable I transposon insertions that could serve as activation tags in plant genomes. The activation tag construct shown in Figure 1 consists of three main components: (a) the En (Spm) transposase coding sequence under control of the CaMV 35S promoter and terminator sequences; (b) a mobile, nonautonomous I (dSpm) component harboring a tetramer of the CaMV 35S enhancer and the BAR gene between the terminal-inverted repeats, denominated activating I element (AIE); and (c) the negatively selectable marker SU1, adjacent to the transposon components within the T-DNA (Tissier et al., 1999).
Figure 1.
Schematic representation of the construct used for plant transformation. Relevant EcoRI sites used for the molecular analysis are indicated. LB, Left border; RB, right border; 35SP, 35ST, CaMV 35S promoter and terminator, respectively; EnTPase, En immobile transposase source; ILtir, IRtir, I-element left and right terminal-inverted repeat, respectively; 4 Enh., tetramer of the CaMV 35S enhancer; Pnos, Tnos, promoter and terminator sequences from the nopaline synthase gene, respectively; SSU5′, SSU3′, promoter and transit signal peptide to the chloroplast and terminator of the small subunit of Rubisco gene, respectively. The gene specific probes (BAR and SSU3′) used for blot hybridization are indicated as bars above or below the figure.
The transposon construct was introduced by A. tumefaciens-mediated transformation into the Arabidopsis ecotypes Wassilewskija (Ws) and Landsberg erecta (Ler). Twenty-six primary transformants named WAT (Ws) and LAT (Ler) were generated from the transformation experiments. Southern-blot and segregation analysis of a selection of primary transformants revealed that they contained from one to seven T-DNA copies at one or more loci (data not shown).
PCR was performed to analyze excision of AIE from the original construct in the primary transformants. Sequencing of PCR products confirmed the presence of an empty site in the donor T-DNA (data not shown). More than 90% of the first batch of primary transformants showed an empty donor site (EDS) by PCR, due to excision. Individual transformants displayed varying intensities of EDS fragments suggesting that there was enough transposase activity to produce excisions in the majority of transformants and also indicating variation among them.
Preliminary analysis of the efficiency of independent AIE transposition was done by characterizing the kanamycin-resistant progeny (containing the donor T-DNA) of one of the first transformants showing a high-intensity EDS fragment (line WAT2). Among 50 kanamycin-resistant plants analyzed, 17 different insertion bands could be seen in a Southern blot suggesting a high frequency of independent transposition (data not shown).
Because the two I transposon terminal-inverted repeats present in AIE were isolated from different inserts in the maize genome, the left junction from the waxy (Pereira et al., 1985) and the right junction from the a1 (Schwarz-Sommer et al., 1987) mutant loci, they contained different flanking sites that do not form a target site duplication. Because excision of AIE was found to be normal, our results confirm that the presence of a target site duplication is not necessary for excision.
Selection and Evaluation of Active Transposing Lines
To select for stable transposed insertion events (transposants), selfed T2 seeds (progeny seed of primary transformants) were sown at high density in the greenhouse and sprayed with Basta (formulation Finale) and R7402 (DuPont) after germination. Surviving doubly resistant seedlings were counted to estimate the stable transposition frequency (STF, ratio of surviving plants to seeds sown for each family). Genomic DNA from these plants was subjected to Southern analysis to determine different AIE insertions and to estimate the independent transposition frequency (ITF; Aarts et al., 1995). ITF is the ratio of the number of different fragments observed to the total number of plants analyzed from a family. This measure gives an indication of the frequency of independent transposition events and can be used to select families useful for generating new and independent insertions.
Pilot experiments were performed to determine the relationship between the STF and the ITF. For this, seeds from four lines were sown in soil and selected by spraying with Basta and R7402. Doubly resistant plants from each line were individually analyzed by Southern blot to detect different insertions. The results obtained are summarized in Table I. These lines displayed an inverse correlation between the STF and the ITF of each line, as observed previously (Tissier et al., 1999). This can be attributed to the timing of excision: Early excisions produce large clonal sectors in the plant, giving rise to a high number of stable progeny but with a low diversity of AIE insertions. Late excision events give rise to small clonal sectors that are represented as a few resistant progeny seedlings with a high diversity of independent insertions. On the basis of STFs and ITFs, lines WAT2 and WAT5 were identified as potentially suitable parents to generate a population of independent stable insertions.
Table I.
Stable and independent transposition frequencies of sprayed progeny
| Line | Plants | Plants Analyzed | Different Insertions | STFc | ITFd |
|---|---|---|---|---|---|
| n | % | ||||
| Single plant analysisa | |||||
| WAT 1 | 23 | 23 | 2 | 13.33 | 8.9 |
| WAT 2 | 18 | 18 | 9 | 0.23 | 50 |
| WAT 3 | 11 | 11 | 2 | 3.8 | 18.2 |
| WAT 3 | 18 | 18 | 4 | 4.82 | 25 |
| WAT 5 | 18 | 18 | 6 | 2.83 | 31.5 |
| Pooled plantsb | |||||
| WAT 3 | 19 | 6 | 1 | 4.52 | 16.67 |
| WAT 8 | 31 | 7 | 8 | 1.9 | 114 |
| WAT 10 | 25 | 8 | 5 | 1.7 | 62.5 |
| WAT 14 | 29 | 6 | 5 | 1.86 | 83.3 |
| WAT 15 | 8 | 8 | 1 | 0.8 | 12.5 |
| WAT 18 | 29 | 10 | 1 | 2.45 | 10.0 |
| WAT 20 | 8 | 8 | 1 | 0.69 | 12.5 |
| WAT 21 | 9 | 9 | 6 | 1.18 | 66.6 |
| WAT 23 | 10 | 10 | 3 | 0.82 | 30.0 |
| WAT 24 | 32 | 7 | 2 | 1.31 | 28 |
| WAT 29 | 3 | 3 | 3 | 0.92 | 100.0 |
| WAT 30 | 3 | 3 | 2 | 0.68 | 66.7 |
| WAT 32 | 7 | 7 | 3 | 0.39 | 42.8 |
| LAT 2 | 6 | 7 | 4 | 0.83 | 57.1 |
| LAT 3 | 37 | 13 | 2 | 8.2 | 15.4 |
| LAT 26 | 6 | 6 | 1 | 1.43 | 16.7 |
Data obtained with single plant Southern analysis.
Data obtained using pooled samples for the Southern analysis.
The STF was calculated as the ratio between the number of double resistant plants and the number of double-sprayed seeds.
The ITF was estimated as the ratio of the number of different fragments observed to the total number of plants analyzed from a family.
To create a large population, to identify more parental plants useful for insert generation, T2 seeds from the remaining primary transformants were used in double selection experiments on a bigger scale, and the STFs per line were calculated. The surviving seedlings were transferred to new soil, and DNA was extracted from inflorescences in pools of eight to 20 plants from the same transformed line.
