Abstract
Long-chain acyl-coenzyme A (CoA) synthetases (LACSs) activate free fatty acids to acyl-CoA thioesters and as such play critical roles in fatty acid metabolism. This important class of enzymes factors prominently in several fatty acid-derived metabolic pathways, including phospholipid, triacylglycerol, and jasmonate biosynthesis and fatty acid β-oxidation. In an effort to better understand the factors that control fatty acid metabolism in oilseeds, we have sought to identify and characterize genes that encode LACSs in Arabidopsis. Nine cDNAs were identified, cloned, and tested for their ability to complement a LACS-deficient strain of yeast (Saccharomyces cerevisiae). Seven of the nine successfully restored growth, whereas two cDNAs encoding putative peroxisomal isoforms did not. Lysates from yeast cells overexpressing each of the nine cDNAs were active in LACS enzyme assays using oleic acid as a substrate. The substrate specificities of the enzymes were determined after overexpression in LACS-deficient Escherichia coli. Most of the LACS enzymes displayed highest levels of activity with the fatty acids that make up the common structural and storage lipids in Arabidopsis tissues. Analysis of the tissue-specific expression profiles for these genes revealed one flower-specific isoform, whereas all others were expressed in various tissues throughout the plant. These nine cDNAs are thought to constitute the entire LACS family in Arabidopsis, and as such, will serve as powerful tools in the study of acyl-CoA metabolism in oilseeds.
Long-chain acyl-CoA synthetase (LACS, EC 6.2.1.3) occupies a critical position in the biosynthetic pathways of nearly all fatty acid-derived molecules. LACS esterifies free fatty acids to acyl-CoAs, a key activation step that is necessary for the utilization of fatty acids by most lipid metabolic enzymes. LACS catalyzes the formation of acyl-CoA by a two-step mechanism (Groot et al., 1976). In the first step, the free fatty acid is converted to an acyl-AMP intermediate, called an adenylate, through the pyrophosphorolysis of ATP. The activated carbonyl carbon of the adenylate is then coupled to the thiol group of CoA, releasing AMP and the acyl-CoA final product (Groot et al., 1976).
However, this reaction scheme is not exclusive to LACS. The formation of the enzyme-bound adenylate is a common mechanism for acyl activation shared by a wide variety of enzymes in organisms that span the biological spectrum, from algal polyketide synthetases (Bibb et al., 1994) to arthropod luciferases (Conti et al., 1996) to bacterial peptide antibiotic synthetases (Conti et al., 1997). This mechanistic similarity is reflected in the conservation of certain amino acid motifs between all enzymes of this group. One motif in particular (PROSITE PS00455) is very highly conserved and acts as the unifying feature of this large group of enzymes, called the AMP-binding protein (AMPBP) superfamily (Babbitt et al., 1992), to which LACS belongs.
LACS is a particularly challenging and interesting target for molecular analysis, because of its fundamental role in providing activated acyl groups as substrates in various fatty acid metabolic pathways. Eukaryotic organisms contain several isoforms of LACS that participate in a variety of different anabolic and catabolic pathways.
One of the most well-characterized functions of LACS is its role in fatty acid transport. This process has been studied in detail in bacteria, yeast (Saccharomyces cerevisiae), and mammalian cells. Escherichia coli contains a single LACS, encoded by the FadD gene (Black et al., 1992). The seminal observations of Overath et al. (1969) suggested that a portion of the LACS enzyme in E. coli was associated with the cell membrane, and that recruitment of FadD to the membrane facilitated a vectorial esterification process necessary for import and activation of exogenous fatty acids. Intracellular fatty acid transport in eukaryotic cells also depends heavily on LACS activity. Acyl-CoA molecules do not freely diffuse across biological membranes. Transport of activated long-chain fatty acids requires an active transport mechanism of some type, like the carnitine shuttle in mammalian mitochondrial β-oxidation (Eaton et al., 1996) or receptor-mediated import as in yeast peroxisomes (Hettema and Tabak, 2000). In yeast, some evidence suggests that short- and medium-chain free fatty acids enter the peroxisome directly. Passage of these acids through the membrane is driven in part by reactivation to the CoA thioesters by acyl-CoA synthetases (Hettema and Tabak, 2000).
LACS initiates the process of fatty acid β-oxidation. In oilseeds, carbon reserves are stored as triacylglycerol (TAG). With the onset of germination, lipases release free fatty acids from the TAG molecules (Hills and Beevers, 1986; Lin et al., 1986). LACS activates the free fatty acids to acyl-CoAs that enter the β-oxidation pathway in the glyoxysomes of the germinating seedling. The enzymes of the β-oxidation cycle completely degrade fatty acids by the sequential removal of two-carbon units, which are released in the form of acetyl-CoA. The resulting acetyl-CoA pool is essential for production of cellular energy (through the tricarboxylic acid cycle) and for synthesis of sugars and other carbon skeletons (via the glyoxylate cycle and gluconeogenesis; Beevers, 1969; Gerhardt, 1992; Eastmond et al., 2000).
LACS provides the acyl-CoA substrate necessary for protein N-myristoylation, a type of protein modification that alters the properties of target proteins and enzymes, many of which are components of signal transduction pathways. The change in hydrophobicity due to myristoylation causes changes in membrane binding, protein-protein interactions, and/or three-dimensional conformation of the target enzymes (Gordon et al., 1991; Yalovsky et al., 1999; Ishitani et al., 2000; Martin and Busconi, 2000).
Our laboratory focuses on the metabolism of fatty acids and TAG in oilseed crops. From this standpoint, one of the primary roles of LACS is the synthesis of acyl-CoA molecules used as substrates for phospholipid and TAG biosynthesis. In plant cells, fatty acids are synthesized in the plastids. The growing acyl chain is esterified to acyl-carrier protein (ACP), which is cleaved to free fatty acid and ACP by acyl-ACP thioesterase enzymes in the inner envelope when the fatty acid has reached the appropriate length (Ohlrogge and Browse, 1995). The free fatty acid diffuses to the outer envelope of the plastid membrane, where a plastidial LACS activates it to the CoA thioester and releases the acyl-CoA product into the cytosol (Andrews and Keegstra, 1983; Pollard and Ohlrogge, 1999). The resulting acyl-CoAs are utilized by acyltransferases located in the endoplasmic reticulum that catalyze successive acylations of glycerol-3-phosphate. The acylglycerol intermediates formed by these reactions ultimately are converted into the suite of phospholipids necessary for membrane biosynthesis in all tissues of the plant as well as the TAGs synthesized in the developing seeds (Somerville and Browse, 1991). Therefore, the contribution of LACS to glycerolipid biosynthesis by creating and maintaining acyl-CoA pools within the cell is essential to normal growth and development of the organism.
Given the many important roles of LACS just described, one of the goals in our laboratory is to investigate the quantitative and qualitative contributions of LACS to lipid metabolism in plants. Progress in this field has been slow due to the lack of detailed molecular information regarding plant LACS genes. Only two previous reports have described plant LACSs at the molecular level. Pongdontri and Hills (2001) isolated a Brassica napus gene that was active in lipogenic tissues, whereas Fulda et al. (1997) cloned five B. napus cDNAs with homology to known LACSs. Two of these clones produced active LACS enzymes when expressed in E. coli. However, given the variety of LACS-dependent pathways, as well as the reports of LACS activity in many different plant organelles, including peroxisomes (Gerhardt, 1992), chloroplasts (Andrews and Keegstra, 1983), lipid bodies (Olsen and Lusk, 1994), mitochondria (Frentzen et al., 1990), and endoplasmic reticulum (Ichihara et al., 1997), higher plants must contain a much larger number of LACS genes.
This report describes a genome-wide assessment of the LACS gene family in Arabidopsis. Identification of these genes provides the first set of tools necessary for expanding our understanding of the function and control of CoA-dependent fatty acid activation.
