Abstract
The endogenous Δ12-desaturase gene (FAD2) in Arabidopsis was targeted for silencing using seed-specific cosuppression (CS), hairpin (HP) RNA (hpRNA), and intron-spliced HP (iHP) constructs. The iHP construct, incorporating the 120-bp 3′-untranslated region of the FAD2 gene, gave the highest degree of silencing. In some iHP lines Δ12-desaturase activity was reduced to levels as low as those in the null fad2-1 mutant, and every primary transformant showed a pronounced reduction in FAD2 activity. One highly silenced iHP line was propagated for five generations and showed no reversion or diminution in its degree of silencing. About 75% of plants transformed with the HP construct, targeting the FAD2 coding region, gave dramatically reduced Δ12-desaturase activity, whereas approximately 50% of plants transformed with the CS construct, containing the same coding region sequence, showed silencing at a much less profound level. In all three types of constructs, the degree of silencing was increased when the transgenes were homozygous, but this was much more pronounced for the CS constructs. All three types of construct could give a single locus that was capable of effective silencing, but in the one such CS line where this was the case, the locus had a complex insertion pattern. This is consistent with the concept that posttranscriptional gene silencing is induced by double-stranded, or self-complementary, RNA that is formed in cases of CS by complex insertion patterns at a single locus and that the most effective way of generating profoundly silenced plants is by the use of constructs that encode hpRNAs. Furthermore, these results demonstrate for the first time, to our knowledge, that iHP constructs targeted against an endogenous seed-expressed gene are clearly able to generate phenotypic changes that are inherited stably over several generations, making this approach a reliable technique for genetic modification of seed quality and possibly other traits in agricultural plants.
Cosuppression (CS), antisense suppression, and RNA-mediated virus resistance in plants appear to be different manifestations of posttranscriptional gene silencing (PTGS; Waterhouse et al., 1999, 2001; Matzke et al., 2001; Vance and Vaucheret, 2001). The PTGS mechanism is also present in other eukaryotes and termed RNA interference in nematodes (Fire et al., 1998) and fruitfly (Drosophila melanogaster; Kennerdel and Carthew, 1998) and quelling in fungi (Cogoni and Macino, 2000). It operates through dsRNA-directed sequence-specific degradation of ssRNA (Waterhouse et al., 1998; Elbashir et al., 2001), and the components and natural roles of the mechanism have been extensively reviewed (Sharp and Zamore, 2000; Waterhouse et al., 2001).
There are increasing reports of constructs specifically designed to express dsRNA in plants, usually in the form of self-complementary hairpin RNA (hpRNA), eliciting a high degree and frequency of PTGS of invading viruses, reporter transgenes, and endogenous genes (Waterhouse et al., 1998; Chuang and Meyerowitz, 2000; Smith et al., 2000; Wang et al., 2000; Wesley et al., 2001). Such hpRNA constructs have great potential as a tool for gene discovery and validation (Somerville, 2000; Wesley et al., 2001) and to improve crop traits (Wang et al., 2000). In the latter respect, the phenotypic stability of the hpRNA gene silencing over many generations is of crucial importance for the reliable application of gene silencing for crop improvement. The possibility of inactivation of transgenes through promoter methylation (Finnegan and McElroy, 1994) has been raised as a potential cause of phenotypic reversion of the transgene-encoded trait, a phenomenon that may gradually emerge over a number of generations. Although studies in Arabidopsis have shown that dsRNA-mediated gene silencing can be inherited in simple Mendelian fashion in the T2 generation when targeted against either the endogenous CLV3 and API genes (Chuang and Meyerowitz, 2000) or a stably integrated β-glucuronidase reporter transgene (Wang and Waterhouse, 2000), there are no studies reporting on the long-term phenotypic stability of such changes over several generations. In addition, there is relatively little information available comparing alternative hpRNA-encoding construct designs for their efficiency and effectiveness in silencing targeted plant genes of agricultural importance, in particular, for seed-expressed traits.
To examine these issues, we conducted a comparative study involving various gene-silencing constructs, targeted seed specifically, against the FAD2 gene in Arabidopsis. FAD2 encodes the microsomal fatty acid ω6-desaturase enzyme that inserts a double bond at the Δ12 position of oleic acid (C18:1Δ9) bound to phosphatidylcholine to produce linoleic acid (C18:2Δ9, 12) and, for this reason, is also referred to as a Δ12-desaturase. FAD2 is present as a single gene in the Arabidopsis genome, and well-characterized mutants are available that facilitate assessment of the relative degree of silencing achieved using PTGS approaches.
RESULTS
The Effects of Gene-Silencing Constructs on Δ12 Desaturation Levels in Seed from Primary Transformants
Δ12-Desaturase is highly active in developing seeds of nontransgenic Arabidopsis ecotype Columbia 2 (Table I), with 73% of 18:1 being converted to 18:2 and 18:3 for an oleic desaturation proportion (ODP) value of 0.73. In contrast, the fad2-1 mutant has an ODP value of 0.17, indicating about a 75% reduction in Δ12 desaturation. This is reflected in the large accumulation of the 18:1 substrate, up from 17% to 53% (Table I).
Table I.
