Abstract
We posed the question of whether steady-state levels of the higher polyamines spermidine and spermine in plants can be influenced by overexpression of a heterologous cDNA involved in the later steps of the pathway, in the absence of any further manipulation of the two synthases that are also involved in their biosynthesis. Transgenic rice (Oryza sativa) plants engineered with the heterologous Datura stramonium S-adenosylmethionine decarboxylase (samdc) cDNA exhibited accumulation of the transgene steady-state mRNA. Transgene expression did not affect expression of the orthologous samdc gene. Significant increases in SAMDC activity translated to a direct increase in the level of spermidine, but not spermine, in leaves. Seeds recovered from a number of plants exhibited significant increases in spermidine and spermine levels. We demonstrate that overexpression of the D. stramonium samdc cDNA in transgenic rice is sufficient for accumulation of spermidine in leaves and spermidine and spermine in seeds. These findings suggest that increases in enzyme activity in one of the two components of the later parts of the pathway leading to the higher polyamines is sufficient to alter their levels mostly in seeds and, to some extent, in vegetative tissue such as leaves. Implications of our results on the design of rational approaches for the modulation of the polyamine pathway in plants are discussed in the general framework of metabolic pathway engineering.
Relatively few pathways have been elucidated molecularly and biochemically in plants, and an even smaller number are amenable to modulation by molecular approaches. This is because of the complex nature of metabolic networks that are often regulated at different levels, spatially and temporally. In our ongoing efforts to implement rational molecular approaches to modulate plant metabolism, we chose the polyamine pathway as a model to unravel those key factors that still present bottlenecks in pathway engineering. The polyamine pathway is ubiquitous in living organisms (Bagni, 1989). It is a relatively short pathway in terms of the number of enzymes involved, however, it is rather complex because of its impact on crucial physiological, developmental, and regulatory processes in which polyamines are implicated (Malmberg et al., 1998). All enzymes involved in the pathway have been characterized, and corresponding genes/cDNAs have been cloned from different sources (Kumar and Minocha, 1998). As a result, the pathway represents an ideal model to test hypotheses and to answer fundamental biological questions in pathway manipulation using transgenesis.
The polyamine pathway comprises an anabolic phase leading to the elaboration of spermidine and spermine from putrescine. Orn decarboxylase (ODC; EC 4.1.1.19) catalyzes the removal of the carboxyl group from Orn to yield putrescine, whereas S-adenosyl-l-Met (SAM) decarboxylase (SAMDC; EC 4.1.1.50), introduces SAM into the pathway, which is then used in its decarboxylated form (dcSAM) as an aminopropyl donor in the conversion of putrescine to spermidine and subsequently to spermine (Tiburcio et al., 1997). The actual transfer of the aminopropyl moiety is catalyzed by two separate and distinct enzymes, spermidine synthase (SPD SYN; EC. 2.5.1.16) and spermine synthase (EC 2.5.1.16). In bacteria and also in plants, two alternative pathways lead to putrescine formation. In addition to the ODC pathway, decarboxylation of Arg by Arg decarboxylase (ADC; EC 4.1.1.19) also results in putrescine formation via two intermediate steps (Malmberg et al., 1998). The pathway also comprises a catabolic phase. This involves oxidative deamination of putrescine, spermidine, and spermine by the action of amine oxidases, these include the copper diamine oxidase (DAO; EC 1.4.3.6); these enzymes are characterized by their substrate specificity toward diamines. The flavoprotein polyamine oxidases (PAO; EC 1.5.3.3) oxidize spermidine and spermine at their secondary amino groups (Tiburcio et al., 1997). DAO oxidizes the primary amino group of putrescine and spermidine with the formation of pyrroline (from putrescine) and aminopropylpyrroline (from spermidine) along with ammonia and hydrogen peroxide (Smith, 1988). PAO yields pyrroline and aminopropylpyrroline, from spermidine and spermine, respectively, along with 1,3-diaminopropane and hydrogen peroxide. Thus, the pathway ensures the recycling of carbon and nitrogen from putrescine (Flores and Filner, 1985).
SAM is used by plants for the biosynthesis of polyamines and ethylene (Even-Chen et al., 1982). Ethylene is produced from SAM via 1-amino-cyclopropane-1-carboxylic acid by the actions of 1-aminocyclopropane-1-carboxylic synthase and 1-aminocyclopropane-1-carboxylic oxidase (Hedden and Philips, 2000). Interestingly, the functions of polyamines and ethylene in higher plant metabolism differ diametrically. Whereas ethylene is a plant-aging hormone leading to retardation of growth and promotion of senescence (Abeles, 1973), polyamines have been documented to delay senescence (Capell et al., 1993), and they can also inhibit ethylene biosynthesis in several plant tissues (Apelbaum et al., 1981). The mechanism that control these processes have not been elucidated.
SAMDC is a highly regulated enzyme whose levels can fluctuate severalfold depending on the growth state and intracellular polyamine concentration of the cell (Stanley, 1995). Enzyme regulation in vivo can be achieved at the gene expression level, and also post-transcriptionally by polyamines themselves (Pegg, 1986). Transgenic mice harboring a rat samdc gene were generated to study implications of overexpression of polyamine-synthesizing enzymes and their regulation (Heljasvaara et al., 1997). A 2- to 4-fold increase in SAMDC activity was detected in liver and brain tissues of transgenic mice expressing samdc. However, neither these nor hybrid mice overexpressing simultaneously odc and samdc displayed any significant changes in spermidine and spermine levels, but putrescine depletion was measured in these animals. When the human samdc cDNA driven by the cauliflower mosaic virus 35S promoter was transferred into tobacco (Nicotiana tabacum), transgenic plants showed a significant reduction in putrescine levels, whereas spermidine was increased 2- to 3-fold (Noh and Minocha, 1994). Transgenic tissues failed to regenerate when a homologous samdc cDNA driven by the cauliflower mosaic virus 35S promoter was re-introduced into potato (Solanum tuberosum). Using the same cDNA in antisense orientation, plants could be regenerated from transformed tissues with difficulty, although severe phenotypic abnormalities were observed, including stunted phenotypes (Kumar et al., 1996).
Using rice (Oryza sativa) as a model system, we reported previously overexpression or down-regulation of several genes involved in the polyamine pathway (Capell et al., 1998; Bassie et al., 2000a, 2000b; Capell et al., 2000; Noury et al., 2000; Lepri et al., 2001). To investigate later steps in the pathway in terms of how modulation of enzyme activities affect levels of putrescine, spermidine, and spermine, we introduced a heterologous Datura stramonium samdc cDNA (GenBank accession no. Y07768) into regenerable rice tissues.