RESULTS
Cloning Strategy and Sequence Comparisons
Our goal was to identify and clone all Arabidopsis genes that may encode LACS enzyme activities. As described in the introduction, LACS isozymes are involved in a number of important pathways of fatty acid metabolism; therefore, several different LACS genes may directly or indirectly affect the fatty acid composition of the seed TAG. Therefore, it was imperative to identify as many LACS genes as possible for further characterization. Computer-assisted analysis of Arabidopsis sequences found either in cDNA or genomic library screens that we performed or in the public databases revealed 44 genes containing homology to known LACSs from other eukaryotic organisms. Each of these genes contained the AMPBP signature motif. This simple sequence analysis alone was not sufficient for identification of LACS genes. In addition to LACS, the AMPBP superfamily also contains several other classes of genes, some members of which, such as 4-coumarate-CoA ligases and acetyl-CoA synthetases, have been characterized in plants previously (Lee et al., 1995; Ehlting et al., 1999; Ke et al., 2000). Therefore, we sought other LACS-specific sequence determinants with which to identify the more likely candidate LACS genes.
Prior sequence comparisons between two rat (Rattus norvegicus) LACS protein sequences and that of the clickbeetle (Pyrearinus termitillumanans) luciferase (Fujino and Yamamoto, 1992) showed two homologous domains, termed LS1 and LS2. The LS1 and LS2 domains are also conserved in many of the other members of the AMPBP superfamily. The rat LACSs, however, also contained a 45-amino acid residue region not found in the luciferase enzyme. This domain is located directly in between LS1 and LS2 and links the two luciferase-like domains. A linker domain of similar length was also observed in four of the homologous enzymes from B. napus (Fulda et al., 1997). The precise function of the linker domain was unknown, but appeared to be a necessary component of eukaryotic LACS function because removal of the linker domain from the rat brain enzyme eliminated LACS activity in vitro (Iijima et al., 1996). The linker domain was also found in many other eukaryotic LACSs known to activate long-chain (C14–C18) fatty acids (Johnson et al., 1994; Fulda et al., 1997; Kang et al., 1997).
The only known LACS that does not contain the linker domain is the FadD enzyme, the sole LACS in E. coli (Black et al., 1992; Fulda et al., 1994). This prokaryotic enzyme contains reduced sequence similarity to most eukaryotic isoforms and also differs from eukaryotic enzymes in that only a portion of the FadD enzyme pool is membrane bound. FadD is soluble but can be recruited to the cell membrane (Overath et al., 1969). Analysis of the amino acid sequences of several other eukaryotic acyl-CoA synthetases that utilize either short- (<C8), medium- (C8–C12), or very long- (>C22) chain substrates demonstrated that these enzymes also do not contain the linker domain (Steinberg et al., 2000).
The maintenance of the linker domain in LACS enzymes from such evolutionarily distant species as rapeseed and rat, combined with its apparent exclusivity to enzymes that accept only long-chain fatty acids implied that this sequence element might be very useful as a LACS-specific sequence “probe.” Given the large size of the AMPBP superfamily in Arabidopsis, this analysis was an important step in the process of narrowing the number of candidate LACS genes to be chosen for further study. As such, the existence of this sequence element was used to analyze the entire set of 44 Arabidopsis genes that contained the AMPBP signature motif. In addition to the three known 4-coumarate-CoA ligases and one known acetyl-CoA synthetase, we also analyzed 13 previously uncharacterized genes with strong similarity to the 4-coumarate-CoA ligases and two additional genes related to acetyl-CoA synthetases. We also analyzed three additional superfamily members that were not closely associated with any of the larger clades, plus a clade of 14 highly homologous genes of unknown function (data not shown). None of the predicted amino acid sequences for these genes contained the linker domain. These data indicate that the presence of this domain may be a useful tool for the identification of eukaryotic-type LACS genes.
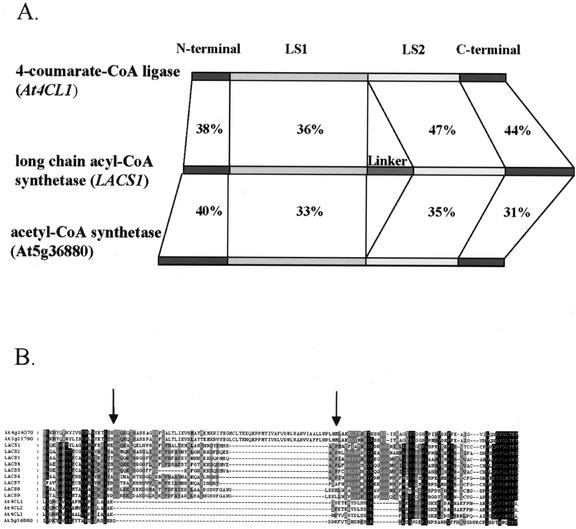
Eleven members of the superfamily contained apparent linker domains near the expected sites within the deduced amino acid sequences. Figure 1A compares the structures and sequences of one candidate LACS enzyme (LACS1) with an acetyl-CoA synthetase and a 4-coumarate-CoA ligase. The four common domains are named N terminal, LS1, LS2, and C terminal, based on the nomenclature of Fujino and Yamamoto (1992). This analysis indicated that each class of AMPBP in Arabidopsis shares similarity across all four domains. This observation was consistent with the crystallographic analysis of firefly luciferase (Conti et al., 1996) and bacterial tyrocidin synthetase (Conti et al., 1997), two members of the AMPBP superfamily, which demonstrated that all four domains contribute to formation of the active sites. However, only the candidate LACS enzymes contained the linker domain.
Figure 1.
Only LACSs contain the eukaryote-type linker domain. A, Schematic comparison of domain structures of various Arabidopsis AMPBP genes. The domain structure of a representative LACS gene (LACS1) is compared with the 4-coumarate-CoA ligase gene At4CL1 and the acetyl-CoA synthetase gene At5g36880. The comparisons were carried out using the GAP program with default parameters (Wisconsin Package Version 10.0, Genetics Computer Group, Madison, WI). The domain nomenclature is described in “Results.” The domain-specific degree of amino acid similarity between LACS1 and the other genes are shown as percentages between each pair of genes. B, Comparison of central regions of a candidate LACS to acetyl-CoA synthetase and 4-coumarate-CoA ligases of Arabidopsis. The amino acid sequences spanning from the C-terminal border of the LS1 domain to the N-terminal border of the LS2 domain were aligned and shaded using the ClustalX and GeneDoc programs. Arrows denote the approximate borders of the linker domain.
The amino acid sequences of the central regions of the 11 candidate LACS enzymes and those of the previously cloned acetyl-CoA synthetase (designated here by its MIPS [Munich Information Center for Protein Sequences] code At5g36880) and 4-coumarate-CoA ligase genes (At4CL1, At4CL2, and At4CL3) are aligned in Figure 1B. Despite strong similarity in the flanking regions, this alignment clearly showed the presence of the linker domain in only the 11 candidate LACSs, with lengths varying between 30 and 70 amino acid residues. The At4g14070 and At3g23790 enzymes contain exceptionally long linker domains, which span approximately 70 residues, compared with the 30 to 40 residue linkers common to most of the other putative LACSs. Sequence comparisons indicated that the shorter linker seen in the other nine candidate genes was also common to most other eukaryotic LACSs, including those of yeast, rat, and human (Abe et al., 1992; Fujino and Yamamoto, 1992; Johnson et al., 1994). The abnormal length of the linker domains in At4g14070 and At3g23790, therefore, immediately raised questions about the identity of these genes. The corresponding enzymes were inactive in in vitro LACS activity assays, as described below. These data and other analyses to be presented in this report led us to decide that the genes at MIPS loci At4g14070 and At3g23790 probably do not encode LACS.
