Abstract
We have genetically modified the fatty acid composition of cottonseed oil using the recently developed technique of hairpin RNA-mediated gene silencing to down-regulate the seed expression of two key fatty acid desaturase genes, ghSAD-1-encoding stearoyl-acyl-carrier protein Δ9-desaturase and ghFAD2-1-encoding oleoyl-phosphatidylcholine ω6-desaturase. Hairpin RNA-encoding gene constructs (HP) targeted against either ghSAD-1 or ghFAD2-1 were transformed into cotton (Gossypium hirsutum cv Coker 315). The resulting down-regulation of the ghSAD-1 gene substantially increased stearic acid from the normal levels of 2% to 3% up to as high as 40%, and silencing of the ghFAD2-1 gene resulted in greatly elevated oleic acid content, up to 77% compared with about 15% in seeds of untransformed plants. In addition, palmitic acid was significantly lowered in both high-stearic and high-oleic lines. Similar fatty acid composition phenotypes were also achieved by transformation with conventional antisense constructs targeted against the same genes, but at much lower frequencies than were achieved with the HP constructs. By intercrossing the high-stearic and high-oleic genotypes, it was possible to simultaneously down-regulate both ghSAD-1 and ghFAD2-1 to the same degree as observed in the individually silenced parental lines, demonstrating for the first time, to our knowledge, that duplex RNA-induced posttranslational gene silencing in independent genes can be stacked without any diminution in the degree of silencing. The silencing of ghSAD-1 and/or ghFAD2-1 to various degrees enables the development of cottonseed oils having novel combinations of palmitic, stearic, oleic, and linoleic contents that can be used in margarines and deep frying without hydrogenation and also potentially in high-value confectionery applications.
Although cotton (Gossypium hirsutum) is primarily grown for fiber production, it is also the world's sixth largest source of vegetable oil. Cottonseed oil is typically composed of about 26% palmitic acid (C16:0), 15% oleic acid (C18:1), and 58% linoleic acid (C18:2). The relatively high level of palmitic acid provides a degree of stability to the oil that makes it suitable for high-temperature frying applications, but is nutritionally undesirable because of the low-density lipoprotein cholesterol-raising properties of this saturated fatty acid (Cox et al., 1995). Although cottonseed oil has recently been shown to lower total serum cholesterol compared with corn (Zea mays) oil (Radcliffe et al., 2001), it did so by lowering the level of the desirable high-density lipoprotein cholesterol without reducing the level of the undesirable low-density lipoprotein cholesterol, presumably because of its significant content of palmitic acid. Furthermore, cottonseed oil is sometimes hydrogenated to achieve the very high stability required in deep-frying food service applications or to provide the solidity required for margarine hard stock. Unfortunately, the hydrogenation process results in the production of trans-fatty acids, which are now recognized as having cholesterol-raising properties equivalent to those of saturated fatty acids (Ascherio and Willett, 1997).
As a result of these factors, there is a growing trend away from the use of oils that are rich in palmitic acid and hydrogenated oils in favor of those that are both nutritionally beneficial and can provide the required functionality without hydrogenation. Oils that are low in palmitic acid and rich in either oleic acid or stearic acid (C18:0) meet these requirements, and such fatty acid profiles have now been developed in several oilseed species through genetic modification of fatty acid synthesis. Selective breeding utilizing natural variants or induced mutations has been used to develop a range of improved oils in the major temperate oilseed crops, including high-stearic (HS) soybean (Glycine max; Graef et al., 1985), high-oleic (HO) rapeseed (Brassica napus; Auld et al., 1992), HO peanut (Arachis hypogaea; Norden et al., 1987), and HS (Osorio et al., 1995) and HO (Soldatov, 1976) sunflower (Helianthus annuus). However, due to a lack of any significant genetic variation for fatty acid composition in cottonseed oil and the allotetraploid nature of cultivated cotton, classical breeding techniques and induced mutagenesis have so far been unsuccessful in developing improved cottonseed oil.
To overcome the limitations of conventional breeding approaches, genetic engineering techniques have now been successfully employed to modify the fatty acid composition in a number of oilseed crops. In particular, posttranslational gene silencing (PTGS) has been used to down-regulate the activity of the desaturase enzymes that control the synthesis of the major seed oil fatty acids, principally stearoyl-acyl-carrier protein (ACP) Δ9-desaturase, which converts stearic acid into oleic acid, and oleoyl-phosphatidylcholine (PC) ω6-desaturase, which converts oleic acid into linoleic acid. For example, stearic acid was raised to around 40% in rapeseed oil through antisense-mediated down-regulation of the stearoyl-ACP Δ9-desaturase activity (Knutzon et al., 1992), and very high levels of oleic acid have been achieved in both soybean and rapeseed through seed-specific cosuppression of oleoyl-PC ω6-desaturase (Kinney, 1996). However, the antisense and cosuppression strategies used in these cases have variable and unpredictable effectiveness and generally require the production of large populations of transgenic plants to obtain an acceptable number of lines exhibiting sufficient degrees of target gene suppression (Knutzon et al., 1992; Kinney, 1996; Hamilton et al., 1998). This presents a particular problem for their application in cottonseed oil improvement because cotton is still relatively difficult to transform and requires long periods of time in tissue culture for regeneration (Cousins et al., 1991; Murray et al., 1999).
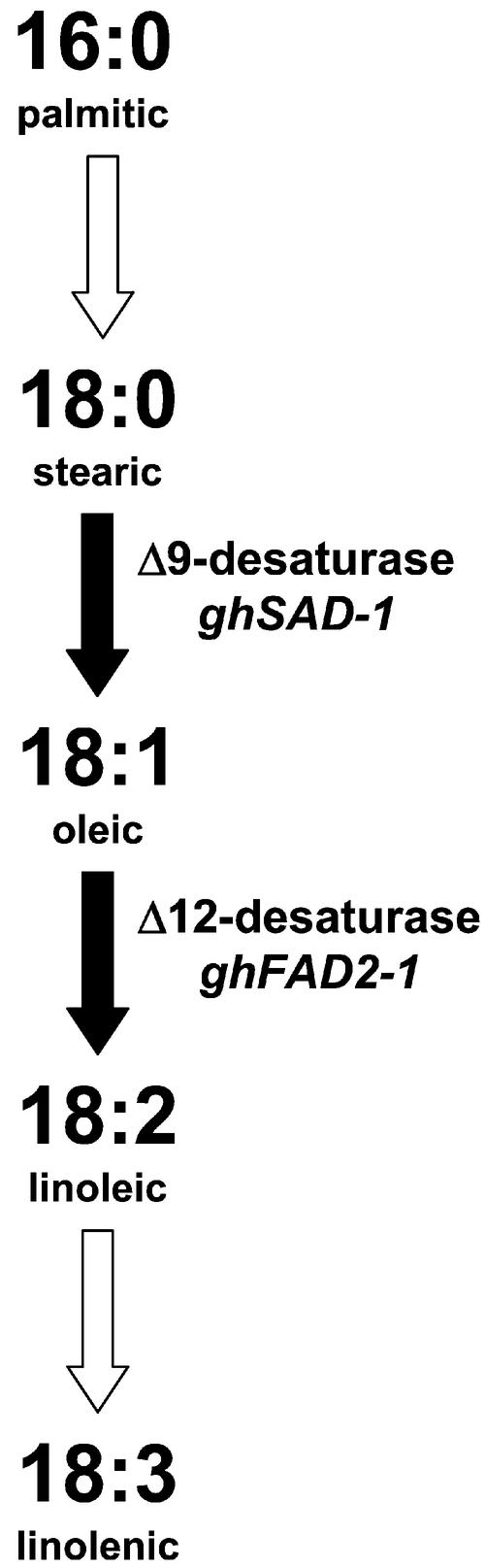
Recently, the discovery that RNA interference occurs in plants and is mediated by sequence-specific degradation of dsRNA has led to the development of highly efficient methods of silencing plant genes (Smith et al., 2000). Specifically designed genetic constructs, such as inverted repeats that encode RNA having regions of self-complementarity, can reliably generate hairpin RNA (hpRNA) transcripts that invoke sequence-specific RNA degradation targeted to the double-stranded region of the hpRNA and to homologous endogenous mRNA molecules. By utilizing a partial sequence of an endogenous gene in the inverted repeat regions of the silencing construct, high-level silencing of the target gene expression can be achieved. We have applied hpRNA-mediated gene-silencing techniques to modify the expression of the Δ12-desaturase gene (FAD2) in Arabidopsis seeds and have demonstrated that they result in much higher efficiency and efficacy of gene silencing than either antisense or cosuppression (Stoutjesdijk et al., 2002). Such high-efficiency gene-silencing techniques have now made it practicable to attempt genetic modification of fatty acid composition of cottonseed oil. The ghSAD-1 and ghFAD2-1 genes in cotton, respectively, encode stearoyl-ACP Δ9-desaturase (Liu et al., 1996) and microsomal ω6-desaturase (Liu et al., 1999b), also referred to as Δ12-desaturase, which are the key enzymes determining the fatty acid composition of cottonseed oil (Fig. 1). Here, we report the use of hpRNA gene-silencing constructs to achieve seed-specific silencing of both ghSAD-1 and ghFAD2-1 resulting in the development of HS and HO cottonseed oils, including molecular analysis of target gene expression, and comparisons of phenotypic patterns observed in a range of independently derived transgenic lines. Furthermore, we demonstrate the stable inheritance of these phenotypes in progeny derived by either selfing or intercrossing, and the generation of further novel fatty acid compositions in hybrid progeny expressing both HO and HS traits.
