Abstract
Huntington's disease (HD) is a fatal inherited neurodegenerative disorder characterized by personality changes, motor impairment, and subcortical dementia. HD is one of a number of diseases caused by expression of an expanded polyglutamine repeat. We have developed several lines of mice that are transgenic for exon 1 of the HD gene containing an expanded CAG sequence. These mice exhibit a defined neurological phenotype along with neuronal changes that are pathognomonic for the disease. We have previously observed the appearance of neuronal intranuclear inclusions, but did not find evidence for neurodegeneration. In this study, we report that all lines of these mice develop a late onset neurodegeneration within the anterior cingulate cortex, dorsal striatum, and of the Purkinje neurons of the cerebellum. Dying neurons characteristically exhibit neuronal intranuclear inclusions, condensation of both the cytoplasm and nucleus, and ruffling of the plasma membrane while maintaining ultrastructural preservation of cellular organelles. These cells do not develop blebbing of the nucleus or cytoplasm, apoptotic bodies, or fragmentation of DNA. Neuronal death occurs over a period of weeks not hours. We also find degenerating cells of similar appearance within these same regions in brains of patients who had died with HD. We therefore suggest that the mechanism of neuronal cell death in both HD and a transgenic mouse model of HD is neither by apoptosis nor by necrosis.
Huntington's disease (HD) is one of a number of inherited, progressive, late onset neurodegenerative disorders that are caused by a polyglutamine expansion. The dominant neuropathological finding is widespread neuronal loss affecting mainly the caudate nucleus, putamen, and frontal lobes (1–4). However, degeneration can eventually appear throughout the brain (2). In cases with juvenile onset (<20 years of age), degeneration can be more widespread with an additional focus for neurodegeneration in the Purkinje cell layer of the cerebellum (3, 4). The striatal changes become more pronounced with disease progression and are associated with a fibrillary reactive astrocytosis and an increased density of oligodendrocytes; there is no inflammatory reaction (3). The majority of remaining neostriatal neurons in HD postmortem brains have normal somatic morphology, but contain more lipofuscin deposited within the cytoplasm, and may be smaller than usually expected (2). In addition, there are scattered atrophic neurons which stain more darkly than the apparently healthy neurons and have been referred to as “neostriatal dark neurons.” These neurons have a scalloped cellular membrane, a finely granular dark cytoplasm, and a nucleus with condensed chromatin (2).
We have previously generated a mouse model of HD that is transgenic for exon 1 of the human HD gene, containing highly expanded CAG repeats, under the control of the HD promoter (5, 6). Three lines develop a progressive neurological phenotype: R6/1, (CAG)115; R6/2, (CAG)145; and R6/5, (CAG)128–156, the symptoms of which include a movement disorder and weight loss with similarities to HD. The R6/2 line has an age of onset for symptoms of ≈2 mo and the disease progresses rapidly such that mice are rarely kept beyond 14 weeks of age (5). In all three lines, the onset of symptoms is preceded by the appearance of a distinct temporal sequence of neuropathological change proceeding from the formation of filamentous neuronal intranuclear inclusions (NIIs) and similar filamentous inclusions within dystrophic neurites (6, 7). Both types of inclusion are principally composed of the transgenic protein in a ubiquitinated form in combination with many components of the proteasome/molecular chaperone complex (ref. 7 and S.W.D., unpublished observations). In vitro studies have shown that the polyglutamine-containing proteins form filaments, adopting a cross β-pleated sheet conformation (8, 9). Perutz (10) have proposed that expanded polyglutamine repeats can self-associate into aggregates with this β-pleated sheet structure.
Both NII and dystrophic neurites are found in postmortem brain from patients who had died with HD (11–13), whereas NII in the absence of dystrophic neurites have now been found in dentatorubral-pallidoluysian atrophy (12, 14, 15), spinobulbar muscular atrophy (16), and spinocerebellar ataxias types 1 (17), 3 (18), and 6 and 7 (19, 20).
In this report, we investigate the ultrastructural features of neuronal cell death in the R6 lines of HD transgenic mice and compare these changes with those found in degenerating neurons in postmortem brain from patients dying with HD. We suggest that the ultrastructural features of cell death in both the human disease (HD) and the transgenic mouse model are unlike those originally used to define the process of apoptotic cell death (21).
