Abstract
We transposed Dissociation (Ds) elements from three start loci on chromosome 5 in Arabidopsis (Nossen ecotype) by using a local transposition system. We determined partial genomic sequences flanking the Ds elements and mapped the elements' insertion sites in 1,173 transposed lines by comparison with the published genomic sequence. Most of the lines contained a single copy of the Ds element. One-half of the lines contained Ds on chromosome 5; in particular, insertion “hot spots” near the three start loci were clearly observed. In the other lines, the Ds elements were transposed across chromosomes. We found other insertion hot spots at the tops of chromosomes 2 and 4, near nucleolus organizer regions 2 and 4, respectively. Another characteristic feature was that the Ds elements tended to transpose near the chromosome ends and rarely transposed near centromeres. The distribution patterns differed among the three start loci, even though they possessed the same Ds construct. More than one-half of the Ds elements were inserted irregularly into the genome; that is, they did not retain the perfect inverted repeat sequence of Ds nor leave perfect target site duplications. This precise analysis of distribution patterns will contribute to a comprehensive understanding of the transposing mechanism. From these Ds insertion sites, we have constructed a database for screening gene-knockout mutants in silico. In 583 of the 1,173 lines, the Ds elements were inserted into protein-coding genes, which suggests that these lines are gene-knockout mutants. The database and individual lines will be available freely for academic use from the RIKEN Bio-Resource Center (http://www.brc.riken.go.jp/Eng/index.html).
The whole Arabidopsis genome sequence was determined in December 2000 (The Arabidopsis Genome Initiative, 2000). However, the functions of a large number of genes remain uncharacterized. A reverse-genetic approach will, therefore, become important in characterizing gene functions. One of the best strategies for reverse genetics is based on insertional mutagenesis. A random insertional mutagenesis approach using a transposon or T-DNA (an element of the tumor inducing [Ti] plasmid of Agrobacterium tumefaciens) as a mutagen offers a viable method for obtaining insertion mutants of genes of interest. The inserted foreign DNA not only introduces a mutation but also “tags” the responsible gene (Maes et al., 1999). Having the whole genome sequence enables us to map these tags precisely by simply reading sequences flanking the tags. At the same time, we can reveal the precise distribution pattern of the transposable elements on the Arabidopsis genome when those elements are used as mutagens. The distribution pattern will become valuable for elucidation of the transposing mechanism.
We have been generating Dissociation (Ds)-inserted lines on the Arabidopsis genome (Nossen ecotype; Ito et al., 1999). Ds is a nonautonomous transposable element of maize (Zea mays). We are using a local transposition system (Fedoroff and Smith, 1993) and have selected three parental lines whose initial loci of the Ds elements have been mapped on chromosome 5 (Fig. 1). By using the Ds-transposed lines, we have already reported two individual mutants (Ito et al., 2000; Motohashi et al., 2001). Here, we identify the insertion position of the moved Ds element by using the thermal asymmetric interlaced (TAIL)-PCR technique (Liu and Whittier, 1995). On the basis of our results, we describe the genome distribution pattern of the Ds elements of 1,173 independent transposed lines and the construction of a database for searching gene-knockout lines.
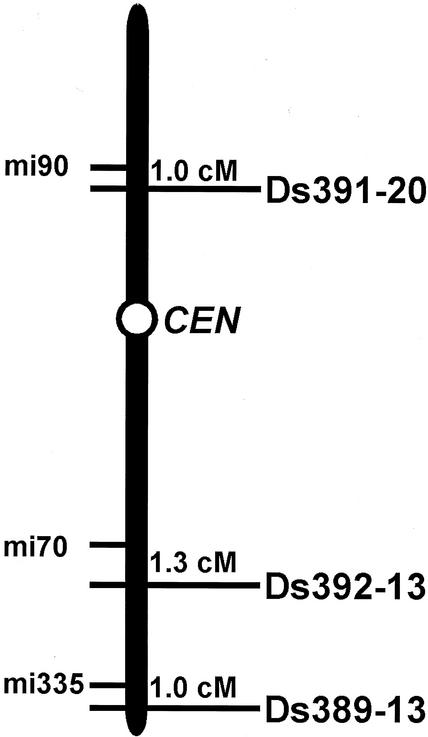
Figure 1.