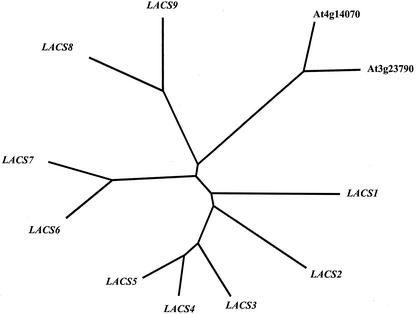
The other nine genes were named LACS and numbered consecutively starting with the number 1. A phylogenetic tree was constructed to visually compare the relationship between each of the candidate LACSs. This tree is shown in Figure 2. As a group, the family of genes is clearly delineated into several distinct clades. The LACS6/LACS7 clade shares 74% amino acid identity, whereas the LACS3/LACS4/LACS5 enzymes are 72% to 80% identical. The enzymes of the LACS8/LACS9 and At4g14070/At3g23790 clades are 67% and 77% identical, respectively. The presence of multiple genes on some branches of the phylogenetic tree may represent certain levels of overlap in function and/or subcellular location of the enzymes from each branch. However, as a group, this family of enzymes was only 30% identical. The genes in this family are also distributed widely throughout the genome. All five chromosomes contain at least one LACS, and those genes that do exist on the same chromosome (LACS2, 3, and 9 on chromosome 1, LACS1 and 8 on chromosome 2, and LACS4 and 5 on chromosome 4) are physically separated by at least 5.3 Mbp, suggesting that none of these genes have arisen from recent gene duplication events. This level of divergence suggested that these enzymes have evolved to fulfill a variety of specific functions within the plant cell.
Figure 2.
Phylogenetic comparison of 11 candidate LACS genes. The deduced full-length amino acid sequences of the genes that contained both the AMPBP signature motif and the linker domain were aligned and displayed as an unrooted nearest neighbor phylogenetic tree using the TreeView program.
A summary of the information pertaining to each of the LACS genes, including the numbers and tissue-specificities of the corresponding expressed sequence tag (EST) sequences, is shown in Table I. The number of ESTs associated with each of the LACS genes varied considerably, with some genes represented by numerous ESTs and others not represented at all. The results of the sequence analysis summarized in Figure 2 and Table I indicated that this family of LACS genes represents a very diverse group. The heterogeneity is also reflected in the length of the encoded enzymes. One-half of the members were nearly identical in length (approximately 665 amino acids; LACS1, 2, 3, 4, and 5), whereas the other one-half were significantly longer, primarily due to the presence of N-terminal extensions of between 30 and 60 amino acid residues (LACS6, 7, 8, 9, and At4g14070 and At3g23790; data not shown). The N-terminal extensions of these proteins were analyzed for the presence of chloroplast transit peptides or mitochondrial targeting peptides using the ChloroP and TargetP servers (Emanuelsson et al., 1999, 2000). Based on a necessary minimum predictive score of 0.50, ChloroP and TargetP both predicted chloroplastic targeting for LACS6, At4g14070, and At3g23790, whereas LACS8 and LACS9 did not receive an acceptable predictive score for any compartment from either program. However, these predictions should be judged with considerable caution and the targeting of these proteins must be confirmed experimentally. For example, studies of fusions between the LACS6 and LACS7 proteins and green fluorescent protein indicate that both of these enzymes are not targeted to chloroplasts as predicted for LACS6, but instead are targeted to peroxisomes (Fulda et al., 2002). LACS9, on the other hand, is targeted to the chloroplasts (Schnurr et al., 2002) despite the negative chloroplast predictions by both programs.
Table I.
Summary of Genbank accession nos., chromosomal locations, and EST profiles of cloned LACS genes
| Gene | Genbank Accession No. | Chromosome/MIPS Code | Corresponding ESTs |
|---|---|---|---|
| LACS1 | AF503751 | Chromosome 2, At2g47240 | Developing seed: eight |
| Green siliques: seven | |||
| Roots: two | |||
| Flower buds/inflorescences: two | |||
| LACS2 | AF503752 | Chromosome 1, At1g49430 | Developing seed: one |
| Green siliques: four | |||
| Roots: one | |||
| Flower buds/inflorescences: one | |||
| LACS3 | AF503753 | Chromosome 1, At1g64400 | Rosette leaves: one |
| Liquid-cultured seedlings: one | |||
| LACS4 | AF503754 | Chromosome 4, At4g23850 | Green siliques: two |
| LACS5 | AF503755 | Chromosome 4, At4g11030 | None |
| LACS6 | AF503756 | Chromosome 3, At3g05970 | Green siliques: one |
| Roots: one | |||
| LACS7 | AF503757 | Chromosome 5, At5g27600 | Developing seed: two |
| Roots: one | |||
| LACS8 | AF503758 | Chromosome 2, At2g04350 | Roots: one |
| Etiolated hypocotyls: one | |||
| LACS9 | AF503759 | Chromosome 1, At1g77590 | Developing seed: two |
| Green siliques: two | |||
| Roots: three |
Complementation of a Yeast Mutant Deficient in LACS
The ultimate focus of the present work was to determine which of the candidate LACS cDNAs encode enzymes capable of activating the C14 to C18 fatty acids commonly found in Arabidopsis glycerolipids. To address this question, the 11 candidate LACS cDNAs were cloned into the yeast expression vectors pYES2 or pRS426. These constructs were tested for their ability to complement the phenotype of yeast strain YB525 (kindly provided by Prof. J.I. Gordon, Washington University, St. Louis). This strain contains insertional disruptions in two of its LACS genes, FAA1 and FAA4 (Knoll et al., 1995), which activate essentially all exogenous fatty acids in yeast (Fargeman et al., 2001). Growth of YB525 is completely dependent on complementation with an active LACS when grown on media containing long-chain fatty acids as a sole carbon source and cerulenin, which inhibits endogenous fatty acid synthesis by the fatty acid synthase complex. Under these conditions, the YB525 mutant is unable to produce necessary levels of long-chain acyl-CoAs that are needed to acylate various essential cellular enzymes and proteins. The lack of protein acylation compromises the viability of the mutant yeast cells (Knoll et al., 1995). Yeast colonies containing each of the LACS expression constructs were selected and cultured as described in “Materials and Methods.” After 4 d at 30°C, seven of the 11 candidate cDNAs had complemented the mutant phenotype and restored growth rates to wild-type levels, as compared with the wild-type strain Invisc (Invitrogen, Carlsbad, CA) that was used as a positive control. Only LACS6, LACS7, At4g14070, and At3g23790 did not complement the mutant phenotype (data not shown). Ten other cDNAs of the Arabidopsis AMPBP superfamily were also cloned and tested. These cDNAs represent 10 of the 14 members of the unique clade of sequences of unknown function described above. The encoded proteins did not contain the linker domain, but did display approximately 25% amino acid identity to known LACS enzymes. None of these cDNAs successfully complemented the mutant yeast (data not shown).
In general, however, the results of the complementation experiment suggested that most of the candidate cDNA sequences were in fact LACSs. Successful phenotypic restoration in YB525 is dependent on subcellular targeting, compatible substrate specificity, and other factors. Some LACS genes may not complement the phenotype of YB525, as evidenced by the inability of the remaining endogenous yeast LACS enzymes, Faa2p and Faa3p, to support cell growth under these conditions. Faa2p is targeted to peroxisomes. The subcellular targeting of Faa3p is not known, but the enzyme apparently has access only to endogenously synthesized fatty acids (Knoll et al., 1995). To definitively establish the identity of the candidate cDNAs, the yeast expression constructs were used to directly test the encoded enzymatic activities.
Measurement of Acyl-CoA Synthetase Enzyme Activity by in Vitro Assays
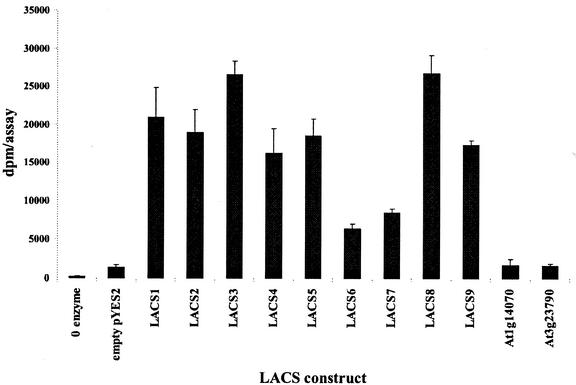
Cell-free lysates were prepared from yeast YB525 cells expressing each of the 11 candidate LACS genes, as described in “Materials and Methods.” These lysates served as enzyme sources in LACS activity assays, using 1-[14C]oleic acid as a substrate. The results of these assays are shown in Figure 3. LACS6 and LACS7, in contrast to the complementation study, did produce active enzymes. These results proved unequivocally that the other seven members of this family are LACSs as well, and that the linker domain described above is a reliable tool for distinguishing LACS genes from other related AMPBP genes. Therefore, this family represents the largest LACS gene family yet described in a single species, surpassing even that of humans, currently known to contain six genes that encode LACS or VLCS (very LACS; Steinberg et al., 2000). Consistent with their inability to complement yeast YB525, all 10 representative cDNAs from the unique clade of AMPBP genes described above did not produce measurable levels of enzyme activity (data not shown).