In the current investigation, we describe and characterize transgenic rice germplasm expressing the D. stramonium samdc cDNA and discuss how changes in the activity of this key enzyme influence (a) steady-state levels of polyamines in vegetative (leaf) and storage (seeds) tissues; (b) activities of other enzymes involved in the pathway; (c) whether transcription and/or translation of the rice ortholog is affected; and (d) whether transcription of the rice spd syn gene is affected.
RESULTS
Recovery of Primary Transformants
The transformation vector containing the D. stramonium samdc cDNA was constructed as described in “Materials and Methods.” Mature rice (var. EYI 105) embryos were cobombarded with plasmid Ubi::Dsamdc (Fig. 1A) containing the D. stramonium samdc cDNA driven by the Ubi-1 promoter and a plasmid containing the hygromycin phosphotransferase (hpt) gene as a selectable marker (Valdez et al., 1998; Sudhakar et al., 1998). We analyzed 20 independently derived transgenic rice plants.
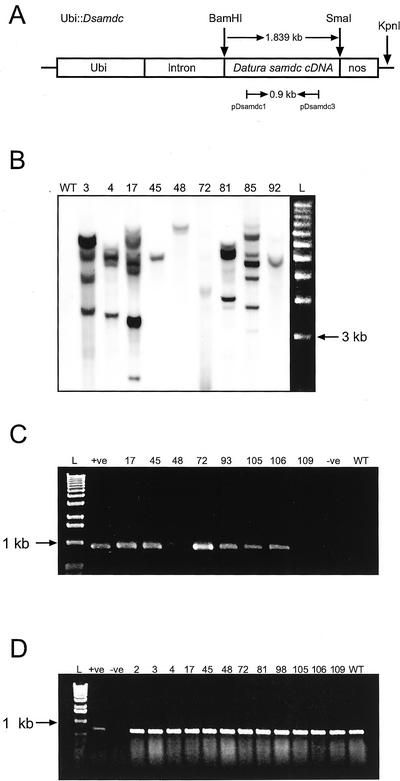
Figure 1.
Generation and molecular characterization of transgenic rice plants expressing the D. stramonium samdc cDNA. A, Map of Ubi::Dsamdc showing transcription unit, relevant restriction sites, and primers used for PCR and RT-PCR analyses. The D. stramonium samdc cDNA is 1.839 kb in size. KpnI has a single restriction site in the plasmid. Nos, Nopaline synthase. Arrows represent primers and length of amplified fragment. B, DNA gel-blot analysis of transgenic rice plants. Genomic DNA (10 μg) was digested with KpnI and probed with the 0.9-kb DIG-labeled PCR product from Ubi::Dsamdc. Exposure time was 10 min; wt, wild type; numbers represent putative transgenic plants; L, molecular size marker (1-kb DNA ladder, Invitrogen, Carlsbad, CA). C, RT-PCR analysis of D. stramonium samdc cDNA (0.9 kb) from total RNA extracted from controls and plants transformed with Ubi::Dsamdc. L, Molecular size marker (1-kb DNA ladder, Invitrogen); +ve, positive control, plasmid Ubi::Dsamdc; −ve, negative control (water); numbers indicate independent transgenic plants; wt, wild type. D, RT-PCR analysis of rice samdc from total RNA extracted from controls and plants transformed with Ubi::Dsamdc. L, Molecular size marker (1-kb DNA ladder, Invitrogen); +ve, positive control, plasmid Ubi::Dsamdc; −ve, negative control (water); numbers represent indicate independent transgenic plants; wt, wild type.
Molecular Characterization of Transgenic Rice Plants
All regenerated plants were screened by PCR (see “Materials and Methods”) to amplify a 0.9-kb fragment of the Ubi::Dsamdc. Genomic DNA gel-blot analysis further confirmed integration of the transgene in the genome of these plants. Genomic DNA-blot analysis of a representative sample of plants is shown in Figure 1B. Digestion was carried out with KpnI, which cuts once within the backbone sequence of the transforming plasmid (Fig. 1A). A 0.9-kb PCR-labeled probe was used to detect the transgene in 19 of the 20 lines we analyzed. The remaining line only had the hpt-selectable marker. Transformed lines exhibited unique integration patterns confirming their independent origin. Twelve of the 19 transgenic lines expressed the D. stramonium samdc mRNA. Reverse transcription (RT)-PCR analysis of total RNA was performed using the pair of primers pDsamdc-1/pDsamdc-3 (Fig. 1C). We studied expression of the rice samdc ortholog using the pair of primers pRsamdc-1/pRsamdc-2. We detected no differences in the level of expression of the endogenous rice samdc in Ubi::Dsamdc-transformed lines, hpt-transformants, and wild-type controls (Fig. 1D). RNA gel-blot analysis demonstrated expression of steady-state D. stramonium samdc mRNA in 10 of the 12 lines that expressed the gene by RT-PCR (Figs. 1C and 2, A and B). The remaining two lines (45 and 105) that did not show expression of the D. stramonium samdc mRNA in northern blots, expressed the RNA at a low level, which was only detectable by RT-PCR. When the same membrane was reprobed using the rice samdc and the rice spd syt DIG-labeled probes, we observed comparable levels of steady-state rice samdc mRNA in all lines (Fig. 2C). Similar results were observed when the membrane was reprobed with the rice spd syt DIG-labeled probe (Fig. 2, C and D).
Figure 2.
Transcript accumulation in rice leaves. A, Normalization of hybridization signals in leaf tissue after densitometric analysis of autoradiographs. D. stramonium samdc mRNA levels were quantified, and the resulting values were normalized using values obtained from RNA loading levels. Column size represents the relative D. stramonium samdc mRNA level generated by comparing the normalized values of each lane with that of the highest expressing sample. B, Gel-blot analyses of total RNA from transgenic leaf tissue (WT, wild type 2, 3, 4, 17, 34, 45, 72, 75, 81, 98, 105, and 106). A 0.9-kb DIG-labeled PCR probe from D. stramonium samdc cDNA was used. Exposure time was 10 min. C, Gel-blot analyses of total RNA from transgenic leaf tissue (WT, wild type 2, 3, 4, 17, 34, 45, 72, 75, 81, 98, 105, and 106). A 0.7-kb DIG-labeled PCR probe from rice samdc cDNA was used. Exposure time was 20 min. D, Gel-blot analyses of total RNA from transgenic leaf tissue (WT, wild type 2, 3, 4, 17, 34, 45, 72, 75, 81, 98, 105, and 106). A 0.9-kb DIG-labeled PCR probe from rice spd syn cDNA was used. Exposure time was 30 min. E, UV fluorescence of ethidium bromide-stained gel showing equal amount of total RNA loading from plants used for the hybridization shown above.