Genetic map position on chromosome 5 of the three start lines used in this study. The nearest RFLP marker and genetic distance are shown. CEN, Centromere. This map is derived from Smith et al. (1996).
RESULTS AND DISCUSSION
We have generated transposed lines from three start loci on chromosome 5 (Fig. 1). Most of the lines contained a single copy of the Ds element per plant. Because this transposition system detects both linked and unlinked transposition events with the start loci (Fedoroff and Smith, 1993; Ito and Shinozaki, 2002), we can obtain all of the transposed elements irrespective of the distance from the start loci. The engineered Ds construct in a T-DNA vector (Ds-GUS-T-DNA construct) had been introduced into the Arabidopsis genome by A. tumefaciens-mediated transformation (Smith et al., 1996). By determining the genomic sequences flanking the T-DNA, we identified the precise insertion positions of the three Ds-GUS-T-DNAs at the nucleotide level (Table I). To examine the distribution pattern of the transposable elements and to construct a database for reverse genetics, we recovered genomic DNA fragments that flank the 5′ and 3′ ends of the Ds elements by using TAIL-PCR (Liu and Whittier, 1995) and read the partial sequences. We identified the insertion positions of 37% of the lines from both flanking sequences and of 49% from either the 5′- or the 3′-flanking sequence. In total, we identified the insertion sites in 1,173 lines. The position of the remaining 14% was identical to the initial position; therefore, we excluded these lines from the transposed collection. Our pilot examination indicates that approximately 8% of the collection is spurious information derived from PCR or seed cross-contamination. These lines will be excluded when the seeds are distributed from RIKEN Bio-Resource Center.
Table I.
Physical map position of the donor Ds-GUS-T-DNA constructs used in this study
| Donor Linea | Start Locus
|
|
|---|---|---|
| P1 cloneb | Inserted positionc | |
| Ds391-20 | MQM1 (10178221) | 52,533 |
| Ds392-13 | MGO3 (3869070) | 36,740 |
| Ds389-13d | K2A18 (2924651) | 75,249d |
These donor lines are described by Smith et al. (1996).
P1 clone in which Ds-GUS-T-DNA is inserted. Parentheses indicate GenBank GI no. of the P1 clone sequence.
Coordinates of the P1 sequence (GenBank entry) flanking the left border of the Ds-GUS-T-DNA.
Ds-GUS-T-DNA of this line was inserted into a protein-coding gene (protein entry code, At5g66210; annotation, calcium-dependent protein kinase; MAtDB), and the homozygous line showed a semi-dwarf phenotype (data not shown).
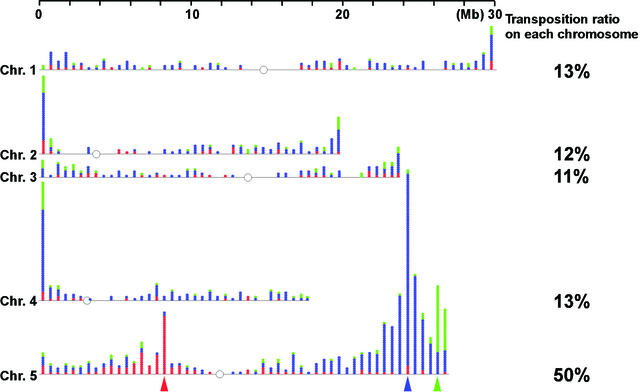
We plotted the insertion sites across the genome at 500-kb intervals (Fig. 2). The distribution pattern indicates that 50% of the Ds elements were transposed within chromosome 5, particularly near the initial loci. The other 50% were transposed to different chromosomes. This result indicates that Ds moves preferentially to linked sites, but it also moves to unlinked position at a relatively high frequency. Two other insertion “hot spots” were observed at the tops of chromosomes 2 and 4, adjacent to nucleolus organizer region (NOR) 2 and NOR4, respectively. The insertion sites were clustered over a 500-kb region (Fig. 3). NOR consists of tandemly repeated ribosomal RNA gene clusters and forms a loosened chromatin structure, because the ribosomal RNA is highly transcribed. These hot spots also were observed in another Activator (Ac)/Ds system in Arabidopsis (Parinov et al., 1999) but not in a Suppressor-mutator/defective Suppressor-mutator system in Arabidopsis (Tissier et al., 1999). Because Ac/Ds and Suppressor-mutator/defective Suppressor-mutator are different classes of transposable elements, this finding suggests that the tops of chromosomes 2 and 4 are hot spots not simply because transposase is physically accessible to these regions. In addition, we found that transpositions were rare around centromeres but more frequent near chromosome ends (Fig. 2). These results indicate that the Ds element is useful for disrupting genes on chromosome ends, especially the tops of chromosomes 2 and 4, and genes near a start locus.