Figure 3.
LACS enzyme activity measurements from cell-free lysates of overexpressing yeast strains. Gal-induced liquid cultures for each LACS construct were harvested, and spheroplasts prepared and lysed by sonication. Cell-free extracts were used as enzyme sources in in vitro LACS enzyme assays, using 1-[14C]oleic acid as a substrate. Levels of activity were measured as the numbers of aqueous-soluble counts converted per assay. Each construct was assayed in triplicate. The error bar represents the sd.
Determination of Fatty Acid Substrate Specificities for LACS Enzymes
One possible explanation for the abundance of LACS genes in Arabidopsis could be that some or all of the encoded enzymes possess distinct fatty acid substrate specificities. We addressed this possibility by cloning each of the LACS cDNAs in prokaryotic expression vectors and overexpressing the enzymes in E. coli. This organism was chosen instead of yeast because a mutant strain that completely lacks endogenous acyl-CoA synthetase activity is available. This mutant, called K27 (Overath et al., 1969), can be obtained from the American Type Culture Collection. After induction with isopropylthio-β-galactoside, the membrane fractions from lysed cells were isolated by ultracentrifugation. The membranes were used in in vitro enzyme assays using eight different radioactive fatty acid substrates, ranging in length from 14 to 20 carbons, and spanning a range of desaturation, from zero to three double bonds.
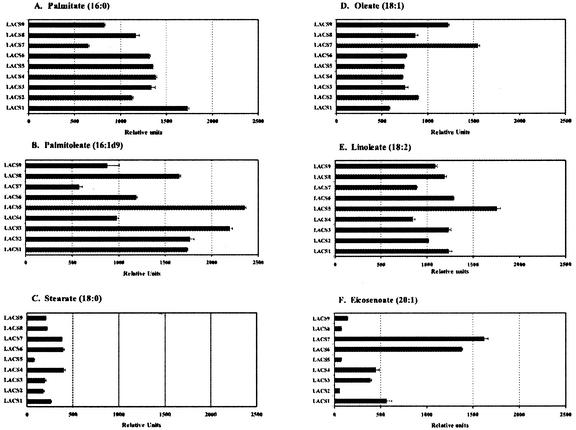
A summary of the specificities of the enzymes toward six of the eight fatty acids is shown in Figure 4. The remaining two fatty acids (myristate [14:0] and linolenate [18:3]) were activated at very similar rates by most of the enzymes and therefore were omitted for clarity. The putative peroxisomal enzymes LACS6 and LACS7 activated all the fatty acids tested at relatively high rates. Especially noteworthy was the strong activity by LACS6 and LACS7 toward eicosenoic acid, a 20-carbon fatty acid found only in the seed storage lipids of Arabidopsis. Peroxisomal LACSs participate in β-oxidation, and therefore would be expected to effectively utilize all fatty acids stored in the seed TAGs. These two enzymes and the genes that encode them are described in additional detail in a separate report (Fulda et al., 2002). The other seven LACS enzymes showed very similar patterns of substrate preference, as shown in Figure 4. Each enzyme activated all of the substrates tested, with highest levels of activity observed with both the saturated and monounsaturated 16-carbon fatty acids and the monounsaturated and polyunsaturated 18-carbon fatty acids. LACS9 preferred oleic acid slightly more than any of the other fatty acids. This enzyme is the major plastidial isoform (Schnurr et al., 2002), and as such should effectively activate oleate, the most abundant fatty acid produced by the plastid fatty acid synthase complex in Arabidopsis. For most of these enzymes, stearate (18:0) and eicosenoate (20:1) were poor substrates. These data correlate very strongly with the fatty acid profiles seen in Arabidopsis leaf lipids, which consist mostly of monounsaturated and polyunsaturated 16- and 18-carbon acyl groups (Ohlrogge and Browse, 1995).
Figure 4.
Substrate specificity analysis of individual LACS enzymes. All nine Arabidopsis LACS genes were expressed in LACS-deficient E. coli. Membrane fractions were isolated and used as enzyme sources in in vitro enzyme assays using six different radioactive fatty acids. Enzyme levels were normalized as described in “Materials and Methods.” Enzyme activities were measured by liquid scintillation counting and converted into relative units, as described in “Materials and Methods,” to compensate for the differences in the expression levels of each enzyme and specific activities of each fatty acid. Each assay was performed in triplicate. The error bars represent the sds.
Analysis of LACS Tissue-Specific Expression Patterns
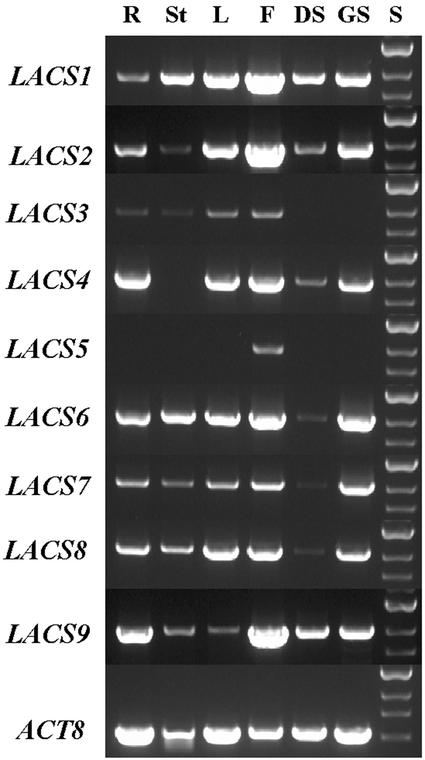
Our interest in the role of LACS in fatty acid metabolism in oilseeds would necessitate expression of these genes in the developing seeds. Northern-blot analysis indicated that some of the LACS genes were expressed at relatively low levels, making sensitive comparisons between them difficult (data not shown). Therefore, we examined the tissue-specific RNA expression profiles of each of the LACS genes by semiquantitative reverse transcription (RT)-PCR (Kong et al., 1999). Each gene was analyzed using RNA from tissue culture-grown roots, leaves, stems, flowers, developing seeds, and 2 d-old germinating seedlings, as described in “Materials and Methods.” A composite of the agarose gel ethidium bromide fluorescence images is shown in Figure 5. The Arabidopsis actin ACT8 gene (An et al., 1996) was used as a control. As seen in Figure 5, most of the LACS genes are expressed in a variety of tissues at widely varying levels. Close inspection of the data presented in Figure 5 reveals several interesting phenomena. First, several LACS genes are expressed in the germinating seedlings. This observation is consistent with a strong demand for the enzymes of β-oxidation and membrane lipid biosynthesis in the emerging seedling, in which rapid cell expansion and division occurs. The second interesting pattern observed is the strength of expression of all 11 genes in flowers. A strong demand for LACS activities in floral tissues correlates well with the strong expression levels of various other lipid metabolic genes in flowers (Engeseth et al., 1996; Dormann et al., 2000), which suggests that this organ is very active with respect to fatty acid metabolism. LACS5 is expressed strongly in flowers, and is unique within this set of genes in that its transcript is undetectable in any other tissue. Also, several of the genes are expressed in developing seeds. LACS1, LACS2, LACS4, and LACS9 showed highest levels of expression, whereas LACS6, LACS7, and LACS8 were expressed at much lower levels. This observation suggests that many genes in this family may participate in glycerolipid synthesis in the developing seed.
Figure 5.
Comparison of tissue-specific expression levels of LACS genes. Aliquots of total RNA from each organ or tissue were analyzed for expression of each LACS gene, as well as the actin ACT8 gene as a positive control, by semiquantitative RT-PCR using gene-specific primer pairs. The yield of each product was measured while still in the linear range. R, Root; St, stem; L; leaf; F, flower; DS, developing seed; GS, germinating seedling; S, size marker.