Transgene Transcript Accumulation Results in Increases in SAMDC Activity
SAMDC activity was analyzed in leaf tissue simultaneously with mRNA and polyamine measurements. Background SAMDC activity in control plants was on the order of 0.40 to 0.50 nKat mg−1 protein. Six of the 12 transgenic lines that expressed the D. stramonium samdc at the mRNA level (3, 4, 72, 81, 98, and 105) had a significant increase in SAMDC activity (Fig. 3A). Plant 3 had a maximum 3-fold increase in SAMDC activity (1.42 nKat mg−1 protein; P < 0.001) when compared with control lines (Fig. 3A). This plant also accumulated the D. stramonium samdc transcript at the highest level (Fig. 2, A and B). The D. stramonium samdc transcript levels in plant 105 were significantly lower compared with the other expressing plants (Fig. 1C). However, SAMDC activity in this clone was of the same order as in the other clones (Fig. 3A).
Figure 3.
Biochemical characterization of transgenic rice plants expressing Ubi::Dsamdc. A, SAMDC enzyme activity in different transgenic lines compared with appropriated controls. Values are means ± se for control lines (n = 6) and means ± se in transgenic lines (n = 4). SAMDC activity in clones 4, 72, 81, 98, and 105 was significantly different from controls at P < 0.01; for clone 3 at P < 0.001. Remaining values were not significantly different from control levels at P > 0.05. B, Cellular polyamine levels in controls and 12 representative transgenic plants. Values are means ± se in control lines (n = 36) and means ± se in transgenic lines (n = 9). Putrescine levels were significantly different from controls at P < 0.01 for clone 98 and P < 0.05 for clone 105. Spermidine levels were significantly different from controls at P < 0.05 for all clones. Spermine levels were not significantly different from controls at P > 0.05.
The Triamine Spermidine Accumulates in Leaf Tissue as a Result of Increases in SAMDC Activity
Spermidine and spermine levels were analyzed in transgenic leaf tissue simultaneously with enzyme activity measurements (ADC, ODC, SAMDC, DAO, and PAO). Spermidine levels were increased significantly in all six lines that showed changes in SAMDC activity in leaves. Increases varied from 1.5-fold in plant 3 (370 nmol g−1 fresh weight; P < 0.05) to 2.5-fold in plant 98 (540 nmol g−1 fresh weight; P < 0.05) compared with wild-type or hpt-transformed controls (175–250 nmol g−1 fresh weight; Fig. 3B). No significant variation (P > 0.05) in spermine levels in leaves was observed in any of the plants analyzed when compared with controls (Fig. 3B).
Putrescine Accumulation in Leaf Tissue Is Related to Increases in ADC and ODC Activities
Two of the 12 lines analyzed had a significant increase in putrescine levels. A 2-fold increase was detected in plants 98 and 105 [1,220 (P < 0.01) and 1,169 (P < 0.05) nmol g−1 fresh weight, respectively] when compared with wild-type and hpt-transformed controls (470–560 nmol g−1 fresh weight, Fig. 3B). When we measured activities of early enzymes in the pathway, ADC and ODC, a surprising result was observed in plants 98 and 105. A significant increase in ADC and ODC activity was detected. A maximum 1.6-fold increase in ADC activity (5.4 nKat mg−1 protein; P < 0.01) and a 5.7-fold increase in ODC activity (6.9 nKat mg−1 protein; P < 0.001) were detected in plant 98 when compared with controls (3.27 nKat mg−1 protein and 1.21 nKat mg−1 protein for ADC and ODC activities, respectively; Fig. 4). We then measured activity of the two enzymes involved in the catabolism of polyamines. We did not detect any significant variation in DAO or PAO activities in any of the plants we analyzed (Fig. 4).
Figure 4.
Rice ADC, ODC, DAO, and PAO activities in leaf tissue. Values are means ± se in control lines (n = 4) and means ± se in transgenic lines (n = 4). ADC activity was significantly different from control at P < 0.01 for clone 98 and at P < 0.05 for clone 105. ODC activity was significantly different from control at P < 0.01 for clones 98 and 105. ADC, ODC, DAO, and PAO activities were not significantly different from controls at P > 0.05 in any of the remaining lines.
Spermidine and Spermine Accumulate in R1 Seeds of Plants Expressing Ubi::Dsamdc
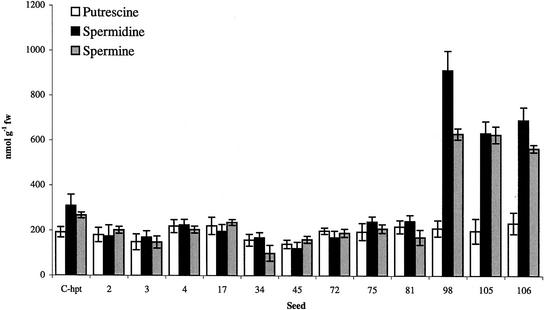
The polyamine content of seeds harvested from primary transformants was determined after collection of mature seeds and desiccation. Seeds from plants 98, 105, and 106 showed a significant increase in spermidine (2.5-fold increase, 700–900 nmol g−1 fresh weight; P < 0.01) and spermine levels (2-fold increase, 600 nmol g−1 fresh weight; P < 0.05) when compared with wild-type and hpt controls (300 and 30 nmol spermine g−1 fresh weight; Fig. 5). No significant variation was detected among the remaining lines and controls in spermidine and spermine levels. No significant variation in putrescine levels was detected in any of the lines (Fig. 5).
Figure 5.
Polyamine levels in controls and Ubi::Dsamdc-containing seeds. Values are means ± se in control lines (n = 36) and means ± se in transgenic lines (n = 3). Putrescine levels were significantly different from controls at P < 0.05 for clones 98 and 106. Spermidine levels were significantly different from controls at P < 0.01 and P < 0.05 for clones 98 and 106, respectively. Spermine levels were significantly different from control at P < 0.05 for clones 98 and 106. Remaining values were not significantly different from control levels at P > 0.05.