Figure 1.
Diagramatic representation of fatty acid biosynthetic pathway in cottonseed.
RESULTS
Identification of Transgenic Plants
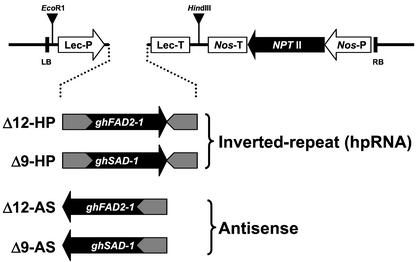
Cotton cv Coker 315 was transformed with gene-silencing constructs consisting of the ghSAD-1 or ghFAD2-1 cDNA clone in either inverted repeat or antisense configurations driven seed specifically by a soybean lectin promoter (Fig. 2). Thirty-four and 25 fertile plants were established from kanamycin-resistant calli for the Δ9-HP and Δ9-AS constructs, respectively. Thirty-six and 27 fertile plants were obtained by transformation with constructs Δ12-HP and Δ12-AS, respectively. No obvious difference was observed in terms of callus induction, somatic embryogenesis, and establishment of fertile plants among the four transformations. Transgenic status of the regenerants was confirmed by PCR amplification of the soybean lectin 3′ terminator DNA fragment from genomic DNA.
Figure 2.
Schematic diagram of the chimeric silencing constructs transformed into cotton. The ghSAD-1 and ghFAD2-1 genes in either inverted repeat (HP) or antisense (AS) orientation were placed under the control of the seed-specific soybean lectin promoter (Lec-P) and terminator (Lec-T). The neomycin phosphotransferase selectable marker gene (NptII) was driven by the Nos promoter (Nos-P). LB and RB correspond to the T-DNA left and right borders, respectively. The positions of EcoRI and HindIII restriction enzyme sites are indicated.
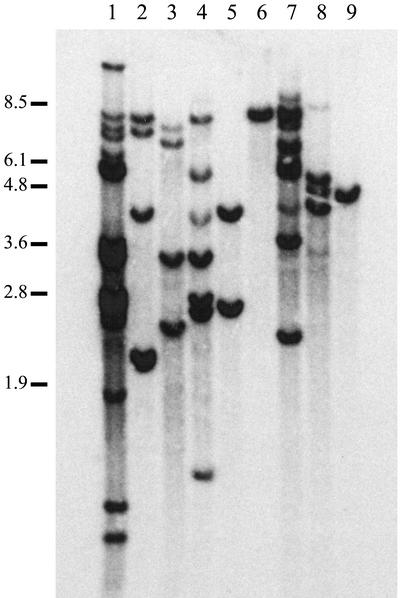
Southern-blot analysis of genomic DNA was used to confirm the integration of the transgene into the cotton genome and to estimate the transgene copy number. The Southern blots were performed with HindIII-digested genomic DNA from the leaf tissue of transgenic plants, and probed with a radiolabeled DNA fragment consisting of the promoter region of the soybean lectin gene. HindIII cuts once within the T-DNA of the Δ12 constructs and twice within the Δ9 constructs, in each case downstream of the lectin promoter. Probing of the Southern blots with the lectin promoter identified uniquely sized fragments that spanned the junction of the T-DNA to the adjacent genomic DNA in each transformation event, verifying that the individual transgenic lines each originated from independent transformation events. A number of representative transgenic lines are shown in Figure 3. Bands of different sizes were interpreted to represent different transgene insertions and demonstrated that several transgenic lines contained multiple numbers of insertions (Fig. 3). In the case of transgenic lines with multiple 5′ lectin bands, different signal intensities were frequently observed among these bands on the same lane. The high-intensity bands may harbor multiple fragments of similar sizes. Although highly variable among the four individual transformations, in each case it is clear that there are a significantly high proportion of transgenic lines containing only a single insertion locus (Table I).
Figure 3.
Southern hybridization analysis of nine representative T1 transgenic lines harboring silencing transgenes. From left: lane 1, Δ9-HP*6; lane 2, Δ9-HP*51; lane 3, Δ9-HP*62; lane 4, Δ9-HP*150; lane 5, Δ9-AS*118; lane 6, Δ12-HP*23; lane 7, Δ12-HP*83; lane 8, Δ12-HP*128; and lane 9, Δ12-HP*124. Genomic DNA from each line was digested with HindIII and probed with the whole promoter fragment of the soybean lectin gene. The bands represent the transgene insertions. The migration of DNA size markers is shown on the left in kb.
Table I.
Nos. of transgenic plants produced using gene-silencing constructs and estimated nos. of transgene insertions
| Transgene Construct | No. of Confirmed Transgenic | No. of Transgenic Plants Having Transgene Copy No.
|
||
|---|---|---|---|---|
| 1 | 2 | >2 | ||
| Δ9-HP | 29 | 13 | 3 | 13 |
| Δ9-AS | 25 | 9 | 4 | 12 |
| Δ12-HP | 34 | 16 | 7 | 11 |
| Δ12-AS | 27 | 10 | 4 | 13 |
Silencing of Δ12-Desaturase Results in HO Cottonseed Oil
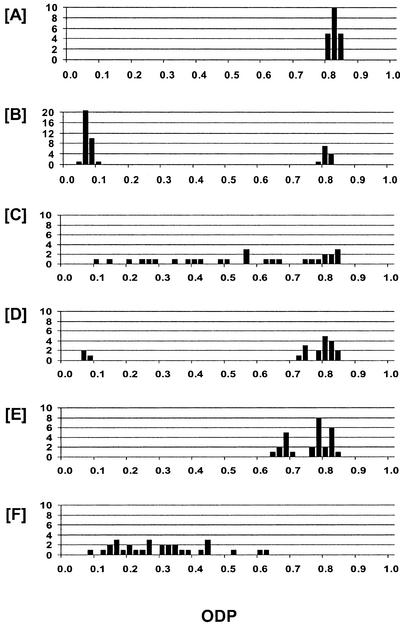
The parental variety Coker 315 has a consistently HO desaturation proportion (ODP) value ranging from 0.80 to 0.85 (Fig. 4A), meaning that over 80% of oleic acid produced in the developing seed is subsequently converted to linoleic acid by the action of the oleoyl-PC Δ12-desaturase enzyme. Many of the T1 plants carrying the Δ12-HP and Δ12-AS constructs showed a considerable reduction in ODP value, down to as low as 0.04, indicating a profound (95%) down-regulation of Δ12-desaturase activity. The HP construct was more effective than the AS construct in achieving the down-regulation of Δ12-desaturase activity, with 18 of 34 (53%) of the Δ12-HP T1 plants showing a reduction in ODP, compared with 10 of 27 (37%) for the Δ12-AS T1 plants.
Figure 4.