Figure 2.
Distribution of Ds insertions on the Arabidopsis genome. Inserted sites are plotted at 500-kb intervals. Red, blue, and green dots indicate transpositions from Ds391-20, Ds392-13, and Ds389-13, respectively. Red, blue, and green arrowheads represent the respective start positions. The circle represents the centromere.
Figure 3.
Distribution of Ds insertions in hot spots near NORs on chromosomes 2 and 4. Arrows on the chromosomes show insertions. Positions of the sequenced BACs are also indicated.
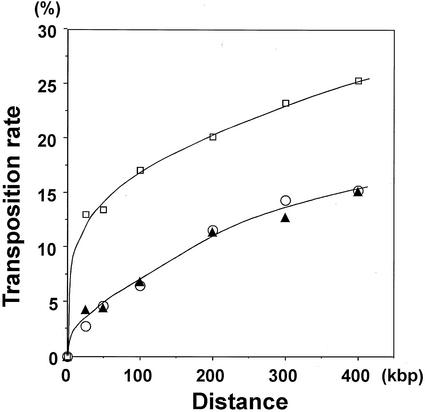
Although Ds tended to transpose to linked sites (Fig. 2), the local distribution patterns differed among the three start lines (Fig. 4). Transpositions from Ds391-20 and Ds392-13 showed a similar pattern, but that from Ds389-13 showed a narrower distribution. Fifteen percent of the total Ds elements from Ds391-20 and Ds392-13 transposed within 400 kb, whereas 15% from Ds389-13 were transposed within 70 kb (Fig. 4). These Ds elements possess the same construct. Therefore, the different patterns are derived from differences in the start positions. One possible determinant may be the chromatin structure around the start position.
Figure 4.
Transposition rate within a given distance from a start site. Circles, triangles, and rectangles represent transposition rates from Ds391-20, Ds392-13, and Ds389-13, respectively.
To screen gene-knockout mutants in silico, we constructed a database of Ds insertion sites. Ds elements of 583 (50%) of the 1,173 lines were inserted into protein-coding genes. These are probably null mutants. Those of 110 lines (9%) lay within 500 bp upstream from the start codon of a gene, and those of 75 lines (6%) lay within 500 bp downstream from the stop codon. Gene expression of these mutants may be disordered. Ds elements of the other 405 lines (35%) were inserted into intergenic regions more than 500 bp from protein-coding genes. The insertion sites of 1,124 of the 1,173 lines are listed in the supplemental materials (they can be viewed at http://www.plantphysiol.org); all will be freely obtained for academic use from RIKEN Bio-Resource Center (http://www.brc.riken.go.jp/Eng/index.html).
These Ds lines are useful in both forward and reverse genetics. We are screening mutants showing the albino or pale-green phenotype to isolate nuclear genes functioning in chloroplasts (Motohashi et al., 2001). We isolated 20 albino mutants from about 4,000 lines. Albino phenotypes of 15 of the mutants were cosegregated with Ds insertions and were probably tagged (R. Motohashi, T. Ito, and K. Shinozaki, unpublished data). Azpiroz-Leehan and Feldmann (1997) estimated that only 35% to 40% of mutants from T-DNA-mutagenized populations are tagged. Thus, our Ds population is more efficient for gene isolation than is a T-DNA population.