Fatty acid profiles of selfed seed from T1 plants of Arabidopsis (ecotype Columbia 2) transformed with cosuppression (CS), hpRNA (HP), and intron-spliced hpRNA (iHP) constructs designed to silence FAD2
| Line | Fatty Acid Composition
|
ODP | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 16:0 | 18:0 | 18:1 | 18:2 | 18:3 | 20:0 | 20:1 | 22:1 | ||
| % | |||||||||
| Columbia 2 | 7.4 | 3.1 | 17.0 | 28.7 | 18.0 | 2.4 | 20.0 | 2.5 | 0.73 |
| fad2-1 | 5.6 | 2.9 | 52.9 | 3.0 | 7.8 | 1.6 | 23.4 | 1.7 | 0.17 |
| CS1 | 7.7 | 3.9 | 36.0 | 16.8 | 15.4 | 1.9 | 15.4 | 1.8 | 0.47 |
| CS2 | 7.4 | 3.6 | 42.2 | 13.3 | 13.3 | 1.8 | 15.2 | 1.6 | 0.39 |
| HP1 | 6.1 | 3.4 | 51.7 | 9.8 | 8.0 | 1.7 | 16.0 | 2.0 | 0.26 |
| HP2 | 5.2 | 3.3 | 54.8 | 5.5 | 7.8 | 1.8 | 18.5 | 1.8 | 0.19 |
| HP3 | 5.5 | 3.6 | 57.3 | 4.6 | 5.9 | 1.8 | 18.0 | 1.9 | 0.16 |
| iHP | 5.6 | 2.8 | 54.2 | 4.8 | 6.8 | 1.3 | 22.3 | 1.4 | 0.18 |
Profiles for Columbia 2 and the fad2-1 mutant are also shown. Minor fatty acids are not reported.
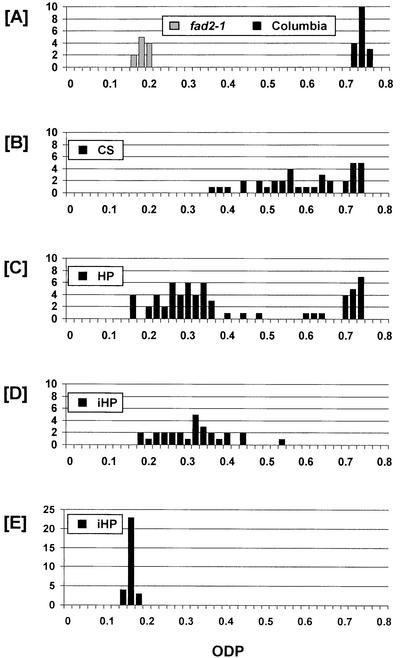
Arabidopsis plants were transformed with CS, hpRNA (HP), or intron-spliced hpRNA (iHP) constructs (Fig. 1) targeted at the FAD2 Δ12-desaturase gene. All three constructs used the rapeseed (Brassica napus) napin promoter (Stalberg et al., 1993), which directs high-level transcription of the transgenes specifically in the embryo and endosperm of the developing Arabidopsis seed. A proportion of plants transformed with each construct produced seed with significantly reduced Δ12 desaturation levels, and there were noticeable differences in effectiveness among the three constructs (Fig. 2). The greatest degree of Δ12-desaturase silencing was found with the iHP construct. Seed from all 28 iHP primary transformants (T1 plants) had considerable reductions in ODP values, ranging from 0.54 down to 0.18 (Table I; see Figs. 2D and 6). The most silenced iHP line had a 75% reduction in Δ12 desaturation, equivalent to that of the fad2-1 mutant. The most silenced HP line achieved a similar reduction of Δ12 desaturation, but only three-quarters (47 of 63) of the HP lines had reductions in ODP. The CS construct was the least effective in Δ12-desaturase silencing. Although a substantial proportion (24 of 36; 67%) of CS T1 plants showed reduced ODP, the degree of the reduction was considerably less than that of the HP constructs. The most silenced CS T1 line had an ODP of 0.37, indicating only a 50% reduction in Δ12 desaturation.
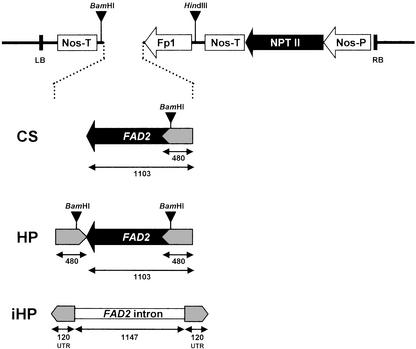
Figure 1.
Diagrammatic representation (not to scale) of FAD2-silencing constructs and associated NPTII selectable marker gene used to transform Arabidopsis. The CS construct has the FAD2 (1,103-nucleotide) sequence in sense orientation downstream from the rapeseed napin promoter (Fp1). The HP construct is the same as the CS construct but with the addition of an inverted-repeat of the 480-nucleotide 5′ region of the FAD2 sequence. The iHP construct consists of an inverted repeat of the 120-nucleotide FAD2 3′-UTR separated by intron 1 of FAD2 (1,147 nucleotide) in the correct (spliceable) orientation. All of these constructs were cloned into the HindIII/EcoRI site of pBI121 (CLONTECH Laboratories, Palo Alto, CA). The location of HindIII and BamHI restriction sites is shown.
Figure 2.
Frequency distribution (no. of plants) of ODP in seed of Columbia and fad2-1 mutant controls (A), independent T1 plants carrying the CS (B), HP (C), or iHP (D) constructs, and T5 plants carrying the iHP construct (E).
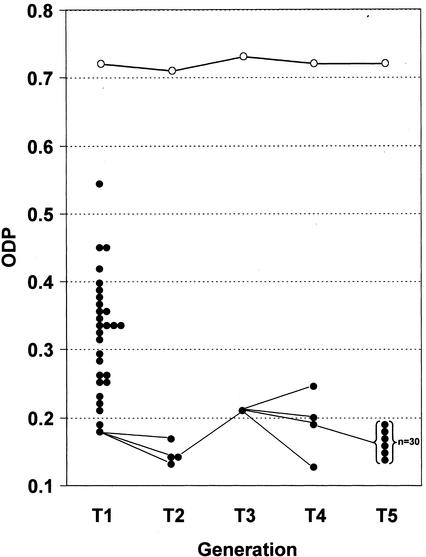
Figure 6.
Distribution of ODP values for iHP (●) T1 plant population and the T2, T3, T4, and T5 progeny of a selected highly silenced T1 plant, in comparison with Columbia (○) control plants grown at the same time.