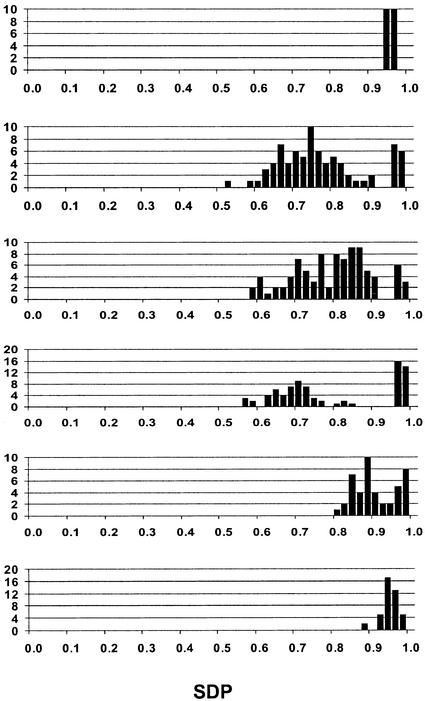
Frequency distribution for ODP in individual seeds of cotton cv Coker 315 (A), and T2 seeds from Δ12-desaturase-silenced lines Δ12-HP*23 (B), Δ12-HP*72 (C), Δ12-AS*132 (D), Δ12-AS*126 (E), and Δ12-AS*86 (F).
The pattern of segregation for the Δ12-silencing trait was examined by analyzing fatty acid composition of individual selfed T2 seeds borne on 15 of the T1 plants. An average of 35 T2 seeds were analyzed from each of the selected T1 plants carrying the Δ12-HP construct. Although Southern-blot analysis (Fig. 3) showed the presence of single transgene insertions in some lines (for example, Δ12-HP*23 and Δ12-HP*124) and multiple insertions in others (for example, Δ12-HP*83 and Δ12-HP*128), in all cases the ratio of low ODP (Δ12 silenced) to normal ODP fitted a 3:1 ratio when tested using the Chi-squared goodness of fit parameter, indicating a single gene inheritance. However, two distinct segregation patterns were evident and are typified by the examples shown in Figure 4, B and C. In 11 of 15 lines examined, typified by line Δ12-HP*23 (Fig. 4B), there was a very high level of silencing of the Δ12-desaturase in all T2 seeds carrying the transgene, indicating that the low ODP phenotype was inherited as a completely dominant trait. In contrast to this pattern, the distribution for ODP in the T2 seeds from the other four T1 plants, as typified by line Δ12-HP*72 (Fig. 4C), revealed a more intermediate level of silencing and a continuous distribution between the wild-type value and that of the most silenced seed. The very wide spread of ODP values in the silenced class suggests that either the expression of this silencing was highly variable, or that there were phenotypic differences between transgene hemizygotes and homozygotes. Progeny testing would be necessary to resolve these possibilities.
T2 seeds were also analyzed from three self-fertilized Δ12-AS lines that showed distinct patterns of Δ12-desaturase down-regulation. For line Δ12-AS*132 (Fig. 4D), about one-quarter of the T2 seeds showed very high-level silencing of Δ12-desaturase, equivalent to the best silencing from the Δ12-HP lines, with the remaining seeds displaying the wild-type phenotype. Such a 1:3 ratio suggests that silencing was only effective when the transgene was in the homozygous state, with hemizygotes showing little or no silencing. This type of segregation pattern was also evident in line Δ12-AS*126 (Fig. 4E), but the degree of silencing was much lower in this case. In the third line, Δ12-AS*86 (Fig. 4F), there was moderate to high silencing in all 30 T2 seeds with no wild-type phenotypes being observed. This pattern would be consistent with the control of silencing by multiple independent copies of the transgene, where the population size may have been insufficient to recover the expected low frequency of null genotypes.
As expected, the pronounced silencing of Δ12-desaturase resulted in large reductions in linoleic acid and concomitant increases in oleic acid (Table II), regardless of whether the transgene was Δ12-HP or Δ12-AS. In the most extreme case of silencing using the Δ12-HP construct (line Δ12-HP*23), oleic acid was increased from the normal level of 13% up to 78%, and correspondingly the level of linoleic acid reduced from normally 59% down to 4%. Interestingly, palmitic acid was also significantly reduced, down from 26% to 15%. Stearic acid is only present in very small amounts and was unchanged in the Δ12-desaturase-silenced lines.
Table II.
Fatty acid composition, SDP, and ODP values for Coker 315, the highest stearic acid seed obtained by silencing ghSAD-1 (from line Δ9-HP*150), the highest oleic acid seed obtained by silencing ghFAD2-1 (from line Δ12-HP*23), and putative homozygous recombinant F2 seed from the cross Δ9-HP*150 × Δ12-HP*23
| Line | Fatty Acid Composition
|
SDP | ODP | |||||
|---|---|---|---|---|---|---|---|---|
| Palmitic (16:0) | Stearic (18:0) | Oleic (18:1) | Linoleic (18:2) | Linolenic (18:3) | Arachidic (20:0) | |||
| % | ||||||||
| Coker 315 | 25.6 | 2.3 | 13.2 | 58.5 | 0.1 | 0.3 | 0.97 | 0.82 |
| Δ9-HP*150 | 14.9 | 39.8 | 3.8 | 38.8 | 0.2 | 2.4 | 0.52 | 0.91 |
| Δ12-HP*23 | 15.3 | 2.3 | 78.2 | 3.7 | 0.1 | 0.3 | 0.97 | 0.05 |
| F2 | 13.7 | 39.9 | 37.4 | 6.0 | 0.6 | 2.4 | 0.52 | 0.15 |
Silencing of Δ9-Desaturase Results in HS Cottonseed Oil
Δ9-desaturation is extremely active in developing cotton seeds, with around 96% of stearic acid formed during seed lipid synthesis being desaturated, initially to oleic acid, in cotton cv Coker 315 and only 2% to 3% of stearic acid remaining in the seed oil at maturity. As was the case for Δ12-desaturase, the Δ9-HP construct was more effective than the Δ9-AS construct in silencing the target Δ9-desaturase gene. For the Δ9-HP construct, 18 of 29 T1 plants (62%) showed reductions in stearic desaturation proportion (SDP), whereas only 6 of 25 (24%) of the Δ9-AS lines were reduced. However, the degree of silencing achieved was considerably less for Δ9-desaturase than was the case for Δ12-desaturase. SDP was reduced from 0.96 in cotton cv Coker 315 down to 0.52 in the most silenced individual seed, representing only a halving of Δ9-desaturase activity compared with the over 95% reduction that was achieved for Δ12-desaturase using the same hpRNA strategy.
The pattern of segregation for the modified Δ9-desaturation trait was examined by analyzing fatty acid composition of individual T2 seeds borne on a range of self-fertilized T1 plants. An average of 48 T2 seeds were analyzed from each of 10 T1 plants carrying the Δ9-HP construct. Segregation patterns for five of these lines that typify the overall results are shown in Figure 5. Three of the T1 plants had individual T2 seeds that showed relatively high degrees of silencing. In each of these lines, the class of seed having reduced Δ9-desaturation (mid-SDP) was distinct from that having the wild-type (high-SDP) phenotype. In the case of line Δ9-HP*150 (Fig. 5B), the ratio of mid-SDP:high-SDP fitted a 3:1 ratio expected for the segregation of a single locus trait, even though Southern-blot analysis revealed the presence of at least six copies of the transgene (Fig. 3, lane 4). The distribution of SDP for line Δ9-HP*62 (Fig. 5C) closely fitted a 15:1 ratio, suggesting the segregation of two independent silencing loci, with Southern-blot analysis indicating three to four copies of the transgene (Fig. 3, lane 3). In contrast, the distribution of SDP in line Δ9-HP*37 (Fig. 5D) did not fit any predicted ratio, there being an excess of wild-type individuals compared with that expected with even a single locus segregation. The distribution within the mid-SDP classes was continuous in each of these three lines and did not enable any putative discrimination of transgene hemizygotes and homozygotes.
Figure 5.
Frequency distribution for SDP in individual seeds of cotton cv Coker 315 (A), and T2 seeds from Δ9-desaturase-silenced lines Δ9-HP*150 (B), Δ9-HP*62 (C), Δ9-HP*37 (D), Δ9-HP*6 (E), and Δ9-HP*72 (F).