DISCUSSION
A limited number of plant metabolic pathways have been studied in depth, primarily because of the complexity of metabolic networks and how these are regulated. An even smaller number of pathways have been manipulated using molecular, genetic, or biochemical tools. Examples of such pathways include flavonoid biosynthesis (Van der Krol et al., 1990), lignins (Guo et al., 2001), carotenoids (Römer et al., 2000; Ye et al., 2000), fatty acids (Kinney, 1998), and some secondary metabolic pathways (Yun and Hashimoto, 1992; Nessler, 1994). Different approaches have been used to understand metabolic processes in plants. These include the use of inhibitors (Malmberg and McIndoo, 1983; Hiatt et al., 1986), mutants that lack particular enzymatic steps in a given pathway (Somerville and Browse, 1991; Watson et al., 1998), and homologous or heterologous genes in molecular studies (Capell et al., 2000; Halpin et al., 2001). As our appreciation of the complexity of biosynthetic pathways in plants increases, it becomes necessary to develop a knowledge base in molecular and biochemical terms to understand how such pathways control vital physiological, developmental, and metabolic processes (Capell et al., 2000). A reductionist approach to simplify the complexity of biosynthetic pathways in plants will help unravel biochemical components that play a crucial role in determining levels of end-products and intermediates. Such an approach needs to be validated on a well-characterized pathway in terms of enzymology and biochemistry.
The polyamine biosynthetic pathway in higher plants provides such an example. The pathway comprises anabolic and catabolic components (Malmberg et al., 1998). The two higher polyamines spermidine and spermine are synthesized from the diamine putrescine by a sequential addition of aminopropyl moieties from dcSAM by SAMDC (Pegg, 1986). There are two alternative pathways for the biosynthesis of putrescine in plants. All enzymes in the polyamine pathway have been characterized, and corresponding cDNAs have been cloned from different organisms (Kumar and Minocha, 1998). As a consequence, we have in place all components that are necessary to test and validate such an approach using a relatively short, yet complex pathway. The pathway is remarkable for its biochemical diversity, and for the number of regulatory and physiological processes in which it has been implicated (Malmberg and Rose, 1987). A transgenic approach to answer such fundamental biological questions has distinct advantages. By introducing appropriate transgenes into plants and analyzing the effects transgene products have on end-product accumulation, we may begin to understand how individual components of the pathway(s) contribute toward their concerted regulation (Kumar and Minocha, 1998).
For the past several years, we have been studying molecular, biochemical, and genetic components of polyamine metabolism in plants (Capell et al., 1998; Bassie et al., 2000a, 2000b; Capell et al., 2000; Noury et al., 2000; Lepri et al., 2001). Through comparison of ADC and ODC activities and the corresponding polyamine profiles in transgenic rice overexpressing the oat (Avena sativa) adc or the human (Homo sapiens) odc cDNAs, we found strong evidence that ODC is most likely the enzyme responsible for regulating the formation of putrescine in plants (Lepri et al., 2001). We showed that the tight regulation of the polyamine pathway at the end-product level can be overcome by overexpressing key enzymes involved in the pathway. Thus, by screening transgenic plants expressing ADC or ODC, we were able to identify populations with substantial changes in polyamine levels (Capell et al., 1998; Noury et al., 2000; Lepri et al., 2001). We also investigated the effect of shutting down ADC by antisense approaches (Capell et al., 2000) and the consequences of down-regulating DAO (Bassie et al., 2000b). Such experiments allowed us to develop an understanding of how early steps in the polyamine pathway control levels of the parent polyamine, putrescine, in plants and how this compound is further converted into the higher polyamines spermidine and spermine. Having investigated the role of early enzymes in the pathway, we wished to elucidate the contribution of enzymes involved in later parts of the pathway, particularly the role of SAMDC.
We posed the question of whether levels of the higher polyamines spermidine and spermine could be modulated in plants by overexpressing SAMDC without any involvement from SPD SYN or spermine synthase, which are also involved in their biosynthesis. We had previously demonstrated that levels of these higher polyamines could be altered by expressing early enzymes involved in the pathway and also by down-regulating DAO (Bassie et al., 2000b).
Accumulation of the D. stramonium samdc Transcript Results in an Increase in SAMDC Activity and Spermidine Accumulation in Leaves
We introduced the Ubi::Dsamdc (Fig. 1A) into rice, and we recovered transgenic plants that integrated the transgene stably. DNA gel blots demonstrated the independent origin of all transgenic plants we recovered (Fig. 1B). Transcription of the D. stramonium samdc was confirmed by RNA gel-blot analysis (Figs. 1C and 2, A and B). Leaf extracts from transgenic rice plants, exhibited significant increases in SAMDC activity (Fig. 3A). As a result of this increase in enzyme activity, we measured a 1.5- to 2.5-fold increase in the levels of spermidine in leaves, confirming that the D. stramonium SAMDC enzyme was functional and that the dicotyledonous enzyme was correctly processed in monocotyledonous plants.
RNA gel-blot analysis indicated no changes in the steady-state rice samdc mRNA (Fig. 2C) in transgenic plants expressing the D. stramonium gene. This demonstrates clearly that the heterologous transgene operates independently of its rice ortholog. Increases in spermidine levels in leaves were attributable to expression of the D. stramonium samdc alone, because we did not detect any changes in the endogenous spd syn transcript (Fig. 2C). We did not detect any increases in spermine in leaves (Fig. 3). The question arises then as to why expression of SAMDC affects levels of spermidine but not spermine in leaves of the transgenic rice plants we generated, because the same enzyme is responsible for the generation of spermine from spermidine by a second transfer of an aminopropyl group from dcSAM. Noh and Minocha (1994) overexpressed the human samdc cDNA in transgenic tobacco plants resulting in a 2- to 3-fold increase in spermidine but no significant variation in spermine levels. Similar results were observed when the homologous samdc cDNA was re-introduced into potato driven by the tuber-specific patatin promoter (Rafart-Pedros et al., 1999). Spermidine concentration was significantly higher in tubers, whereas no variation was observed in spermine levels. This pattern indicates a tighter regulation of cellular spermine metabolism, compared with putrescine (Noury et al., 2000) or spermidine (Bassie et al., 2000a). Although spermine is ubiquitous in eukaryotic cells at high levels, the physiological roles of spermine are unclear (Hamasaki-Katagiri et al., 1998). It is possible that the reason we do not see any changes in spermine levels in leaves of these plants is because the spermidine pool is not large enough to permit conversion of excess spermidine to spermine. We had previously proposed a similar threshold model in terms of the size of the putrescine pool to explain why rice tissues expressing the oat adc cDNA driven by a very strong constitutive promoter were able to accumulate higher levels of spermidine and spermine (Bassie et al., 2000a) compared with plants engineered with the same transgene driven by a weaker promoter that did not show any changes in the levels of the higher polyamines (Capell et al., 1998).