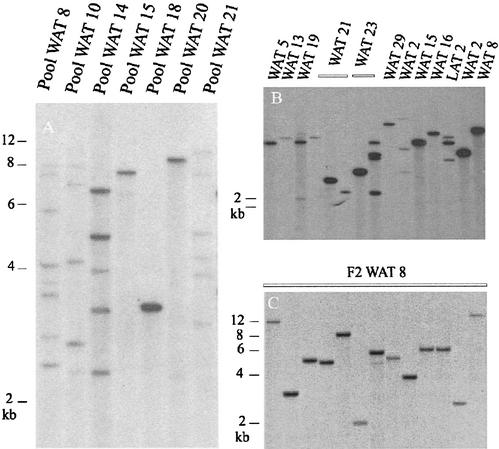
The DNA from the pools was analyzed by Southern-blot analysis and transposon insertion display (TID; Tissier et al., 1999; Yephremov and Saedler, 2000) to identify independent insertions. Southern analysis proved to be sensitive enough to estimate single AIE inserts in the pool based on reconstruction experiments (data not shown). The autoradiogram shown in Figure 2A displays the variation in insert number from the pooled DNA from stable insertion lines derived from different transformants.
Figure 2.
Southern blot of double-resistant plants revealing stable inserts with a BAR gene probe. A, Double-resistant progeny pools from different first transformants (WATs) are displayed. The bands show independent insertions. A ladder showing the size in kilobase pairs is indicated on the left side. The numbers of plants per pool are: WAT 8, seven plants; WAT 10, eight plants; WAT 14, six plants; WAT 15, eight plants; WAT 18, 10 plants; WAT 20, eight plants; and WAT 21, nine plants. B, Southern blot of random individual plants derived form different first transformants (population 1). C, Plants selected from spraying F2 (obtained by outcrossing T3 plants) seeds of line WAT8, obtained in population 5.
The ITF ranges from less than 5% to 114% (where at least one of the sampled plants had more than one independent AIE) among the different lines tested. For example, eight different bands can be seen in the Figure 2A in the pool of plants derived from WAT8, which included seven plants. In contrast, only one band is visible in a pool of 10 plants from line WAT18. The small sample of plants used in this analysis did not allow a precise comparison with the STFs but supported the selection of optimal lines. Table I shows the ITF and STF for most primary transformants.
This larger double selection experiment resulted in the generation of an initial population of 1,300 doubly resistant plants (denominated population 1). The total number of independent inserts in population 1 was estimated to range between 350 and 500.
Preliminary analysis of individual stable plants revealed that the number of AIE insertions was low, ranging from one to four per plant. In a random sample of 17 doubly resistant plants from population 1 obtained from different primary transformants, 76.4% contained one AIE, 17.6% contained three AIE inserts, and 5.8% contained four AIE inserts (Fig. 2B). In two additional independent samples of plants derived from the same primary transformant (WAT2), the majority of the plants contained one insert (60% and 73% in each group), and the rest had two (13% and 33%) or three insertions (13% and 8% in each group).
TID (Tissier et al., 1999), using representative pools of plants, provided data on the distribution of insertions in population 1. Table II shows the chromosomal distribution of 69 insertions obtained from DNA of plant pools derived from different lines. These results show that insertions in most Arabidopsis chromosomes can be obtained in the progeny from a single line and that the progeny of different lines produces an even distribution of insertions in the Arabidopsis genome (Table II). This suggests that the diversity of insertions in populations generated from these lines will be large and most probably should be able to saturate the genome when an adequate number of plants is used.
Table II.
Number of inserts in each chromosome (Chr.)
| Line | Chr. I | Chr. II | Chr. III | Chr. IV | Chr. V | Total Insertionsa |
|---|---|---|---|---|---|---|
| WAT2 | 1 | 2 | 7 | 6 | 2 | 18 |
| WAT3 | 1 | 1 | 2 | |||
| WAT8 | 2 | 5 | 4 | 1 | 12 | |
| WAT10 | 2 | 2 | 3 | 7 | ||
| WAT13 | 1 | 2 | 1 | 2 | 6 | |
| WAT14 | 1 | 4 | 1 | 6 | ||
| WAT17 | 2 | 1 | 1 | 1 | 5 | |
| WAT19 | 2 | 1 | 2 | 1 | 6 | |
| WAT21 | 2 | 1 | 3 | |||
| WAT24 | 1 | 1 | 2 | |||
| WAT30 | 1 | 1 | ||||
| WAT32 | 1 | 1 | ||||
| Total | 11 | 14 | 20 | 17 | 7 | 69 |
Inserts in representative plants from the first population.
Lines showing the highest ITF (more than 30%–40%) and an acceptable STF (less than 3%, like in lines WAT 2, 5, 8, 10, 14, 21, 23, 29, 30, 32, LAT2, and 26) were chosen for further double selections to increase the insert collection (populations 2–5). The unselected T2 and T3 plants from the chosen lines were next used to generate more stable insertions with the double-spray selection method. To check the ITF of the following generation, plants resistant to the double selection were analyzed by Southern blot. Line WAT8 still showed an ITF of 50% with mainly single inserts, whereas line WAT10 had an ITF of less than 30% independent insertions, indicating that the calculated frequencies were adequate enough to make predictions for the population (data not shown).
The AIE number per plant remained consistent in different generations for the analyzed lines (WAT8, WAT10, and WAT14), with four being the highest number of inserts found per individual. For example, from 20 T3 WAT8 plants, 14 have one (70%), five have two (25%), and one has three inserts (5%). Progeny derived from out-crossing T3 WAT8 plants with the wild type still bear a low number of insertions, as can be observed in Figure 2C.
Mutant Identification and Characterization
Population 1 was selected from among the original (T2) progeny of the transformants and was expected to have mainly heterozygous insertions. Phenotypic screens for obvious morphological mutants in the greenhouse revealed some new and previously observed phenotypes within this population. Inheritance tests were done using selfed or back-crossed seed. Though some mutant phenotypes did not reappear in the progeny, eight phenotypes were shown to be heritable. After segregation analysis, two mutants were found to be recessive and four dominant, and the inheritance of the remaining two remained unclear, but they were apparently dominant.
The recessive mutants included the well-described mutant fiddlehead (fdh) with fused inflorescences that originally did not display a leaf fusion phenotype, although the progeny showed a phenotype similar to previously isolated mutant alleles (Yephremov et al., 1999; Pruitt et al., 2000). Variegated leaves and highly variable flower phenotypes characterized the second recessive phenotype.
The dominant mutants observed included thread, a sterile plant with pronounced apical dominance and reduced and epinastic leaves; empty siliques, which showed developed but seedless siliques (see below); a mutant displaying slightly upward folded rosette leaves with decreased fertility, reduced size, and in late development increased shoot number; and a fourth mutant showing wrinkled leaves of increased size, late flowering time, and vertically oriented siliques.