Materials and Methods
Transgenic Mice.
Three symptomatic lines of mice that are transgenic for the HD mutation were studied (5, 6). Neurological symptoms first appear at ≈2 mo for R6/2 hemizygotes, 5 mo for R6/1 hemizygotes, and 9 months for R6/5 homozygotes. Mice were studied up to 17 weeks (R6/2), 13 months (R6/1 hemizygotes), and 17 months (R6/5 homozygotes), respectively. The ultrastructural analysis of the central nervous system in R6 transgenic mice was always compared with age-matched material from nontransgenic littermate controls, from the transgenic R6/0 line which does not express the transgene (21 mo), and from the HDex6 and HDex27 transgenic lines expressing a similar construct carrying a (CAG)18 tract and studied for up to 21 mo of age.
Human Postmortem Tissue.
We studied postmortem samples of anterior cingulate cortex (Brodmann area 24), caudate nucleus, putamen, and cerebellar vermis from two HD patients and two controls. CAG repeat sizes within the HD gene were 17/101 (HD1), 17/69 (HD2), 17/23 (C1), and 16/19 (C2), respectively. These juvenile HD patients had an onset of disease at 5 years and 14 years of age and died 6 and 7 years later, respectively.
Light Microscopy.
Animals were anesthetized with an overdose of sodium pentobarbitone (Sagatal, 100 mg/kg i.p.) and perfused through the left cardiac ventricle with 35–50 ml of a 2% paraformaldehyde/lysine/periodate fixative in phosphate buffer (pH 7.4). Brains were carefully removed and placed into fixative for 4–6 h before being transferred to 30% sucrose in 0.1 M Tris (pH 7.4) for 48 h at 4°C. Brains were mounted in Tissue-Tek OCT compound (Miles), frozen with powdered solid CO2, and sectioned in the coronal plane at 40 μm on a sledge microtome. Sections were mounted onto gelatinized glass slides and allowed to dry overnight. Alternate sections were stained for Nissl substance with thionin before all sections were rapidly dehydrated through ethanols, cleared in Histoclear (National Diagnostics), and coverslipped.
Electron Microscopy.
Animals were anesthetized as described above and then perfused through the left cardiac ventricle with 35–50 ml of 4% paraformaldehyde and either 0.5% glutaraldehyde or 0.1% glutaraldehyde in 0.1 M Millonig's phosphate buffer (pH 7.4). The brain was removed from the skull and placed in fresh fixative overnight at 4°C. Coronal sections through the cerebellum, striatum, and cerebral cortex (50–200 μm) were cut on an Oxford vibratome (Lancer) and collected in serial order in 0.1 M phosphate buffer. After being osmicated (30 min in 1% OsO4 in 0.1 M phosphate buffer), the sections were stained for 15 min in 0.1% uranyl acetate in sodium acetate buffer at 4°C, dehydrated in ethanols, cleared in propylene oxide, and embedded in araldite between two sheets of Melanex (ICI). Semithin (1 μm) sections were cut with glass knives and stained with toluidine blue adjacent to thin sections cut with a diamond knife on a Reichert Ultracut ultramicrotome. The sections were collected on mesh grids coated with a thin Formvar film, counterstained with lead citrate, and viewed in a JEOL 1010 electron microscope.
Terminal Deoxynucelotidyltransferase-Mediated UTP End Labeling (TUNEL).
Ten to 12 sections from mice of the R6/2 line at 10–17 weeks of age (three mice per age group) were used for terminal deoxynucleotidyltransferase catalyzed incorporation of either digoxigenin-11-dUTP or biotin-16-dUTP into the free 3′-OH terminals of fragmented nuclear DNA (Roche Molecular Biochemicals or Oncor). The incorporated digoxigenin-dUTP was detected with a horseradish peroxidase conjugated anti-digoxigenin antibody. Incorporated biotin was detected with an avidin–biotin–peroxidase complex (Vector Laboratories). Sites of enzymatic activity were visualized with H2O2 and 3,3′-diaminobenzidine as chromagen. Positive controls were sections through the neocortex, striatum, and subventricular zone beneath the cortex of 5- to 7-day-old mice, whereas negative controls were sections from 14.5- to 17-week-old nontransgenic littermates.