In the six other T1 lines, exemplified by Δ9-HP*6 (Fig. 5E) and Δ9-HP*72 (Fig. 5F), the degree of Δ9-desaturase silencing was relatively minor. Although in each case the distribution of SDP was continuous, cleavage at an SDP value of 0.95, based on the cotton cv Coker 315 wild type having a consistently higher SDP value, resulted in the segregation of silenced to wild-type individuals that fitted a 3:1 ratio in five of the six lines. For the remaining line, the distribution of SDP was continuous with only very minor reductions in SDP evident. None of the Δ9-AS lines were examined for segregation of the trait among T2 seeds.
The partial silencing of Δ9-desaturase resulted in significant increases in stearic acid and concomitant decreases in both oleic and linoleic acids (Table II) in the Δ9-HP and Δ9-AS lines. In line Δ9-HP*150, one of the most extreme cases of silencing using the Δ9-HP construct, stearic acid was raised from normal levels of 2% up to 40%, with oleic acid being reduced from 13% to 4% and linoleic acid down from 59% to 39%. As was the case with Δ12-desaturase silencing, palmitic acid was also significantly reduced, down from 26% to 15%.
Reduced ODP and SDP Is Seed Specific and Associated with Reduced Desaturase mRNA Levels
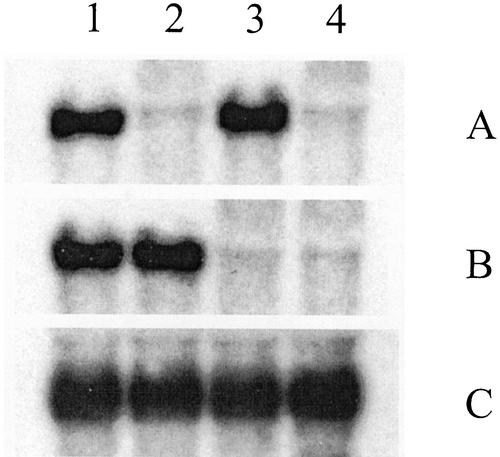
Expression levels of the targeted ghSAD-1 and ghFAD2-1 genes were examined by northern-blot analysis of RNA isolated from developing seeds. We have previously demonstrated that ghSAD-1 (Liu et al., 1996) and ghFAD2-1 (Liu et al., 1999b) are highly expressed in the developing cotton embryos, concomitant to the accumulation of storage lipids, with the highest expression at mid-maturation stages. RNAs originating from pooled samples of developing T2 seeds from Δ9-HP*62 and Δ12-HP*23 (Fig. 6, lanes 2 and 3, respectively) were probed with the ghSAD-1 or ghFAD2-1 cDNA clones at high stringency. The Δ9- and Δ12-silenced lines showed drastic reductions in ghSAD-1 and ghFAD2-1 mRNA levels, respectively, compared with the Coker 315 control (Fig. 6, lane 1), but each line had normal levels of mRNA for the desaturase that was not targeted by the silencing transgene. The sharp contrast in expression levels of the respective targeted desaturase genes compared with those in the untransformed cotton cv Coker 315 clearly indicates that the drastic reduction in SDP and ODP levels in the transgenic lines is associated with substantially lowered levels of each transcript. Furthermore, analysis of leaf lipids in a number of Δ9-HP and Δ12-HP lines (data not shown) demonstrated that they were identical to those of Coker 315, suggesting that the gene silencing was restricted to the developing seeds as expected from reporter gene studies using the soybean lectin promoter (Townsend and Llewellyn, 2002).
Figure 6.
Northern-blot analyses of ghSAD-1- (A), ghFAD2-1- (B), and ghKASII- (C) specific RNAs in developing embryos of transgenic cotton and control. Samples were extracted from mid-maturation embryos (approximately 30 d after fertilization) from untransformed cotton cv Coker 315 (lane 1), T2 seeds of Δ9-HP*62 (lane 2) and Δ12-HP*23 (lane 3), and F2 seeds of the cross Δ9-HP*150 × Δ12-HP*23 (lane 4).
Combination of HO and HS Traits by Crossing Δ12-HP and Δ9-HP Lines
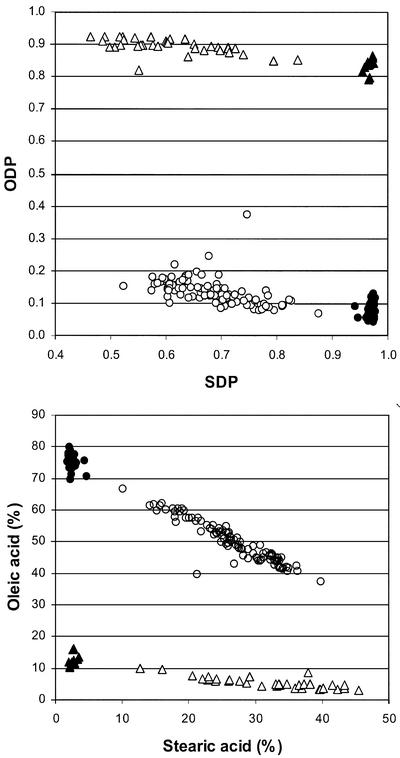
The possibility of combining the silencing of the Δ9-desaturase and Δ12-desaturase genes into a single line to generate further variability for fatty acid composition was examined initially by intercrossing the Δ9-HP*150 and Δ12-HP*23 T1 plants. Based on Southern-blot analysis and within-line trait segregation, the Δ12-HP*23 parent contains only one copy of the Δ12-silencing transgene, whereas the Δ9-HP*150 parent appears to contain at least six copies of the Δ9-silencing transgene (Fig. 3), of which only one copy seems to be contributing to the Δ9-desaturase silencing. Because the hybridization was performed using hemizygous T1 plants as parents, the F1 seeds are expected to segregate for the transgenes. Among 36 F1 seeds analyzed nondestructively for fatty acid composition, 12 seeds had levels of stearic and oleic acids similar to wild type, 10 seeds had elevated stearic acid alone, seven seeds had elevated oleic acid alone, and seven seeds had elevated levels of both stearic and oleic acids, indicating that they carried both Δ9- and Δ12-silencing transgenes. One F1 plant was established from this latter class and 199 individual F2 seeds borne on this plant were analyzed for fatty acid composition to determine the phenotypes of the transgene recombinants. The plots of SDP against ODP (Fig. 7A) and stearic acid against oleic acid content (Fig. 7B) clearly show the presence of four classes representing the combinations of wild-type and silenced phenotypes for each of the two targeted desaturases. The ratio of silenced to wild type for the Δ12-desaturase (140:59) and for the Δ9-desaturase (152:47) in each case fitted a 3:1 ratio expected for the segregation of a single dominant gene (χ21 = 2.29 and 0.20, respectively). The joint segregation of the two traits also closely fitted the pattern expected for two independently segregating genes (χ21 = 1.15).
Figure 7.
Joint segregation of ODP with SDP (A), and oleic acid with stearic acid (B), in F2 seed populations from the cross Δ9-HP*150 × Δ12-HP*23. Seeds are classified as typical of the Δ9-HP*150 HS parent (▵), the Δ12-HP*23 HO parent (●), recombinant HO and HS (○), and wild-type cotton cv Coker 315 (▴).