The expression patterns of the LACS gene family was also analyzed in another way by taking advantage of the large collection of EST sequences that are available in the Arabidopsis databases. These data sets can be used to investigate the expression levels of a particular gene or many genes by comparing the relative abundance of ESTs for each gene. This type of analysis, often called “digital northerns,” may also reveal characteristics of the flux rates through a pathway or clues regarding the regulation of certain enzymes (Ohlrogge and Benning, 2000). The data in Figure 5 clearly show that none of the LACS genes in Arabidopsis is seed specific. However, we felt that analysis of the EST populations, especially those derived from green siliques (Asamizu et al., 2000) and developing seeds of Arabidopsis (White et al., 2000), might provide additional insight regarding which LACS genes play roles in lipid synthesis during seed development. The last column in Table I provides a summary of the tissue specificity of the LACS ESTs. Nearly all the genes are represented in the green silique and/or developing seed EST populations, with the exceptions of LACS3, LACS5, and LACS8. These data confirmed those observed for these three genes in Figure 5. Quantitatively, LACS1 was by far the predominantly expressed LACS gene. LACS1 was represented by 15 ESTs from green siliques or developing seeds, compared with five ESTs for LACS2 and four ESTs for LACS9. As a whole, both the digital and RT-PCR data confirm that LACS1, LACS2, and LACS9 are the quantitatively significant genes in developing seeds, and as such, may be of particular interest with regard to their roles in fatty acyl-CoA metabolism in this tissue.
DISCUSSION
A greater understanding of the control of fatty acid metabolism in higher plants has long been confounded by the lack of detailed molecular information regarding the enzymes that catalyze many of the known anabolic and catabolic reactions that utilize fatty acids and their derivatives. Genomics studies have illuminated the complexity of these pathways. For example, systematic analysis of the annotated Arabidopsis genome sequence has identified more than 40 genes that encode putative lipase or hydrolase enzymes that are likely involved in the production of free fatty acids. Therefore, LACS plays a pivotal role in fatty acid utilization by activating free fatty acids to the corresponding CoA thioesters, which are the preferred substrates for most fatty acid metabolizing enzymes. We describe here the cloning and initial characterization of nine cDNAs from Arabidopsis that encode active LACS enzymes, plus an additional two cDNAs that possess strong sequence similarity to the others yet do not display LACS activity under the conditions used in our experiments.
This subset of sequences was selected from a much larger group of genes known as the AMPBP gene superfamily, which share a mechanism step in carboxylic acid activation via adenylation (Babbitt et al., 1992). The initially daunting task of cloning and characterizing 44 genes was simplified via sequence analysis of the predicted amino acid sequences of the genes in this family. Eukaryotic long-chain LACS genes are unique within the AMPBP superfamily in that only this subfamily contains a linker domain, a motif of between 30 and 70 amino acid residues that links two larger domains that are broadly conserved across most members of the superfamily. This linker domain is present in many other eukaryotic LACSs, but not in the short-chain, medium-chain, or very LACS enzymes from eukaryotic organisms. Eleven of the 44 genes in the AMPBP superfamily contained the linker domain and were chosen for further characterization. Phylogenetic comparisons of the candidate LACS sequences indicated a great deal of heterogeneity. The central and C-terminal regions of the proteins were generally more conserved, whereas the N-terminal domains varied considerably in length, primarily due to the presence of N-terminal extensions of 30 to 60 residues in some of the amino acid sequences. These extensions may serve to target the relevant LACS enzymes to specific subcellular compartments within the cell. These data correlate very well with the body of biochemical literature from plants, which suggests that many organelles contain one or more LACS activities.
To confirm the enzymatic activity of the 11 candidate cDNAs, each was expressed in yeast strain YB525. This strain lacks the LACS activities necessary to activate exogenous fatty acids. When grown on media containing long-chain (C14–C18) fatty acids as a sole carbon source and cerulenin (which inhibits endogenous fatty acid synthesis), complementation with active LACS genes is necessary to restore growth to the organism (Knoll et al., 1995). Seven of the LACS genes effectively complemented the growth phenotype of the mutant yeast, suggesting that these genes were in fact LACSs. Only LACS6, LACS7, and the At4g14070 and At3g23790 genes did not restore growth. All 11 constructs were also directly tested for their ability to produce active LACS enzymes by performing in vitro LACS assays, using cell-free lysates from the overexpressing yeast strains. Nine of the 11 genes, including all the genes that complemented the yeast mutant, and LACS6 and LACS7, produced high levels of LACS enzyme activity using 1-[14C]oleate as a substrate, confirming that all nine genes encode LACS enzymes. LACS6 and LACS7 contain PTS2 and PTS1 peroxisome targeting sequences, respectively, thus possibly explaining their inability to complement the YB525 phenotype. Targeting of a LACS to the peroxisome may render the enzyme inaccessible to the pool of exogenous fatty acids, as evidenced by the inability of Faa2p, the endogenous yeast peroxisomal LACS (Johnson et al., 1994; Knoll et al., 1995), to support growth under the conditions used in this experiment.
Only the At4g14070 and At3g23790 genes were ineffective in both the yeast complementation and in vitro enzyme assay experiments. The results for these two genes were consistent with the unsuccessful E. coli expression studies using a B. napus homolog of these genes (Fulda et al., 1997), and provide further support for the hypothesis that the enzymes encoded by At4g14070 and At3g23790 are somehow different from the other nine LACS genes. These genes may encode LACSs that activate specialized substrates, or they may encode a different type of enzyme related to LACS. It is also possible that these genes are LACSs whose protein products are inactive under the conditions used in our experiments due to special protein folding or subunit multimer formation requirements, or the need for posttranslational modifications not met by the cellular machinery of yeast or E. coli.
The fatty acid substrate specificity for each of the LACS enzymes was also determined. All nine LACSs were active in the membrane fraction from E. coli cells. Analysis of the relative activities of each enzyme was tested using a variety of fatty acid substrates. The specificity profiles were quite similar for many of the LACS enzymes, as shown in Figure 4. Saturated and unsaturated 16-carbon and unsaturated 18-carbon fatty acids were the preferred substrates. Eicosenoic acid (20:1) and stearic acid (18:0) were poor substrates for most of the enzymes. Two notable exceptions to this general profile were LACS6 and LACS7, which showed much stronger levels of activity toward eicosenoate, a seed-specific fatty acid, while still maintaining high activity toward all other fatty acids as well. These two isoforms are targeted to the peroxisome, and may be important in β-oxidation (Fulda et al., 2002). In general, the fatty acid preferences for these enzymes correlate very well with the observed fatty acid compositions of Arabidopsis membrane and seed storage lipids, which are made up primarily of 16:0, 18:0, 18:1, 18:2, 18:3, and 20:1. The lack of striking substrate specificity differences between the different isoforms suggests that the specific roles fulfilled by each enzyme are not determined by substrate preference but by other factors such as subcellular targeting, or differences in temporal or cell-type expression.
The tissue-specific expression patterns of the LACS genes were also investigated by semiquantitative RT-PCR. With few exceptions, each member of the gene family was expressed in numerous tissues, often at widely varying levels. Analysis of gene expression at the cell-type level and enzyme targeting at the subcellular level may be necessary to determine if different isoforms are expressed in distinct parts of the same tissues or organs. Only then will we gain a better understanding of the precise roles that each LACS gene fulfills. Only one gene, LACS5, was expressed in a tissue-specific manner. This gene was expressed at very high levels in flowers. Every other member of the gene family was also expressed in flowers, suggesting a high level of acyl-CoA metabolism in floral tissues. We were particularly interested in the expression patterns in the developing seeds and germinating seedlings. Germinating seedlings depend heavily on β-oxidation. The peroxisomal β-oxidation pathway utilizes fatty acids from TAG reserves to provide cellular energy and carbon skeletons for the emerging seedling during germination. One or more LACS isoforms may be needed to fulfill this role. Two peroxisomal genes, LACS6 and LACS7, are currently under investigation in this respect (Fulda et al., 2002). Developing seeds are the site of TAG deposition and as such have a strong demand for fatty acid biosynthesis to supply the acyltransferases that produce TAG. Therefore, the plastidial isoform(s) of LACS will play an important role in activating the fatty acids synthesized de novo in the developing seeds. LACS9 has been identified as the major plastidial isoform. LACS9 is characterized in more detail in a separate report (Schnurr et al., 2002).