Results
Ultrastructural Features of Neuronal Cell Death.
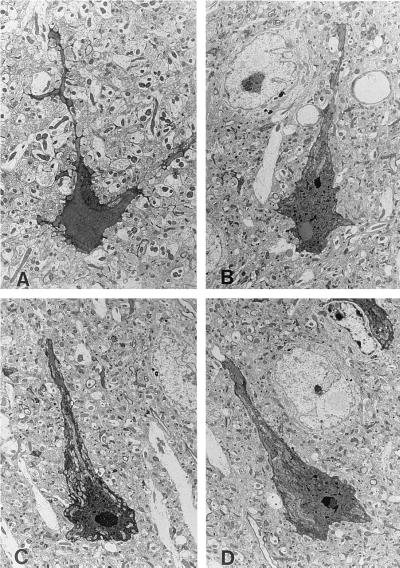
We have now studied in great detail R6/2 mice kept for up to 17 weeks of age. However, observations similar in nature to those that we are reporting can also be found in the other two older symptomatic lines, but not in any of the asymptomatic lines of mice studied for up to at least 2 years of age (see Materials and Methods for full details of the mouse lines). Detailed examination throughout the brains of these mice using both light and transmission electron microscopy detected degenerating neurons in the anterior cingulate cortex, in the dorsal striatum, and within the vermis of the cerebellum. We did not find degenerating neurons in any other areas of the central nervous system, but are currently performing a detailed investigation using the ultrastructural criteria defined for degenerating neurons in this study. Degenerating neurons can be detected by their enhanced affinity for staining with either toluidine blue or osmium, which delineates both perikarya and proximal dendrites (Fig. 1). These neurons exhibit nuclear and cytoplasmic condensation and clumping of chromatin, while subcellular organelles, including mitochondria, appear to be intact. However, during the process of degeneration and cellular darkening, both mitochondria and the Golgi network undergo a transient swelling before condensation in the final stages of cell death. The cellular plasma membrane is ruffled, giving a scalloped appearance to the darkened cell. All degenerating cells contain a distinctive NII and exhibit extensive invagination of the nuclear membrane. There is no fragmentation or “blebbing” of either nucleus or cytoplasm. Adjacent neurons are of normal appearance (Fig. 1 B and D) despite the presence of a distinct NII and invariably contain markedly increased amounts of lipofuscin.
Figure 1.
Ultrastructural appearance of neurons undergoing dark cell degeneration in the striatum (A) and anterior cingulate cortex (B–D) of 17-week-old R6/2 transgenic mice. Note the juxtaposition of neurons containing NII, but without any of the characteristic features of dark cell degeneration (arrows in B and D) (magnification ×650).
Pathological Changes in the Neuronal Nucleus.
We have repeatedly tried to identify DNA fragmentation within “dark neurons” by labeling with the terminal deoxynucleotidyltransferase-mediated dUTP–biotin nick end labeling (TUNEL) method for the in situ determination of DNA strand breaks (22–24), but we were unable to find any labeling of these neurons. We can however routinely demonstrate apoptotic profiles with this method during the development of both the cortex and striatum in the early postnatal period (5–7 days postnatal mouse, data not shown).
Glial Response.
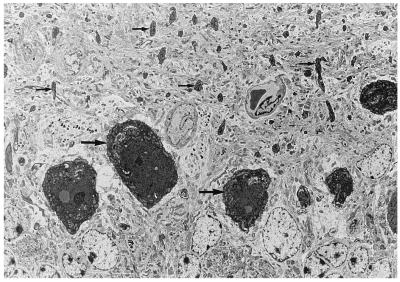
There is no accompanying inflammatory response to neurodegeneration, with no appearance of either macrophages or microglia in the adjacent neuropil (see Figs. 1, 3, and 4). However, we do find a discrete glial response with one or two pale immature reactive astrocytes, without cytoplasmic glial fibrillary acidic protein filaments, in the immediate vicinity of degenerating neurons. These astrocytes frequently have processes that surround the degenerating neuron, but in no case do we see engulfment of the neuron by either macrophages or glial cells.