Inheritance studies of the other two mutants have not been conclusive but strongly point to dominance. For example, one phenotype could not be recovered when Columbia (Col) wild-type plants were used as female parents for the cross (data not shown), but was recovered when Ws plants were used. One of these two mutants was found to be extremely small in size, with a profusion of small, serrated and wide leaves, reduced shoots, and very little seed set. In the second one (ecotype Ler), two different phenotypes were segregating in its progeny: one sterile with trichomes and the other fertile without trichomes. Both plant types also had very short, thick, crooked siliques and curved leaves.
The phenotype of the empty siliques mutant was characterized further to describe the nature of the lack of seed set. Crosses to the wild type in both (male and female) directions resulted in the formation of viable seed. Fertility is lower than in the wild type in both cases, with the mutant being even less fertile as a male. When pollinated by wild-type pollen, the mutant silique grows further and finally becomes larger than unpollinated mutant siliques and wider than wild-type ones. The stem grows in an undulating fashion, and the F1 seeds (produced by the female mutant) are larger than the wild-type seeds, whereas ovule and embryo sac development are normal (C.-M. Liu, personal communication).
Molecular Analysis of Mutants
To characterize the nature of the insertions conferring the different mutant phenotypes, DNA flanking the inserts was sequenced and used to position the insert in the genome and to determine its context with respect to adjacent annotated genes. The fiddlehead mutant had the most recognizable phenotype, and isolation of flanking DNA confirmed the mutant to be a knockout in the FDH gene (Yephremov et al., 1999).
For the dominant mutants, however, the AIE was not found to be present inside a predicted or experimentally defined coding region. The predicted/annotated genes in the region around the insertion were then identified, and primers were designed to clone them and analyze their expression. Genes closer to the enhancer of the AIE were regarded as primary candidates to be overexpressed and to account for the observed phenotype.
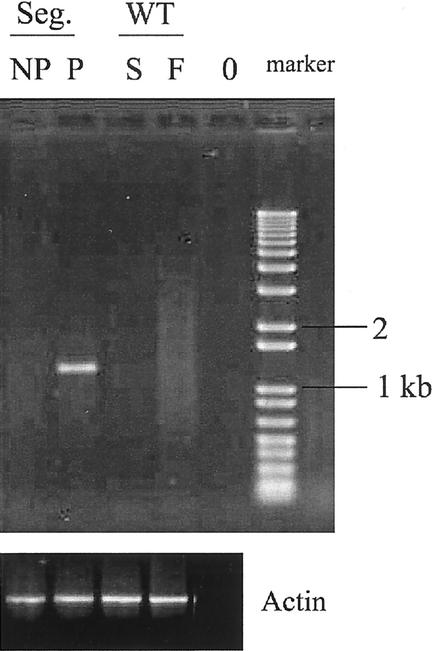
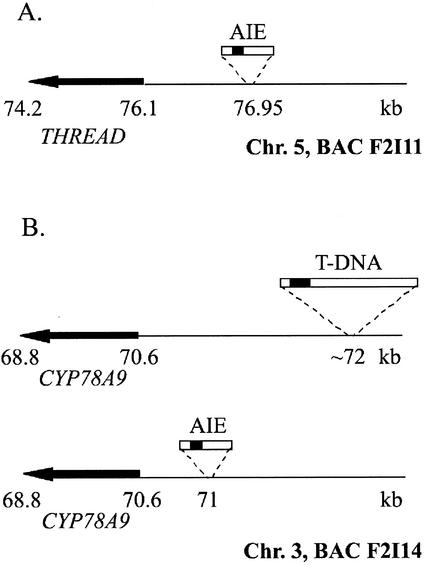
The expression level of the candidate mutant genes was first examined by semiquantitative reverse transcriptase (RT)-PCR analysis in mutant and nonmutant siblings (wild-type control) among the progeny of the original mutant. In all cases, increased levels of RT-PCR products from a gene near the insertion site were obtained from the plants showing the phenotype compared with the wild type. This analysis is shown in Figure 3 for the thread mutant. This mutant has an AIE insertion 828 bp upstream of the predicted translation start codon of the THREAD gene (F2I11_210; Fig. 4A). A clear PCR fragment is visible only in the RT-PCR samples of segregating seedlings showing the mutant phenotype, but not in wild-type plants, suggesting overexpression of the gene in the mutant and its possible role in causing the phenotype. No PCR product could be visually detected after 30 amplification cycles for wild-type flowers, but after 38 cycles a fragment of low intensity could be observed (data not shown).
Figure 3.
Semiquantitative RT-PCR using thread (top) or actin (bottom) primers. The lanes display the following samples: segregant (Seg) seedlings not showing (NP) and showing (P) the mutant phenotype; WT, Ws wild-type; s, seedlings; and f, flowers. Negative control (0), no RNA.
Figure 4.
Position of the AIE and T-DNA insertions in some mutants. A, AIE insertion in the thread mutant. B, AIE and T-DNA (Ito et al., 2000) insertions affecting the CYP78A9 gene (empty siliques phenotype). The dark box inside the “insertions” represents the enhancer tetramer.
To confirm that the observed phenotype was due to gene activation, a gene construct was made in which the THREAD coding sequence (from Ws) was placed under control of the CaMV 35S promoter and transformed to wild-type ecotypes Col, Ler, and Ws (the original ecotype) plants. Figure 5, E and F, shows the overexpression phenotype obtained by transforming the THREAD gene compared with the original thread mutant phenotype (Fig. 5B). The phenotype was recovered in the three ecotypes transformed, although with some differences. From seven Ws transgenic lines, all had epinastic leaves and short siliques, showing a very similar phenotype to the original mutant except one that had elongated siliques. These results confirm that the phenotype observed in the activation mutant is due to the enhanced transcription of the THREAD gene.
Figure 5.
thread, empty siliques, and other phenotypes. A, thread mutant flowering. B through D, thread mutant, wild-type Ws, and Col seedlings, respectively. E and F, 35S overexpression transformant seedlings in Col and Ler ecotypes, respectively. G, empty siliques mutant and wild-type siliques are indicated in the picture. H and I, Miniature bushy mutant with a yellow tip (approximately 5 cm) for comparison (H) and a few weeks later (I). J, Mutant showing serrated leaves.
From 13 Col transformants, 11 showed the epinastic phenotype in the leaves, with varying degrees of severity. The original mutant in the Ws ecotype had short siliques, but in some of the Col transformants the siliques were elongated, even though the epinastic leaf phenotype was evident. From 10 Ler transformants, all displayed the leaf phenotype, but the siliques, with one exception, were elongated. This illustrates the variation in phenotype not only among transgenic lines, but also among ecotypes.
The THREAD gene (F2I11_120) has been annotated and predicted to encode a flavin mono-oxygenase-like enzyme, although no phenotype or function has been experimentally determined previously. However, the predicted THREAD protein shares 66% amino acid identity with YUCCA, encoded by a cytochrome P450 gene that has recently been characterized by activation tagging (Zhao et al., 2001). The YUCCA gene family has nine members, and the overexpression of some of them leads to the overproduction of auxin, which results in the yucca phenotype (Zhao et al., 2001). The THREAD gene phenotype described here is, thus, a new member of the family that displays a similar phenotype, suggesting similar functions.