Among the F2 seeds, the pattern of silencing for the individual Δ9 and Δ12 desaturases was similar to that observed among the T2 seeds of the parental lines. Within the Δ12-silenced genotypes, variation for ODP was relatively narrow, ranging only between 0.04 and 0.13 in the Δ9 wild-type class and mainly between 0.07 and 0.24 in the Δ9-silenced class. In contrast, variation for SDP within the Δ9-silenced genotypes was very broad, ranging from 0.46 to 0.84 in the Δ12 wild-type class and between 0.52 and 0.88 in the Δ12-silenced class. This was consistent with the relative variation observed for ODP and SDP, respectively, in the T2 seeds of the parental lines, Δ12-HP*23 (Fig. 4B) and Δ9-HP*150 (Fig. 5B). The combined effects of the Δ9- and Δ12-desaturase silencing resulted in a wide range of novel combinations of stearic acid and oleic acid levels in the F2 seeds carrying both transgenes. There was a strong negative correlation between oleic acid and stearic acid levels, with the extreme types having on the one hand 10% stearic and 65% oleic, and on the other hand 40% stearic and 38% oleic, reflecting the precursor/product relationship of these two fatty acids. Because the two silencing transgenes are segregating independently, it is expected that 1 of 16 of the F2 seeds are homozygous for both the Δ9- and Δ12-silencing transgenes. These are most likely to be the seeds having around 30% to 35% stearic and 40% to 45% oleic acid, although progeny testing would be necessary to confirm the genotypes of the individual seeds. As was the case in both parental lines, palmitic acid was around 10% lower in the lines carrying both silencing constructs compared with the wild-type genotypes, being as low as 12% of total fatty acids in the putative group of double homozygous F2 individuals. Northern-blot analysis using the RNA extracted from the developing F2 embryos confirmed the simultaneous silencing of ghSAD-1 and ghFAD2-1 (Fig. 6, A and B, lane 4).
In some of the initial transgenic lines and recombinants expressing the HS trait, we observed that some HS seeds, although germinating well at room temperature on water-soaked filter paper, were slow to establish and grow when transferred to soil. By analyzing fatty acid composition of a small proportion of the seed and leaving the rest to germinate in soil, we observed a correlation between high levels of stearic acid and reduced survival ability of seedlings (data not shown). However, germination and growth of HS seeds was increased significantly when they were germinated and grown at approximately 5°C higher temperature. Furthermore, all HS seeds gave rise to viable plants when they were germinated and established on Suc-supplemented tissue culture media. No germination difficulties were observed with any of the seeds expressing only the HO trait.
DISCUSSION
We have recently demonstrated in Arabidopsis that inverted repeat gene-silencing constructs are more efficient than either antisense or cosuppression in down-regulating expression of the FAD2 gene encoding Δ12-desaturase, both in terms of higher frequency of transgenics showing silencing, and generally higher degrees of silencing, particularly in heterozygotes (Stoutjesdijk et al., 2002). Those observations are now further supported by the current results from cotton where inverted repeat constructs targeted against either the FAD2-1 or the SAD-1 gene resulted in substantially higher recovery of silenced individuals than did the corresponding antisense constructs, and by our recent demonstration that the inclusion of an intron in the inverted repeat constructs targeted against the FAD2 gene results in 100% efficiency of silencing (Wesley et al., 2001). The high efficiency of gene silencing obtainable through the use of hpRNA-mediated PTGS makes this technique a valuable contribution to practical trait modification in agricultural plants. This is particularly important for those plants, such as cotton, that have relatively low transformation efficiency, or where it is required to have larger transgenic populations to obtain selectable marker free plants by segregation.
Our results demonstrate that hpRNA constructs targeted specifically against the ghFAD2-1 gene can almost completely silence Δ12-desaturase activity in developing cottonseed (ODP reduced to approximately 6% of normal levels), apparently without impairing normal seed development or subsequent germination and plant growth. Because of its tetraploid origin, two highly homologous copies (98% DNA sequence similarity) of the ghFAD2-1 gene exist in cotton, one in each of the A and D subgenomes, and appear to contribute equally to the abundance of ghFAD2-1 mRNA in the developing cottonseeds (Liu et al., 1999b). Northern-blot analysis showed the presence of only very low levels of ghFAD2-1 mRNA during the period of rapid fatty acid synthesis in developing seeds of the most highly silenced Δ12-HP transgenic line, confirming the expectation that mRNA transcripts from both subgenomic copies of FAD2-1 would be targeted for degradation. Interestingly, the ghFAD2-1-silencing trait was inherited as a single dominant gene in several of the highly silenced inverted repeat lines, indicating that a single copy of the silencing transgene can be sufficient to achieve maximum suppression of the target ghFAD2-1 gene. This feature was more pronounced in the current cotton experiments than was the case with FAD2 silencing in Arabidopsis (Stoutjesdijk et al., 2002), where hemizygotes for highly effective silencing transgenes approached, but generally did not equal, the degree of silencing achieved in homozygotes. This difference may indicate that the timing and degree of expression of the lectin-driven silencing transgene was a more effective fit to the expression pattern of the targeted endogenous FAD2-1 gene in cotton than was the case with the napin-driven transgene in Arabidopsis. The profound reduction in Δ12-desaturase activity achieved through hpRNA-mediated silencing of the ghFAD2-1 gene is considerably greater than that recently reported in cottonseed by Chapman et al. (2001), where transformation with a full-length, but nonfunctional, rapeseed FAD2 sequence in sense orientation achieved only a halving of Δ12-desaturation, with substantial amounts of linoleic acid (approximately 28%) remaining in the most extreme lines. In that case, the introduced sequence has relatively low homology to the cotton FAD2 genes and it seems probable that the reduction of Δ12-desaturation is the consequence of a phenomenon other than PTGS.
It is notable that even in the most highly Δ12-silenced lines, there is still a small amount of linoleic acid (approximately 3%) that accumulates in the oil, reflecting a low but significant residual level of Δ12-desaturase activity. This is almost identical to our experience with silencing of the equivalent FAD2 gene in Arabidopsis using hpRNA-mediated PTGS (Stoutjesdijk et al., 2002), where it seemed likely that the residual Δ12-desaturase activity is encoded by other divergent Δ12-desaturase genes that are not effectively targeted by the silencing construct, presumably the FAD6 gene encoding a Δ12-desaturase that acts on oleic acid bound to glycerolipid substrates in the plastid. It is possible that such FAD6-mediated Δ12-desaturation may also account for some of the residual linoleic acid present in the FAD2-silenced cotton lines. In addition, whereas Arabidopsis has only a single FAD2 gene, cotton has been shown by Southern-blot analysis to have at least five copies (Liu et al., 1999b). Although the ghFAD2-1 member of this gene family appeared to be the major contributor to the desaturation of oleic acid in the seed oil, it is not known how much contribution the other members make. We have isolated one other member, ghFAD2-2 (Liu et al., 1999a), which appears to be expressed only at low levels throughout the plant, including in the seeds. Similar to the situation in soybean (Heppard et al., 1996), this constitutively expressed ghFAD2-2 sequence has only about 70% identity with ghFAD2-1, which is probably a sufficient mismatch for ghFAD2-2 mRNA to escape significant homology-dependent silencing in the ghFAD2-1-targeted lines and to perhaps contribute to the residual Δ12-desaturase activity in their seeds.
In contrast to the very high-level silencing of Δ12-desaturase, only intermediate down-regulation of Δ9-desaturase was achieved in the current study. The most highly silenced Δ9-HP line recovered still accumulated about 43% of C18 unsaturated fatty acids, indicating that Δ9-desaturation had only been halved by the ghSAD-1-silencing transgene. The absence of high-level reductions in Δ9-desaturase activity in cottonseed, even with inverted repeat gene-silencing techniques, accords with previous attempts to down-regulate this enzyme in rapeseed using antisense techniques (Knutzon et al., 1992), where stearic acid was also raised only up to 40%. This common experience possibly reflects the essential role that Δ9-desaturase plays in cellular lipid synthesis. All C18 unsaturated fatty acids present in plant cells, whether in plastidic or microsomal membranes or deposited as triacylglycerols in oleosomes, originate from the desaturation of stearoyl-ACP in the plastid by the stearoyl-ACP Δ9-desaturase. Complete removal of this enzyme activity would leave cells without the ability to synthesize any C18 unsaturated fatty acids and impair their ability to appropriately manipulate membrane fluidity (Lightner et al., 1994). This essential nature of stearoyl-ACP Δ9-desaturase may have been a factor favoring the evolutionary development of the multigene families that encode it in many plant species. For example, even though the small genome of Arabidopsis has only one FAD2 gene encoding Δ12-desaturase, it contains at least five genes encoding stearoyl-ACP Δ9-desaturase (Ohlrogge and Jaworski, 1997). Similarly, Southern-blot analysis in cotton has shown that the Δ9-desaturase multigene family consists of six to eight members per diploid genome (Liu et al., 1996; Liu, 1998) and we have recently isolated at least four cDNA clones with unique 3′-untranslated regions (UTRs) from a cottonseed cDNA library (Q. Liu, unpublished data).