Analysis of T2 and T3 Progeny Plants Transformed with HP and CS Constructs
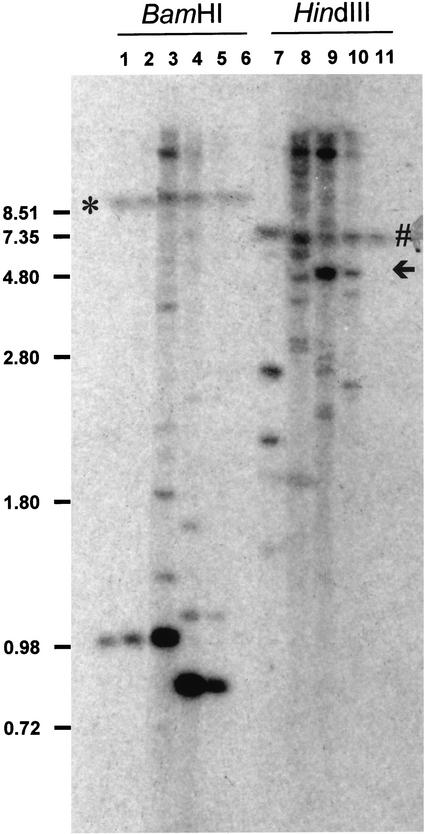
Primary transformant (T1) plants are hemizygous for transgene insertions. Therefore, the fatty acid analysis of the bulk seed from self-pollinated T1 plants will give an average value for each population of segregating genotypes, which will include some seeds that do not carry the transgene. To determine the inheritance of the silencing engendered by the constructs, the three most silenced HP lines (HP1, HP2, and HP3) and the two most silenced CS lines (CS1 and CS2) were examined by fatty acid analysis of seed borne on T2 plants and by Southern blot for transgene copy number on T3 plants. BamHI digestion of the genomic DNA of the HP and CS lines, as expected, released a 1- and a 0.8-kb internal FAD2 fragment from the HP and CS constructs, respectively. On the other hand, HindIII, which cuts once within the HP and CS constructs, gave bands that were greater than 2 kb in size (Figs. 1 and 3). A comparison of the intensities of the 1- and 0.8-kb fragments to that of the endogenous BamHI-FAD2 and HindIII-FAD2 fragments allows the estimation of copy number of the HP and CS constructs. It is estimated that HP1 contains two transgene inserts at a single locus (data not shown for HindIII digest) and HP2 contains two inserts at separate loci, whereas HP3, CS1, and CS2 contain multiple transgenes in complex loci. Also noteworthy is the strongly hybridizing band in the HindIII digest of CS1, suggestive of tandem, possibly inverted, repeat insertions of the CS transgene at some loci (Fig. 3).
Figure 3.
Southern-blot analysis of HP and CS transgenic Arabidopsis lines. Five micrograms of genomic DNA was digested with either BamHI or HindIII, and the blot was probed with a FAD2 gene probe. Lane 1, HP1; lanes 2 and 7, HP2; lanes 3 and 8, HP3; lanes 4 and 9, CS1; lanes 5 and 10, CS2; and lanes 6 and 11, untransformed Columbia. * and # indicate endogenous FAD2-hybridizing bands and the arrow indicates a possible tandem-repeat arrangement of the transgene. DNA size markers in kilobases are indicated on the left.
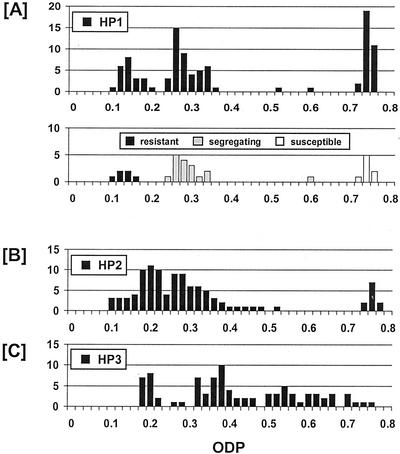
A large population (>80) of T2 plants was established for each of the five HP and CS lines and an ODP measured for a seed sample from each plant. As expected, the T2 plants from HP1 showed a trimodal distribution for ODP (Fig. 4A). The relative frequencies of T2 plants having ODP values that were high (32 plants), medium (45 plants), and low (22 plants) are consistent with the expected ratio of 1:2:1 for a single-locus segregation (χ22 = 2.84 ns). The high ODP class represents null homozygotes, the intermediate ODP class are putative hemizygotes (with the same ODP value as the hemizygous T1 plants), and the very low ODP class are putative homozygotes for the transgene. To confirm the genotypes of each group, T3 seed from a randomly chosen sample of 33 T2 plants was screened for kanamycin resistance (Fig. 4A). The trimodal distribution of these T2 plants into fully resistant (six plants), segregating (17 plants), and susceptible (eight plants) was again consistent with the expected ratio for a single-locus trait (χ22 = 0.55 ns), and the classes accorded fully with those inferred for ODP in the full T2 population.
Figure 4.
Frequency distribution (no. of plants) of ODP in seed of T2 plants from three lines of Arabidopsis transformed with HP1 (A; n = 99), HP2 (B; n = 94), and HP3 (C; n = 88). A also includes the frequency distribution of ODP in a subsample of 31 T2 plants scored for the status of the NPTII selectable marker gene.
The relative frequency of HP2 T2 plants having seed with suppressed FAD2 activity compared with those with wild-type activity was 94:11 (Fig. 4B), which is closer to the 15:1 ratio expected for a two-locus segregation than the 3:1 ratio expected for inheritance of a single-locus trait. A two-locus segregation pattern of 1 (four copies): 4 (three copies): 6 (two copies): 4 (one copy): 1 (null) can be envisaged in the distribution of ODP values, on the assumption that increased copy number gives increased silencing. The imperfect fit with a 15:1 ratio may be due to some degree of linkage between the two transgene loci. The ODP distribution (Fig. 4C) in seed from T2 plants of HP3 appears to form a continuum, which is consistent with the multiple complex transgene loci (Fig. 3) randomly assorting to provide additive effects on the degree of inhibition of FAD2 activity.
