Abstract
To learn more about the role of the CER6 condensing enzyme in Arabidopsis surface wax production, we determined CER6 transcription domains and the timing of CER6 transcription in vegetative and reproductive structures from juvenile, mature, and senescing tissues. We found that CER6 is highly transcribed throughout development, exclusively in the epidermal cells in all tissues examined. The only exception to the epidermal expression was observed in anthers nearing maturity, in which CER6 mRNA was localized in the tapetum. To determine if environmental factors such as light and water deficit, which are known to stimulate wax accumulation, induce CER6 transcription, we examined the effects of these factors on CER6 transcript abundance. Our results demonstrate that light is essential for CER6 transcription, and that osmotic stress and the presence of abscisic acid enhance CER6 transcript accumulation. CER6 promoter-directed expression of the β-glucuronidase reporter gene in transgenic plants demonstrated that the CER6 promoter was highly effective in directing epidermis-specific expression in Arabidopsis and tobacco (Nicotiana tabacum). Furthermore, CER6 promoter-driven CER6 overexpression resulted in increased wax deposition in Arabidopsis stems. These experiments indicate that the expression level of CER6 in the epidermis is one of the factors controlling wax accumulation on Arabidopsis stems.
Epidermal cells of all aerial plant organs are covered with a cuticle, a polyester matrix of hydroxy and hydroxy-epoxy fatty acids, C16- and C18-long (cutin), embedded and overlaid with a mixture of very long chain lipids, commonly referred to as cuticular waxes (Walton, 1990). The primary role of cuticular waxes is to protect the plant from desiccation (Reed and Tukey, 1982), but they also play roles in protection from UV light and frost damage (Blum, 1975; Reicosky and Hanover, 1978; Richards et al., 1986; Grant et al., 1995; Barnes et al., 1996). Furthermore, waxes are believed to be one of the factors involved in plant defense against bacterial and fungal pathogens (Jenks et al., 1994), and they may contribute to a variety of plant-insect interactions (Eigenbrode and Espelie, 1995). The wax-related differences in plant resistance/susceptibility to environmental stresses, pathogens, or insects have been linked to wax accumulation (load) and wax composition, which vary among plant species (Eigenbrode and Espelie, 1995; Post-Beittenmiller, 1996). Therefore, the mechanisms by which plants control wax accumulation and composition are of considerable interest.
The predominant cuticular wax constituents, such as primary and secondary alcohols, aldehydes, alkanes, ketones, and esters, are derived from saturated very-long-chain fatty acids (VLCFAs; chain length C20–C34; von Wettstein-Knowles, 1995; Post-Beittenmiller, 1996). Thus, the first step in wax biosynthesis is the elongation of C18:0 fatty acid produced in the plastid to generate VLCFA wax precursors up to 34 carbons in length (Baker, 1982; Lemieux et al., 1994; Post-Beittenmiller, 1996). Fatty acid elongation (FAE) involves four enzymatic reactions. The initial condensation reaction, which adds two carbon units from malonyl-coenzyme A (CoA) to a C18 fatty acid substrate, is catalyzed by a β-ketoacyl-CoA synthase. Experimental evidence suggests that this is the substrate-specific step and the rate-limiting step of FAE (Lassner et al., 1996; Millar and Kunst, 1997). Subsequent reactions include a reduction of the β-ketoacyl-CoA to β-hydroxy-CoA, dehydration to an enoyl-CoA, and a second reduction to yield acyl-CoA extended by two carbons. Enzymes catalyzing these last three reactions are thought to be constitutively expressed throughout the plant and used with condensing enzymes present in the same cell (Millar and Kunst, 1997).
To date, several VLCFA condensing enzymes have been studied from Arabidopsis. A single condensing enzyme, FAE1, catalyzes VLCFA synthesis in seeds (Kunst et al., 1992; James et al., 1995), whereas three condensing enzymes, KCS1 (Todd et al., 1999), FDH (Yephremov et al., 1999; Pruitt et al., 2000), and CER6 (Millar et al., 1999; Fiebig et al., 2000), have been implicated in the synthesis of VLCFA precursors for wax production in shoots. It is interesting that a major reduction of CER6 activity in cer6 mutants and sense suppressed CER6 plants nearly abolished stem wax accumulation, and resulted in conditional male sterility (Millar et al., 1999), suggesting that there is no significant functional overlap of CER6 with KCS1 and FDH activities in the stem and anther of Arabidopsis. Furthermore, a recently identified VLCFA condensing enzyme, CER60 (Fiebig et al., 2000), with high amino acid sequence identity to CER6, does not appear to significantly contribute to the synthesis of stem and pollen surface lipids. This could be due to a very low level of CER60 expression or a different tissue specificity of CER60. Taken together, these data implicate CER6 as the major condensing enzyme for stem wax and pollen coat lipid biosynthesis.
To improve our understanding of the role that CER6 plays in stem wax production, we determined the site and the timing of CER6 transcription during Arabidopsis development by reverse transcriptase (RT)-PCR, RNA-blot hybridization analysis, and in situ hybridization. We also isolated a 5′-CER6 promoter fragment, fused it to the β-glucuronidase (GUS) reporter gene, and examined its tissue specificity and activity in transgenic Arabidopsis and tobacco (Nicotiana tabacum).
Because condensing enzymes are rate-limiting activities of VLCFA biosynthesis, and because CER6 is a major condensing enzyme providing VLCFA precursors for stem wax and pollen lipid biosynthesis, we were interested if wax accumulation is, in part, regulated by the level of CER6 transcription. Thus, we examined the effects of light and osmotic stress, known to induce wax synthesis in higher plants (Thomas and Barber, 1974; Bengtson et al., 1979; von Wettstein-Knowles et al., 1979; Hadley, 1989), on CER6 transcript accumulation. In addition, we generated a series of transgenic Arabidopsis lines overexpressing CER6 in the epidermis. Our results provide evidence that the transcription level of CER6 is one of the factors controlling wax accumulation on Arabidopsis stems.
RESULTS
Expression Analyses of the CER6 Gene
RNA-blot hybridization was used to investigate the transcription profile of the CER6 gene in developing roots, leaves, stems, siliques, and flower buds of Arabidopsis. The presence of a CER6-like gene, CER60, in the Arabidopsis genome that shares 79% and 85% nucleotide identity with CER6 in exons 1 and 2, respectively (Fiebig et al., 2000), prompted us to carefully evaluate the probes used for the RNA-blot analyses. DNA-blot analyses originally performed by Millar et al. (1999) indicated that CER6 and CER60 do not crosshybridize at high stringency. Using the same full-length CER6 coding region as a probe (Millar et al., 1999), we confirmed that this was the case. To further ensure the specificity of hybridization, we generated CER6 and CER60 gene-specific probes originating from the 5′-untranslated region (UTR) of these two genes and used them to hybridize the corresponding RNA blots. A CER6-specific probe detected CER6 transcript in all shoot tissues assayed, but not in roots (data not shown). Surprisingly, we could not detect the CER60 transcript using the CER60 5′-UTR probe. Thus, CER60 was not transcribed in the selected tissues at the time when the tissues were harvested, or the level of CER60 transcription was low.
In an attempt to examine CER60 transcript levels and compare them with CER6, and to determine the tissue specificity of CER60 transcription in Arabidopsis shoots, we designed gene-specific primers and performed a quantitative RT-PCR experiment using the histone H1 sequence as an internal standard (Fig. 1). Cycle and template optimization revealed that a PCR protocol using 22 cycles and 25 ng of cDNA template gave a result within the linear range for all the sequences amplified. The RT-PCR assay showed consistently high CER6 transcript levels throughout the shoot, but no significant CER6 mRNA accumulation in roots (Fig. 1). In contrast, CER60 transcript levels were low in all the mature shoot tissues examined, and almost undetectable in roots and stems (Fig. 1). CER60 mRNA levels were somewhat higher only in floral tissues and the shoots of developing 8-d-old seedlings. This pattern of transcription of CER60 suggests that it also may have a role in the synthesis of wax precursors. However, low levels of CER60 transcription throughout the mature shoot, together with increased transcript accumulation during early shoot morphogenesis and flowering (pollen production), are consistent with an accessory role in wax deposition at certain stages of development that require higher levels of wax production.
Figure 1.
CER6 expression is limited to shoots and is much higher than that of CER60. SYBRGreen I-stained agarose gel showing RT-PCR products of CER6 (700 bp), CER60 (600 bp), and histone H1 (800 bp) from different tissues of Arabidopsis.