At present, neither the degree of sequence diversity between the individual SAD genes, nor their relative contribution to the overall Δ9-desaturation activity in the developing cottonseed storage lipids, are known. Therefore, it is not possible to predict how much the expression of other SAD genes is likely to be affected by the ghSAD-1-silencing construct. It might be the case that the SAD gene family has high levels of sequence homology and is capable of being globally silenced by a ghSAD-1-silencing construct. If so, highly expressed ghSAD-1-silencing transgenics accumulating very high levels of stearic acid might not be recovered due to lethality resulting from either inadequate levels of C18 unsaturated fatty acids in their membrane lipids or inability of transgenic embryoids to mobilize the HS lipids to support germination. In such a case, it might be expected that only those transgenics with weakly expressing silencing transgenes, and consequently intermediate levels of stearic acid and essential C18 unsaturated fatty acids, might be recovered. However, in the current study we did not observe any substantially lower recovery rates from transformations involving the Δ9-desaturase-silencing constructs compared with those involving the Δ12-desaturase constructs. Furthermore, the highest stearic line had a dramatic reduction in ghSAD-1 mRNA, indicative of high-level silencing of that gene. Therefore, it appears more likely that the significant remaining Δ9-desaturation activity in the HS lines is due to other SAD genes escaping silencing, rather than a global but weak silencing of all members of this gene family.
Assuming that it is possible to use PTGS to achieve greater reductions in Δ9-desaturase activity in cottonseed than were obtained in the current study, there may still be physiological limitations to the level of stearic acid that can ultimately be attained. In the present study, seed germination and seedling establishment were impaired in some of the HS cotton lines, but not in any of the HO lines, even though initial indications are that both types had apparently normal oil content (data not shown). This problem was readily overcome by germinating the HS seeds on Suc-containing medium, suggesting that it was caused by poor ability to mobilize the altered seed oil as an energy source. Similar germination problems have been reported in HS mutants of soybean (Rahman et al., 1997) and HS genotypes of Brassica spp. produced by antisense-mediated down-regulation of stearoyl-ACP Δ9 desaturase (Knutzon et al., 1992). The inability of cottonseeds to effectively utilize HS seed oils during germination could operate through a number of mechanisms. Future studies comparing germination of cottonseeds having various fatty acid modifications, and analyzing the fatty acid structure of triglycerides and other cellular lipids in the HS cottonseed lines, should provide further insight into this issue.
As expected, the principal effect of silencing Δ9- and Δ12-desaturase was to alter the relative proportions of the C18 fatty acids—stearic, oleic, and linoleic acids—by decreasing the levels of the fatty acids downstream of the relevant enzyme and increasing the levels of the immediate fatty acid substrate. However, palmitic acid was also significantly reduced in the HS and HO lines, as well as in the lines carrying both traits. One possible explanation for this may be that the accumulation of oleic acid in the Δ12-silenced lines and stearic acid in the Δ9-silenced lines results in increased levels of oleoyl-CoA and stearoyl-CoA, respectively, in the cytoplasm and that this alters the relative selectivity of acyl-transferases responsible for the movement of the fatty acids into triglycerides in a manner that reduces the incorporation of palmitate. The concentrations of minor fatty acids were relatively unaltered in the Δ9- and Δ12-silenced lines. In particular, the levels of the cyclopropenoid fatty acids, malvalic acid and sterculic acid, remained low (<1%) in oil extracts taken from bulked HS or HO lines (data not shown). Thus, despite the large increase in oleic acid in the Δ12-silenced lines, there was no evidence of increased production of the cyclopropenoid fatty acids that are synthesized from oleic acid. This observation is consistent with the theory that cyclopropenoid fatty acid synthesis occurs mainly in the embryo axis of cottonseed (Wood, 1986), where it would be spatially separated from the highly enriched cotyledonary oleic acid pool in the Δ12-silenced lines. Ultimately, it will be interesting to examine the levels of cyclopropenoid fatty acids in the embryo axis lipids of the very HO lines to determine whether they are substantially altered as a result of the increased level of oleic acid.
The alterations to fatty acid composition of cottonseed oil achieved using PTGS should enable the development of a range of cottonseed oils that better match current end-use requirements. In particular, the HO cottonseed oil containing predominantly oleic acid (75%) and palmitic acid (15%) is expected to be even more stable than the HO forms of other oilseeds such as soybean, rapeseed, and sunflower, and should be usable in long-life deep-frying applications, such as in the food service and snack food sectors, without the need for hydrogenation and the associated production of nutritionally undesirable trans-fatty acids. Similarly, the HS cottonseed oil should prove suitable for solid fat applications, such as margarines and shortenings, without hydrogenation. Although it may be possible to further reduce linoleic acid below the 4% present in the best HO lines, such a change may not be advantageous because there is evidence from studies with midoleic genotypes of rapeseed (Xu et al., 2000) that a modest level of linoleic acid in the oil is desirable from a flavor standpoint, and should not be sacrificed for the minimal further improvement in stability that would result from its complete removal. In fact, it may even be desirable to select HO lines that have slightly higher levels of linoleic acid to produce an optimal cooking oil.
The relatively high level of palmitic acid naturally present in cottonseed oil has been an important contributor to the stability of the oil and to the solidity of its hydrogenated derivatives, but is nutritionally undesirable. Because the increases in oleic acid or stearic acid are able to impart the required functional properties on the modified oils, it should now be possible to dramatically lower palmitic acid in cottonseed oil without compromising performance. Both the HO and HS oils developed in the current study already have a significant reduction in palmitic acid, and thereby enhanced nutritional value. Further reductions in palmitic acid should be possible in both the HO and HS oils through genetic manipulation of the enzymes controlling palmitic acid synthesis, in particular palmitoyl-ACP thioesterase and keto-acyl synthase II.
The HO and HS characteristics behaved as independent traits that were able to be brought together in recombinant genotypes having elevated levels of both fatty acids. The degree of silencing of both Δ9- and Δ12-desaturase in these recombinant genotypes was equivalent in its magnitude to that observed in the individually silenced parental lines, demonstrating for the first time, to our knowledge, that hpRNA-induced PTGS in independent genes can be stacked without any diminution in the degree of silencing. Because stearic acid and oleic acid percentages are negatively correlated due to their precursor/product relationship, the recombinant genotypes show intermediate levels of both fatty acids. Furthermore, because it was possible to obtain various degrees of elevation of oleic acid and stearic acid in individually silenced transgenic lines, the opportunity exists to develop a wide range of alternative palmitic, stearic, and oleic acid combinations through recombination of appropriately chosen parental lines. This should enable the development of cottonseed fats and oils that satisfy different application requirements, in particular lines with fatty acid profiles matching those of valuable specialty confectionery fats such as cocoa (Theobroma cocoa) butter.
MATERIALS AND METHODS
Gene-Silencing Constructs
Gene-silencing constructs designed to target the endogenous cotton (Gossypium hirsutum) genes encoding either stearoyl-ACP Δ9-desaturase or microsomal Δ12-desaturase, using the ghSAD-1 or ghFAD2-1 cDNA sequences, respectively, in either antisense or inverted repeat configurations, are shown in Figure 2. The silencing constructs were each driven by the soybean (Glycine max) lectin promoter, which has been shown to direct seed-specific expression of a β-glucuronidase reporter gene in cotton (Townsend and Llewellyn, 2002). A HindIII/EcoRI fragment containing the soybean lectin promoter and terminator sequences was excised from the pGLe-10 plasmid (Cho et al., 1995) and engineered into the same restriction sites of binary vector pBI121 (CLONTECH) from which the cauliflower mosaic virus 35S-Gus-Nos chimeric gene had been removed. This formed a pBI-Lec binary vector that was subsequently used to carry all the gene-silencing constructs described in this paper. The Δ9-desaturase antisense construct (Δ9-AS) consisted of the entire ghSAD-1 cDNA (Liu et al., 1996) cloned behind the lectin promoter in an antisense orientation. For the Δ9-desaturase inverted repeat construct (Δ9-HP), a 514-bp fragment was PCR amplified from the 5′ end of ghSAD-1 using oligonucleotides Δ9s1 (5′-TTTTAATGCCATCGCCTCG-3′) and Δ9a1 (5′-CTTCAGCAGTCCAAGCCCTG-3′) and inserted at the 3′ end of the ghSAD-1 sequence to form an inverted repeat. This ghSAD-1 inverted repeat construct was then ligated behind the lectin promoter in the sense orientation in relation to the full-length ghSAD-1 sequence. A 1,351-bp fragment of ghFAD2-1 (Liu et al., 1999b) was PCR amplified using oligonucleotides Δ12s1 (5′-CCTGGCGTTAAACTG CTTTC-3′) and Δ12a1 (5′-CCATATAGTTTATTAATATAACAC-3′) and consisted of the entire Δ12-desaturase coding region, the full 3′-UTR, and a partial 5′-UTR. This ghFAD2-1 fragment was cloned behind the lectin promoter in an antisense orientation to make the antisense construct (Δ12-AS). For the Δ12-desaturase inverted repeat construct (Δ12-HP), an 853-bp fragment was PCR amplified from the 5′ end of the ghFAD2-1 with oligonucleotides Δ12s1 and Δ12a2 (5′-TATGTTGCCGTAGGTGATC-3′) and ligated at the 3′ end of ghFAD2-1 to form an inverted repeat. The whole ghFAD2-1 inverted repeat unit was then ligated behind the lectin promoter in pBI-Lec binary vector in a sense orientation in relation to the full-length ghFAD2-1 sequence.