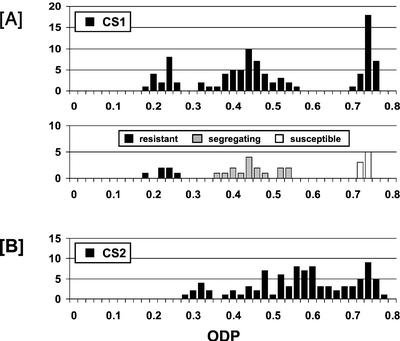
A similar ODP continuum, although for a more modest FAD2 silencing (Fig. 5B), is also found in seed from the multitransgene-containing CS2 line. However, seed lots from CS1 T2 plants show a trimodal ODP distribution (Fig. 5A), suggesting the presence of a single effective transgene insertion, despite this line containing multiple complex transgene loci. The relative frequencies of T2 plants having ODP values that were high (30 plants), medium (47 plants), and low (17 plants) departed slightly but not significantly from the expected ratio of 1:2:1 for a single-locus segregation (χ2 = 3.60 ns), due to a deficiency in transgene homozygotes and an excess of null homozygotes. Seed from a randomly chosen subsample of 30 CS1 T2 plants was examined for kanamycin resistance. The results (Fig. 5A) showed that the population segregated as eight susceptible, high ODP; 16 segregating resistant, medium ODP; and six resistant, low ODP. This distribution correlates more closely with a 1:2:1 ratio (χ2 = 0.40 ns) and indicates a cosegregation of effective kanamycin resistance with FAD2 silencing. However, most of the kanamycin-resistant plants grew slowly on kanamycin selection media with partial to full bleaching of the first true leaves and cotyledons suggesting that the NPTII gene(s) were not operating optimally. Perhaps this reflects some degree of PTGS impacting on NPTII transgene expression as a result of the possible existence of double-stranded NPTII RNA molecules emanating from some of the T-DNAs inserted as inverted-repeat structures in CS1.
Figure 5.
Frequency distribution (no. of plants) of ODP in seed of T2 plants from two lines of Arabidopsis transformed with CS1 (A; n = 94) and CS2 (B; n = 87). A also includes the frequency distribution of ODP in a subsample of 30 T2 plants scored for the status of the NPTII selectable marker gene.
Analysis of Multiple Generations of Seed from a Selected iHP Line
To examine the stability of inherited silencing, the progeny of a single highly silenced iHP line was carried through to T5 plants and analyzed at each generation for ODP level (Fig. 6). The wide variation for ODP in the hemizygous T1 plant population reflects the different silencing effectiveness of various transgene insertion events. A T1 plant having a single-transgene insertion (based on the pattern of selectable marker segregation) and a very low ODP value was selected and four T2 progeny plants grown. ODP varied from 0.13 to 0.17 among these T2 plants. A T2 plant having ODP of 0.14 was used to derive progeny in the T3 and T4 generations. ODP averaged 0.19 among four T4 plants, indicating that the high degree of FAD2 silencing was being maintained. A T4 plant with average ODP (0.19) was used to generate an expanded population of 30 T5 plants, which showed consistently low ODP values ranging from 0.14 to 0.19 (Fig. 2E). Control Columbia plants grown alongside the iHP progeny populations maintained normal high levels of ODP in the range of 0.71 to 0.73 throughout the generations. These results demonstrate that genotypes homozygous for a single insertion of an iHP transgene breed true and maintain the high level of target gene silencing throughout five successive generations.
DISCUSSION
It is becoming increasingly apparent that PTGS occurs via a mechanism involving the production of double-stranded or self-complementary hpRNA (Waterhouse et al., 1998, 2001; Matzke et al., 2001; Vance and Vaucheret, 2001) and that constructs designed to express such transcripts are an efficient way of inducing targeted gene silencing (Waterhouse et al., 1998; Chuang and Meyerowitz, 2000; Smith et al., 2000; Levin et al., 2001; Wesley et al., 2001). Our results further extend this growing body of evidence, with HP and iHP gene-silencing constructs in this study being highly efficient in down-regulating endogenous FAD2 Δ12-desaturase activity in Arabidopsis. Furthermore, this high degree of gene silencing achieved with the iHP construct was stably inherited over five generations.
The highest silencing efficiency was obtained with the iHP construct targeted against the relatively short 120-bp 3′-untranslated region (UTR) of the FAD2 gene, with all transformants showing significant silencing. The HP construct, which was targeted against the FAD2 coding region and had part of the FAD2 gene as the spacer region rather than a spliceable intron, was 25% less efficient than the iHP construct, but still produced a higher frequency of silencing than the CS construct. It is notable that the 66% efficiency of FAD2 silencing achieved with the CS construct in the current study is much higher than the 17% previously reported by Cartea et al. (1998). This enhancement is possibly related to the different constructs used in the two studies. The CS construct in the current work consisted of the rapeseed napin promoter (Fp1) driving a truncated FAD2 coding sequence that was missing the 5′-UTR and a small portion of the 5′ coding region, whereas the construct used by Cartea et al. (1998) consisted of the full-length FAD2 coding sequence driven by rapeseed napin promoter. It is possible that this latter construct may result in expression of an effective Δ12-desaturase in some transformants and, thereby, reduce the efficiency of silencing. In this regard, it is relevant that Cartea et al. (1998) reported significantly higher efficiency of silencing (57%) using FAD2 antisense constructs. Conventional CS and antisense constructs often appear to induce PTGS by producing dsRNA when they are integrated into the genome in particular orientations, such as inverted-repeat arrangement. It is pertinent to point out that Southern-blot data for one of the most silenced CS lines (CS1) suggests the presence of tandem-repeat arrangement of inserted T-DNA. Such tandem-repeat, and especially inverted-repeat, structures have been associated with the production of double-stranded RNA molecules, which can trigger PTGS (Wang and Waterhouse, 2000). The likely low frequency of such integration events may explain the lower recovery of gene silencing in CS and antisense plants compared with plants containing HP constructs.