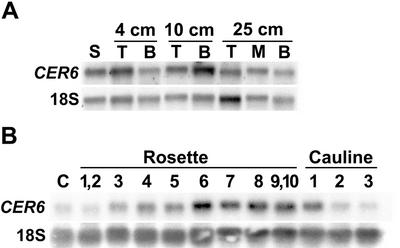
Deposition of cuticular waxes is known to begin very early in plant development, likely as soon as epidermal cells are exposed to air (Jeffree, 1996), and continues during subsequent organ expansion. To investigate the involvement of CER6 in cuticular wax biosynthesis during stem and leaf development, we analyzed CER6 transcript accumulation in these organs by RNA-blot hybridization (Fig. 2). CER6 mRNA levels were already high in young 8-d-old seedlings, as well as in 4-cm-tall bolting stems, and increased further in 10-cm stems. It is surprising that similar CER6 transcript levels were detected in tops and bases of stems of the same height, even in old 25- to 30-cm-tall stems, suggesting that CER6 transcription did not cease once the stems had finished elongating (Fig. 2A). In leaves, CER6 mRNA was present at every developmental stage from cotyledons to the youngest cauline leaves, but was the most abundant in rosette leaves numbers 6 through 10 (Fig. 2B).
Figure 2.
RNA-blot hybridization analysis of CER6 expression in developing Arabidopsis stems and leaves. In each case, 10 μg lane−1 of total RNA was probed with CER6 coding region and 18S rRNA (loading control). A, RNA extracted from 8-d-old seedlings (S) and from 2-cm segments of the tops (T) and bottoms (B) of bolting stems 4, 10, and 25 cm (when flower production had ceased) tall. RNA was also extracted from the 2-cm section at the center (M) of 25-cm-tall stems. B, RNA extracted from cotyledons (C), rosette (in sequence; first initiated, 1; last initiated, 10), and cauline leaves (also in sequence; numbering as with rosette leaves). Leaves were harvested when the plants had bolting stems 10 cm tall.
CER6 Is Transcribed in a Cell-Specific Manner
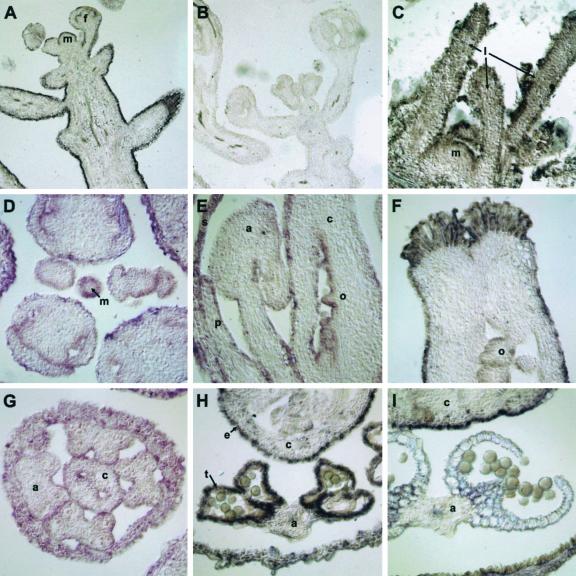
Spatial distribution of CER6 mRNA in Arabidopsis shoots was analyzed in more detail in an in situ hybridization experiment (Fig. 3). In all of the plant organs tested, reproductive and vegetative meristematic regions (Fig. 3, A and C), floral primordia, developing carpels, ovules, and stamens (Fig. 3, D–I), the CER6 transcript was exclusively present in the epidermal cell layer. The only exception to the epidermis-specific transcription was found in the anthers. Even in the anthers, CER6 was transcribed in the epidermis from the primordial stage until the time when the sporogenous tissue underwent meiosis. However, shortly after young microspores were released from the tetrads, CER6 transcripts accumulated only in tapetal cells, but not in other anther tissues (Fig. 3, E, G, and H). Transcription of CER6 in the tapetum during microsporogenesis could be expected of a condensing enzyme required for production of pollen coat lipids, as the tapetum is responsible for the production of the lipidic components of the pollen coat (Piffanelli et al., 1998). CER6 mRNA persisted in the tapetum until the breakdown of the tapetal layer, after which CER6 transcripts were undetectable in the anthers (Fig. 3I).
Figure 3.
In situ localization of CER6 mRNA. Sections of Arabidopsis tissues hybridized to antisense CER6 RNA probe (A, C–I) and sense CER6 RNA probe (B). Hybridization is indicated by a purple precipitate produced as a result of an alkaline phosphatase reaction with the nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate substrate. A and B, Longitudinal sections of the inflorescence meristem (m) and developing flower buds (f), showing epidermal staining (hybridization) with the antisense probe (A) that is absent with the sense probe (B; control; 10×). C, Vegetative meristem of an 8-d-old seedling showing epidermal hybridization over the meristem (m) and in the leaf primordia (l; 40×). D, Cross section through the inflorescence meristem (m) and surrounding flower buds showing epidermal hybridization (40×). E, Longitudinal section through a flower bud showing the developing sepal (s), petal (p), stamen (a), and carpel (c) with the ovule primordia (o) beginning to develop in the carpel. Epidermal hybridization is detected in the carpel, including ovule primordia, stamen, and petal, but not in the sepal at this stage (40×). F, Longitudinal section through the nearly mature carpel, exhibiting epidermal staining on the outside of the carpel and on the ovules (o; 40×). G, Cross section of a flower bud with developing carpel (c) and anthers (a) with microspores in meiosis (40×). H, Cross section through a flower bud with anther (a) showing tapetal staining (t) during pollen development and epidermal staining (e) of the carpel (c; 40×). I, Cross section through a flower bud with degraded tapetum and dehiscing anthers (a) that do not show any hybridization with CER6; hybridization is still apparent in the carpel (c) epidermis (40×).
Isolation of the CER6 Promoter and Expression of CER6 Promoter-GUS Fusions in Transgenic Plants
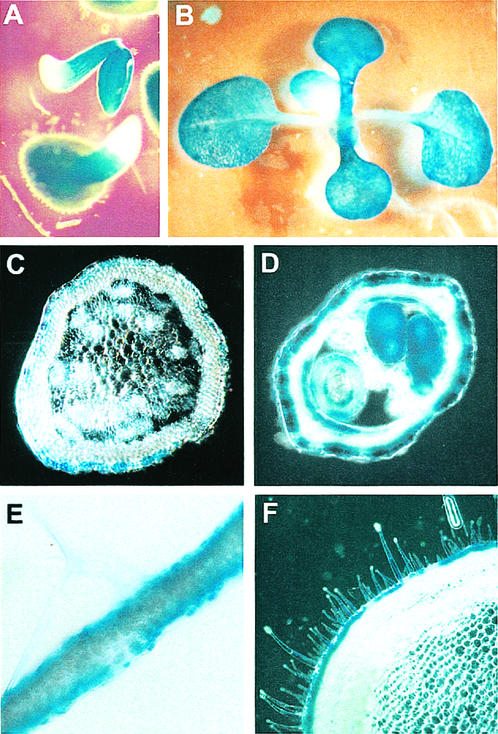
To further investigate the tissue specificity and timing of expression of the CER6 gene, the 1,208-bp genomic fragment immediately upstream of the CER6 coding region was fused to a promoterless bacterial uidA gene encoding GUS, and was used to transform Arabidopsis and tobacco. Tissue samples of five independent transgenic Arabidopsis lines were stained for GUS activity. GUS activity was found in all aerial parts of the plants, but never in roots (Fig. 4, A and B), consistent with the RT-PCR data (Fig. 1). Free-hand cross sections of stems, leaves, and siliques from all five lines showed a localization of GUS activity exclusively in the epidermal cells (Fig. 4, C–E), mirroring the result of the in situ experiments (Fig. 3). Histochemical GUS staining of stem and leaf cross sections of five independent transgenic tobacco plants demonstrated that the epidermal specificity of the CER6 promoter was retained even in plant species unrelated to Arabidopsis (Fig. 4F). Thus, it appears that the CER6 promoter will be a useful tool for targeting the expression of genes of interest to the epidermis in transgenic plants.
Figure 4.