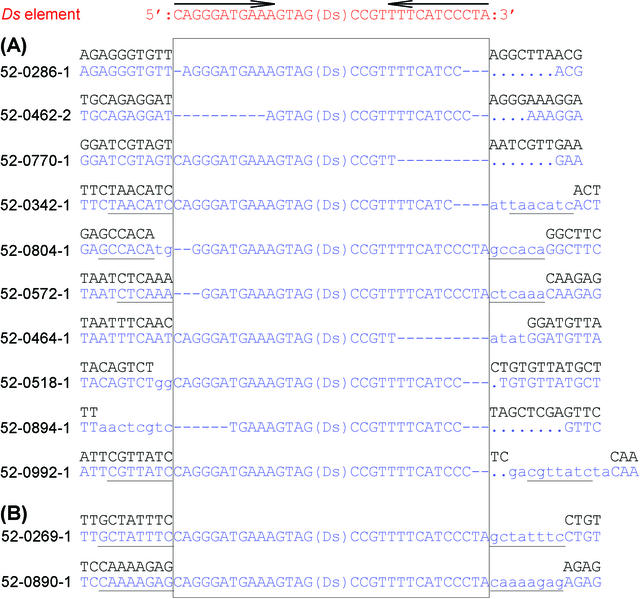
One of the advantages of transposable elements is that they can be remobilized. Mapped insertion lines will become useful resources for the targeted mutagenesis of closely linked genes. If one finds an insertion near a gene of interest, a gene disruptant may be obtained by a secondary transposition procedure. For efficient transposition, it is necessary for the Ds to possess perfect inverted repeat (IR) sequences in the 5′ and 3′ ends, because IR sequences are a major requirement for transposition. To assess a strategy for the secondary transposition, we examined 54 junction sequences of the transposed Ds end and its flanking genome. Thirty transposed lines (56%) possessed neither a perfect IR element nor a perfect 8-bp target site duplication sequence (irregularly inserted patterns; Fig. 5A). This result indicates that it is inefficient to use these lines as launching pads for transposition. Only 24 lines (44%) contained both a perfect IR element and a perfect 8-bp target site duplication (authentic patterns; Fig. 5B). It would be efficient to use these lines as launching pads for transposition. We conclude that we have to check the junction sequence of the transposed Ds end and its flanking genome when we use a transposed line as a Ds-donor line. These abnormal, and presumably no longer transposition-competent Ds elements, could explain many of the stable mutants that frequently arise in Ac or Ds mobilization experiments in many plants.
Figure 5.
Nucleotide sequences around Ds insertion sites. A, Examples of irregularly inserted patterns. B, Examples of authentic patterns. Arrows indicate IR sequences of a Ds element necessary for transposition. Ds elements in the insertion lines are surrounded by a rectangle. Sequences duplicated by transposition are underlined. Sequences shown as red and blue indicate those of the original Ds and the insertion line, respectively. The sequence shown as black is the Nossen wild-type sequence. Hyphens and dots show deleted nucleotides in the insertion lines compared with original Ds element (--) and wild-type genomic DNA (·). Additional nucleotides in the insertion lines are indicated in lowercase.
MATERIALS AND METHODS
Extraction of Genomic DNA, TAIL-PCR, and Sequencing of PCR Products
Extraction of genomic DNA, TAIL-PCR, and sequencing of PCR products were carried out as described by Ito et al. (2001).
Determination of Ds Insertion Sites
The flanking sequences were used in a BLASTN search of the Munich Information Center for Protein Sequences Arabidopsis Database (v.040701; http://mips.gsf.de/proj/thal/db/). The position showing the highest similarity score was defined as an insertion site. When Ds was inserted into a protein-coding gene, we picked out the protein entry code (e.g. At5g23670) of the gene. When Ds was inserted into an intergenic region between protein-coding genes, we picked out the protein entry codes of the two genes on either side (see supplemental data at http://www.plantphysiol.org).
Supplementary Material
ACKNOWLEDGMENTS
We thank Y. Notoshi for information about donor locus Ds392-13 and his useful comments, J. Ishida and M. Nakajima for sequencing, and T. Hirayama and K. Akiyama for constructing the database. We also thank S. Kawamura for maintaining the greenhouse.