The significantly higher proportion of silenced transformants obtained by the iHP construct compared with the HP construct might be explained by these constructs differing in their ability to produce steady-state levels of duplex RNA in excess of the threshold levels considered necessary to activate PTGS in plants (Waterhouse et al., 2001). Thus, when inserted into poorly transcribed regions of the genome, as no doubt occurs in a proportion of random insertion events, transgenes inherently capable of producing higher levels of duplex RNA might activate silencing, whereas those producing lower levels might not. A number of attributes of iHP constructs could make them capable of producing higher levels of duplex RNA than HP constructs. First, the iHP construct is expected to be transcribed into a mature hpRNA comprising a 120-bp stem and only a 21-bp loop, whereas the HP construct is expected to give a mature hpRNA comprising a 480-bp stem and a 623-bp loop. This means that the iHP construct gives a hpRNA with a considerably tighter loop and is, hence, less prone to nuclease attack than that produced by the HP construct, which could result in the iHP construct having higher steady-state levels of duplex RNA. Second, the process of intron splicing of the ihpRNA could be responsible for more efficient duplex formation through alignment of the complimentary arms in the spliceosome complex, whereas the complimentary arms of hpRNA would have to hybridize by the less-efficient random but tethered collisions. Third, the mere presence of the intron in the iHP construct may result in increased or more-stabilized transcript levels than in the nonintron-containing hpRNA (Callis et al., 1987; Tanaka et al., 1990). At this stage, we would favor the third explanation for the greater efficiency of the iHP construct to elicit PTGS because Wesley et al. (2001) have presented evidence that may discount the first two explanations. In addition to the significant differences in the rate of recovery of FAD2-suppressed lines, it is clear that the maximum degree of silencing occurred with HP constructs. For both the HP and iHP constructs, it was possible to generate transformants that had ODP values around 0.10, which indicates equivalent lowering of Δ12-desaturase activity to that of the fad2-1 ethyl methanesulphonate (EMS) mutant. Thus, although significantly enhancing the efficiency of silencing, the presence of a spliceable intron did not alter the maximum degree of silencing achievable with HP constructs.
Transgenic expression of either CS or HP constructs targeted against the FAD2 gene resulted in a reduction of up to 85% of the wild-type levels of Δ12 desaturation, which is roughly equivalent to that which occurs with the fad2-1 mutant. It is, therefore, interesting to consider whether this reduction could represent a complete silencing of FAD2-mediated Δ12 desaturation. C18 polyunsaturated fatty acids present in Arabidopsis seed lipids can be formed by either of two distinct pathways. In the “eukaryotic” pathway that operates on the endoplasmic reticulum membranes of the cytoplasm, 18:1 esterified to the sn1 and sn2 positions of phosphatidylcholine is desaturated at the Δ12 position by the action of a membrane-bound ω6-desaturase that in Arabidopsis is encoded by the single-copy FAD2 gene (Okuley et al., 1994). This pathway provides the great majority of lipid precursors for synthesis of triacylglycerols in the oil-accumulating tissues of the developing seeds (Browse and Somerville, 1991). In addition, there is a “prokaryotic” pathway operating within the plastids of leaves and seeds in which 18:1 esterified to the sn-1 position of the major plastidic lipids (phosphatidic acid, galactolipids, and sulfolipids) is desaturated to 18:2 by a similar membrane-bound ω6-desaturase encoded by a different gene, the FAD6 gene in Arabidopsis (Browse and Somerville, 1991). Detailed analysis using mutants in the FAD2 and FAD6 genes has revealed that there is considerable metabolic flexibility in determining the final complement of fatty acids in the cell. It appears that significant interplay occurs between the eukaryotic and prokaryotic pathways through a reversible exchange of fatty acids between plastidic and endoplasmic reticulum membranes (Miquel and Browse, 1994) enabling each pathway to partially compensate for mutations in the other pathway to maintain adequate levels of polyunsaturated fatty acids. The EMS-induced fad2-1 mutant, for example, although producing apparently normal levels of FAD2 mRNA transcript, has been shown biochemically to have negligible microsomal Δ12-desaturase activity, suggesting that the mutation results in the translation of a defective FAD2 desaturase protein (Miquel and Browse, 1994). However, seeds of the fad2-1 mutant still accumulate significant levels of 18:2 and 18:3, approximately 8% in total, presumably as a result of the FAD6-encoded plastidic Δ12-desaturase. Polyunsaturated fatty acids resulting from FAD6 activity either remain located in the plastid itself or are exported and subsequently incorporated in the endoplasmic reticulum lipids and triacylglycerols. The Arabidopsis FAD6 gene has 41% identity to the FAD2 gene at the DNA level and, therefore, should be sufficiently divergent to escape cross-silencing by the FAD2-targeted gene-silencing constructs used in the present study. Our observation that transformation of the fad2-1 mutant with either of the highly effective HP and iHP FAD2-silencing constructs did not bring about a further lowering of 18:2 and 18:3 content in any of nine independently derived transgenic plants (data not shown) suggests that PTGS-mediated removal of any FAD2 mRNA transcripts remaining in the fad2-1 plants achieves no additional reduction in Δ12 desaturation. This is consistent with the fad2-1 protein being enzymatically ineffective and, thus, with the remaining Δ12 desaturation being mediated by FAD6.