Figure 3.
Cerebellar Purkinje cells undergoing dark cell degeneration, neuronal soma (large arrows), and dendritic processes (small arrows) both show darkened features (magnification ×720).
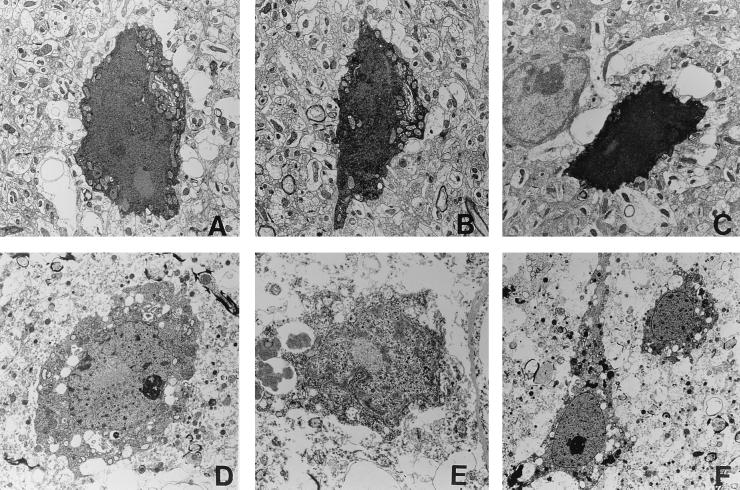
Figure 4.
Examples of degenerating neurons in the right anterior cingulate cortex from an R6/2 transgenic mouse (A–C; 17 weeks old, 141 CAG repeats) and an HD patient (D–E; 11 years old, 100 CAG repeats). NII are present within all of these neurons. (Magnifications: A–C, ×1,250; D–E, ×2,250; F, ×1,000.)
Late Onset Progressive Neurodegeneration.
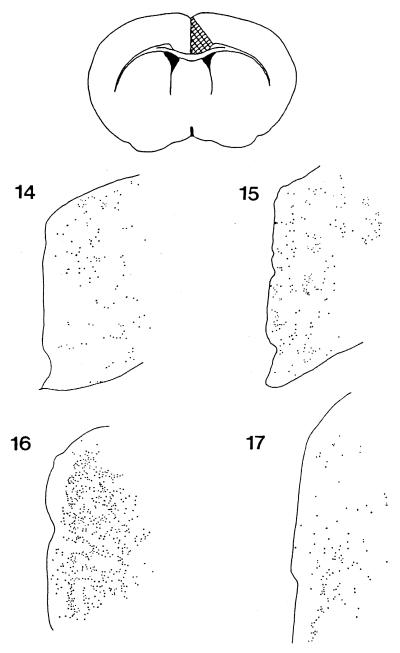
Within the anterior cingulate cortex, dark neurons are found throughout cortical layers II–VI and are always more prevalent within the right hemisphere than the left. These degenerating neurons can first be detected at 14–14.5 weeks of age in the R6/2 line and increase in number until 16 weeks of age, declining thereafter (Fig. 2). Analysis of 1 μm of toluidine blue-stained sections suggests that dark neurons represent a significant proportion of cells within this cortical region and that degenerating neurons persist for several weeks within this area. We can additionally find dark-staining neurons of similar appearance at approximately the same time (14–15 weeks) within the dorsal pericallosal region of the anterior striatum (Fig. 1A) and within the Purkinje cell layer of the cerebellar vermis (Fig. 3). After 16 weeks, darkened neurons can additionally be found within more lateral areas of the frontal cortex (data not shown).
Figure 2.
Camera Lucida drawings of 1-μm toluidine blue-stained araldite-embedded sections through the anterior cingulate cortex showing the distribution of darkened, degenerating neurons. Sections are from R6/2 mice of 14 weeks, 15 weeks, 16 weeks, and 17 weeks.
Neurodegeneration in the Postmortem HD Brain.