We feel that based on the preceding criteria, the genes described in this report represent the entire LACS gene family from Arabidopsis. Other classes of uncharacterized genes belonging to the AMPBP superfamily were also tested but failed to produce active LACS enzymes. Cloning of the entire family of LACS genes from Arabidopsis opens up avenues of study that were previously impossible. Future experiments will be designed to investigate the cell type-specific expression and subcellular targeting of each of the LACS genes. The contributions of individual LACS genes to specific fatty acid metabolic pathways will be also be investigated through reverse genetic studies and antisense suppression analyses. This family of LACS genes will serve as powerful tools that will allow us to gain a greater understanding of glycerolipid biosynthesis and fatty acid metabolism as a whole.
MATERIALS AND METHODS
Sequencing and Sequence Homology Analysis
All DNA sequencing was conducted in the Macromolecular Analysis Laboratory (Washington State University) using automated sequencing equipment (Applied Biosystems, Foster City, CA). Sequences were assembled and modified using the GCG suite of programs (Wisconsin Package Version 10.0, Genetics Computer Group). Database homology searches were conducted against the AtDB Illustra database (http://genome-www.stanford.edu/Arabidopsis/), and its successor at The Arabidopsis Information Resource (http://www.Arabidopsis.org/). For subcellular targeting predictions, protein sequences were analyzed using either TargetP (http://www. cbs.dtu.dk/services/TargetP/; Emanuelsson et al., 2000) or ChloroP (http://www.cbs.dtu.dk/services/ChloroP/; Emanuelsson et al., 1999). MIPS designations refer to the nomenclature used at the Munich Information Center for Protein Sequences Arabidopsis database (http://mips.gsf.de/proj/thal/db/search/search_frame.html). Protein sequence alignments were conducted using the ClustalX program (Thompson et al., 1997) and phylogenetic trees drawn from the alignments using TREEVIEW (Page, 1996).
Identification and Cloning of LACS Genes
Full-length LACS clones were isolated by first screening the EST databases (Newman et al., 1994) to identify partial cDNA clones with homology to known LACSs. The inserts from these clones were used to screen for full-length clones present in any of various cDNA libraries available from the Arabidopsis Biological Resource Center (Weigel et al., 1992; Kieber et al., 1993). When full-length clones could not be identified using this approach, the missing portions of the genes were identified by isolation of genomic clones from an Arabidopsis genomic DNA library (Voytas et al., 1990). Once the initiator codon of each gene had been determined, a new gene-specific oligonucleotide primer pair was used to isolate RT-PCR products spanning the full-length open reading frame.
Cloning of Arabidopsis LACS Genes in Escherichia coli and Yeast (Saccharomyces cerevisiae)
For expression in yeast, one of two methods was used to reamplify the open reading frames of the Arabidopsis cDNAs for recloning. Some genes were amplified from the original plasmids using new oligonucleotide primer pairs that introduced restriction sites compatible for insertion into the multiple cloning site of the yeast-inducible expression vector pYES2 (Invitrogen). The PCR products were restricted and ligated to appropriately digested pYES2 DNA (all LACS genes except LACS4) or pRS426 DNA (LACS4) and transformed into competent E. coli. Plasmid DNA from the resulting bacterial colonies was transformed into yeast YB525 cells (generously provided by Prof. J.I. Gordon, Washington University, St. Louis; Knoll et al., 1995) that had been made competent for chemical transformation using the S.c. EasyComp kit (Invitrogen). Alternatively, PCR products for some of the LACS cDNAs were generated using the sticky-end PCR technique (Zeng, 1998). These products were ligated and transformed as described above.
The LACS genes were also cloned into pET24 vectors for expression in E. coli. PCR products were cloned, either by restriction or sticky-end ligation, into either pET24c or pET24d (Novagen, Madison, WI). Each plasmid was then transformed into competent cells prepared from the K27 mutant of E. coli (Overath et al., 1969). This mutant had first been made to express the T7 RNA polymerase gene from λDE3 by directed integration of the prophage into the E coli chromosome using the DE3 lysogenization kit (Novagen).
Yeast Complementation
The expression constructs were transformed into chemically competent YB525 cells and uracil auxotrophs selected on Dropout base agar-uracil (DOBA)-plates (DOBA: 2% [w/v] yeast nitrogen base, 2% [w/v] dextrose, 0.1% [w/v] complete supplement mixture lacking uracil, and 17g L−1 agar; Bio 101, Vista, CA). Representative colonies were chosen at random and grown until mid- to late-log phase in Dropout base liquid medium (DOBA minus agar). Gal was added to a concentration of 2% (w/v) to induce high-level expression of the transgenes from the GAL1 promoter of the vector. The cultures were then grown for an additional 2 to 4 h. Aliquots of each culture were diluted 1:1 (v/v) with 2 m sorbitol and 5-μL aliquots plated on DOBA plates containing Gal plus 500 μm myristic acid and 25 μm cerulenin, followed by incubation at 30°C for 3 to 4 d.
Enzyme Overproduction in Yeast
Transformed YB525 cells were selected, grown in liquid medium, and induced as in the previous section. The cells were harvested by centrifugation, washed once with distilled water, and harvested again for spheroplast production. Spheroplasts were generated from intact cells using lytic enzyme (ICN Pharmaceuticals, Aurora, OH) following the manufacturer's protocol. The spheroplasts were lysed by sonication on ice (2 × 1 min) followed by removal of solid debris by centrifugation at 8,000g for 15 min at 4°C. The resulting supernatants were used as enzyme sources for the LACS assay.
Enzyme Overproduction in E. coli
For production of the LACS enzymes in E. coli, starter cultures for each of the LACS-pET24 constructs were grown overnight at 37°C in LB media containing 50 μg mL−1 kanamycin. Aliquots from these cultures were used to inoculate 100-mL cultures that were grown at 22°C until mid-log phase had been reached. The cultures were induced to express the LACS enzymes with 1 mm isopropylthio-β-galactoside and growth was continued for 16 h. The cells were harvested, and lysed by sonication for 1 min. The cellular debris was removed by centrifugation at 4,300g for 15 min. The membrane fraction was isolated from the low-speed supernatant by ultracentrifugation at 100,000g for 1 h. The membrane pellet was resuspended in 50 mm Tris-HCl (pH 8.0) containing 20% (v/v) glycerol.
To normalize the levels of enzyme activity, each membrane fraction was used in trial enzyme assays to determine the amount necessary to utilize approximately 100,000 dpm of oleic acid substrate in a 10-min assay. The level of activity of each enzyme for each fatty acid was then multiplied by the relevant conversion factor, where a conversion factor of 1.00 equaled 100,000-dpm activity using oleic acid. For conversion of the oleic acid activity data to relative units, it was normalized against the palmitic acid data.
LACS in Vitro Enzyme Assay
The LACS enzyme assay was conducted in 1.5-mL Eppendorf tubes (Eppendorf Scientific, Westbury, NY) in a volume of 100 μL. The assay mixture contained 100 mm Bis-Tris-propane (pH 7.6), 10 mm MgCl2, 5 mm ATP, 2.5 mm dithiothreitol, 1 mm CoA, 30 μm 1-[14C] fatty acid (specific activity 50–57 mCi mmol−1, PerkinElmer Life Sciences, Boston, MA), or 9,10-[3H] fatty acid (specific activity 60 Ci mmol−1, American Radiolabeled Chemicals, St. Louis), and 20 μg of crude yeast cell lysate protein or 0.02 to 15 μg E. coli membrane protein. The assay was initiated by addition of the fatty acid and incubated at room temperature for 10 min for the E. coli assays or 15 min for the yeast assays. The reactions were stopped by addition of 100 μL of 10% (v/v) acetic acid in isopropanol and extracted twice with 900 μL of hexane (previously saturated with 50% [v/v] isopropanol). Enzyme activity was measured by analyzing aliquots of the aqueous phase by liquid scintillation counting. Lysates from yeast cells or E. coli cells bearing the appropriate empty vector served as negative controls, whereas commercial ACS enzyme from Pseudomonas sp. (Sigma, St. Louis) served as the positive control.