Against this background, the fact that the most highly FAD2-silenced lines of Arabidopsis in the current study have residual levels of 18:2 and 18:3 equivalent to those attributable to FAD6 activity in the fad2-1 mutant line, most likely indicates that FAD2 expression has been completely blocked in these silenced lines. In wild-type Arabidopsis, it is considered that FAD2 expression is essentially under translational and posttranslational control, with the FAD2 mRNA transcript being present in substantial excess of the amount needed to account for the level of FAD2-mediated Δ12 desaturation. This is based on the observation that the T-DNA insertional mutant fad2-5 has barely detectable levels of FAD2 mRNA transcript, but is nevertheless able to effect more than one-half of the Δ12 desaturation attributable to FAD2 in the wild type (Okuley et al., 1994). Thus, the level of mRNA transcripts in the most highly silenced lines in the current study, which show considerably greater reduction in Δ12 desaturation than does the fad2-5 mutant, must be very low and indicative of highly effective mRNA degradation.
The extremely high efficiency and efficacy of endogenous gene silencing obtainable using ihpRNA constructs provides several substantial enhancements in the utility of PTGS in genetic modification. First, such high-level silencing enables down-regulation of endogenous genes to be attempted in difficult species that have relatively low-transformation rates. For example, we have already used the efficiency of hpRNA constructs to separately down-regulate both Δ9-desaturase and Δ12-desaturase in cotton, a poorly transformable species, to produce novel high-stearic and high-oleic cottonseed oils (Liu et al., 2002). Second, by reducing the population size needed to recover a good silencing transformation, the use of ihpRNA constructs will facilitate the removal by segregation of selectable marker genes that have been inserted at separate loci to the silencing transgene, such as by using twin T-DNA vectors (Matthews et al., 2001). Third, the observation that almost maximal levels of gene silencing were obtainable in plants that were heterozygous for the HP or iHP constructs but not for CS heterozygotes suggests that HP constructs may be effectively deployed for trait modification in F1 hybrid varieties. Finally, it should be possible to use the ability of HP constructs to target small UTRs (as little as 120 bp) of a gene to selectively silence individual members of multigene families if there is sufficient divergence in the UTR sequences of the various members of the gene family. Targeting to highly conserved regions should conversely enable simultaneous silencing of the complete gene family. These features and the demonstrated multigenerational stability make hpRNA-mediated gene silencing a valuable technique for trait modification in plant improvement programs.
MATERIALS AND METHODS
Gene-Silencing Constructs
HP and CS constructs targeted against the Arabidopsis Δ12-desaturase gene (FAD2) were made using a 1,103-bp fragment of the coding region of the gene. This fragment is a 5′-truncated version of the full FAD2 coding region and was used to prevent any overexpression that might otherwise result from the sense transcription and translation of the full coding region. Expression was directed to the developing seeds by driving the constructs with the truncated version (Fp1) of the seed-specific napin promoter from rapeseed (Brassica napus; Stalberg et al., 1993). Three types of gene-silencing constructs were evaluated: a CS construct consisting of the 1,103-bp coding region fragment and its complete 3′-UTR (Fig. 1A); an HP construct consisting of the CS construct with a 480-bp fragment of the 5′ end of the FAD2 sequence inserted in an antisense orientation immediately behind the 3′ end (Fig. 1B); and an iHP construct in which the inverted-repeat regions consisted of just the 120-bp 3′-UTR of the FAD2 gene and were separated by intron 1 of the FAD2 gene in spliceable orientation (Fig. 1C). In the iHP construct, the inverted-repeat arms were oriented in the antisense and sense directions at the 5′ and 3′ ends of the construct, respectively. The NPTII kanamycin resistance gene driven by the nopaline synthase promoter was placed upstream of each of the silencing constructs to facilitate selection of transgenic plants.
The gene-silencing constructs were produced as follows. All constructs were assembled in the plasmid vector pBluescript SK− (Stratagene, La Jolla, CA) before cloning into the binary vector pBI121 (CLONTECH Laboratories) as HindIII-SacI fragments, replacing the β-glucuronidase-containing HindIII-SacI region of pBI121. The truncated Fp1 promoter, containing sequences between −309 and +1 (Stalberg et al., 1993), was cloned into the HindIII-EcoRV site of pBluescript. For the CS construct, a 1,103-bp fragment was amplified from the FAD2 cDNA clone (Okuley et al., 1994) using PCR primers At1 and At2 and cloned into the EcoRV-EcoRI site behind the Fp1 promoter. For the HP construct, a 480-bp FAD2 fragment was amplified using PCR primers At3 and At4 and cloned into the PstI-XbaI site of the CS construct. For the iHP construct, a 1,147-bp FAD2 intron 1 fragment was amplified from genomic DNA isolated from Columbia ecotype of Arabidopsis using PCR primers At5 and At6. This intron fragment also contained 17 bp of exon I and 4 bp of exon II to ensure the inclusion of the 5′ and 3′ splice sites (Okuley et al., 1994). The FAD2 intron 1 fragment was then cloned in a 5′-3′ orientation behind the Fp1 promoter in the EcoRI site. A 120-bp 3′-UTR fragment was amplified from the FAD2 cDNA clone using PCR primers At7 and At8 and cloned into the BamHI-SacII sites behind the intron 1 sequence. Similarly, to complete the inverted repeat structure of the 3′-UTR, the same 120-bp 3′-UTR fragment was amplified using PCR primers At9 and AT10 and cloned into the EcoRV site between the Fp1 promoter and the FAD2 intron 1. Nucleotide sequences of the PCR primers mentioned above are: At1, EcoRV ATCATTATAGCCTCATGCTTC; At2, EcoRI AACATAATGAGCAGCCAAAATG; At3, PstI TCGGTCATTGTAGATGGGAGC; At4, same as At1 but with XbaI site; At5, EcoRI GTCAGCTCCATCTCCAGGTCC; At6, EcoRI GTTCTGCAGAAAACCAAAAGC; At7, BamHI GAGCATGATGGTGAAGAAATT; At8, SacII GCAGCCAAAATGTCATAACAC; At9, same as At7 but with SmaI site; and At10, same as At8 but with EcoRV site.