The CER6 promoter directs epidermis-specific expression of GUS in Arabidopsis throughout the shoot from very early stages of development, as well as in tobacco. Seedlings (A and B) and tissue sections (D and E) of Arabidopsis and tobacco (F) plants transformed with the CER6 promoter-GUS construct, and incubated in 5-bromo-4-chloro-3-indolyl-β-d-glucuronide assay buffer. GUS activity is indicated by a blue precipitate. A, One-day-old seedlings; B, 8-d-old seedlings; C, free-hand cross section of the top of a bolting stem; D, silique cross section; E, leaf cross section; F, tobacco stem cross section.
The onset of gene expression directed by the CER6 promoter was also examined in young germinating CER6 promoter-GUS seedlings. GUS expression was detected in the cotyledons and hypocotyl as early as the beginning of radicle emergence from the seed, 1 d after transfer of stratified seeds to 20°C for germination (Fig. 4A). In agreement with the RNA-blot hybridization data (Fig. 2), high levels of GUS staining persisted throughout the aerial parts of the seedlings (assayed on d 3, 5, 8, and 14 postgermination), and in rosette leaves and bolting mature plants (assayed on d 21 postgermination; Fig. 4 and data not shown).
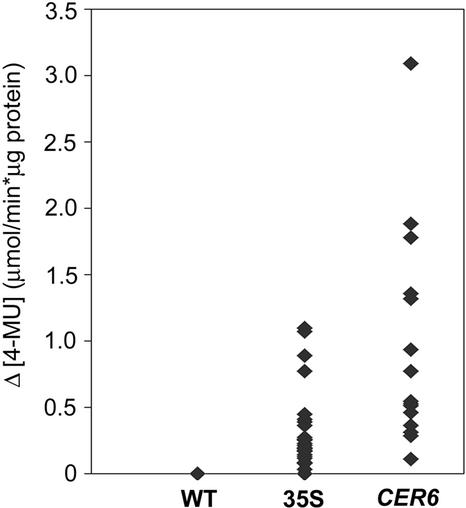
To evaluate the strength of the CER6 promoter, crude protein extracts of 8-d-old seedlings of independent CER6 promoter-GUS lines were assayed for GUS activity using a quantitative fluorometric assay. The GUS activity measured in CER6 promoter-GUS lines was compared with that of transgenic lines carrying a 35S promoter-GUS construct. The average rate of hydrolysis of the 4-methylumbelliferone-glucuronide (MUG) substrate in the CER6 promoter-GUS lines (894 nmol 4-methylumbelliferone [4-MU] min−1 μg−1 protein) was considerably higher than that in the 35S promoter-GUS plants (325 nmol 4-MU min−1 μg−1 protein; Fig. 5). Furthermore, the highest rate of MUG hydrolysis in CER6 promoter-GUS plants (3,089 nmol 4-MU min−1 μg−1 protein) was also about 3-fold higher than that of the best 35S promoter-GUS line (1,097 nmol 4-MU min−1 μg−1 protein). Thus, the CER6 promoter is highly active in transgenic Arabidopsis.
Figure 5.
GUS activity in plants transformed with CER6 promoter-GUS is higher than that found in plants transformed with the cauliflower mosaic virus (CaMV)35S promoter-GUS. Activity of GUS from protein extracts of 8-d-old Arabidopsis seedlings, measured by accumulation of 4-MU. Wild-type (Columbia-2) and lines transformed with CaMV35S promoter-GUS and with CER6 promoter-GUS (CER6). Each point represents activity from a separate transformed line.
Environmental Effects on CER6 Transcription
The presence of a number of putative regulatory elements in the CER6 promoter, such as the I-box and the GT1-binding site found in light-inducible promoters (Terzaghi and Cashmore, 1995), and the abscisic acid (ABA)-responsive cis-acting elements (ABRE; Guiltinan et al., 1990) involved in ABA-regulated gene expression of a number of drought- and cold-inducible genes (Fig. 6) suggested that light and osmotic stress may influence CER6 expression. Therefore, we analyzed the effects of these factors on CER6 transcript abundance by RNA-blot hybridization.
Figure 6.
The CER6 promoter region has numerous consensus sequences for light and ABRE. Genomic sequence 5′ to the CER6 coding region, showing ABRE (light shading), I-boxes (single border), and GT1 binding sites (double border). The ATG initiating translation is marked with dark shading.
The effect of light was monitored in 8-d-old seedlings and in bolting stem tops (Fig. 7). CER6 transcripts were undetectable in etiolated seedling shoots, but accumulated to significant levels 2 d after the seedlings were transferred to light (Fig. 7A). Light was also required for CER6 transcription in developing bolting stems, as the absence of light for 24 h resulted in a significant reduction in CER6 mRNA levels. CER6 mRNA was undetectable in stems of plants deprived of light for 96 h (Fig. 7B). We also examined CER6 transcript accumulation in the det1 and det2 mutants, which develop as light-grown plants even when grown in darkness due to defects in the light-regulated signal transduction pathways (Chory and Susek, 1994). As in wild-type plants, CER6 was not transcribed in the det1 mutant in the absence of light (Fig. 7A). Thus, repression of CER6 in the dark does not depend on the DET1 pathway. However, CER6 transcription in the det2 mutant was not completely repressed by darkness, suggesting that the DET2 pathway may be involved in CER6 dark repression.
Figure 7.
Light is required for CER6 expression in seedlings and in bolting stems, as shown by RNA-blot hybridization. Ten micrograms of total RNA was loaded into each lane. A, RNA extracted from wild-type (Columbia-2), det1, and det2 mutant 8-d-old seedlings germinated on agar plates in continuous light (L), continuous darkness (D), and dark for 6 d followed by transfer to light for the last 2 d (6D + 2L; wild type only). The blot was probed with the CER6 coding region and 18S rRNA (loading control). B, RNA extracted from stem tops of plants incubated in continuous light (L) or placed in darkness for 8, 24, 48, or 96 h. The blot was hybridized to the CER6 5′-UTR and 18S rRNA probes.
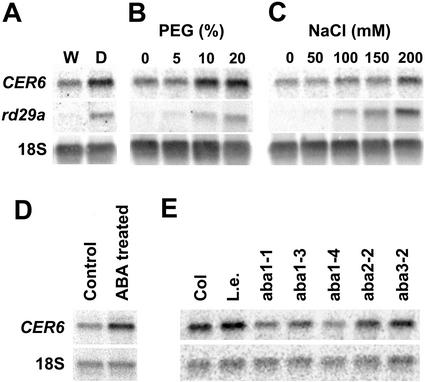
CER6 transcript accumulation in response to osmotic stress caused by water deficit, polyethylene glycol (PEG), or salt was examined in comparison with rd29A (Yamaguchi-Shinozaki and Shinozaki, 1993), an extensively studied drought-inducible gene (Fig. 8). In bolting stems, water stress resulted in 2-fold higher CER6 mRNA levels than in well-watered control plants (Fig. 8A). Application of PEG or salt caused a similar increase in CER6 transcript levels in seedlings, with higher concentrations of PEG and salt resulting in greater accumulations of CER6 mRNA (Fig. 8, B and C). Changes in the CER6 mRNA levels under osmotic stress are obviously not as dramatic as those seen for rd29A. This is probably because, unlike rd29A, CER6 transcription is relatively high even in unstressed tissue.
Figure 8.
Water deficit and ABA treatment increase CER6 transcript accumulation in seedlings and in bolting stems of Arabidopsis. RNA-blot hybridization of total RNA (10 μg lane−1) probed with CER6 5′-UTR, rd29A unique sequence, and 18S rRNA (loading control). A, RNA extracted from the tops of 10 cm tall bolting stems of plants grown in well-watered (W) pots and in pots allowed to dry out immediately after transplanting (D). B, RNA extracted from shoots of 14-d-old seedlings germinated on agar plates, then transferred to and incubated for 10 h in liquid AT medium containing PEG (0%–20%) prior to harvest. C, RNA extracted from shoots of 14-d-old seedlings germinated on agar plates, then transferred to and incubated for 10 h in liquid AT medium containing NaCl (0–200 mm) prior to harvest. D, RNA extracted from shoots of 14-d-old seedlings germinated on agar plates, then transferred to and incubated for 10 h in liquid AT medium containing ABA in methanol (10−4 m) or an equal volume of methanol (control). E, RNA extracted from the tops of 10 cm tall bolting stems of wild-type (Columbia-2 and Landsberg erecta) and ABA synthesis mutants (aba1-1, aba1-3, aba1-4, aba2-2, and aba3-2).