Cotton Transformation
Transgenic cotton cv Coker 315 plants were generated by Agrobacterium tumefaciens-mediated transformation, and selection on medium containing kanamycin sulfate, by a modification of the method described by Cousins et al. (1991). Cotton seedlings were germinated aseptically on Murashige and Skoog medium (Murashige and Skoog, 1962) solidified using phytagel (Sigma, St. Louis). Seedlings were maintained under low-light conditions at 28°C. Cotyledon explants from 10- to 14-d-old seedlings were cocultivated with A. tumefaciens strain AGL1 containing the relevant gene construct for 2 d on the medium containing Murashige and Skoog macro- and micro-elements and B5 vitamins (Gamborg et al., 1968), 100 mg L−1 myo-inositol, 30 g L−1 Glc, 0.2 mg L−1 2,4-dichlorophenoxyacetic acid, 0.1 mg L−1 kinetin, and 0.93 g L−1 magnesium chloride, and solidified using 2 g L−1 phytagel. The callus was induced on the same medium but supplemented with 50 mg L−1 kanamycin sulfate and 250 mg L−1 cefotaxime at 28°C for 6 weeks. Healthy calli were then transferred to Murashige and Skoog medium containing 5 mg L−1 6-(γ,γ-dimethylallylamino)-purine, 0.1 mg L−1 naphthalene acetic acid, 25 mg L−1 kanamycin, and 250 mg L−1 cefotaxime for a second selection period of 6 weeks at 28°C. Somatic embryogenesis was initiated on the solidified Murashige and Skoog medium, without added phytohormone or antibiotic, and the embryoids formed were then germinated on Stewart and Hsu medium (Stewart and Hsu, 1977) solidified with phytagel to produce transgenic cotton plantlets. Primary transgenic cotton plantlets (herein referred to as the T1 generation) were transferred to soil and maintained in a greenhouse once leaves and roots developed.
Identification of Transgenic Plants by PCR
The presence of the transgenes in each regenerated cotton plant was initially determined by PCR, using cotton genomic DNA as a template, and the following oligonucleotides: 3Lec-s1, 5′-CATGTGACAGATCGAAGGAA-3′; and 3Lec-a1, 5′-ATCTAATTATTCTATTCAGAC-3′.
This process amplifies an approximately 300-bp DNA fragment comprising the transcriptional terminator of the soybean lectin gene. Accordingly, amplification only occurs from plant DNA containing the introduced chimeric genes. Further confirmation of transgenic status of the regenerated cotton plants was obtained by Southern-blot analysis as described below.
DNA Isolation and Southern-Blot Analysis
Cotton genomic DNA was isolated according to Paterson et al. (1993) and further purified using the CsCl gradient method as described by Sambrook et al. (1989). Approximately 10 μg of DNA was digested by HindIII and the restriction fragments were separated on a 0.7% (w/v) agarose gel by electrophoresis and transferred onto a Hybond N+ nylon membrane (Amersham, Buckinghamshire, UK) using 0.4 m NaCl for 4 h. Southern-blot analyses were carried out by hybridizing with an [α-32P]dCTP-labeled DNA fragment consisting of the promoter region of the soybean lectin gene that is specific for the transgene. The hybridization and subsequent washing were carried out as previously described (Liu et al., 1999b).
RNA Isolation and Northern-Blot Analysis
Cotton embryos at mid-maturation, approximately 30 d after fertilization, were harvested and RNA was isolated using the RNeasy Plant Mini Kit (Qiagen USA, Valencia, CA). RNA was separated on a denaturing formaldehyde gel and transferred onto a Hybond N+ nylon membrane according to Sambrook et al. (1989). The entire coding regions of the ghSAD-1 and ghFAD2-1 genes were used as probes. A cDNA fragment containing the entire coding region of the cotton keto-acyl synthase II gene (ghKASII) was obtained (Q. Liu, unpublished data) and used to probe the blot as a control, indicating the level of expression of nontargeted lipid synthesis genes in each line. The hybridization and the after washing was essentially the same as previously described (Liu et al., 1999b).
Fatty Acid Analysis
Self-pollinated seeds were harvested from each primary transgenic (T1) plant and analyzed for fatty acid composition. As an initial screen, the total lipids were extracted from pooled three-seed samples from each T1 plant by the method of Bligh and Dyer (1959) and used for fatty acid analysis. Subsequently, interesting lines were examined in more detail by performing fatty acid analysis on a number of individual T2 seeds borne on each T1 plant. In addition, some of the T1 plants carrying the ghSAD-1 or ghFAD2-1-silencing constructs were intercrossed to combine the traits. F1 seeds from these crosses were analyzed nondestructively by cutting off approximately a one-sixth portion of the seed distal to the embryonic axis and crushing this onto filter paper discs to provide expressed oil for analysis. The remaining larger portion of each seed, containing the embryonic axis, was planted directly into soil to establish F1 plants. F2 seeds were analyzed individually for fatty acids by methylation of oil expressed from individual seeds.
Fatty acid methyl esters were prepared by alkaline transmethylation. Samples of solvent-extracted or -expressed oil were loaded onto filter paper discs and methylated in 2 mL of 0.02 m sodium methoxide for 1 h at 90°C, followed by addition of 1.5 mL of hexane and 2 mL of water. After vortexing and phase separation, the upper hexane layer containing the fatty acid methyl esters was transferred to a microvial. Fatty acid methyl esters were analyzed by gas-liquid chromatography as previously described (Stoutjesdijk et al., 2002). Cyclopropenoid fatty acids were not routinely determined on all lines. Relative fatty acid compositions were calculated as the percentage that each fatty acid represented of the total measured fatty acids. An indirect method of assessing the cumulative effects of Δ9-desaturase and Δ12-desaturase activity during seed fatty acid synthesis is through the SDP and ODP parameters, derived by the following formulae: SDP = (% oleic + % linoleic)/(% stearic + % oleic + % linoleic) and ODP = (% linoleic)/(% oleic + % linoleic), respectively. These parameters represent the ratio of the total fatty acid products of desaturation to the amount of fatty acid substrate that was available, and are useful in illustrating the effects of gene silencing on the activities of the target enzymes. Cottonseed oil typically has an SDP value of around 0.97 and an ODP value of around 0.80, indicating that about 97% of stearic acid formed during fatty acid synthesis is subsequently desaturated to oleic acid and about 80% of this is further desaturated to linoleic acid. Phenotypic distributions for fatty acid composition, SDP, and ODP in T2 seed populations were compared with expected segregation ratios using the Chi-squared goodness of fit test at P = 0.05.
ACKNOWLEDGMENTS
We are grateful to Clive Hurlstone and Rhenzong Liu for technical assistance and to Dr. Danny Llewellyn and Dr. Belinda Townsend for advice on cotton transformation. We also thank Dr. Lorraine Tonnet, Lorraine Mason, and Richard Philips for analyzing fatty acid composition.