The AIE in the mutant empty siliques (Fig. 5G) is adjacent to a gene that has been previously identified by T-DNA activation tagging (Ito and Meyerowitz, 2000). This mutant has a transposon insertion around 500 bp upstream of the cytochrome P450 gene homolog classified as CYP78A9, as shown in Figure 4. The other two dominant mutants analyzed had insertions near genes not previously reported and will be described elsewhere.
In general, in the dominant mutants, the insert was placed upstream of the overexpressed gene, in a range of 105 bp to 1.9 kb from the translation start codon. The enhancer tetramer is present next to the left border of the mobile element. However, because the AIE is small, the enhancer sequences can act on genes that are on either side of it. In three cases, the overexpressed gene was positioned closer to the AIE left border (thus, near the enhancer). In the fourth case, the overexpressed gene was 1.9 kb to the right side of the AIE and 3.9 kb away from the enhancer, showing the furthest distance between the enhancer and the overexpressed gene among the sampled mutants.
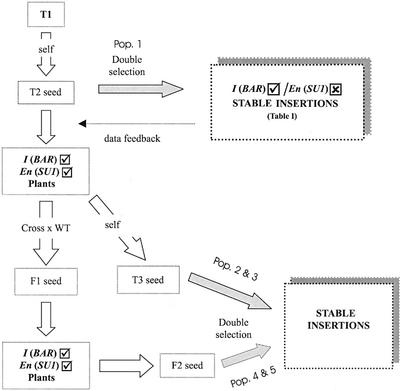
Generation of an En-I Activation Tag Population
Given the encouraging results obtained with the initial batch of transposants, the experiment was scaled up. The transformed genotypes that revealed optimal frequencies of stable transposants and ITFs on the basis of molecular genetic analysis were selected to generate a larger population of inserts. Initially the selfed T2 and T3 progeny were used as shown in Figure 6 for construction of populations 1 to 3. These lines often gave high numbers of SU1 homozygous plants and could harbor fixed insertions that occurred early in the transformed lines. To avoid these potential problems, the T2/T3 progeny of the primary transformants was crossed to wild-type and the F2 progeny used for selection of stable inserts to obtain populations 4 and 5 (Fig. 6).
Figure 6.
Steps followed to generate the population of stable insertions. The process starts from the first transformants (T1) containing the construct shown in Figure 1. Several rounds of stable insertion selections were carried out using either self or F2 seed as parental lines. The resulting population number reported in Table III is indicated for each case.
Molecular analysis of sets of plants from selected populations using the TID technique and Southern blots revealed the ITFs of different lines and allowed an estimate of the frequency of stable insertions. Lines with very high numbers (more than 20 stable plants of 1,000 tested seeds) were regarded as producing redundant or dependent inserts. On the other hand, lines that produced a few inserts were considered to be mostly independent. Based on this data, low frequency of stable transposition (fewer than 20 plants per 1,000 seeds) was used as an indicator for selection of lines for generating new transposant populations.
In populations 1 and 5, doubly resistant plants were grown in batches of about eight plants per pot allowing convenient observation of mutants. In populations 2 to 4, the objective was to obtain large populations in a small space and plants were grown in 96-well trays. However, this latter growing condition did not aid in the selection of mutant phenotypes.
Because population 5 was grown under optimal conditions for mutant identification, this population is, therefore, ideal to estimate the dominant mutant frequency. Some mutants could not produce seed because of sterility or weakness and, thus, were not used further in the analysis. An example of a sterile, miniature, bushy mutant is shown in Figure 5, H and I. Among 26 putative mutants from population 5 for which the progeny could be analyzed, 25 showed a dominant or semidominant inheritance of the phenotype. These 25 mutants from 2,500 estimated stable activation tags show that the frequency of observed dominant mutants is about 1%. This result is consistent with the data generated obtained in the pilot batch of stable insertions (population 1). These mutants showed diverse phenotypes with alterations in a wide range of characteristics such as fertility, height, flowering time, branching, flower structure, plant architecture etc. An example of a developmental mutant with a serrated leaf structure is shown in Figure 5J, indicating that developmental mutants can also be obtained using activation tagging with this transposon system.
In the resulting total population (consisting of five subpopulations) of about 13,000 plants containing an estimated of 8,300 inserts, a variety of aerial morphological mutants have already been identified as shown in Table III.
Table III.
Number of stable transposants and estimated insertions in different populations
| Population No. | Parental Genotypesa | Selected Transposantsb | Insertions Estimatedc | Mutants (Dominant)d |
|---|---|---|---|---|
| n | ||||
| 1e | 54 | 1346 | 400 | 8 (4) |
| 2 | 188 | 2,688 | 800 | 1 n.d. |
| 3 | 246 | 1,248 | 600 | 4 (3) |
| 4 | 1,250 | 4,960 | 4,000 | 8 (8) |
| 5e | 726 | 3,000 | 2,500 | 35 (25) |
| Total | 13,242 | 8,300 | 56 (40) | |
n.d., Not determined.
Number of segregating T2/T3 or F2 genotypes used.
Plants with stable insertions.
Estimated on basis of molecular analysis and stable insertion frequency.
Number of confirmed dominant mutants based on segregation.
Plants screened in pots at low density.
DISCUSSION AND CONCLUSIONS
Analysis of an En-I Transposon Activation Tagging Population
We describe here the use of the En/I (Spm-dSpm) maize transposon system for activation tagging in Arabidopsis. To position the 35S enhancer tetramer in the I element close to adjacent genes, fragments of 200 and 400 bp of the I-element terminal-inverted repeats were used. These fragments are much smaller than those described previously (Tissier et al., 1999). Excisions were observed in most transformants that contained a complete T-DNA, indicating that these short transposon terminal-inverted repeats are still functional for efficient transposition. The total size of the AIE (Fig. 1) is 3.7 kb, and the enhancer tetramer is only 200 bp away from the left end and around 2 kb from the right end of the element.
It has been previously reported (Weigel et al., 2000) that genes further than 3 kb from the enhancer can be activated to give a novel phenotype. In one case, we found that in the 35S enhancers could activate genes on both sides of the AIE (data not shown) requiring confirmation of the gene responsible for the observed phenotype.
To visualize the different stable insertions in the doubly resistant progeny of the primary transformants, Southern-blot analysis was done on individual and pooled plants. An inverse correlation was observed between the frequency of stable insertions and the ITFs in progenies from different transformants. Analysis of these parameters in different families of transformants allowed the selection of suitable parents with high ITFs that were useful for making a large population of insertions. For this purpose, optimal parental lines were chosen as starting lines on the basis of their ITF, as described in “Results.” Either selfed T2/T3 or F2 seed from crosses with the wild type were sown and then sprayed with both R7402 and Basta to select new stable insertions. Selfed seed has the disadvantage that a number of parents homozygous for the T-DNA or containing multiple T-DNA inserts will not be able to produce stable transposants. Moreover, insertions that are “fixed” as early transpositions will eventually be obtained in the next generation at a high frequency. In view of this, segregating F2 seed was used for the generation of the latter batches of the population. The parental F1s were obtained by crossing T2 or T3 plants with a wild-type Ws plant as the female parent to facilitate identification of F1 progeny compared with self-pollinated escapes, lacking the SU1 phenotype.