The phytohormone ABA plays an important role in mediating the transcription of a large number of genes that respond to drought and salt stress. These stresses trigger the production of ABA, which in turn induces gene expression. Such genes are also induced by exogenous application of ABA and typically contain ABRE (Guiltinan et al., 1990) in their promoter. The presence of the ABREs in the CER6 promoter (Fig. 6) led us to investigate whether CER6 transcription is under ABA control. CER6 transcript abundance was determined in 14-d-old seedlings floated on an AT (Arabidopsis thaliana) medium (Somerville and Ogren, 1982) supplemented with 10−4 m ABA for 10h and compared with controls floated on the same AT medium without ABA. ABA treatment resulted in 2.5- to 3-fold greater CER6 mRNA accumulation in Arabidopsis seedlings (Fig. 8D), suggesting that ABA can induce CER6 transcription. To further test the role of ABA in CER6 expression, we examined CER6 transcript accumulation in stems of ABA-deficient mutants under a normal watering regime. In all the mutant plants tested, CER6 transcription was substantially reduced (Fig. 8E), with aba1 mutants accumulating less than 50% of the wild-type levels of CER6 mRNA. Thus, even under normal watering conditions, ABA appears to be involved in the regulation CER6 transcription.
Overexpression of CER6 Can Increase Surface Wax Accumulation
To more directly assess whether the extent of wax accumulation can be affected by changing the level of CER6 transcription, we overexpressed the CER6 gene in Arabidopsis using the strong constitutive CaMV 35S promoter. RNA-blot analysis revealed high levels of CER6 transcript in a number of transgenic lines. However, none of these lines had a significantly greater wax load (Millar et al., 1999 and data not shown). These experiments suggested that higher levels of CER6 transcription in the epidermis might be required to impact wax production. Therefore, we transformed Arabidopsis plants with an extra copy of the CER6 gene under the control of its native CER6 promoter (1× cassette). In addition, we investigated if introducing two copies in tandem of CER6 per T-DNA copy (2× cassette) or three copies (3× cassette) would result in a greater accumulation of wax or would generate a high-wax phenotype at a higher frequency. Seventy-four kanamycin-resistant lines transformed with a 1× cassette (35 waxy and 39 waxless), 66 plants transformed with a 2× cassette (29 waxy and 37 waxless), and 67 plants transformed with a 3× cassette (37 waxy and 30 waxless) were recovered.
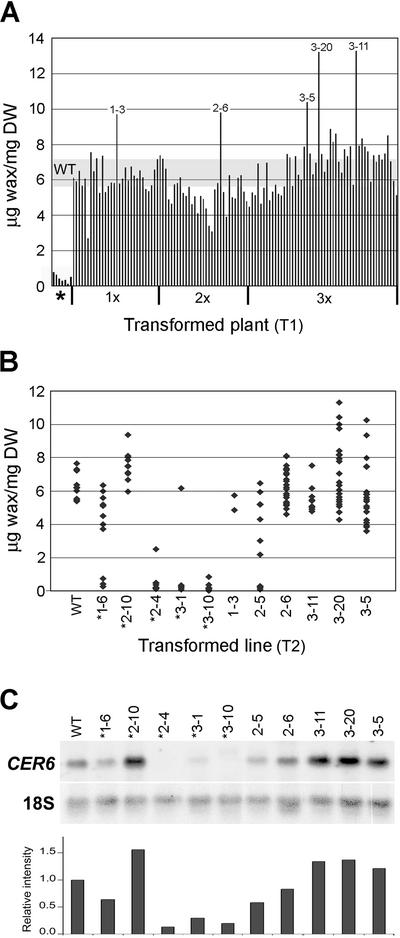
The wax load in transgenic plants with visible surface wax receiving all three types of cassettes ranged between 3 and approximately 13 μg mg−1 dry wt, with an average value similar to that of the wild type (5.6–7.2 μg mg−1 dry weight; Fig. 9A). However, a few lines had a wax load almost 100% greater than that measured for wild-type plants. It is interesting that the highest wax loads achieved with 1× and 2× cassettes were similar, but those generated with three copies of the CER6 transgene per insert (3× cassette) were considerably higher. However, the frequency with which transgenic lines containing 3× cassettes produced a high-wax phenotype was comparable with that obtained with 1× and 2× cassettes. A large proportion of transgenic plants receiving 1×, 2×, and 3× cassettes exhibited a waxless phenotype, presumably due to sense suppression (Fig. 9A and data not shown). Gas chromatographic analysis of surface wax extracted from stems of seven randomly selected waxless plants showed that they all had less than 10% of the wax load measured in wild-type Arabidopsis (ecotype Columbia-2).
Figure 9.
Wax load and CER6 expression in stems of plants transformed with the 1× cassette, 2× cassette, and 3× cassette of the CER6 promoter-CER6. CER6 transcript accumulation correlates with wax loads of plants transformed with CER6 promoter-CER6. A, T1 generation; each bar represents the wax load of a single transgenic plant. Wax loads of seven transgenic lines with no visible wax (first seven bars; indicated with an asterisk), and all the transgenic plants recovered that had visible surface wax are shown. The type of cassette introduced is indicated. Plants with wax loads significantly increased over that of the wild type are marked with their line numbers. The range of wax loads of six wild-type control plants is shaded across the graph. B, T2 generation; each point represents the wax load of a single plant. Each column represents progeny from one transgenic line. Lines marked with asterisks were waxless in the T1 generation. The first number in the transformed line indicates the type of cassette used (1×, 2×, or 3×). C, RNA-blot hybridization of total RNA (10 μg lane−1) extracted from the tops of 10 cm tall bolting stems of T2 progeny of lines transformed with 1×, 2×, or 3× cassettes. The blot was probed with CER6 coding region and 18S rRNA (loading control). The relative intensity of CER6 transcript accumulation was calculated and standardized according to that of the untransformed wild-type plants.
We also analyzed the composition of wax on the stems of transgenic Arabidopsis lines with greater wax loads. We found that wax composition of all lines examined was very similar to that of the Columbia-2 wild type grown under the same conditions (data not shown), and in good agreement with wax composition previously reported for the Columbia-2 ecotype (Millar et al., 1999).
To follow the wax phenotype in the progeny of plants overproducing wax, seed was collected from the T1 plants that had more wax than the untransformed controls, as well as from a few of the waxless plants. T2 seeds from these primary transformants were planted, and wax load (Fig. 9B) and CER6 mRNA accumulation (Fig. 9C) were determined for 10 to 20 kanamycin-resistant T2 progeny. Line 3-20 had the individual T2 progeny with the highest wax accumulation (11.3 μg wax mg−1 dry weight; Fig. 9B). Similarly, there were several individuals originating from line 3-5 with a substantially greater wax load than that of the wild type. It is interesting to note that line 2-10, which was waxless in the T1 generation, also had several T2 progeny with higher than wild-type wax loads. However, on average, the wax loads of the T2 progeny of the wax overproducing lines fell within the wild-type range.
CER6 transcript accumulation in the T2 generation of the selected lines of transformants (Fig. 9C) correlated with the wax loads found on the bolting stems (Fig. 9B). Lines 2-10, 3-11, 3-20, and 3-5 all had greater accumulations of CER6 transcript than the wild type. Similarly, the lines with many waxless individuals, 2-4, 3-1, and 3-10 showed extremely low levels of the CER6 transcript.
Stems of two to three randomly chosen transgenic plants (T2 generation) descended from wax overproducers or waxless T1 individuals were examined by scanning electron microscopy. This analysis revealed that the transgenic plants often had waxless sectors on an otherwise waxy stem (data not shown).
DISCUSSION
Spatial and Temporal Pattern of CER6 Transcription
VLCFAs with chain lengths greater than 18 carbon atoms are used as substrates for the production of cuticular waxes, suberin seed storage lipids, and ceramides, minor but important structural components of cellular membranes. Their synthesis is controlled by the activity of the β-ketoacyl-CoA synthase enzymes (condensing enzymes) of the fatty acid elongase, which determine the amounts and the overall chain lengths of fatty acid products of the elongation process (Lassner et al., 1996; Millar and Kunst, 1997).