Plant Transformation
All transformation experiments were conducted with the Columbia ecotype of Arabidopsis using the vacuum infiltration technique (Bechtold et al., 1993). Seed was first placed in small pots for 2 d at 4°C to synchronize germination and seedlings were subsequently transferred to a 16-h-light (23°C)/8-h-dark (17°C) regime. After approximately 3 weeks, primary bolts were cut back, and the plants were allowed to grow for another 2 weeks or until the bolts were between 3 and 10 cm high. Plants were then vacuum infiltrated with Agrobacterium tumefaciens strain AglI and grown under plastic wrap for 2 d to maintain high humidity. Approximately 72 plants were vacuum infiltrated for each silencing construct. Seed was collected from the treated (T0) plants at maturity. Primary transformants (T1 plants) were established by screening selfed seed from T0 plants for kanamycin resistance on Murashige and Skoog plates (Murashige and Skoog, 1962) supplemented with 1.5% (w/v) Suc and 50 mg L−1 kanamycin sulfate. Kanamycin-resistant plantlets were transferred to soil and allowed to mature. Care was taken to rescue any plant that grew past the cotyledon stage or resisted bleaching during the selection phase.
All transgenic plants were grown in a greenhouse under natural daylength with a controlled temperature of 24°C in the daylight hours and 18°C in the evening, with Columbia wild type and its fad2-1 mutant grown along side as controls. Seed was harvested from individual T1 and control plants, and 100-seed samples were analyzed for fatty acid composition. T2 plant populations were subsequently established for selected families and grown alongside Columbia and fad2-1 control plants.
DNA Blotting Analysis
DNA was extracted from six T3 plants per line using the procedure of Shure et al. (1983). After freezing and grinding of leaf tissue in liquid N2, approximately 0.5 g of leaf tissue was added to 750 μL of extraction buffer (0.3 m NaCl, 50 mm Tris-HCl, pH 7.5, 20 mm EDTA, 2% [w/v] Sarkosyl, 0.5% [w/v] SDS, 5 m urea, and 5% [w/v] phenol added just before use). After mixing, an equal volume of phenol chloroform (1:1) was added and gently agitated for 5 min. The samples were centrifuged at 14,000 rpm for 5 min, and the supernatant was transferred to a new tube where DNA was precipitated by the addition of an equal volume of isopropanol. After centrifugation at 14,000 rpm, the pellet was washed twice with 70% (w/v) ethanol, air dried, and resuspended in Tris-EDTA buffer. After RNase treatment, the DNA was digested with HindIII or BamHI. Five micrograms of digested DNA from each of the T3 lines was separated by electrophoresis (0.8% [w/v] agarose). Size-fractionated DNA was transferred to Hybond N+ membrane by standard Southern-blot techniques (Maniatis et al., 1982). Radiolabeled probes were made using the FAD2 coding sequence and a Megaprime DNA labeling system (Amersham-Pharmacia Biotech, Uppsala). After overnight hybridization (4× SSC, 0.1% [w/v] SDS, 5× Denhardt's solution, and 100 μg mL−1 denatured salmon sperm DNA) at 65°C, filters were washed once with 2× SSC at room temperature and twice with 0.2× SSC and 0.1% (w/v) SDS at 65°C for 20 min. Hybridization patterns were recorded on a PhosphoImager (Molecular Dynamics, Sunnyvale, CA).
Fatty Acid Analysis
Fatty acid methyl esters were prepared by an acidic methylation method (Christie, 1982). Approximately 100 seeds per plant were pooled and crushed onto filter paper discs. The samples were methylated in 2 mL of 5% (v/v) HCl in methanol for 90 min at 80°C, followed by the addition of 3 mL of petroleum spirit and 1 mL of water. After vortexing and phase separation, the upper petroleum spirit layer containing the fatty acid methyl esters was transferred to a microvial. One gram of 10% (w/w) K2CO3/Na2SO4 was added to the vial and mixed by vortexing. Fatty acid methyl esters were separated on a SGE BPX70 column (0.25-mm diameter, 60-m length, and 2.5-μm film thickness) in a gas chromatograph (model 3400, Varian, Palo Alto, CA) using helium as the carrier gas. The initial temperature of the column was 170°C and was programmed to increase at 3°C per minute until the final temperature of 220°C was reached and maintained until the completion of the analysis. Relative fatty acid compositions were calculated as the percentage that each fatty acid represented of the total fatty acid profile. Alterations to the activity of the Δ12-desaturase caused by the action of introduced transgenes could be seen as changes in the amounts of oleic acid and in the seed oil profiles. An additional indirect method of assessing the cumulative effects of Δ12-desaturase activity during seed fatty acid synthesis is through the ODP parameter, derived by the following formula:
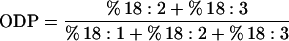 |
ODP represents the ratio of the total fatty acids accounting for the products of 18:1 desaturation (i.e. 18:2 and 18:3) to the total amount of 18:1 substrate that was available, i.e. these products of 18:1 modification plus the remaining 18:1. Arabidopsis typically has an ODP value of around 0.70 to 0.72, indicating that around 70% to 72% of 18:1 formed during fatty acid synthesis is subsequently converted to the polyunsaturated C18 fatty acids initially via the action of Δ12-desaturase. This parameter is useful in illustrating the effects of the FAD2 gene-silencing transgenes on the level of endogenous Δ12-desaturase activity.
ACKNOWLEDGMENTS
We thank Sue McKinney and Diana Hall for their excellent technical assistance, and Lorraine Tonnet, Lorraine Mason, and Richard Philips for conducting the fatty acid analysis.
Footnotes
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.006353.