The variety of independent inserts can be increased in this way over several rounds of selection as outlined in Figure 6. The progeny of the selected lines can behave differently than the original progenitors or their siblings (data not shown). Because it was not possible to analyze molecularly each new stable transposant seedling to decide whether to include it in the collection, the inverse correlation between the STF and ITF provided a parameter to aid selection. To avoid the selection of stable insertions originating from plants with very low ITF and early clonal sectors that give a high percentage of stable transposants, the seeds from crosses of different progenitors were handled separately. In this way, seeds that give rise to a very high number of double resistant seedlings (and suspected to have a low ITF) can easily be identified, and only one or a few seedlings are then included in the population. Thus, individual families having a low to moderate frequency of stable transposants were used to select plants and to build up the population of independent inserts. This strategy is different from that used previously (Tissier et al., 1999) where the best lines were bulked and used for transposant generation.
The selection of progenitors whose seed would give rise to stable insertions, was done by spraying them with Basta to ensure the presence of the I element. Most conveniently, the SU1 gene serves as a visual marker because it confers a dwarf and dark-green phenotype that allows the selection of transposase-containing plants for use as parents for the double-spraying experiments. Each step of stable transposant selection resulted in batches of 1,000 to nearly 5,000 individuals (as shown in Table III). The total number of plants obtained to date is around 13,000, with 8,300 estimated independent inserts (Table III). Mutant identification in separate batches shows a considerable variation due to differences in plant handling and screening. Subpopulations 1 and 5 were obtained in optimal conditions and are the most representative, having a frequency of dominant aerial morphological mutants of 1%.
One advantage of using populations with low number of insertions per plant is that they allow the rapid identification of genes that are responsible for a phenotype of interest. Analysis of individual stable plants from different subpopulations, derived either from single or from a number of starting lines showed that they contain one (the majority) or just few transposon inserts. In addition, the inserts seem to be well distributed throughout the genome, as observed in a random sample of stable plants (Table II), suggesting that genome saturation of insertions within these genotypes is possible. In addition, the seeds of each transposant plant were collected separately in all subpopulations, enabling the screening of families and the correlation of phenotypes within a line. This can be particularly useful in cases where screenings for sensitive mutants to certain compound or environmental condition are used. It also avoids the problem of losing phenotypes that have been identified (as occurs when pooled seed is used). However, to facilitate large screens, pools of seed can also be made and seed from the original individual can be traced back once an interesting phenotype is identified.
Characterization of Transposon Activation-Tagged Mutants
In the pilot experiment on population generation, a number of dominant mutants were identified. Characterization of these mutants revealed that two were novel, and two coincided with previously reported genes or gene families isolated by means of T-DNA activation tagging. The recovery of activation-tagged genes similar to those obtained with T-DNA insertions indicates that the mechanisms involved in activation are similar for the transposon system, and validates the use of this system for gene activation.
One of the genes identified in our screen is the THREAD gene encoding a flavin mono-oxygenase-like enzyme, which belongs to the family of the YUCCA gene (Zhao et al., 2001). YUCCA was shown to be an enzyme involved in the Trp-dependent auxin biosynthesis pathway. This family has nine members, and the overexpression of some of them leads to the overproduction of auxin that results in the yucca phenotype (Zhao et al., 2001). THREAD shares a 66% amino acid sequence identity with YUCCA. The overexpression phenotype that THREAD produces is remarkably similar to the one described for the yucca mutant: long hypocotyl, enhanced apical dominance, narrow and epinastic leaves, and sterility. The auxin content in the thread mutant was not analyzed. However, its phenotype suggests the effects of elevated levels of this hormone.
RT-PCR experiments reveal a low level of expression of THREAD transcript in wild-type flowers. This is supported by the presence of an EST corresponding to the THREAD gene in a flower bud cDNA library (Asamizu et al., 2000). It is unclear whether other family members are expressed similarly or differ in their regulation. Interestingly, the transformants obtained with the overexpression construct display some differences between ecotypes. This might be due to the different genetic backgrounds that give rise to different gene interactions in each ecotype.
Another mutant is empty siliques in which overexpression of a cytochrome P450 (CYP78A9) gene causes a seedless phenotype. The CYP78A9 mutant was originally identified in a T-DNA activation-tagged population by screening for suppressors of the apetala2-1 phenotype. The insertion in the T-DNA mutant line was located around 2 kb away from the CYP78A9 gene (Fig. 4; Ito and Meyerowitz, 2000). In the transposon mutant line described here, the seedless phenotype (shown in Fig. 5G) was also observed but in a wild-type Ws background. Other phenotypic features besides the lack of seed, such as silique length and increased width when pollinated, were also similar. Some observations suggest that the lack of seed formation is caused by failure of fertilization with no zygotic and endosperm development in the ovules and could be due to a signal missing from the ovule or at other stages in the fertilization process (C.-M. Liu, personal communication).
It is intriguing to know whether the occurrence of these similar mutants in different activation-tagged populations (Ito and Meyerowitz, 2000; Zhao et al., 2001) is just a coincidence or indicates that the likelihood to overexpress particular genes or members of certain families is higher.
Our analysis of a number of tagged genes reveals that in many cases there is an ectopic overexpression of the tagged gene, although the presence of a low level of expression of the wild-type gene in certain tissues cannot be ruled out.
Transposons as Activation Tags
The identification of at least four gain-of-function activation mutants in the first batch of 400 activation-tagged inserts and 25 in an independent subpopulation containing about 2,500 inserts indicates a high frequency of activation tagging using the En-I transposon-based activation tagging system. Using the T-DNA activation tag system, one mutant per thousand individuals was reported (Weigel et al., 2000). Because it is likely that in the reported T-DNA populations there are several T-DNA inserts per individual, the frequency of activation mutants identified in our transposon tag collection is considerably higher. A higher frequency was also observed in a transposon tag population with a 35S promoter containing Ds element (Wilson et al., 1996). This would support the notion that activation tagging is more effective with transposons than with T-DNA.
The lower frequency of T-DNA activation mutants could be caused by a high incidence of complex and repeated loci in the T-DNA tagging population. Such complex loci occur often in T-DNA integration and are associated with silencing (Matzke et al., 2000). They may interfere with enhancer action by inactivating the enhancer through RNA silencing-like mechanisms and/or methylation (Mette et al., 2000; Sijen et al., 2001). In complex loci, the enhancer could also become positioned too far away to activate plant genes efficiently.