The CER6 condensing enzyme has been previously shown to be essential for wax production in the bolting stems of Arabidopsis, and in the anthers for the synthesis of pollen lipids (Millar et al., 1999; Fiebig et al., 2000). In this study, our aim was to more precisely define the transcription domains of the CER6 gene, determine the timing of CER6 transcription in Arabidopsis shoots during development, and examine if CER6 expression is induced under environmental conditions that increase wax accumulation. RT-PCR, RNA-blot analyses, in situ hybridization, and CER6-promoter driven GUS activity assays demonstrated that CER6 was expressed not only in the stems, but in all the aerial parts of the plant examined, including leaves, flowers, and siliques (Figs. 1–4). Furthermore, in all tissues, CER6 expression was restricted to the epidermal cell layer (Figs. 3 and 4), except in the anthers during later stages of microsporogenesis. At that time, CER6 mRNA was localized only in the tapetal cells of the anthers. The monolayer tapetum surrounds the maturing microspores and produces abundant lipids. At the end of microspore development, tapetal cells disintegrate, thereby releasing lipids that get deposited on the pollen surface. Thus, CER6 transcription in the tapetum is consistent with its role in the production of pollen coat lipids (Millar et al., 1999; Fiebig et al., 2000).
CER6 expression throughout the shoot and at all stages of stem and leaf development, starting from 1-d-old seedlings (Figs. 2 and 4), supports the idea that CER6 is the key condensing enzyme dedicated to wax biosynthesis in Arabidopsis. Additional condensing enzymes reported to be involved in wax production, such as CER60, FDH, or KCS1, appear to be expressed only in certain tissues or during a specific developmental stage, perhaps to boost the overall wax production when necessary. Relatively high levels of CER6 mRNA were detected even in mature stems. This is surprising because the wax bloom that is mechanically removed from older parts of the stem does not regenerate (L. Samuels, unpublished data). It may be that older stems still produce VLCFA precursors for wax regeneration, but wax synthesis does not take place or wax composition is altered. The microcrystalline structure of epicuticular wax seems to be related to its chemical composition, and perhaps the way and rate by which the wax is exuded through the cuticle (Hall et al., 1965; von Wettstein-Knowles, 1974). Therefore, if wax produced later in development has a different composition, it may not form the rod- and tube-like microcrystals, but rather take on a more amorphous form that would not be visible as a wax bloom.
Light and Osmotic Stress Influence CER6 Transcription Levels
Wax accumulation in higher plants is known to be influenced by a variety of environmental factors, including light (von Wettstein-Knowles et al., 1979) and water deficit due to lack of soil water or freezing temperatures (Thomas and Barber, 1974; Bengtson et al., 1979; Hadley, 1989). However, the molecular events underlying the observed phenomenon have not been established. One possibility is that these environmental signals enhance wax synthesis by up-regulating the production of key wax biosynthetic enzymes like CER6. The presence in the CER6 promoter of I-box-like and GT1-like sequences previously found in light-responsive genes (Terzaghi and Cashmore, 1995), and ABRE elements (Guiltinan et al., 1990) identified in drought- and cold-inducible genes responsive to ABA suggested that CER6 transcription may be induced by these stimuli. Our results presented here clearly demonstrate that light plays a pivotal role in the transcription of CER6 (Fig. 7), and that in the absence of light, CER6 transcript levels quickly decline. However, a complete repression of CER6 transcription in the dark did not occur in the det2 mutant, suggesting that this response may be mediated by a DET2-dependent signal transduction pathway. CER6 transcript also accumulates in response to osmotic stress (Fig. 8), suggesting that dehydration may enhance wax deposition by increasing VLCFA production. Furthermore, considerably greater CER6 transcript accumulation in ABA-treated seedlings and reduced CER6 transcript levels in ABA-deficient mutants implicate ABA in the induction of CER6 transcription in response to drought.
The CER6 Promoter Directs High Levels of Gene Expression in the Shoot Epidermis
The accumulation of the CER6 transcript exclusively in the epidermis of Arabidopsis shoots suggested that this gene might be controlled by an interesting promoter of potential value for genetic engineering applications that require epidermis-specific expression of genes. To evaluate the CER6 promoter, a 1.2-kb fragment immediately upstream of the CER6 coding region was fused to the GUS reporter gene and was transformed into Arabidopsis and tobacco. In both transgenic systems, GUS activity was restricted to the shoot epidermis, demonstrating that the 1.2-kb promoter fragment used contained all the regulatory elements required to direct epidermis-specific expression, and that the same regulatory elements were recognized in tobacco. The GUS histochemical assay of plants at different stages of development also revealed an early and strong GUS activity that persisted throughout shoot development. The high level of GUS expression directed by the CER6 promoter was confirmed by comparison with the CaMV 35S promoter (Fig. 5). Data showing that the CER6 promoter was comparable with, if not stronger than, the 35S promoter are striking in view of the fact that the 35S promoter is considered a constitutive promoter, and its expression is not restricted to the epidermal cells. Taken together, these experiments demonstrate that the CER6 promoter is very effective in directing high levels of gene expression in the plant epidermis not only in Arabidopsis, but also in unrelated plant species like tobacco. Thus, it should be a useful tool for the modification of surface characteristics of crop plants.
The Effect of CER6 Overexpression on Stem Wax Accumulation
If the level of CER6 transcription is one of the factors controlling wax deposition in Arabidopsis, overexpression of CER6 should increase wax accumulation. Overexpression of the CER6 gene using the strong constitutive CaMV 35S promoter resulted in high levels of CER6 mRNA, but failed to promote greater wax deposition (Millar et al., 1999). In contrast, high levels of CER6 expression in the epidermis using the native CER6 promoter resulted in appreciably greater wax accumulation in a number of transgenic lines. Thus, similar to lignin modification experiments, which require accurate temporal expression of genes specifically in cells undergoing lignification (Meyer et al., 1998), effective manipulation of surface wax accumulation appears to require correctly timed epidermis-specific expression of relevant genes. Furthermore, because wax composition in the wax-overproducing lines was unchanged from that of the wild type, this increase in the wax load is likely due to increased carbon flux via both the decarbonylation and the acyl-reduction pathways of wax synthesis.
The relatively wide range of wax load values observed in the T2 progeny of plants with increased wax accumulation in the T1 generation may be due to a number of reasons. For example, if the high wax phenotype in primary transformants (T1) is caused by insertions of multiple copies of the CER6 transgene, segregation of transgenes in the T2 generation would result in a reduced transgene copy number in a number of individuals. In addition, the remaining transgenes in each T2 individual would be present at a variety of chromosomal locations. Genomic location affects the level of expression of the transgene, which in turn influences wax accumulation. Another explanation for reduced wax accumulation in T2 plants is offered by the presence of waxless sectors on otherwise waxy stems. Wax synthesis and deposition has been shown to be cell-autonomous (von Wettstein-Knowles and Netting, 1976). Thus, individual cells could overexpress or silence the CER6 transgene, generating a mosaic of waxy and waxless sectors. The overall wax load of a particular T2 plant would depend on the number and size of waxless sectors. The fact that the sectoring effect was also present in the T1 generation could account for reduced wax loads measured in many of these plants in comparison with the wild type.
Our results demonstrate that wax accumulation in Arabidopsis is, in part, regulated by the level of CER6 transcription. The CER6 gene is highly transcribed in the epidermis of all the shoot tissues throughout Arabidopsis development. The CER6 promoter is very effective in directing epidermal expression of genes in transgenic plants. The responsiveness of CER6 transcription to light and water deficit, as well as to the phytohormone ABA, suggests that these factors cause changes in wax production by altering the abundance of key wax biosynthetic enzymes, including CER6, in the epidermal cells. The evaluation of this hypothesis requires the development of an appropriate enzymatic assay and a CER6-specific antibody to monitor CER6 protein levels.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Seeds of Arabidopsis (ecotype Columbia-2), a gift from Dr. Shauna Somerville (Carnegie Institution of Washington, Stanford, CA), and aba mutants obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus) were stratified for 3 d at 4°C, and were then germinated on AT-agar plates (Somerville and Ogren, 1982; containing 50 μg mL−1 kanamycin [w/v] for transgenic plants) at 20°C under continuous light (100 μE m−2 s−1 photosynthetically active radiation). Seven-day-old seedlings were transplanted to soil (Terralite Redi-Earth, W.R. Grace and Co., Canada Ltd., Ajax, Ontario, Canada) in 12-cm pots (nine seedlings/pot) and were returned to continuous light at 20°C until maturity. For PEG, NaCl, and ABA treatments, seedlings were grown on AT-agar plates for 14 d, transferred to liquid AT medium containing the treatment solution, and were incubated for 10 h prior to harvest.