SAMDC Expression Results in Spermidine and Spermine Accumulation in Storage Tissue
We had previously observed a hierarchical accumulation of polyamines in different tissues/organs (Lepri et al., 2001; P. Trung-Nghia et al., personal communication). The general picture that emerges from these studies strongly demonstrates that less metabolically active tissues, such as seeds, accumulate higher levels of polyamines. This was the case in transgenic rice plants expressing the human odc or the oat adc cDNAs (Noury et al., 2000; Lepri et al., 2001). There are no reports in any other transgenic plant system describing the accumulation of any polyamines in storage tissues. In transgenic rice expressing the Ubi::Dsamdc, spermidine and spermine levels were significantly increased, whereas putrescine levels remained unchanged. Our results are in line with experiments in which metabolites such as vitamin A and pharmaceutical antibodies accumulate at high levels in seeds of rice (Ye et al., 2000; Torres et al., 2001), wheat (Triticum aestivum; Stöger et al., 2000), and pea (Pisum sativum; Perrin et al., 2000). It is reasonable to assume that dormant or less metabolically active tissue provides a conducive environment for the accumulation of transgenic products. In extreme cases, the formation of recombinant proteins in the form of paracrystalline structures in cereal endosperm, is easily observed by optical microscopy (Stöger et al., 2001).
Activities of Early Enzymes in the Pathway and Those Responsible for Polyamine Catabolism Are Rarely Altered in Transgenic Plants Expressing Ubi::Dsamdc
Manipulation of a particular enzyme involved in a metabolic pathway may result in pleotropic changes in other enzymes in the pathway. This may be the result of a compensation mechanism through which plants adjust their metabolism to maintain steady-state pools of key metabolites. Changes in the concentration of metabolites or end-products may also affect other enzyme activities, because certain compounds appear to feedback inhibit or regulate enzymes in different ways. When spermidine and spermine were applied to tobacco cell cultures, a significant reduction in ADC and SAMDC activity was measured. These polyamines did not affect ODC activity (Hiatt et al., 1986). In mammalian systems, an increase in the intracellular content of polyamines reduces the activity of ODC (Kameji and Pegg, 1987). This reduction occurs as a result of the loss of enzyme protein (Persson et al., 1984). The decline in enzyme protein occurs partly by means of an increased degradation rate (Murakami et al., 1985) and partly by a reduced rate of synthesis (Höltta and Pohjanpelto, 1986).
A majority of the plants that we generated that expressed the Ubi::Dsamdc did not have any changes in the activities of the rice ADC, ODC, DAO, or PAO in leaf tissue (Fig. 4). However, two transgenic plants that accumulated high levels of spermidine (up to 2.5-fold) also exhibited increases in putrescine levels (2-fold) as a result of increase in ADC and ODC activity (Figs. 3B and 4). These two plants (98 and 105) exhibited the most dramatic increases in rice ODC activity (up to 5.7-fold) compared with all of the clones we recovered (Fig. 4). Rice ADC activity was increased a maximum of 1.6-fold in these lines as well. These results are indeed consistent with data we published previously that demonstrated that ODC rather than ADC is responsible for changes in putrescine levels in plants (Lepri et al., 2001). Our results suggest that application of exogenous polyamines and their generation in situ in plant cells as a result of heterologous transgene expression appear to result in different responses at the biochemical level. This may be explained by the fact that the two systems are physiologically very different and as such, endogenous enzyme activities respond in different ways to what appears to be the same stimulus. It may be that changes in the endogenous enzyme activities in some of these transgenic plants is an exception that may be a result of differential regulation of enzymes in different transformants.
CONCLUSION
We have demonstrated that expression of a heterologous samdc cDNA in plants is adequate to increase enzyme activity and end-product levels in a tissue-dependent manner and is uncoupled from the endogenous polyamine biosynthetic machinery. Our results further suggest that it is possible to modulate complex pathways in plants by overexpression of appropriate heterologous transgenes, even in situations in which a particular product or intermediate requires input from additional components of the pathway. We can, thus, envisage strategies for the manipulation of other pathways in plants by applying findings we obtained as a result of experiments involving the polyamine pathway.
MATERIALS AND METHODS
Plasmid Construction, Rice (Oryza sativa) Transformation, and Plant Regeneration
The 1.8-kb Datura stramonium samdc cDNA (GenBank accession no. Y07768) containing the 5′-untranslated sequence and the cDNA coding region, was excised as an XhoI fragment from pBluescript, blunt ended, and digested again with BamHI. The BamHI/blunt-end fragment was subcloned into the BamHI/SmaI site of the plasmid pAL76 (Christensen and Quail, 1996), which contains the maize (Zea mays) 1 ubiquitin (Ubi-1) promoter and first intron, and a Nos transcriptional termination. This plasmid was subsequently referred to as Ubi::Dsamdc.
Rice transformation, selection, and plant regeneration procedures were as described previously (Sudhakar et al., 1998; Valdez et al., 1998).
PCR and RT-PCR Analyses
Genomic DNA was extracted from leaf tissue according to the method of Edwards et al. (1991). Genomic PCR amplification to detect D. stramonium samdc cDNA was carried out in a total volume of 25 μL comprising 50 ng of genomic DNA, 1× PCR buffer (50 mm KCl, 10 mm Tris-HCl, pH 8.3, and 1.5 mm MgCl2, Roche Molecular Biochemicals, Mannheim, Germany), 200 μm each dNTP, 50 nm of each primer (the forward sequence primer started from position 630 in the D. stramonium samdc open reading frame and consisted of 5′-CGGACCTGCTGAGTGCACCATTGT-3′, primer pDsamdc-1; reverse primer, 5′-CCAGCAGCCCTTCAGAACGG-3′, primer pDsamdc-3) and 1.25 units of Taq DNA polymerase (Roche Molecular Biochemicals). We carried out 35 amplification cycles: denaturation (94°C, 40 s), annealing (64°C, 1 min), and extension (72°C, 2 min). The 0.9-kb product was visualized by agarose gel electrophoresis (0.8% [w/v] Tris-borate/EDTA [TBE]).