The double-selection system that we used to obtain stable transposon insertions assures, in most cases, the elimination of the original T-DNA from where the transposon excised. Therefore, in contrast to complex T-DNA insertions that are often generated during plant transformation, the nonautonomous enhancer-carrying transposons produce insertions containing a single copy of the intact element that could, therefore, be less susceptible to silencing phenomena. Another factor that could affect activation efficiency is that transposon insertions have a greater bias for insertion near transcriptionally active sites.
In addition, the use of a transposon as the carrier of the enhancer offers further advantages. Once an activated gene has been identified, the transposon could be remobilized to activate related genes or copies near the original, if present, and to produce knock-out insertion phenotypes (Wilson et al., 1996). Unstable transposons could also be helpful, particularly in recovering insertions that could confer embryo lethal activation phenotypes. Somatic insertions would then potentially display aberrant phenotypes only in specific sectors of the plant, from where tissue can be obtained to culture or to isolate DNA and identify the genes involved.
In conclusion, we have described an alternative strategy to efficiently produce populations of stable transposon-based activation tags in the Arabidopsis genome. We validate the En/I activation tag system by describing some of the mutants obtained. Moreover, the principles developed here are applicable to many plants that do not have efficient transformation systems (only a small number of primary transgenic lines is required to generate the activation population) or where somaclonal variation is high, a disturbing feature in forward mutant screens. The efficiency of the transposon activation tags, suggests that this system could be employed to saturate the Arabidopsis and other more complex genomes with inserts.
MATERIALS AND METHODS
Construction of En-I Activation Tag Vector
The construct for activation tagging was assembled in a single six-point ligation of multiple fragments excised from specific subclones. The fragments used were: (a) NotI-HindIII linker, (b) NotI-XhoI immobile 35S-En transposase, (c) a XhoI/XbaI left end of the I element with terminal-inverted repeat and a CaMV 35S enhancer tetramer, (d) a BamHI-SpeI fragment containing the right end of the I terminal-inverted repeat fused to the Nos promoter-BAR-Nos terminator, (e) an EcoRI-BamHI fragment containing the SSU-SU12 gene (O'Keefe et al., 1994), and (f) a HindIII-EcoRI-digested pBinplus vector (Engelen et al., 1995). The transposase fragment was derived from an immobile En transposase gene driven by a 35S promoter (Aarts et al., 1995). The nonautonomous I element contains a tetramer of the CaMV 35S enhancer (twice −392 to −90 and −526 to −90; Odell et al., 1985) and the BAR gene (De Block et al., 1987; Thompson et al., 1987).
Arabidopsis Transformation
Arabidopsis ecotypes Ws-3 and Ler-1, were grown in a climate chamber for 4 weeks under short days (8 h light, 16°C) and 2 weeks under long-day conditions (16 h of light, 22°C) at about 70% relative humidity.
The constructs were introduced into the plants using the floral dipping transformation method (Clough and Bent, 1998). The seeds were plated on one-half-strength Murashige and Skoog medium (Murashige and Skoog, 1962) in petri dishes with 1% (w/v) Suc and 50 mg L−1 kanamycin. After a 4°C treatment for three nights, they were transferred to a growth chamber (16 h of light, 23°C) for 9 d. The resistant seedlings were then transferred to soil in the greenhouse at around 22°C. Primary transformants (T1) were further confirmed by PCR for the presence of the marker genes and the correct position of the main elements.
Parental Lines and Transposant Selection
Selections for parental lines and transposants were done in the greenhouse. For transposant selection, progeny seedlings of first transformants or parental lines were sprayed with 0.7 mL L−1 Finale (commercial formulation, Aventis, which contains 150 g L−1 glufosinate ammonium) three times, and with 100 μg L−1 R7402 (DuPont) daily after germination for 8 d. Resistant plants were transferred to new soil 5 to 7 d after the last spray, and phenotypes were scored and allowed to set seed. Populations 2, 3, and 4 were transferred to new soil in 96-well trays, with one single plant per well. Populations 1 (8–20 plants per pot) and 5 (8–10 plants per pot) were transferred to new soil in 14-cm-diameter pots. Phenotypes were prescored during plantlet transfer for all subpopulations and at least once more during the life of the plant for subpopulations 1 and 5. T2/T3 and F1 plants to be used as parental lines were handled as for transposant selection, but were sprayed only twice with 0.7 mL L−1 Finale, and were visually evaluated (dwarf, dark, reduced apical dominance plants were selected). Their seeds were collected and used for transposant selection.
Molecular Analysis of Excision
DNA was isolated (Pereira and Aarts, 1998) from two leaves or young flower buds, either in Eppendorf tubes or in a 96 tube-rack. Genomic DNA (approximately 20 ng) isolated from the primary transformants was used in a PCR reaction to reveal EDSs. The following primers were employed: 35S-T2, 5′-CCA AAA TCC AGT GGG TAC CGA GC-3′; SSU-301-TF, 5′-GTT GGT TGA GAG TCT TGT GGC CT-3′. The PCR reaction comprised a denaturing step of 95°C for 3 min, followed by 35 cycles of 1 min at 95°C, 1 min at 60°C, and 1 min at 72°C, ending with an elongation step of 5 min at 72°C.
Transposition Analysis using Southern Hybridization
Approximately 500 ng for individual plants or 1 μg for genomic DNA from pooled plant tissue was digested with EcoRI and electrophoresed in a 0.8% (w/v) agarose gel in 1× TAE (40 mm Tris-acetate and 1 mm EDTA) and transferred to Hybond N+ membranes (Amersham, Buckinghamshire, UK). Digestion with EcoRI distinguishes between I elements in the original full donor site and EDS T-DNAs. To visualize the EDS and full donor site, labeled probes were used either containing an approximately 1.2-kb BamHI/PstI SU1 fragment or an approximately 620-bp BamHI/PstI BAR fragment (shown in Fig. 1). The BAR probe also allowed visualization of different I transpositions in the genome and was used to estimate the frequency of independent insertions. TID (Tissier et al., 1999) was used to support the results of Southern hybridization and to provide data on the frequency of insertions in the population.
Genetic Analysis of Mutants
The first and fifth subpopulations of stable transposed elements were screened visually for easily distinguishable morphological phenotypes in the greenhouse. The putative mutants were crossed to the wild-type ecotype Ws as female or male. A few crosses to other ecotypes, or Ler, were also carried out in some cases. The F1 and/or selfed progeny were grown to confirm the heritability and check dominance of the mutants.
Isolation of Flanking Sequences
To characterize the insertions in different mutants, genomic DNA was used to isolate fragments adjacent to the AIEs by thermal asymmetric interlaced-PCR (Liu et al., 1995; Liu and Whittier, 1995; Tsugeki et al., 1996) with the following modifications. The specific nested primers for the I element were: Int2, 5′-CAGGGTAGCTTACTGATGTGCG-3′; Irj-201, 5′-CATAAGAGTGTCGGTTGCTTGTTG-3′; and DSpm1 (Tissier et al., 1999), 5′-CTTATTTCAGTAAGAGTGTGGGGTTTTGG-3′ for the first, second, and third thermal asymmetric interlaced-PCRs, respectively. A fourth primer, I terminal-inverted repeat 3 (5′-CTTACCTTTTTTCTTGTAGTG-3′) was used to directly sequence the obtained PCR products with high specificity.