Tobacco (Nicotiana tabacum cv Xanthi SR1) seed was obtained from Dr. Carl Douglas (Department of Botany, University of British Columbia, Vancouver). Tobacco plants were grown in growth chambers at 23°C under an 8-h dark/16-h light regime. Tobacco transformation was carried out as described (Millar and Kunst, 1997).
RNA Gel-Blot Analysis
Arabidopsis stem segments 2 cm long from just below the inflorescence (stem tops) or just above the rosette (stem bases), whole leaves, unopened flower buds, opened flowers, whole siliques less than 1 cm long, whole seedling shoots, or whole roots were harvested and immediately frozen in liquid nitrogen. Total RNA was extracted using Trizol Reagent (Invitrogen, Carlsbad, CA) and isopropanol precipitation, according to the manufacturer's protocol. Ten micrograms of each sample was separated by electrophoresis on a 1% (w/v) MOPS-agarose gel containing formaldehyde. Separated RNA was downward-blotted (Koetsier et al., 1993) onto a Hybond XL membrane (Amersham Pharmacia Biotech, Piscataway, NJ) and was fixed to the membrane by baking at 80°C for 2 h. Hybridization was carried out in modified Church buffer (0.5 m Na-phosphate buffer, pH 7, 7% [w/v] SDS, and 1 mm EDTA) overnight at 65°C using a PCR-generated 32P-labeled DNA probe. A CER6 full-length coding sequence probe was made as in Millar et al. (1999), where CER6 was designated as “CUT1. ” A CER6 5′-UTR probe was made using Taq polymerase (Invitrogen) and the oligonucleotides 5′-ATATCCTTCACCTTCCC-3′ and 5′-CTCTGGCATCGGTGC-3′. Amplification conditions were 94°C for 2 min, 30 cycles of denaturation at 94°C for 15 s, annealing at 50°C for 15 s, and extension at 72°C for 30 s, followed by a final extension at 72°C for 5 min in a DNA Thermal Cycler 480 (PerkinElmer Instruments, Norwalk, CT). An rd29A (Yamaguchi-Shinozaki and Shinozaki, 1993) unique sequence probe used as a control for water deficit response corresponded to a part of the coding sequence for this gene that did not produce a significant score when BLASTed against the Arabidopsis genome database. Primers used to amplify it from genomic DNA were 5′-GATCAGAAGCCAGGACAATTTG-3′ and 5′-TCCAGCTCAGCTCCTGACTC-3′. Amplification conditions were the same as above, except that annealing temperature was 59°C. Arabidopsis 18S rRNA (Unfried et al., 1989) was used as a loading control. It was amplified from genomic DNA using the oligonucleotide primers 5′-CTGCCAGTAGTCATATGC-3′ and 5′ATGGATCCTCGTTAAGGG-3′, and the same amplification program as for the CER6 5′-UTR probe. Blots were washed in 2× SSC and 0.1% (w/v) SDS for 2 × 5 min; 1× SSC and 0.1% (w/v) SDS for 15 min; and 0.1× SSC and 0.1% (w/v) SDS for 2 × 10 min at 65°C, and were then autoradiographed overnight at −80°C using film (Kodak XAR-5; Eastman-Kodak, Rochester, NY), or they were exposed to a phosphor screen that was then scanned with a STORM 860 phosphor imager (Amersham Pharmacia Biotech). Each blot was sequentially hybridized with a probe corresponding to the CER6 coding region or the 5′-UTR, followed by the 18S rRNA probe. The intensity of the bands on the autoradiogram was quantified by densitometry using a digital camera (AlphaImager 1220; Alpha Innotech Corporation, San Leandro, CA) and densitometry software (Alpha Innotech Corporation).
Quantitative RT-PCR
Total RNA isolated as described above for RNA gel-blot analysis was used for cDNA synthesis by C. therm. polymerase (from Carboxydothermus hydrogenoformans; Roche Diagnostics, Laval, QC, Canada) following the manufacturer's protocol. Gene-specific primers were designed to amplify 600- and 700-bp fragments of CER6 and CER60, respectively, spanning the introns (CER6, 400 bp; CER60, 200 bp) to differentiate products amplified from cDNA from any product amplified from contaminating genomic DNA. An 800-bp fragment of histone H1 was also reverse-transcribed in the same reaction, and amplified separately.
PCR cycle number and template amounts were optimized for all fragments amplified to yield products in the linear range of the reaction. Primer sequences were as follows: CER6 sense 5′-ATCTAGCCCGCGACTTGCTC-3′, CER6 antisense 5′-CACTTGAAACCACTCCCAAACG-3′; CER60 sense 5′-TCTAACCGCAGATCCGACAGG-3′, CER60 antisense 5′-ACAATCCGACCACGCTCCATC-3′; and histone H1 sense 5′-CCGGAATTCCGGGGTTAAAGTCAAAGCTTCTTTTAAGA-3′, histone H1 antisense 5′-CCGCTCGAGCGGGAGTGAAGAAACCATCACATTATA-3′. PCR conditions used for comparison of CER6 and CER60 transcription levels were: 25 ng of template cDNA denatured at 94°C for 2 min, followed by 22 cycles of 94°C for 15 s, 58°C for 30 s, and 72°C for 60 s. Reactions were maintained at 72°C for 7 min before separation of PCR products by electrophoresis in a 1.2% (w/v) agarose-Tris-acetate EDTA (TAE) gel. PCR products were visualized by SYBRGreen I (Molecular Probes, Eugene, OR) staining of the gel and were quantified by densitometry using the ImageQuant 5.2 software (Molecular Dynamics, Sunnyvale, CA) after fluorescence scanning by the STORM 860 fluorescence imager (Amersham Pharmacia Biotech) at 450-nm excitation and 520-nm emission wavelengths.
In Situ Hybridization
In situ hybridization of Arabidopsis inflorescences including 0.5 cm of the stem adjacent to the apex and 8-d-old seedling shoots was carried out according to the protocol of Samach et al. (1997) using digoxygenin-11-UTP-labeled ssRNA probes in a sense (negative control) and antisense orientation with respect to the CER6 coding region.
To synthesize the probes, DNA templates were amplified by PCR with Pwo polymerase (Roche Diagnostics) from cloned CER6 cDNA using primers incorporating the T7 RNA polymerase binding site. For the antisense probe, the primer sequences were: 5′-ATGCCTCAGGCACCG-3′ and 5′-GATAATACGACTCACTATAGGGTTATTTGAGTACACC-3′. For the sense probe, the primer sequences were: 5′-TTATTTGAGTACACC-3′ and 5′-GATAATACGACTCACTATAGGATGCCTCAGGCACCG-3′. Amplification conditions were 94°C for 2 min, 10 cycles of denaturation at 94°C for 15 s, annealing at 55°C for 30 s, and extension at 72°C for 90 s; 20 cycles of the same conditions but increasing the extension time by 20 s/cycle; followed by a final extension at 72°C for 7 min. RNA probes were transcribed from the PCR-generated DNA templates using T7 RNA polymerase and digoxygenin-11-UTP-labeled nucleotide mix (Roche Diagnostics), according to the manufacturer's directions. The probes were then cleaved to approximately 150 bp in length by alkaline hydrolysis for 55 min at 60°C in 0.2 m sodium carbonate buffer (pH 10.4).
Isolation of CER6 Promoter Fragment and Generation of the CER6 Promoter::GUS Construct
A genomic library of Arabidopsis (ecotype Columbia-2) generated in the vector λGEM11 (Promega, Madison, WI) by John Mulligan and Ronald Davis (Stanford University, Stanford, CA) was screened using standard procedures (Sambrook et al., 1989) with a full-length CER6 cDNA as a probe (Millar et al., 1999). Eleven genomic clones were isolated, and DNA gel-blot analysis revealed that a single clone when digested with SalI resulted in two restriction fragments of 11 and 2.8 kb, as expected from the restriction map of the CER6 cDNA. The 2.8-kb SalI fragment was subcloned into the SalI site of pT7T3 18U (Amersham Pharmacia Biotech) and its nucleotide sequence was determined. The first 551 nucleotides of this fragment corresponded to the CER6 cDNA, confirming that we isolated the correct genomic clone.