LITERATURE CITED
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. CR Acad Sci (Paris) 1993;316:1194–1199. [Google Scholar]
- Browse J, Somerville CR. Glycerolipid synthesis: biochemistry and regulation. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:467–506. [Google Scholar]
- Callis J, Fromm M, Walbot V. Introns increase gene expression in cultured maize cells. Genes Dev. 1987;1:1183–1200. doi: 10.1101/gad.1.10.1183. [DOI] [PubMed] [Google Scholar]
- Cartea ME, Migdal M, Galle AM, Pelletier G, Guerche P. Comparison of sense and antisense methodologies for modifying the fatty acid composition of Arabidopsis thaliana oilseed. Plant Sci. 1998;136:181–194. [Google Scholar]
- Christie WW. Lipid Analysis. Isolation, Separation, Identification and Structural Analysis of Lipids. Oxford: Pergamon Press; 1982. [Google Scholar]
- Chuang CH, Meyerowitz EM. Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2000;97:4985–4990. doi: 10.1073/pnas.060034297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni C, Macino G. Gene silencing in Neurospora crassa requires a protein homologous to RNA dependent RNA polymerase. Nature. 2000;399:166–169. doi: 10.1038/20215. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan J, McElroy D. Transgene inactivation: plants fight back. Bio-Technology. 1994;12:883–888. [Google Scholar]
- Fire A, Xu SQ, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Kennerdel JR, Carthew RW. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell. 1998;95:1017–1026. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- Levin JZ, de Framond AJ, Tuttle A, Bauer MW, Heifetz PB. Methods of double stranded RNA-mediated gene inactivation in Arabidopsis and their use to define an essential gene in methionine biosynthesis. Plant Mol Biol. 2001;44:759–775. doi: 10.1023/a:1026584607941. [DOI] [PubMed] [Google Scholar]
- Liu Q, Singh S, Green A (2002) High-stearic and high-oleic cottonseed oils produced by hpRNA-mediated posttranscriptional gene silencing. Plant Physiol (in press) [DOI] [PMC free article] [PubMed]
- Maniatis T, Fritsch E, Sambrook J. Molecular Cloning. A Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- Matthews PR, Wang MB, Waterhouse PM, Thornton S, Fieg SJ, Gubler F, Jacobsen JV. Marker gene elimination from transgenic barley, using co-transformation with adjacent ‘twin T-DNAs’ on a standard Agrobacterium transformation vector. Mol Breeding. 2001;7:195–202. [Google Scholar]
- Matzke MA, Matzke AJ, Pruss GJ, Vance VB. RNA-based silencing strategies in plants. Curr Opin Genet Dev. 2001;2:221–227. doi: 10.1016/s0959-437x(00)00183-0. [DOI] [PubMed] [Google Scholar]
- Miquel MF, Browse JA. High-oleate oilseeds fail to develop at low temperature. Plant Physiol. 1994;106:421–427. doi: 10.1104/pp.106.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Okuley J, Lightner J, Feldmann K, Yadav N, Lark E, Browse J. Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell. 1994;6:147–158. doi: 10.1105/tpc.6.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PA, Zamore PD. RNA interference. Science. 2000;287:2431–2433. doi: 10.1126/science.287.5462.2431. [DOI] [PubMed] [Google Scholar]
- Shure M, Wessler S, Fedoroff N. Molecular identification and isolation of the waxy locus in maize. Cell. 1983;35:225–233. doi: 10.1016/0092-8674(83)90225-8. [DOI] [PubMed] [Google Scholar]
- Smith NA, Singh SP, Wang MB, Stoutjesdijk PA, Green AG, Waterhouse PM. Total silencing by intron-spliced hairpin RNAs. Nature. 2000;407:319–320. doi: 10.1038/35030305. [DOI] [PubMed] [Google Scholar]
- Somerville C. Genomics: plant biology 2010. Science. 2000;290:2077–2078. doi: 10.1126/science.290.5499.2077. [DOI] [PubMed] [Google Scholar]
- Stalberg K, Ellerstrom M, Josefsson LG, Rask L. Deletion analysis of a 2S seed storage protein promoter of Brassica napus in transgenic tobacco. Plant Mol Biol. 1993;23:671–683. doi: 10.1007/BF00021523. [DOI] [PubMed] [Google Scholar]
- Tanaka A, Mita S, Ohta S, Kyozuka J, Shimamoto K, Nakamura K. Enhancement of foreign gene expression by a dicot intron in rice but not in tobacco is correlated with an increased level of mRNA and an efficient splicing of the intron. Nucleic Acids Res. 1990;18:6767–6770. doi: 10.1093/nar/18.23.6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance V, Vaucheret H. RNA silencing in plants: defense and counterdefense. Science. 2001;292:2277–2280. doi: 10.1126/science.1061334. [DOI] [PubMed] [Google Scholar]
- Wang MB, Abbott D, Waterhouse PM. A single copy of a virus derived transgene encoding hairpin RNA gives immunity to barley yellow dwarf virus. Mol Plant Pathol. 2000;1:401–410. doi: 10.1046/j.1364-3703.2000.00038.x. [DOI] [PubMed] [Google Scholar]
- Wang MB, Waterhouse PM. High efficiency silencing of a β-glucuronidase gene in rice is correlated with repetitive transgene structure but is independent of DNA methylation. Plant Mol Biol. 2000;43:67–82. doi: 10.1023/a:1006490331303. [DOI] [PubMed] [Google Scholar]
- Waterhouse PM, Graham HW, Wang MB. Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc Natl Acad Sci USA. 1998;95:13959–13964. doi: 10.1073/pnas.95.23.13959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse PM, Smith NA, Wang MB. Virus resistance and gene silencing: killing the messenger. Trends Plant Sci. 1999;4:452–457. doi: 10.1016/s1360-1385(99)01493-4. [DOI] [PubMed] [Google Scholar]
- Waterhouse PM, Wang MB, Lough T. Gene silencing as an adaptive defence against viruses. Nature. 2001;411:834–842. doi: 10.1038/35081168. [DOI] [PubMed] [Google Scholar]
- Wesley V, Helliwell C, Smith N, Wang MB, Rouse D, Liu Q, Gooding P, Singh S, Abbott D, Stoutjesdijk P et al. Construct design for efficient, effective and high throughput gene silencing in plants. Plant J. 2001;27:581–590. doi: 10.1046/j.1365-313x.2001.01105.x. [DOI] [PubMed] [Google Scholar]