Footnotes
This work was supported by the Australian Cotton Research and Development Corporation (grant no. CSP–78C).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.001933.
LITERATURE CITED
- Ascherio A, Willett WC. Health effects on trans fatty acids. Am J Clin Nutr. 1997;66:1006S–1010S. doi: 10.1093/ajcn/66.4.1006S. [DOI] [PubMed] [Google Scholar]
- Auld DL, Heikkinen MK, Erickson DA, Sernyk JL, Romero JE. Rapeseed mutants with reduced levels of polyunsaturated fatty acids and increased levels of oleic acid. Crop Sci. 1992;32:657–662. [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Chapman KD, Austin-Brown S, Sparace SA, Kinney AJ, Ripp KG, Pirtle IL, Pirtle RM. Transgenic cotton plants with increased seed oleic acid content. J Am Oil Chem Soc. 2001;78:941–947. [Google Scholar]
- Cho MJ, Widholm JM, Vodkin LO. Cassettes for seed-specific expression tested in transformed embryogenic cultures of soybean. Plant Mol Biol Rep. 1995;13:255–269. [Google Scholar]
- Cousins YL, Lyon BR, Llewellyn DJ. Transformation of an Australian cotton cultivar: prospects for cotton improvement through genetic engineering. Aust J Plant Physiol. 1991;18:481–494. [Google Scholar]
- Cox C, Mann J, Sutherland W, Chisholm A, Skeaff M. Effects of coconut oil, and safflower oil on lipids and lipoproteins in persons with moderately elevated cholesterol levels. J Lipid Res. 1995;36:1787–1795. [PubMed] [Google Scholar]
- Gamborg OL, Miller RA, Ojima K. Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res. 1968;50:151–158. doi: 10.1016/0014-4827(68)90403-5. [DOI] [PubMed] [Google Scholar]
- Graef GL, Miller LA, Fehr WR, Hammond EG. Fatty acid development in a soybean mutant with high stearic acid. J Am Oil Chem Soc. 1985;62:773–775. [Google Scholar]
- Hamilton AJ, Brown S, Yuanhai H, Ishizuka M, Lowe A, Solis A-GA, Gierson D. A transgene with repeated DNA causes high frequency, post-transcriptional suppression of ACC-oxidase gene expression in tomato. Plant J. 1998;15:737–746. doi: 10.1046/j.1365-313X.1998.00251.x. [DOI] [PubMed] [Google Scholar]
- Heppard EP, Kinney AJ, Stecca KL, Miao GH. Developmental and growth temperature relation of two different microsomal ω-6 desaturase genes in soybeans. Plant Physiol. 1996;110:311–319. doi: 10.1104/pp.110.1.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney AJ. Improving soybean seed quality. Nature Biotechnol. 1996;14:946. doi: 10.1038/nbt0896-946. [DOI] [PubMed] [Google Scholar]
- Knutzon DS, Thompson GA, Radke SE, Johnson WB, Knauf VC, Kridl JC. Modification of Brassica seed oil by antisense expression of a stearoyl-acyl carrier protein desaturase gene. Proc Natl Acad Sci USA. 1992;89:2624–2628. doi: 10.1073/pnas.89.7.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightner J, Wu J, Browse J. A mutant of Arabidopsis with increased levels of stearic acid. Plant Physiol. 1994;106:1443–1451. doi: 10.1104/pp.106.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q. The isolation and characterisation of fatty acid desaturase genes in cotton. PhD thesis. Sydney University; 1998. [Google Scholar]
- Liu Q, Singh SP, Brubaker CL, Green AG. Cloning and sequence analysis of a novel member (accession No. Y10112) of the microsomal ω-6 fatty acid desaturase family from cotton (Gossypium hirsutum) Plant Physiol. 1999a;120:339. [Google Scholar]
- Liu Q, Singh SP, Brubaker CL, Sharp PJ, Green AG, Marshall DR. Molecular cloning and expression of a cDNA encoding a microsomal ω-6 fatty acid desaturase in cotton (Gossypium hirsutum L.) Aust J Plant Physiol. 1999b;26:101–106. [Google Scholar]
- Liu Q, Singh SP, Sharp PJ, Green AG, Marshall DR. Nucleotide sequence of a cDNA from Gossypium hirsutum encoding a stearoyl-acyl carrier protein desaturase (accession No. X95988) (OGR 96-012) Plant Physiol. 1996;110:1435. [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Murray F, Llewellyn D, McFadden H, Last D, Dennis ES, Peacock WJ. Expression of the Talaromyces flavus glucose oxidase gene in cotton and tobacco reduced fungal infection, but is also phytotoxic. Mol Breed. 1999;5:219–232. [Google Scholar]
- Norden AJ, Gorbet DW, Knauft DA, Young CT. Variability in oil quality among peanut genotypes on the Florida breeding program. Peanut Sci. 1987;14:7–11. [Google Scholar]
- Ohlrogge JB, Jaworski JG. Regulation of fatty acid synthesis. Ann Rev Plant Physiol Plant Mol Bio. 1997;48:109–136. doi: 10.1146/annurev.arplant.48.1.109. [DOI] [PubMed] [Google Scholar]
- Osorio J, Fernandez-Martinez J, Mancha M, Garces R. Mutant sunflower with high concentration of saturated fatty acids in their oil. Crop Sci. 1995;35:739–742. [Google Scholar]
- Paterson AH, Brubaker CL, Wendel JF. A rapid method for extraction of cotton (Gossypium spp.) genomic DNA suitable for RFLP or PCR analysis. Plant Mol Biol Rep. 1993;11:122–127. [Google Scholar]
- Radcliffe JD, King CC, Czajka-Narins DM, Imrhan V. Serum and liver lipids in rats fed diets containing corn oil, cottonseed oil, or a mixture of corn and cottonseed oils. Plant Foods Human Nutr. 2001;56:51–60. doi: 10.1023/a:1008189503099. [DOI] [PubMed] [Google Scholar]
- Rahman SM, Takagi Y, Knoshita T. Genetic control of high stearic acid content in seed oil of two soybean mutants. Theor Appl Genet. 1997;95:772–776. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Plainview, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Smith NA, Singh SP, Wang MB, Stoutjesdijk PA, Green AG, Waterhouse PM. Totally silencing by intron-spliced hairpin RNAs. Nature. 2000;407:319–320. doi: 10.1038/35030305. [DOI] [PubMed] [Google Scholar]
- Soldatov KI. Proceedings of the Seventh International Sunflower Association, Vlaardingen, The Netherlands. Toowoomba, Australia: International Sunflower Association; 1976. Chemical mutagenesis in sunflower breeding; pp. 352–357. [Google Scholar]
- Stewart JM, Hsu CL. In ovulo embryo culture and seedling development of cotton (Gossypium hirsutum L.) Planta. 1977;137:113–117. doi: 10.1007/BF00387547. [DOI] [PubMed] [Google Scholar]
- Stoutjesdijk P, Singh SP, Liu Q, Hurlstone C, Waterhouse P, Green A. hpRNA-mediated targeting of the Arabidopsis FAD2 gene gives highly efficient and stable silencing. Plant Physiol. 2002;129:1723–1731. doi: 10.1104/pp.006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend BJ, Llewellyn DJ (2002) Spatial and temporal regulation of a soybean (Glycine max) lectin promoter in transgenic cotton (Gossypium hirsutum). Funct Plant Biol (in press) [DOI] [PubMed]
- Wesley V, Helliwell C, Smith N, Wang MB, Rouse D, Liu Q, Gooding P, Singh S, Abbott D, Stoutjesdijk P et al. Construct design for efficient, effective and high throughput gene silencing in plants. Plant J. 2001;27:581–590. doi: 10.1046/j.1365-313x.2001.01105.x. [DOI] [PubMed] [Google Scholar]
- Wood R. Comparison of the cyclopropene fatty acid content of cottonseed varieties, glanded and glandless seeds, and various seed structures. Biochem Arch. 1996;2:73–80. [Google Scholar]
- Xu XQ, Tran VH, Palmer MV, White K, Salisbury P. Chemical, physical and sensory properties of Monola oil, palm olein and their blends in deep frying trials. Food Aust. 2000;52:77–82. [Google Scholar]