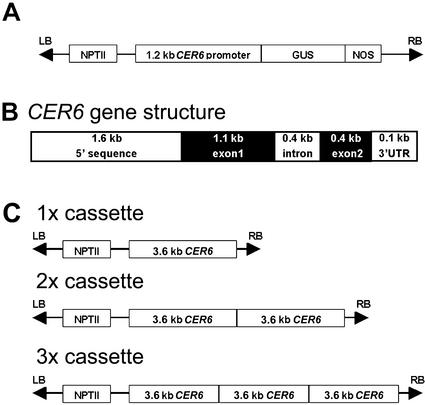
The 1,208 nucleotides immediately upstream of the CER6 coding region were amplified by PCR from genomic Arabidopsis DNA using the oligonucleotide primers 5′-CGTCGGAGAGTTTTAATG-3′ and 5′-CTTCGATATCGGTTGTTG-3′ and high-fidelity Pfu polymerase (Stratagene, La Jolla, CA). The 1.2-kb PCR fragment obtained was blunt-end cloned into the HincII site of pT7T3 U18 (Amersham Pharmacia Biotech), resulting in the plasmid pCER6PRO. Sequencing confirmed that the PCR product corresponded to the upstream of the CER6 gene in the Arabidopsis database. The product was oriented such that the 5′ end was near the HindIII site, whereas the 3′ end was adjacent to the XbaI site in the polylinker of pT7T3 18U. The HindIII/XbaI fragment was then cleaved from pCER6PRO and was subcloned into the HindIII/XbaI sites of pBI101 (CLONTECH Laboratories, Palo Alto, CA), resulting in the binary vector pCER6-GUS (Fig. 10A). pCER6-GUS was introduced into the Agrobacterium tumefaciens strain GV3101 (pMP90; Koncz and Schell, 1986) that was used in the transformation of Arabidopsis and tobacco as previously described (Millar and Kunst, 1997).
Figure 10.
Diagram of the constructs used in plant transformation experiments. A, T-DNA containing the CER6 promoter-GUS gene fusion used to evaluate the tissue specificity of CER6 expression in Arabidopsis and tobacco plants. B, Structure of the 3.6-kb CER6 genomic clone used to construct the 1×, 2×, and 3× expression cassettes. C, T-DNA containing one, two, and three copies (1×, 2×, and 3× expression cassettes, respectively) of the CER6 genomic clone used to transform Arabidopsis plants.
Transformation of Arabidopsis with Additional Copies of CER6 promoter::CER6
A 3.6-kb fragment, including 1.6 kb of the CER6 promoter and the CER6 coding region, was amplified from Arabidopsis ecotype Columbia-2 genomic DNA by PCR using the Expand High Fidelity thermostable DNA polymerase (Roche Diagnostics) and primers 5′-CAAATGACACAATTGTTC-3′ (forward) and 5′-CCCAAATGAAAAGCAGAG-3′ (reverse). The amplified fragment was ligated into the SmaI site of pGEM-7Zf+ (Promega). The genomic fragment was then directionally subcloned into pRD400 (Datla et al., 1992) using the XbaI and BamHI sites, to produce the 1× expression cassette. To introduce the second and third copies of the genomic fragment, thus producing the 2× and 3× cassettes, the original PCR-amplified fragment was ligated into the HincII site of pBluescriptII KS+ (Stratagene). This fragment was then excised and subcloned into the XbaI and XhoI sites of the vector pGEM-7Zf+ to introduce a BamHI site at either end of the fragment. The second and third copies of the genomic CER6 fragment were added to the 1× cassette by subcloning into the BamHI site. The orientation and number of CER6 fragments in each of the cassettes was confirmed by restriction analysis. Constructs containing a single copy of CER6 with its native promoter, or two and three tandem copies of the gene (Fig. 10, B and C) were used to transform A. tumefaciens strain GV3101 (pMP90; Koncz and Schell, 1986) by electroporation. A. tumefaciens lines harboring the binary vectors were used to transform Arabidopsis (Columbia-2 ecotype) by the floral dip method (Clough and Bent, 1998). Screening for transformed seed was performed on 50 μg mL−1 kanamycin as described previously (Katavic et al., 1994).
Wax Analysis
Wax load was determined on individual T1 transformants and on T2 progeny of selected T1 plants. Wax was extracted from 5-cm-long stem bases of senesced, dry plants and was analyzed by gas chromatography as described in Millar et al. (1999).
GUS Histochemical Assay
Tissues of pCER6-GUS transformed Arabidopsis plants and transgenic plants harboring the 35SCaMV-GUS construct were incubated in GUS assay buffer containing 100 mm phosphate buffer, pH 7.0, 10 mm EDTA, 0.1% (w/v) Triton X-100, 1 mm potassium ferricyanide, 1 mm potassium ferrocyanide, and 1 mg mL−1 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (Jefferson, 1987) at 37°C for 0.5 h to overnight. The reaction was stopped by removal of the assay buffer and the addition of 95% (v/v) ethanol. Samples were cleared by incubation in 95% (v/v) ethanol overnight.
GUS Fluorometric Assay
Wild-type and pCER6-GUS transgenic seedlings were harvested and homogenized with sand in protein extraction buffer (50 mm phosphate buffer, pH 7.0, 10 mm β-mercaptoethanol, 10 mm Na2EDTA, 0.1% [w/v] sodium lauryl sarcosine, and 0.1% [w/v] Triton X-100; Federle et al., 2000). Cell debris was removed by centrifugation, and protein concentration of the supernatants was quantified against a bovine serum albumin standard curve using the Bradford assay (Federle et al., 2000). The GUS assay was started by addition of 20 to 40 μL of this crude extract to a solution of 2 mm MUG in extraction buffer at 37°C. For each transgenic line, aliquots were removed at several time points and were added to 0.2 m Na2CO3 to stop the reaction. The concentration of 4-MU in each aliquot was measured by fluorometer (excitation at 365 ± 7 nm; emission at 460 ± 15 nm). To determine the rates of MUG hydrolysis for different transgenic lines, the slopes of the linear portions of the curves were determined; these values were standardized to the protein concentrations of the extracts.
ACKNOWLEDGMENTS
We thank Gangamma Chowrira, Lacey Samuels, Mark Smith, Tamara Western, and Huanquan Zheng for helpful discussions, and two anonymous reviewers for valuable comments during preparation of this manuscript. We also thank Sabine Clemens for her skilled technical assistance. We thank the Arabidopsis Biological Resource Center for providing the aba mutant seeds.
Footnotes
This work was supported by the Natural Sciences and Engineering Research Council of Canada (grant to L.K.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.003707.