Total RNA was extracted from leaves of transgenic plants using the Trizol-Reagent (Invitrogen). RNA samples were treated with RQ1 RNase-free DNase (Promega, Madison, WI) as recommended by manufacturer. RT was carried out using the Access RT-PCR system (Promega) in 25-μL reaction volumes containing 100 ng of total RNA. The primer pairs used for RT-PCR were pDsamdc-1/pDsamdc-3 to study D. stramonium samdc cDNA expression and pRsamdc-1/pRsamdc-2 to study rice samdc gene expression. The primer sequences for rice samdc cDNA were as follows: the forward sequence primer started from position 1,000 in the rice samdc open reading frame and consisted of 5′-GGAGATCCAGCAAAGCCTGGCC-3′ (pRsamdc-1), and the reverse sequence consisted of 5′-CCCAGGGGAGAAGATTGC-CCAG-3′ (pRsamdc-2). D. stramonium samdc cDNA and rice samdc cDNA were amplified for 40 cycles: denaturation (94°C, 40 s), annealing (65°C, 1 min), and extension (68°C, 2 min). The 0.9-kb D. stramonium samdc and the 0.7-kb rice samdc were visualized on a 1% (w/v) TBE agarose gel.
DNA and RNA Gel-Blot Analysis
Genomic DNA from leaf tissue was extracted as described by Edwards et al. (1991). DNA was digested with KpnI, fractionated by 0.8% (w/v) TBE agarose gel electrophoresis (Sambrook et al., 1989), and transferred to a positively charged nitrocellulose membrane (Roche Molecular Biochemicals). Nucleic acids were fixed by baking at 80°C for 2 h. Filters were washed in 2× SSC for 30 min and subsequently prehybridized at 42°C for 1 h using the DIG-easy hybridization solution (Roche Molecular Biochemicals). The 0.9-kb D. stramonium samdc, the 0.7-kb rice samdc, and the 0.9-kb rice spd syn were labeled using the PCR DIG probe synthesis kit (Roche Molecular Biochemicals). The primer sequences for rice spd syt cDNA were as follows: The forward sequence primer started from position 196 bp in the rice spd syn open reading frame and consisted of 5′-GGATGGTTCTCCGAGATTAG-3′ (pRspdsyn-1), and the reverse sequence consisted of 5′-GATCTAGTT-GGCCTTGGATC-3′ (pRspdsyn-2). Alkali-labile DIG-11-dUTP was incorporated into the probe in a final volume of 50 μL comprising 4 μm dATP, 4 μm dCTP, 4 μm dGTP, 3.2 μm dTTP, 0.8 μm DIG-11-dUTP, 1× Roche Molecular Biochemicals PCR buffer (50 mm KCl, 10 mm Tris-HCl, pH 9.0, and 0.1% [w/v] Triton X-100), 2.5 units of Taq DNA polymerase (Roche Molecular Biochemicals), 0.1 mm each of the forward and reverse sequence primers (as PCR above), and 100 ng of the plasmids. After an initial denaturation step for 2 min at 96°C, 35 amplification cycles were carried out, each comprising denaturation at 96°C for 10 s, annealing at 60°C for 10 s, and extension at 72°C for 1 min. The 0.9-, 0.7-, and 0.9-kb-labeled probes were purified using the QIAquick Gel Extraction Kit (QIAGEN, Dorking, Surrey, UK) and denatured at 68°C for 10 min before use. Hybridization was performed at 42°C overnight. The membranes were washed twice for 5 min in 2× SSC, 0.1% (w/v) SDS at room temperature and then twice (15 min) in 0.5× SSC, 0.1% (w/v) SDS at 68°C. Chemiluminescent detection was carried out according to the manufacturer's instructions using the DIG Luminescent Detection Kit. After washing, the membranes were incubated with CSPD Chemiluminescent Substrate (Roche Molecular Biochemicals) and subsequently exposed to x-ray film (Fuji Photo Film, Kanawa, Japan) for 20 min at 37°C.
Total RNA was extracted from leaf tissue using RNeasy Plant Mini Kit (QIAGEN). Denatured RNA (30 μg) was subjected to electrophoresis on 1.2% (w/v) agarose-formaldehyde gel using 1× MOPS buffer (Sambrook et al., 1989). Transfer and hybridization were carried out as described above for DNA procedures. Membranes were exposed to x-ray film for 30 min at 37°C. Stripping and reprobing the membrane with the 0.7-kb rice samdc and 0.9-kb rice spd syn was performed as described by Hloch et al. (2001).
Determination of SAMDC, ADC, ODC, DAO, and PAO Activities
Leaf tissue was used for SAMDC, ADC, ODC, DAO, and PAO activity measurements. Tissue was extracted in buffer (0.1 m Tris, pH 7.5, and 2 mm dithiothreitol) at a ratio of 300 mg ml−1. Polyvinylpyrrolidone (100 mg) was added during grinding. After centrifugation at 12,000g for 20 min, the supernatant was used directly in enzyme activity assays. Enzyme assays were carried out as described in detail in Lepri et al. (2002). Enzyme activity was expressed as nanokatals per milligram of protein.
Polyamine Analysis
Crude extracts from leaves and seeds were dansylated and separated by thin layer chromatography as described (Capell et al., 1998). The dansyl-polyamine bands were identified on the basis of their RF values after visualization under UV light (312 nm) and comparison with dansylated polyamine standards. The image of the chromatogram was captured and analyzed by Quantity One (Quantification Software; Bio-Rad, Hercules, CA). The relative amount of dansyl-polyamine in each sample was determined by calculating the integrated optical density of the bands compared with the integrated optical density of the appropriate dilution of the dansylated control samples. Results were expressed as nanomoles per gram fresh weight.
Statistical Analysis
For molecular and biochemical analyses (enzyme activity and polyamine content) we used hpt-transformed and wild-type controls. The average control for the biochemical analyses was determined by taking three samples from 12 independent lines (six wild type and six hpt-transformants; n = 36). Hygromycin-resistant transformants and wild-type controls were not significantly different (P > 0.05) in terms of enzyme activity and polyamine levels in any of the tissues analyzed (Lepri et al., 2002). Data was analyzed by one-way ANOVA followed by Student's t test using the residual mean square in the ANOVA as the estimate of variability.
ACKNOWLEDGMENTS
We thank Dr. A. Michael for the kind gift of the D. stramonium samdc cDNA, J. Dix for graphic design, and E. Aguado for maintaining plants in the greenhouse.
Footnotes
This work was supported by the Rockefeller Foundation (fellowships to P.T.-H. and P.T.-N.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010966.