Our findings of a uniform ultrastructural appearance of degenerating neurons within the brains of transgenic mice prompted us to look at neurons within these same areas (anterior cingulate cortex, striatum, and cerebellum) in postmortem tissue from two patients with HD. We again found dark neurons of similar ultrastructural appearance to those found in the transgenic mouse within these same regions. These cells, containing NII, were again never found to contain apoptotic bodies nor to show blebbing of the nucleus or cytoplasm (compare neurons in the anterior cingulate cortex of a transgenic mouse Fig. 4 A–C with those found in the same region of an HD patient in Fig. 4 D and E). Intracellular organelles exhibit good ultrastructural preservation in the initial stages of degeneration and then a transient swelling before condensing in the latter stages of neuronal death.
Discussion
These findings clearly suggest that neurons in the brain of transgenic mice, expressing exon 1 of the human HD gene containing an expanded polyglutamine, degenerate within those specific areas of the brain known to be affected in HD and by a process with a well-defined ultrastructural appearance. The defining characteristics of apoptotic cell death were never found. Thus, the fragmentation or blebbing of both nucleus and cytoplasm and the formation of apoptotic bodies, morphologic criteria upon which this process was originally defined, are not present (21, 25). Similarly, we can not find TUNEL-positive neurons, suggesting that within dark neurons there is no internucleosomal DNA fragmentation, a prominent feature of apoptosis. Degenerating neurons of similar ultrastructural appearance can be found in postmortem brain from patients with HD, but to our knowledge have never been reported in other neurodegenerative diseases such as Alzheimer's disease and Parkinson's disease. In HD postmortem striatum, a small percentage of dark neurons have previously been found to be TUNEL-positive, confirming several studies of TUNEL-positive cells (both neurons and glia) in this disease (22–24). However, the demonstration of TUNEL-positive nuclei, without ultrastructural analysis, in postmortem tissue is not currently recognized as definitive of apoptosis (26).
These same ultrastructural features in degenerating neurons have recently been found in an invertebrate model of HD; Drosophila expressing amino-terminal fragments of human huntingtin-containing tracts of 120 glutamine residues develop late onset degeneration of photoreceptor neurons (27). Degenerating neurons are characterized by an enhanced staining with osmium, exhibit condensation of both cytoplasm and nucleus, and contain NII. These cells do not develop apoptotic bodies nor do they show blebbing of cytoplasm or nucleus (27). This neuronal death cannot be blocked by coexpression of the viral antiapoptotic gene P35 (27).
In marked contrast to this late onset nonapoptotic form of neuronal death observed in HD brain, and transgenic mouse and Drosophila models (27), is the rapid apoptotic death observed following expression of high levels of mutant huntingtin in transgenic mice (28) or in transfected cells in culture (29, 30). This latter form of degeneration appears not to be dependent on aggregation of the mutant protein forming intracellular inclusions (either nuclear or neuritic), and the mechanism of neurodegeneration clearly seems to differ from the results obtained in our in vivo investigations.
Our demonstration of such prominent degeneration within the anterior cingulate cortex of HD transgenic mice is somewhat surprising. However, this cortical area has been demonstrated to exhibit hypometabolism in patients with HD, studied by a variety of noninvasive functional imaging methods (31). Similarly, neuropathological studies have highlighted this area as one exhibiting prominent neurite pathology in this disease (32, 33). Our studies, confirmed by results of the studies of postmortem material from HD patients, suggests that further investigation of the anterior cingulate cortex in HD is clearly warranted. The observation of asymmetric degeneration within this area in transgenic mice awaits confirmation in patient material.
These results clearly suggest that the neuronal death found in the brain in HD is by a process that is morphologically and biochemically distinct from apoptosis, and is associated with the presence of intracellular aggregates of mutant protein. This defines a temporal progression of protein aggregation, inclusion formation, appearance of neurological symptoms, and finally neurodegeneration. Potential treatments for HD directed toward prevention of protein aggregation may thus prove more effective than antiapoptotic therapies.