LITERATURE CITED
- Baker EA. Chemistry and morphology of plant epicuticular waxes. In: Cutler DJ, Alvin KL, Price CE, editors. The Plant Cuticle. London: Academic Press; 1982. pp. 139–165. [Google Scholar]
- Barnes JD, Percy KE, Paul ND, Jones P, McLaughlin CK, Mullineaux PM, Creissen G, Wellburn AR. The influence of UV-B radiation on the physicochemical nature of tobacco (Nicotiana tabacum L.) leaf surfaces. J Exp Bot. 1996;47:99–109. [Google Scholar]
- Bengtson C, Larsson S, Liljenberg C. Water stress, epicuticular wax and cuticular transpiration rate. In: Appelqvist LA, Liljenberg C, editors. Advances in the Biochemistry and Physiology of Plant Lipids. New York: Elsevier/North Holland Biomedical Press; 1979. pp. 269–274. [Google Scholar]
- Blum A. Effect of the Bm gene on epicuticular wax and the water relations of Sorghum bicolor L. (Moench) Isr J Bot. 1975;24:50–51. [Google Scholar]
- Chory J, Susek RE. Light signal transduction and the control of seedling development. In: Meyerowitz EM, Somerville CR, editors. Arabidopsis. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. pp. 579–614. [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Datla RS, Hammerlindl JK, Panchuk B, Pelcher LE, Keller W. Modified binary plant transformation vectors with the wild type gene encoding NPTII. Gene. 1992;211:383–384. doi: 10.1016/0378-1119(92)90232-e. [DOI] [PubMed] [Google Scholar]
- Eigenbrode SD, Espelie KE. Effects of plant epicuticular lipids on insect herbivores. Annu Rev Entomol. 1995;40:171–194. [Google Scholar]
- Federle W, Rohrseitz K, Holldobler B. Attachment forces of ants measured with a centrifuge: Better “wax-runners” have a poorer attachment to a smooth surface. J Exp Biol. 2000;203:505–512. doi: 10.1242/jeb.203.3.505. [DOI] [PubMed] [Google Scholar]
- Fiebig A, Mayfield JA, Miley NL, Chau S, Fischer RL, Preuss D. Alterations in CER6, a gene identical to CUT1, differentially affect long-chain lipid content on the surface of pollen and stems. Plant Cell. 2000;12:2001–2008. doi: 10.1105/tpc.12.10.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant RH, Jenks MA, Rich PJ, Peters PJ, Ashworth EN. Scattering of ultraviolet and photosynthetically active radiation by sorghum bicolor: influence of epicuticular wax. Agric Forest Meteorol. 1995;75:263–281. [Google Scholar]
- Guiltinan MJ, Marcotte WR, Jr, Quatrano RS. A plant leucine zipper protein that recognizes an abscisic acid response element. Science. 1990;250:267–270. doi: 10.1126/science.2145628. [DOI] [PubMed] [Google Scholar]
- Hadley NF. Lipid water barriers in biological systems. Prog Lipid Res. 1989;28:1–33. doi: 10.1016/0163-7827(89)90005-2. [DOI] [PubMed] [Google Scholar]
- Hall DM, Matus AI, Lamberton JA, Barber HN. Intraspecific variation in wax on leaf surfaces. Aust J Biol Sci. 1965;18:323–332. [Google Scholar]
- James DW, Lim E, Keller J, Plooy I, Ralston E, Dooner HK. Directed tagging of the Arabidopsis FATTY ACID ELONGATION1 (FAE1) gene with the maize transposon activator. Plant Cell. 1995;7:309–319. doi: 10.1105/tpc.7.3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep. 1987;5:387–405. [Google Scholar]
- Jeffree CE. Structure and ontogeny of plant cuticles. In: Kerstiens G, editor. Plant Cuticles. Oxford: BIOS Scientific Publishers; 1996. pp. 33–82. [Google Scholar]
- Jenks MA, Joly RJ, Peters PJ, Rich PJ, Axtell JD, Ashworth EN. Chemically induced cuticle mutation affecting epidermal conductance to water vapor and disease susceptibility in Sorghum bicolor (L.) Moench. Plant Physiol. 1994;105:1239–1245. doi: 10.1104/pp.105.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katavic V, Haughn GW, Reed D, Martin M, Kunst L. In planta transformation of Arabidopsis thaliana. Mol Gen Genet. 1994;245:363–370. doi: 10.1007/BF00290117. [DOI] [PubMed] [Google Scholar]
- Koetsier PA, Schorr J, Doerfler W. A rapid optimized protocol for downward alkaline Southern blotting of DNA. BioTechniques. 1993;15:260–262. [PubMed] [Google Scholar]
- Koncz C, Schell J. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet. 1986;204:383–396. [Google Scholar]
- Kunst L, Taylor DC, Underhill EW. Fatty acid elongation in developing seeds of Arabidopsis thaliana. Plant Physiol Biochem. 1992;30:425–434. [Google Scholar]
- Lassner MW, Lardizabal K, Metz JG. A jojoba β-ketoacyl-CoA synthase cDNA complements the canola fatty acid elongation mutation in transgenic plants. Plant Cell. 1996;8:281–292. doi: 10.1105/tpc.8.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux B, Koornneef M, Feldmann KA. Epicuticular wax and eceriferum mutants. In: Meyerowitz EM, Somerville CR, editors. Arabidopsis. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. pp. 1031–1047. [Google Scholar]
- Meyer K, Shirley AM, Cusumano JC, Bell-Lelong DA, Chapple C. Lignin monomer composition is determined by the expression of cytochrome P450-dependent monooxygenase in Arabidopsis. Proc Natl Acad Sci USA. 1998;95:6619–6623. doi: 10.1073/pnas.95.12.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AA, Clemens S, Zachgo S, Giblin EM, Taylor DC, Kunst L. CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell. 1999;11:825–838. doi: 10.1105/tpc.11.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AA, Kunst L. Very-long-chain fatty acid biosynthesis is controlled through the expression and specificity of the condensing enzyme. Plant J. 1997;12:121–131. doi: 10.1046/j.1365-313x.1997.12010121.x. [DOI] [PubMed] [Google Scholar]
- Piffanelli P, Ross JHE, Murphy DJ. Biogenesis and function of the lipidic structures of pollen grains. Sex Plant Reprod. 1998;11:65–80. [Google Scholar]
- Post-Beittenmiller D. Biochemistry and molecular biology of wax production in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:405–430. doi: 10.1146/annurev.arplant.47.1.405. [DOI] [PubMed] [Google Scholar]
- Pruitt RE, Vielle-Calzada JP, Ploense SE, Grossniklaus U, Lolle SJ. FIDDLEHEAD, a gene required to suppress epidermal cell interactions in Arabidopsis, encodes a putative lipid biosynthetic enzyme. Proc Natl Acad Sci USA. 2000;97:1311–1316. doi: 10.1073/pnas.97.3.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed DW, Tukey HB. Light intensity and temperature effects on epicuticular wax morphology and internal cuticle ultrastructure of carnation and brussels sprouts leaf cuticles. J Am Soc Hort Sci. 1982;107:417–420. [Google Scholar]
- Reicosky DA, Hanover JW. Physiological effects of surface waxes. Plant Physiol. 1978;62:101–104. doi: 10.1104/pp.62.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards RA, Rawson HM, Johnson DA. Glaucousness in wheat: its development and effect on water-use efficiency, gas exchange and photosynthetic tissue temperatures. Aust J Plant Physiol. 1986;13:465–473. [Google Scholar]
- Samach A, Kohalmi SE, Motte P, Datla R, Haughn GW. Divergence of function and regulation of class B floral organ identity genes. Plant Cell. 1997;9:559–570. doi: 10.1105/tpc.9.4.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Somerville CR, Ogren WL. Isolation of photorespiratory mutants of Arabidopsis. In: Hallick RB, Chua NH, editors. Methods in Chloroplast Molecular Biology. New York: Elsevier; 1982. pp. 129–139. [Google Scholar]
- Terzaghi WB, Cashmore AR. Light-regulated transcription. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:445–474. [Google Scholar]
- Thomas DA, Barber HN. Studies on leaf characteristics of a cline of Eucalyptus urnigera from Mount Wellington, Tasmania: water repellency and the freezing of leaves. Aust J Bot. 1974;22:501–512. [Google Scholar]
- Todd J, Post-Beittenmiller D, Jaworski JG. KCS1 encodes a fatty acid elongase 3-ketoacyl-CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. Plant J. 1999;17:119–130. doi: 10.1046/j.1365-313x.1999.00352.x. [DOI] [PubMed] [Google Scholar]
- Unfried I, Stocker U, Gruendler P. Nucleotide sequence of the 18S rRNA gene from Arabidopsis thaliana Col0. Nucleic Acids Res. 1989;17:7513. doi: 10.1093/nar/17.18.7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wettstein-Knowles P. Ultrastructure and origin of epicuticular wax tubes. J Ultrastruct Res. 1974;46:483–498. doi: 10.1016/s0022-5320(74)90069-0. [DOI] [PubMed] [Google Scholar]
- von Wettstein-Knowles P. Biosynthesis and genetics of waxes. In: Hamilton RJ, editor. Waxes: Chemistry, Molecular Biology and Functions. Dundee, Scotland: Oily Press; 1995. pp. 91–130. [Google Scholar]
- von Wettstein-Knowles P, Avato P, Mikkelsen JD. Light promotes synthesis of the very long chain fatty acyl chains in maize wax. In: Mazliak D, Bennveniste P, Costes C, Douce R, editors. Biogenesis and Function of Plant Lipids. New York: Elsevier/North Holland Biomedical Press; 1979. pp. 271–274. [Google Scholar]
- von Wettstein-Knowles P, Netting AG. Composition of epicuticular waxes on barley spikes. Carlsberg Res Commun. 1976;41:225–235. [Google Scholar]
- Walton TJ. Waxes, cutin and suberin. In: Harwood JL, Bowyer JR, editors. Methods in Plant Biochemistry: Lipids, Membranes and Aspects of Photobiology. San Diego: Academic Press; 1990. pp. 105–158. [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol Gen Genet. 1993;236:331–340. doi: 10.1007/BF00277130. [DOI] [PubMed] [Google Scholar]
- Yephremov A, Wisman E, Huijser P, Huijser C, Wellesen K, Saedler H. Characterization of the FIDDLEHEAD gene of Arabidopsis reveals a link between adhesion response and cell differentiation in the epidermis. Plant Cell. 1999;11:2187–2201. doi: 10.1105/tpc.11.11.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]