LITERATURE CITED
- Abeles FB. Ethylene in Plant Biology. New York: Academic Press; 1973. [Google Scholar]
- Apelbaum A, Burgoon AC, Anderson JD, Lieberman M, Ben-Afir R, Mattoo AK. Polyamines inhibit biosynthesis of ethylene in higher plant tissue and fruit protoplast. Plant Physiol. 1981;68:453–456. doi: 10.1104/pp.68.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagni N. Polyamines in plant growth and development. In: Bachrach U, Heimer YM, editors. The Physiology of Polyamines. II. Boca Raton, FL: CRC Press; 1989. pp. 107–120. [Google Scholar]
- Bassie L, Noury M, Lepri O, Lahaye T, Christou P, Capell T. Promoter strength influences polyamine metabolism and morphogenic capacity in transgenic rice tissues expressing the oat arginine decarboxylase cDNA constitutively. Transgenic Res. 2000a;9:33–42. doi: 10.1023/a:1008997822463. [DOI] [PubMed] [Google Scholar]
- Bassie L, Noury M, Wisniewski JP, Topsom L, Christou P, Capell T. Transgenic cell lines as a useful tool to study the biochemistry of down-regulation of an endogenous rice gene using a heterologous diamine oxidase cDNA. Plant Physiol Biochem. 2000b;38:729–737. [Google Scholar]
- Capell T, Bassie L, Topsom L, Hitchin E, Christou P. Simultaneous down-regulation of two related enzymes in early steps of the polyamine biosynthetic pathway in transgenic rice by a single antisense mRNA species. Mol Gen Genet. 2000;264:470–476. doi: 10.1007/s004380000317. [DOI] [PubMed] [Google Scholar]
- Capell T, Campos JL, Tiburcio AF. Antisenescence properties of guazatine in osmotically stressed oat leaves. Phytochemistry. 1993;32:785–788. [Google Scholar]
- Capell T, Escobar C, Lui H, Burtin D, Lepri O, Christou P. Over-expression of the oat arginine decarboxylase cDNA in transgenic rice (Oryza sativa L.) affects normal development patterns in vitro and results in putrescine accumulation in transgenic plants. Theor Appl Genet. 1998;97:246–254. [Google Scholar]
- Christensen AH, Quail PH. Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res. 1996;5:213–218. doi: 10.1007/BF01969712. [DOI] [PubMed] [Google Scholar]
- Edwards K, Johnstone C, Thompson C. A simple and rapid method for the preparation of plant genomic DNA for PCR. Nucleic Acids Res. 1991;19:1349. doi: 10.1093/nar/19.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Even-Chen Z, Mattoo AK, Goren R. Inhibition of ethylene biosynthesis by aminoethoxyvinylglycine and by polyamines shunts label from 3,4-[14C]methionine into spermidine in aged orange peel disc. Plant Physiol. 1982;69:385–388. doi: 10.1104/pp.69.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores HE, Filner P. Metabolic relationships of putrescine GABA and alkaloids in cell and root cultures. In: Newman KH, Barz W, Reinhard E, editors. Primary and Secondary Metabolism of Plants Cell Cultures. New York: Springer; 1985. pp. 37–42. [Google Scholar]
- Guo D, Chen F, Inoue K, Blount JW, Dixon RA. Downregulation of caffeic acid 3-O-methyltransferase and caffeoyl CoA 3-O-methyltransferase in transgenic alfalfa: impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell. 2001;13:73–88. doi: 10.1105/tpc.13.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpin C, Barakate A, Askari BM, Abbot JC, Ryan MD. Enabling technologies for manipulating multiple genes on complex pathways. Plant Mol Biol. 2001;47:295–310. [PubMed] [Google Scholar]
- Hamasaki-Katagiri N, Katagiri Y, Tabor CW, Tabor H. Spermine is not essential for growth of Saccharomyces cerevisiae: identification of the SPE4 gene (spermine synthase) and characterization of a spe4 deletion mutant. Gene. 1998;210:195–201. doi: 10.1016/s0378-1119(98)00027-4. [DOI] [PubMed] [Google Scholar]
- Hedden P, Philips AL (2000) Manipulation of hormone biosynthetic genes in transgenic plants. Curr Opin Biotechnol 130–137 [DOI] [PubMed]
- Heljasvaara R, Veress I, Halmekytö M, Alhonen L, Jänne J, Laajala P, Pajunen A. Transgenic mice overexpressing ornithine and S-adenosylmethionine decarboxylase maintain a physiological polyamine homeostasis in their tissues. Biochem J. 1997;323:457–462. doi: 10.1042/bj3230457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiatt AC, McIndoo J, Malmberg R. Regulation of polyamine biosynthesis in tobacco: effects of inhibitors and exogenous polyamines on arginine decarboxylase, ornithine decarboxylase, and S-adenosylmethionine decarboxylase. J Biol Chem. 1986;261:1293–1298. [PubMed] [Google Scholar]
- Hloch P, Hoffmann K, Kruchen B, Rueger B. The DIG system: a high sensitive substitute of radioactivity in northern blot analysis. Biochemica. 2001;2:24–25. [Google Scholar]
- Höltta E, Pohjanpelto P. Control of ornithine decarboxylase in Chinese hamster ovary cells by polyamines: translational inhibition of synthesis and acceleration of degradation of the enzyme by putrescine, spermidine and spermine. J Biol Chem. 1986;261:9502–9508. [PubMed] [Google Scholar]
- Kameji T, Pegg AE. Inhibition of translation of mRNAs for ornithine decarboxylase and S-adenosylmethionine decarboxylase by polyamines. J Biol Chem. 1987;262:2427–2430. [PubMed] [Google Scholar]
- Kinney A. Manipulating flux through plant metabolic pathways. Curr Opin Plant Biotechnol. 1998;1:173–178. doi: 10.1016/s1369-5266(98)80021-6. [DOI] [PubMed] [Google Scholar]
- Kumar A, Minocha SC. Transgenic manipulation of polyamine metabolism. In: Lindsey K, editor. Transgenic Research in Plants. London: Harwood Academic Publishers; 1998. pp. 187–199. [Google Scholar]
- Kumar A, Taylor MA, Mad-Arif SA, Davies H. Potato plants expressing antisense and sense S-adenosylmethionine decarboxylase (SAMDC) transgenes show altered levels of polyamine and ethylene: antisense plants display abnormal phenotypes. Plant J. 1996;9:147–158. [Google Scholar]