Acknowledgments
We thank Dr. J. O. Xuereb and R. Hills of the Department of Pathology, Addenbrooke's Hospital (Cambridge) for the generous provision of postmortem human tissue. This work was supported by grants from the Wellcome Trust, Huntington's Disease Society of America, Hereditary Disease Foundation, and the Special Trustees of Guy's Hospital.
Abbreviations
- HD
Huntington's disease
- NII
neuronal intranuclear inclusion
- TUNEL
terminal deoxynucleotidyltransferase-mediated UTP end labeling
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.110078997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.110078997
References
- 1.Vonsattel J-P G, Meyers R H, Stevens T J, Ferrante R J, Bird E D, Richardson E P. J Neuropathol Exp Neurol. 1985;44:559–577. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- 2.Vonsattel J P G, DiFiglia M. J Neuropathol Exp Neurol. 1998;57:369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- 3.Robitaille Y, Lopes-Cendes I, Becher M, Rouleau G, Clark A W. Brain Pathol. 1997;7:877–881. doi: 10.1111/j.1750-3639.1997.tb00893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ross C A, Becher M W, Coloner V, Engelender S, Wood J D, Sharp A H. Brain Pathol. 1997;7:1003–1017. doi: 10.1111/j.1750-3639.1997.tb00898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mangiarini L, Sathasivam K, Seller M, Cozens B A, Harper A, Hetherington C, Lawton M, Trottier Y, Lehrach H, Davies S W, et al. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 6.Davies S W, Turmaine M, Cozens B A, DiFiglia M, Sharp A H, Ross C A, Scherzinger E, Wanker E E, Mangiarini L, Bates G P. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 7.Davies S W, Turmaine M, Cozens B A, Raza A S, Mahal A, Mangiarini L, Bates G P. Philos Trans R Soc London B. 1999;354:971–979. doi: 10.1098/rstb.1999.0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scherzinger E, Lurz R, Turmaine M, Mangiarini L, Hollenbach B, Hasenbank R, Bates G P, Davies S W, Lehrach H, Wanker E E. Cell. 1997;90:549–558. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- 9.Scherzinger E, Sittler A, Schweiger K, Heiser V, Lurz R, Hasenbank R, Bates G P, Lehrach H, Wanker E E. Proc Natl Acad Sci USA. 1999;96:4604–4609. doi: 10.1073/pnas.96.8.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perutz M F. Trends Biochem Sci. 1999;24:58–63. doi: 10.1016/s0968-0004(98)01350-4. [DOI] [PubMed] [Google Scholar]
- 11.DiFiglia M, Sapp E, Chase K O, Davies S W, Bates G P, Vonsattel J P, Aronin N. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 12.Becher M W, Kotzuk J A, Sharp A H, Davies S W, Bates G P, Price D L, Ross C A. Neurobiol Dis. 1998;4:387–398. doi: 10.1006/nbdi.1998.0168. [DOI] [PubMed] [Google Scholar]
- 13.Gourfinkel-An I, Cancel G, Duyckaerts C, Faucheaux B, Hauw J-J, Trottier Y, Brice A, Agid Y, Hirsch E C. NeuroReport. 1998;9:1823–1826. doi: 10.1097/00001756-199806010-00028. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi Y, Kakita A, Yamada M, Koide R, Igarashi S, Takano H, Ikeuchi T, Wakabayashi K, Egawa S, Tsuji S, et al. Acta Neuropathol. 1998;96:547–552. doi: 10.1007/s004010050933. [DOI] [PubMed] [Google Scholar]
- 15.Igarashi S, Koide R, Shimohata T, Yamada M, Hayashi Y, Takano H, Date H, Oyake M, Sato T, Sato A, et al. Nat Genet. 1998;18:111–117. doi: 10.1038/ng0298-111. [DOI] [PubMed] [Google Scholar]