The sequence of the flanking DNA was compared against the Arabidopsis database using BLASTN (Altschul et al., 1997) and enabled positioning of the insert in the genome and its context with adjacent annotated genes.
Expression Analysis
Total RNA from mutant and wild-type whole seedlings, leaves, or flowers was isolated as described by Verwoerd et al. (1989). Approximately 1 μg of total RNA was treated with 1 unit of DNase I as indicated by the supplier (Invitrogen, Carlsbad, CA). cDNA was obtained using the Moloney murine leukemia virus RT as described by the supplier (Invitrogen) in a 20-μL reaction mix that contained 0.1 μg of the DNase-treated RNA and 1 μm oligo(dT) primer (T)23-NN among the components (1× Moloney murine leukemia virus RT buffer, 10 μm dithiothreitol, 1 mm dNTPs, and diethyl pyrocarbonate-treated water). Two microliters of the total RT reaction was used to perform the PCR with the gene-specific primers.
For the flavin mono-oxygenase-like gene (THREAD) the primers used were: Monoox5′, 5′-TTGGTACCCATGGGCACTTGTAGAGAA-3′; and Monoox3′, 5′-′GTGAGCTCTTAGGATTTATTGAAATGAAGATGA-3′. The primers used for Actin were 5′-GCGGTTTTCCCCAGTGTTGTT G-3′ and 5′-TGCCTGGACCTGCTTCATCATACT-3′. The reaction conditions for PCR included a denaturing step of 95°C for 3 min, followed by 32 cycles of 1 min at 95°C, 1 min at 54°C, and 2 min at 72°C, ending with an elongation step of 5 min at 72°C. The PCR to analyze the expression of the gene in wild-type flowers used 38 cycles (data not shown), and control PCR with actin primers comprised only 30 cycles.
Distribution of Materials
The materials developed will be made available to the scientific community in the form of parental lines useful to make subpopulations or pools of stable lines.
ACKNOWLEDGMENTS
We thank Dr. Daniel P. O'Keefe for providing the SU1 gene. We appreciate the assistance of Daan Jaspers, Gerrit Stunnenberg, and Piet de Man in the greenhouse. We also thank Asaph Aharoni, Wim Dirkse, Hanife Firinci, John Franken, Antonio Chalfun Jr., Stefan de Folter, and Jurriaan Mes for diverse kinds of help in different stages of this project; José López Bucio, June Simpson, and Jan Peter Nap for carefully reading the manuscript and for useful suggestions; and Chun-Ming Liu for analysis of the empty siliques phenotype.
Footnotes
This work was supported in part by an internal PRI Strategic expertise development fund (to A.P.) and by the Howard Hughes Medical Institute (grant no. 55003677 to L.H.-E.). N.M.-M. was supported by United Nations Educational, Scientific and Cultural Organization, Consejo Nacional de Ciencia y Tecnología (México), and International Agricultural Center (The Netherlands) fellowships.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.003327.
LITERATURE CITED
- Aarts MGM, Corzaan P, Stiekema WJ, Pereira A. A two-element Enhancer-Inhibitor transposon system in Arabidopsis thaliana. Mol Gen Genet. 1995;247:555–564. doi: 10.1007/BF00290346. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acid Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Asamizu E, Nakamura Y, Sato S, Tabata S. A large scale analysis of cDNA in Arabidopsis thaliana: generation of 12,028 non-redundant expressed sequence tags from normalized and size-selected cDNA libraries. DNA Res. 2000;7:175–180. doi: 10.1093/dnares/7.3.175. [DOI] [PubMed] [Google Scholar]
- Borevitz J, Xia Y, Blount J, Dixon RA, Lamb C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell. 2000;12:2383–2394. doi: 10.1105/tpc.12.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché N, Bouchez D. Arabidopsis gene knockout: phenotypes wanted. Curr Opin Plant Biol. 2001;4:111–117. doi: 10.1016/s1369-5266(00)00145-x. [DOI] [PubMed] [Google Scholar]
- Boyes D, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR. Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell. 2001;13:1499–1510. doi: 10.1105/TPC.010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- De Block M, Botterman J, Vanderwiele M, Dockx J, Thoen C, Gossele V, Movva RN, Thompson C, Van Montagu M, Leemans J. Engineering herbicide resistance in plants by expression of a detoxifying enzyme. EMBO J. 1987;6:2513–2518. doi: 10.1002/j.1460-2075.1987.tb02537.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Neve M, De Buck S, Jacobs A, Van Montagu M, Depicker A. T-DNA integration patterns in co-transformed plant cells suggest that T-DNA repeats originate form co-integration of separate T-DNAs. Plant J. 1997;11:15–29. doi: 10.1046/j.1365-313x.1997.11010015.x. [DOI] [PubMed] [Google Scholar]
- Engelen FA, Molthoff JW, Conner AJ, Nap JP, Pereira A, Stiekema WJ. pBINPLUS: an improved plant transformation vector based on pBIN19. Transgenic Res. 1995;4:288–290. doi: 10.1007/BF01969123. [DOI] [PubMed] [Google Scholar]
- Gheysen G, Herman L, Breyne P, Gielen J, Van Montagu M, Depicker A. Cloning and sequence analysis of truncated T-DNA insertions from Nicotiana tabacum. Gene. 1990;94:155–163. doi: 10.1016/0378-1119(90)90382-2. [DOI] [PubMed] [Google Scholar]
- Huang S, Cerny E, Bhat DS, Brown SM. Cloning of an Arabidopsis patatin-like gene, STURDY, by activation T-DNA tagging. Plant Physiol. 2001;125:573–584. doi: 10.1104/pp.125.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Meyerowitz E. Overexpression of a gene encoding a cytochrome P450, CYP78A9, induces large and seedless fruit in Arabidopsis. Plant Cell. 2000;12:1541–1550. doi: 10.1105/tpc.12.9.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakimoto T. CKI1, a histidine kinase homolog implicated in cytokinin signal transduction. Science. 1996;274:982–985. doi: 10.1126/science.274.5289.982. [DOI] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. Activation tagging of the floral inducer FT. Science. 1999;286:1962–1965. doi: 10.1126/science.286.5446.1962. [DOI] [PubMed] [Google Scholar]
- Lee H, Suh S, Park E, Cho E, Ahn JH, Kim S, Lee JS, Kwon YM, Lee I. The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 2000;14:2366–2376. doi: 10.1101/gad.813600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 1995;8:457–463. doi: 10.1046/j.1365-313x.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- Liu YG, Whittier RF. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics. 1995;25:674–681. doi: 10.1016/0888-7543(95)80010-j. [DOI] [PubMed] [Google Scholar]