Footnotes
This work was supported in part by RIKEN (a grant for Genome Research to K.S.; the Special Fund from the Director of RIKEN Tsukuba Institute to T.I.), by the Ministry of Education, Culture, Sports, Science and Technology of Japan (Grant-in-Aid for Scientific Research on Priority Areas no. 10182101 to K.S. and a Grant-in-Aid for Scientific Research on Priority Areas (C) “Genome Science” to T.I.), and by Bio-Oriented Technology Research Advancement Institution (the Program for Promotion of Basic Research Activities for Innovative Biosciences to K.S.).
The online version of this article contains Web-only data. The supplemental material is available at www.plantphysiol.org.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.002774.
LITERATURE CITED
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Azpiroz-Leehan R, Feldmann KA. T-DNA insertion mutagenesis in Arabidopsis: going back and forth. Trends Genet. 1997;13:152–156. doi: 10.1016/s0168-9525(97)01094-9. [DOI] [PubMed] [Google Scholar]
- Fedoroff NV, Smith DL. A versatile system for detecting transposition in Arabidopsis. Plant J. 1993;3:273–289. doi: 10.1111/j.1365-313x.1993.tb00178.x. [DOI] [PubMed] [Google Scholar]
- Ito T, Kim G-T, Shinozaki K. Disruption of an Arabidopsis cytoplasmic ribosomal protein S13-homologous gene by transposon-mediated mutagenesis causes aberrant growth and development. Plant J. 2000;22:257–264. doi: 10.1046/j.1365-313x.2000.00728.x. [DOI] [PubMed] [Google Scholar]
- Ito T, Motohashi R, Shinozaki K. Preparation of transposon insertion lines and determination of the insertion sites in Arabidopsis genome. In: Braman J, editor. Methods in Molecular Biology. Ed 2. 182, In Vitro Mutagenesis Protocols. Totowa, NJ: Humana Press; 2001. pp. 211–221. [DOI] [PubMed] [Google Scholar]
- Ito T, Seki M, Hayashida N, Shibata D, Shinozaki K. Regional insertional mutagenesis of genes on Arabidopsis thaliana chromosome V using the Ac/Ds transposon in combination with a cDNA scanning method. Plant J. 1999;17:433–444. doi: 10.1046/j.1365-313x.1999.00383.x. [DOI] [PubMed] [Google Scholar]
- Ito T, Shinozaki K. Random insertional mutagenesis in Arabidopsis. In: Jain SM, Brar DS, Ahloowalia BS, editors. Molecular Techniques in Crop Improvement. Dordrecht, The Netherlands: Kluwer Academic Publishers; 2002. pp. 409–425. [Google Scholar]
- Liu Y-G, Whittier RF. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics. 1995;25:674–681. doi: 10.1016/0888-7543(95)80010-j. [DOI] [PubMed] [Google Scholar]
- Maes T, Keukeleire PD, Gerats T. Plant tagnology. Trends Plant Sci. 1999;4:90–96. doi: 10.1016/s1360-1385(99)01375-8. [DOI] [PubMed] [Google Scholar]
- Motohashi R, Nagata N, Ito T, Takahashi S, Hobo T, Yoshida S, Shinozaki K. An essential role of a TatC homologue of a ΔpH-dependent protein transporter in thylakoid membrane formation during chloroplast development in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2001;98:10499–10504. doi: 10.1073/pnas.181304598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parinov S, Sevugan M, Ye D, Yang W-C, Kumaran M, Sundaresan V. Analysis of flanking sequences from Dissociation insertion lines: a database for reverse genetics in Arabidopsis. Plant Cell. 1999;11:2263–2270. doi: 10.1105/tpc.11.12.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D, Yanai Y, Liu Y-G, Ishiguro S, Okada K, Shibata D, Whittier RF, Fedoroff NV. Characterization and mapping of Ds-GUS-T-DNA lines for targeted insertional mutagenesis. Plant J. 1996;10:721–732. doi: 10.1046/j.1365-313x.1996.10040721.x. [DOI] [PubMed] [Google Scholar]
- Tissier AF, Marillonnet S, Klimyuk V, Patel K, Torres MA, Murphy G, Jones JDG. Multiple independent defective Suppressor-mutator transposon insertions in Arabidopsis: a tool for functional genomics. Plant Cell. 1999;11:1841–1852. doi: 10.1105/tpc.11.10.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.