Analysis of Tissue-Specific LACS Gene Expression
The tissue-specific expression patterns for each LACS gene were analyzed by semiquantitative RT-PCR (Kong et al., 1999). RNA preparations from developing seeds, 2-d-old germinating seedlings, roots, young leaves, stems, and flowers were quantified spectrophotometrically and 2-μg aliquots of each used as template for RT, as described above. One microliter of each RT reaction was used as template in a 50-μL PCR reaction containing gene-specific primers. The amplification conditions were as follows: 95°C 3 min, and 30 cycles of 94°C for 15 s, 55°C for 30 s, and 72°C for 1 min. Fifteen percent of each reaction was analyzed by Tris-acetate EDTA-agarose gel electrophoresis and the degree of gene expression correlated to the relative intensity of each band as determined by visual comparison of the ethidium bromide staining intensity when the gels were visualized under UV illumination. The actin gene ACT8 (An et al., 1996) was used as a control.
ACKNOWLEDGMENTS
We thank the National Science Foundation, Dow Chemical Company and Dow AgroSciences, and the Agricultural Research Center at Washington State University for financial support. We also thank Judy Schnurr for her assistance in sequence analysis.
Footnotes
This work was supported in part by the National Science Foundation (postdoctoral fellowship to J.M.S., grant no. BIR–9627559), by Dow Chemical Company/Dow AgroSciences (grant to J.A.B.), by the U.S. Department of Agriculture (grant no. USDA–NRI 2001–35318–10186 to J.A.B.), and by the Agricultural Research Center, Washington State University.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.003269.
LITERATURE CITED
- Abe T, Fujino T, Fukuyama R, Minoshima S, Shimizu N, Toh H, Suzuki H, Yamamoto T. Human long-chain acyl-CoA synthetase: structure and chromosomal location. J Biochem. 1992;111:123–128. doi: 10.1093/oxfordjournals.jbchem.a123707. [DOI] [PubMed] [Google Scholar]
- An YQ, McDowell JM, Huang S, McKinney EC, Chambliss S, Meagher RB. Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J. 1996;10:107–121. doi: 10.1046/j.1365-313x.1996.10010107.x. [DOI] [PubMed] [Google Scholar]
- Andrews J, Keegstra K. Acyl-coenzyme A synthetase is located in the outer membrane and acyl-CoA thioesterase in the inner membranes of pea chloroplast envelopes Pisum sativum. Plant Physiol. 1983;72:735–740. doi: 10.1104/pp.72.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asamizu E, Nakamura Y, Sato S, Tabata S. A large scale analysis of cDNA in Arabidopsis thaliana: generation of 12,028 non-redundant expressed sequence tags from normalized and size-selected cDNA libraries. DNA Res. 2000;7:175–180. doi: 10.1093/dnares/7.3.175. [DOI] [PubMed] [Google Scholar]
- Babbitt PC, Kenyon GL, Martin BM, Charest H, Slyvestre M, Scholten JD, Chang KH, Liang PH, Dunaway-Mariano D. Ancestry of the 4-chlorobenzoate dehalogenase: analysis of amino acid sequence identities among families of acyl:adenyl ligases, enoyl-CoA hydratases/isomerases, and acyl-CoA thioesterases. Biochemistry. 1992;31:5594–5604. doi: 10.1021/bi00139a024. [DOI] [PubMed] [Google Scholar]
- Beevers H. Glyoxysomes of castor bean endosperm and their relation to gluconeogenesis. Ann NY Acad Sci. 1969;168:313–324. doi: 10.1111/j.1749-6632.1969.tb43118.x. [DOI] [PubMed] [Google Scholar]
- Bibb MJ, Sherman DH, Omura S, Hopwood DA. Cloning, sequencing and deduced functions of a cluster of Streptomyces genes probably encoding biosynthesis of the polyketide antibiotic frenolicin. Gene. 1994;142:31–39. doi: 10.1016/0378-1119(94)90351-4. [DOI] [PubMed] [Google Scholar]
- Black PN, DiRusso CC, Metzger AK, Heimert TL. Cloning, sequencing, and expression of the fadD gene of Escherichia coli encoding acyl coenzyme A synthetase. J Biol Chem. 1992;267:25513–25520. [PubMed] [Google Scholar]
- Conti E, Franks NP, Brick P. Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes. Structure. 1996;4:287–298. doi: 10.1016/s0969-2126(96)00033-0. [DOI] [PubMed] [Google Scholar]
- Conti E, Stachelhaus T, Marahiel MA, Brick P. Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidin S. EMBO J. 1997;16:4174–4183. doi: 10.1093/emboj/16.14.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann P, Voelker TA, Ohlrogge JB. Accumulation of palmitate in Arabidopsis mediated by the acyl-acyl carrier protein thioesterase FATB1. Plant Physiol. 2000;123:637–644. doi: 10.1104/pp.123.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond PJ, Germain V, Lange PR, Bryce JH, Smith SM, Graham IA. Postgerminative growth and lipid catabolism in oilseeds lacking the glyoxylate cycle. Proc Natl Acad Sci USA. 2000;97:5669–5674. doi: 10.1073/pnas.97.10.5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton S, Bartlett K, Pourfarzam M. Mammalian mitochondrial beta-oxidation. Biochem J. 1996;320:345–357. doi: 10.1042/bj3200345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlting J, Buttner D, Wang Q, Douglas CJ, Somssich IE, Kombrink E. Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J. 1999;19:9–20. doi: 10.1046/j.1365-313x.1999.00491.x. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, Heijne Gv. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Heijne Gv. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Prot Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engeseth NJ, Pacovsky RS, Newman T, Ohlrogge JB. Characterization of an acyl-CoA-binding protein from Arabidopsis thaliana. Arch Biochem Biophys. 1996;331:55–62. doi: 10.1006/abbi.1996.0282. [DOI] [PubMed] [Google Scholar]
- Fargeman NJ, Black PN, Zhao XD, Knudsen J, DiRusso CC. The acyl-CoA synthetases encoded within FAA1 and FAA4 in Saccharomyces cerevisiae function as components of the fatty acid transport system linking import, activation, and intracellular utilization. J Biol Chem. 2001;276:37051–37059. doi: 10.1074/jbc.M100884200. [DOI] [PubMed] [Google Scholar]
- Frentzen M, Neuburger M, Joyard J, Douce R. Intraorganelle localization and substrate specificities of the mitochondrial acyl-CoA: sn-glycerol-3-phosphate O-acyltransferase and acyl-CoA: 1-acyl-sn-glycerol-3-phosphate O-acyltransferase from potato tubers and pea leaves. Eur J Biochem. 1990;187:395–402. doi: 10.1111/j.1432-1033.1990.tb15317.x. [DOI] [PubMed] [Google Scholar]
- Fujino T, Yamamoto T. Cloning and functional expression of a novel long-chain acyl-CoA synthetase expressed in brain. J Biochem. 1992;111:197–203. doi: 10.1093/oxfordjournals.jbchem.a123737. [DOI] [PubMed] [Google Scholar]
- Fulda M, Heinz E, Wolter FP. The fadD gene of Escherichia coli K12 is located close to rnd at 39.6 min of the chromosomal map and is a new member of the AMP-binding protein family. Mol Gen Genet. 1994;242:241–249. doi: 10.1007/BF00280412. [DOI] [PubMed] [Google Scholar]
- Fulda M, Heinz E, Wolter FP. Brassica napus cDNAs encoding fatty acyl-CoA synthetase. Plant Mol Biol. 1997;33:911–922. doi: 10.1023/a:1005780529307. [DOI] [PubMed] [Google Scholar]
- Fulda M, Shockey J, Werber M, Wolter FP, Heinz E (2002) Two acyl-CoA synthetases from Arabidopsis thaliana involved in peroxisomal fatty acid beta-oxidation. Plant J (in press) [DOI] [PubMed]