- 16.Li M, Miwa S, Kobayshi Y, Merry D E, Yamamoto M, Tanaka F, Doyu F, Hashizume Y, Fischbeck K H, Sobue G. Ann Neurol. 1998;44:249–254. doi: 10.1002/ana.410440216. [DOI] [PubMed] [Google Scholar]
- 17.Skinner P J, Koshy B T, Cummings C J, Klement I A, Helin K, Servadio A, Zoghbi H Y, Orr H T. Nature (London) 1997;389:971–974. doi: 10.1038/40153. [DOI] [PubMed] [Google Scholar]
- 18.Paulson H L, Perez M K, Trottier Y, Trojanowski J Q, Subramony S H, Das S S, Vig P, Mandel J L, Fischbeck K H, Pitmann R N. Neuron. 1997;19:333–344. doi: 10.1016/s0896-6273(00)80943-5. [DOI] [PubMed] [Google Scholar]
- 19.Holmberg M, Duyckaerts C, Dürr A, Cancel G, Gourfinkel-An I, Damier P, Faucheux B, Trottier Y, Hirsch E C, Agid Y, et al. Hum Mol Genet. 1998;7:913–918. doi: 10.1093/hmg/7.5.913. [DOI] [PubMed] [Google Scholar]
- 20.Schmidt T, Landwehrmeyer G B, Schmitt I, Trottier Y, Auberger G, Laccone F, Klockgether T, Völpel M, Epplen J T, Schöls L, et al. Brain Pathol. 1998;8:669–681. doi: 10.1111/j.1750-3639.1998.tb00193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kerr J F R, Wyllie A H, Currie A R. Br J Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Portera-Cailliau C, Hedreen J C, Price D L, Koliatsos V E. J Neurosci. 1995;15:3775–3787. doi: 10.1523/JNEUROSCI.15-05-03775.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dragunow M, Faull R L M, Lawlor P, Beihartz E J, Singleton K, Walker E B, Mee E. Clin Neurosci Neuropathol. 1995;6:1053–1057. doi: 10.1097/00001756-199505090-00026. [DOI] [PubMed] [Google Scholar]
- 24.Brannon Thomas L, Gates D J, Richfield E K, O'Bien T F, Schweitzer J B, Steindler D A. Exp Neurol. 1995;133:265–272. doi: 10.1006/exnr.1995.1029. [DOI] [PubMed] [Google Scholar]
- 25.Clarke P G H. Anat Embryol. 1990;181:195–213. doi: 10.1007/BF00174615. [DOI] [PubMed] [Google Scholar]
- 26.Lucassen P J, Chung W C J, Kamphorst W, Swaab D F. J Neuropathol Exp Neurol. 1997;56:887–900. doi: 10.1097/00005072-199708000-00007. [DOI] [PubMed] [Google Scholar]
- 27.Jackson G R, Salecker I, Dong X, Yao X, Arnheim N, Faber P W, MacDonald M E, Zipursky S L. Cell. 1998;21:633–642. doi: 10.1016/s0896-6273(00)80573-5. [DOI] [PubMed] [Google Scholar]
- 28.Reddy P H, Williams M, Charles C, Garrett L, Pike-Buchanan L, Whetsell W O, Jr, Miller G, Tagle D. Nat Genet. 1998;20:198–202. doi: 10.1038/2510. [DOI] [PubMed] [Google Scholar]
- 29.Saudou F, Finkbinder S, Devys D, Greenberg M E. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 30.Klement I A, Skinner P J, Kayfor M D, Yi H, Hersch S M, Clark H B, Zoghbi H Y, Orr H T. Cell. 1998;95:41–53. doi: 10.1016/s0092-8674(00)81781-x. [DOI] [PubMed] [Google Scholar]
- 31.Mayberg H S, Starkstein S E, Peyser C E, Brandt J, Dannals R F, Folstein S E. Neurology. 1992;42:1791–1797. doi: 10.1212/wnl.42.9.1791. [DOI] [PubMed] [Google Scholar]
- 32.Sotrel A, Paskevitch P A, Kiely D K, Bird E D, Williams R S, Myers R H. Neurology. 1991;41:1117–1123. doi: 10.1212/wnl.41.7.1117. [DOI] [PubMed] [Google Scholar]
- 33.Jackson M, Gentleman S, Lennox G, Ward L, Gray T, Randall K, Morell K, Lowe J. J Neuropathol Appl Neurobiol. 1995;21:18–26. doi: 10.1111/j.1365-2990.1995.tb01024.x. [DOI] [PubMed] [Google Scholar]