- Martienssen RA. Functional genomics: probing plant gene function and expression with transposons. Proc Natl Acad Sci USA. 1998;95:2021–2026. doi: 10.1073/pnas.95.5.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Mette MF, Matzke AJM. Transgene silencing by the host genome defense: implications for the evolution of epigenetic control mechanisms in plants and vertebrates. Plant Mol Biol. 2000;43:401–415. doi: 10.1023/a:1006484806925. [DOI] [PubMed] [Google Scholar]
- Mette MF, Aufsatz W, Van der Winden J, Matzke MA, Matzke AJM. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 2000;19:5194–5201. doi: 10.1093/emboj/19.19.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio-assay with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Nacry P, Camillieri C, Courtial B, Caboche M, Bouchez D. Major chromosomal rearrangements induced by T-DNA transformation in Arabidopsis. Genetics. 1998;149:641–650. doi: 10.1093/genetics/149.2.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff M, Nguyen SM, Malancharuvil EJ, Fujioka S, Noguchi T, Seto H, Tsubuki M, Honda T, Takatsuto S, Yoshida S, Chory J. BAS1: a gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc Natl Acad Sci USA. 1999;96:15316–15323. doi: 10.1073/pnas.96.26.15316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odell JT, Nagy F, Chua N. Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature. 1985;313:810–812. doi: 10.1038/313810a0. [DOI] [PubMed] [Google Scholar]
- O'Keefe DP, Tepperman JM, Dean C, Leto KJ, Erbes DL, Odell JT. Plant expression of a bacterial cytochrome P450 that catalyzes activation of a sulfonylurea pro-herbicide. Plant Physiol. 1994;105:473–482. doi: 10.1104/pp.105.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira A, Aarts MGM. Transposon tagging with the En-I system. In: Martinez-Zapater J, Salinas J, editors. Arabidopsis Protocols. Totowa, NJ: Humana Press; 1998. pp. 329–338. [Google Scholar]
- Pereira A, Schwarz-Sommer Z, Gierl A, Bertram I, Peterson PA, Saedler H. Genetic and molecular analysis of the Enhancer (En) transposable element of Zea mays. EMBO J. 1985;4:17–25. doi: 10.1002/j.1460-2075.1985.tb02311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt RE, Vielle-Calzada JP, Ploense SE, Grossniklaus U, Lolle SJ. FIDDLEHEAD, a gene required to suppress epidermal cell interactions in Arabidopsis, encodes a putative lipid biosynthetic enzyme. Proc Natl Acad Sci USA. 2000;97:1311–1316. doi: 10.1073/pnas.97.3.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Sommer Z, Sheperd N, Tacke E, Gierl A, Rohde W, Leclercq L, Mattes M, Berndtgen R, Peterson PA, Saedler H. Influence of transposable elements on the structure and function of the A1 gene of Zea mays. EMBO J. 1987;6:287–294. doi: 10.1002/j.1460-2075.1987.tb04752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Vijn I, Rebocho A, van Blokland R, Roelofs D, Mol JNM, Kooter JM. Transcriptional and posttranscriptional gene silencing are mechanistically related. Curr Biol. 2001;11:436–440. doi: 10.1016/s0960-9822(01)00116-6. [DOI] [PubMed] [Google Scholar]
- Speulman E, Metz PLJ, Arkel G, Lintel-Hekkert B, Stiekema W, Pereira A. A two-component Enhancer-Inhibitor transposon mutagenesis system for functional analysis of the Arabidopsis genome. Plant Cell. 1999;11:1853–1866. doi: 10.1105/tpc.11.10.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CJ, Movva NR, Tizard R, Crameri R, Davies JE, Lauwereys M, Botterman J. Characterization of the herbicide-resistance gene BAR from Streptomyces hygroscopicus. EMBO J. 1987;6:2519–2523. doi: 10.1002/j.1460-2075.1987.tb02538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissier A, Marillonnet S, Klimyuk V, Patela K, Torres MA, Murphy G, Jones JDG. Multiple independent defective Suppressor-mutator transposon insertions in Arabidopsis: a tool for functional genomics. Plant Cell. 1999;11:1841–1852. doi: 10.1105/tpc.11.10.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugeki R, Kochieva EZ, Fedoroff N. A transposon insertion in the Arabidopsis SSR16 gene causes an embryo-defective lethal mutation. Plant J. 1996;10:479–489. doi: 10.1046/j.1365-313x.1996.10030479.x. [DOI] [PubMed] [Google Scholar]
- Van der Fits L, Memelink J. ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science. 2000;289:295–297. doi: 10.1126/science.289.5477.295. [DOI] [PubMed] [Google Scholar]
- van der Graaff E, Dulk-Ras AD, Hooykaas PJJ, Keller B. Activation tagging of the LEAFY PETIOLE gene affects leaf petiole development in Arabidopsis thaliana. Development. 2000;127:4971–4980. doi: 10.1242/dev.127.22.4971. [DOI] [PubMed] [Google Scholar]
- Verwoerd TC, Dekker BMM, Hoekema A. A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 1989;17:2362. doi: 10.1093/nar/17.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walden R, Fritze K, Hayashi H, Miklashevichs E, Harling H, Schell J. Activation tagging: a means of isolating genes implicated as playing a role in plant growth and development. Plant Mol Biol. 1994;26:1521–1528. doi: 10.1007/BF00016488. [DOI] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blázquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrándiz C, Kardailsky I, Malancharuvil EJ, Neff MM et al. Activation tagging in Arabidopsis. Plant Physiol. 2000;122:1003–1013. doi: 10.1104/pp.122.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K, Long D, Swinburne K, Coupland G. A Dissociation insertion causes a semidominant mutation that increases expression of TINY, an Arabidopsis gene related to APETALA2. Plant Cell. 1996;8:659–671. doi: 10.1105/tpc.8.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisman E, Hartmann U, Sagasser M, Baumann E, Palme K, Hahlbrock K, Saedler H, Weisshaar B. Knock-out mutants from an En-I mutagenized Arabidopsis thaliana population generate phenylpropanoid biosynthesis phenotypes. Proc Natl Acad Sci USA. 1998;95:12432–12437. doi: 10.1073/pnas.95.21.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yephremov A, Saedler H. Technical advance: display and isolation of transposon-flanking sequences starting from genomic DNA or RNA. Plant J. 2000;21:495–505. doi: 10.1046/j.1365-313x.2000.00704.x. [DOI] [PubMed] [Google Scholar]
- Yephremov A, Wisman E, Huijser P, Huijser C, Wellesen K, Saedler H. Characterization of the FIDDLEHEAD gene of Arabidopsis reveals a link between adhesion response and cell differentiation in the epidermis. Plant Cell. 1999;11:2187–2201. doi: 10.1105/tpc.11.11.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science. 2001;291:306–309. doi: 10.1126/science.291.5502.306. [DOI] [PubMed] [Google Scholar]