- Gerhardt B. Fatty acid degradation in plants. Prog Lipid Res. 1992;31:417–446. doi: 10.1016/0163-7827(92)90004-3. [DOI] [PubMed] [Google Scholar]
- Gordon JI, Duronio RJ, Rudnick DA, Adams SP, Gokel GW. Protein N-myristoylation. J Biol Chem. 1991;266:8647–8650. [PubMed] [Google Scholar]
- Groot PH, Scholte HR, Hulsmann WC. Fatty acid activation: specificity, localization, and function. Adv Lipid Res. 1976;14:75–126. doi: 10.1016/b978-0-12-024914-5.50009-7. [DOI] [PubMed] [Google Scholar]
- Hettema EH, Tabak HF. Transport of fatty acids and metabolites across the peroxisomal membrane. Biochim Biophys Acta. 2000;1486:18–27. doi: 10.1016/s1388-1981(00)00045-7. [DOI] [PubMed] [Google Scholar]
- Hills MJ, Beevers H. ATPase in lipid body membranes of castor bean endosperm. Plant Physiol. 1986;82:671–674. doi: 10.1104/pp.82.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihara K, Yamane K, Hirano E. Acyl-CoA synthetase in oilseeds: fatty acid structural requirements for activity and selectivity. Plant Cell Physiol. 1997;38:717–724. [Google Scholar]
- Iijima H, Fujino T, Minekura H, Suzuki H, Kang MJ, Yamamoto T. Biochemical studies of two rat acyl-CoA synthetases, ACS1 and ACS2. Eur J Biochem. 1996;242:186–190. doi: 10.1111/j.1432-1033.1996.0186r.x. [DOI] [PubMed] [Google Scholar]
- Ishitani M, Liu J, Halfter U, Kim CS, Shi W, Zhu JK. SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell. 2000;12:1667–1678. doi: 10.1105/tpc.12.9.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DR, Knoll LJ, Levin DE, Gordon JI. Saccharomyces cerevisiae contains four fatty acid activation (FAA) genes: an assessment of their role in regulating protein N-myristoylation and cellular lipid metabolism. J Cell Biol. 1994;127:751–762. doi: 10.1083/jcb.127.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MJ, Fujino T, Sasano H, Minekura H, Yabuki N, Nagura H, Iijima H, Yamamoto TT. A novel arachidonate-preferring acyl-CoA synthetase is present in steroidogenic cells of the rat adrenal, ovary, and testis. Proc Natl Acad Sci USA. 1997;94:2880–2884. doi: 10.1073/pnas.94.7.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke J, Behal RH, Back SL, Nikolau BJ, Wurtele ES, Oliver DJ. The role of pyruvate dehydrogenase and acetyl-coenzyme A synthetase in fatty acid synthesis in developing Arabidopsis seeds. Plant Physiol. 2000;123:497–508. doi: 10.1104/pp.123.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- Knoll LJ, Johnson DR, Gordon JI. Complementation of Saccharomyces cerevisiae strains containing fatty acid activation gene (FAA) deletions with a mammalian acyl-CoA synthetase. J Biol Chem. 1995;270:10861–10867. doi: 10.1074/jbc.270.18.10861. [DOI] [PubMed] [Google Scholar]
- Kong SE, Hall JC, McCauley RD. Estimation of gene expression within the intestinal mucosa using semiquantitative reverse transcriptase-polymerase chain reaction. Anal Biochem. 1999;271:111–114. doi: 10.1006/abio.1999.4123. [DOI] [PubMed] [Google Scholar]
- Lee D, Ellard M, Wanner LA, Davis KR, Douglas CJ. The Arabidopsis thaliana 4-coumarate:CoA ligase (4CL) gene: stress and developmentally regulated expression and nucleotide sequence of its cDNA. Plant Mol Biol. 1995;28:871–884. doi: 10.1007/BF00042072. [DOI] [PubMed] [Google Scholar]
- Lin YH, Yu C, Huang AHC. Substrate specificities of lipases from corn and other seeds. Arch Biochem Biophys. 1986;244:346–356. doi: 10.1016/0003-9861(86)90123-2. [DOI] [PubMed] [Google Scholar]
- Martin ML, Busconi L. Membrane localization of a rice calcium-dependent protein kinase (CDPK) is mediated by myristoylation and palmitoylation. Plant J. 2000;24:429–435. doi: 10.1046/j.1365-313x.2000.00889.x. [DOI] [PubMed] [Google Scholar]
- Newman T, de Bruijn FJ, Green P, Keegstra K, Kende H, McIntosh L, Ohlrogge J, Raikhel N, Somerville S, Thomashow M et al. Genes galore: a summary of methods for accessing results from large-scale partial sequencing of anonymous Arabidopsis cDNA clones. Plant Physiol. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge J, Benning C. Unraveling plant metabolism by EST analysis. Curr Opin Plant Biol. 2000;3:224–228. [PubMed] [Google Scholar]
- Ohlrogge J, Browse J. Lipid biosynthesis. Plant Cell. 1995;7:957–970. doi: 10.1105/tpc.7.7.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JA, Lusk KR. Acyl-CoA synthetase activity associated with rapeseed lipid body membranes. Phytochemistry. 1994;36:7–9. [Google Scholar]
- Overath P, Pauli G, Schairer HU. Fatty acid degradation in Escherichia coli. An inducible acyl-CoA synthetase, the mapping of old-mutations, and the isolation of regulatory mutants. Eur J Biochem. 1969;7:559–574. [PubMed] [Google Scholar]
- Page R. TREEVIEW: an application to display phylogenetic trees on personal computers. Comp Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Pollard M, Ohlrogge J. Testing models of fatty acid transfer and lipid synthesis in spinach leaf using in vivo oxygen-18 labeling. Plant Physiol. 1999;121:1217–1226. doi: 10.1104/pp.121.4.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongdontri P, Hills M. Characterization of a novel plant acyl-coA synthetase that is expressed in lipogenic tissues of Brassica napus L. Plant Mol Biol. 2001;47:717–726. doi: 10.1023/a:1013652014744. [DOI] [PubMed] [Google Scholar]
- Schnurr JA, Shockey JM, de Boer G-J, Browse JA. Fatty acid export from the chloroplast: molecular characterization of a major plastidial acyl-coenzymeA synthetase from Arabidopsis. Plant Physiol. 2002;129:1700–1709. doi: 10.1104/pp.003251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C, Browse J. Plant lipids: metabolism, mutants, and membranes. Science. 1991;252:80–87. doi: 10.1126/science.252.5002.80. [DOI] [PubMed] [Google Scholar]
- Steinberg SJ, Morgenthaler J, Heinzer AK, Smith KD, Watkins PA. Very long-chain acyl-CoA synthetases: human “bubblegum” represents a new family of proteins capable of activating very long-chain fatty acids. J Biol Chem. 2000;275:35162–35169. doi: 10.1074/jbc.M006403200. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytas DF, Konieczny A, Cummings MP, Ausubel FM. The structure, distribution and evolution of the Ta1 retrotransposable element family of Arabidopsis thaliana. Genetics. 1990;126:713–721. doi: 10.1093/genetics/126.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM. LEAFY controls floral meristem identity in Arabidopsis. Cell. 1992;69:843–859. doi: 10.1016/0092-8674(92)90295-n. [DOI] [PubMed] [Google Scholar]
- White JA, Todd J, Newman T, Focks N, Girke T, de Ilarduya OM, Jaworski JG, Ohlrogge JB, Benning C. A new set of Arabidopsis expressed sequence tags from developing seeds. The metabolic pathway from carbohydrates to seed oil. Plant Physiol. 2000;124:1582–1594. doi: 10.1104/pp.124.4.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalovsky S, Rodriguez-Concepcion M, Gruissem W. Lipid modifications of proteins-slipping in and out of membranes. Trends Plant Sci. 1999;4:439–445. doi: 10.1016/s1360-1385(99)01492-2. [DOI] [PubMed] [Google Scholar]
- Zeng G. Sticky-end PCR: new method for subcloning. Biotechniques. 1998;25:206–208. doi: 10.2144/98252bm05. [DOI] [PubMed] [Google Scholar]