- Lepri O, Bassie L, Safwat G, Thu-Hang P, Trung-Nghia P, Hölttä E, Christou P, Capell T. Over-expression of the human ornithine decarboxylase cDNA in transgenic rice plants alters the polyamine pool in a tissue-specific manner. Mol Gen Genet. 2001;266:303–312. doi: 10.1007/s004380100557. [DOI] [PubMed] [Google Scholar]
- Lepri O, Bassie L, Thu-Hang P, Christou P, Capell T (2002) Endogenous enzyme activities and polyamine levels in diverse rice cultivars depend on genetic background and are not affected by the presence of the hygromycin phosphotransferase selectable marker. Theor Appl Genet (in press) [DOI] [PubMed]
- Malmberg RL, McIndoo J. Abnormal floral development of a tobacco mutant with elevated polyamine levels. Nature. 1983;305:623–625. [Google Scholar]
- Malmberg RL, Rose DJ. Biochemical genetics of resistance to MGBG in tobacco: mutants that alter SAM decarboxylase or polyamine ratios, and floral morphology. Mol Gen Genet. 1987;201:9–14. [Google Scholar]
- Malmberg RL, Watson MB, Galloway GL, Yu W. Molecular genetic analysis of plant polyamines. Crit Rev Plant Sci. 1998;17:199–224. [Google Scholar]
- Murakami Y, Fujita K, Kameji T, Hayashi S. Accumulation of ornithine decarboxylase-antizyme complex in HMOA cells. Biochem J. 1985;225:669–697. doi: 10.1042/bj2250689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nessler C. Metabolic engineering of plant secondary products. Transgenic Res. 1994;3:109–115. doi: 10.1007/BF01974088. [DOI] [PubMed] [Google Scholar]
- Noh EW, Minocha SC. Expression of a human S-adenosylmethionine decarboxylase cDNA in transgenic tobacco and its effects on polyamine biosynthesis. Transgenic Res. 1994;3:25–53. doi: 10.1007/BF01976024. [DOI] [PubMed] [Google Scholar]
- Noury M, Bassie L, Lepri O, Kurek I, Christou P, Capell T. A transgenic rice cell lineage expressing the oat arginine decarboxylase (adc) cDNA constitutively accumulates putrescine in callus and seeds but not in vegetative tissues. Plant Mol Biol. 2000;43:357–544. doi: 10.1023/a:1006480304879. [DOI] [PubMed] [Google Scholar]
- Pegg AE. Recent advances in the biochemistry of polyamines of eukaryotes. Biochem J. 1986;234:249–262. doi: 10.1042/bj2340249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin Y, Vaquero C, Gerrad I, Sack M, Drossard J, Stöger E, Christou P, Fisher R. Transgenic pea seeds as bioreactors for the production of a single-chain Fv fragment (scFV) antibody used in cancer diagnosis and therapy. Mol Breed. 2000;6:345–352. [Google Scholar]
- Persson L, Seely JE, Pegg AE. Investigation of structure and rate of synthesis of ornithine decarboxylase protein in mouse kidney. Biochemistry. 1984;23:3777–3783. doi: 10.1021/bi00311a033. [DOI] [PubMed] [Google Scholar]
- Rafart-Pedros A, MacLeod MR, Ross HA, McRae D, Tiburcio AF, Davies HV, Taylor MA. Manipulation of S-adenosylmethionine decarboxylase activity in potato tubers. Planta. 1999;209:153–160. doi: 10.1007/s004250050617. [DOI] [PubMed] [Google Scholar]
- Römer S, Fraser PD, Kiano JW, Shipton CA, Misawa N, Schuch W, Bramley PM. Elevation of the provitamin A content of transgenic tomato plants. Nat Biotechnol. 2000;18:666–669. doi: 10.1038/76523. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Smith TA. The di- and poly-amine oxidases of higher plants. Biochem Soc Trans. 1988;13:319–322. doi: 10.1042/bst0130319. [DOI] [PubMed] [Google Scholar]
- Somerville C, Browse J. Plant lipids: metabolism, mutants and membranes. Science. 1991;252:80–87. doi: 10.1126/science.252.5002.80. [DOI] [PubMed] [Google Scholar]
- Stanley BA. Mammalian S-adenosylmethionine decarboxylase regulation and processing. In: Casero R, editor. Polyamines: Regulation and Molecular Interaction. Vol. 3. R.G. Landes Company; 1995. pp. 27–75. [Google Scholar]
- Stöger E, Parker M, Christou P, Casey R. Pea legumin over-expressed in wheat endosperm assembles into an ordered paracrystalline matrix. Plant Physiol. 2001;125:1732–1742. doi: 10.1104/pp.125.4.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöger E, Vaquero C, Torres E, Sack M, Nicholson L, Drossard J, Williams S, Keen D, Perrin Y, Christou P et al. Cereal crops as viable production and storage systems for pharmaceutical scFv antibodies. Plant Mol Biol. 2000;42:583–590. doi: 10.1023/a:1006301519427. [DOI] [PubMed] [Google Scholar]
- Sudhakar D, Duc LT, Bong BB, Tinjuangjun P, Maqbool SB, Valdez M, Jefferson R, Christou P. An efficient rice transformation system utilizing mature seed-derived explants and a portable, inexpensive particle bombardment device. Transgenic Res. 1998;7:289–294. [Google Scholar]
- Tiburcio AF, Altabella T, Borrel A, Masgrau C. Polyamine metabolism and its regulation. Physiol Plant. 1997;100:664–674. [Google Scholar]
- Torres E, Gonzales-Melendi P, Stöger E, Shaw P, Twyman RM, Nicholson L, Vaquero C, Fischer R, Christou P, Perrin Y. Native and artificial reticuloplasmins co-accumulate in distinct domains of the endoplasmic reticulum (ER) and in post-ER vesicles. Plant Physiol. 2001;127:1212–1223. [PMC free article] [PubMed] [Google Scholar]
- Valdez M, Cabrera-Ponce JL, Sudhakhar D, Herrera-Estrella L, Christou P. Transgenic Central American, West African and Asian elite rice varieties resulting from particle bombardment of foreign DNA into mature seed-derived explants utilizing three different bombardment devices. Ann Bot. 1998;82:795–801. [Google Scholar]
- Van der Krol AR, Mur LA, Beld M, Mol JNM, Stuitje A. Flavonoid genes in petunia: Addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell. 1990;2:291–299. doi: 10.1105/tpc.2.4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson MB, Emory KK, Piatak RM, Malmberg RL. Arginine decarboxylase (polyamine synthesis) mutants of Arabidopsis thaliana with altered root growth. Plant J. 1998;13:231–239. doi: 10.1046/j.1365-313x.1998.00027.x. [DOI] [PubMed] [Google Scholar]
- Ye X, Al-Babili S, Klöti A, Zhang J, Lucca P, Beyer P, Potricus I. Engineering the provitamin A (β-carotene) biosynthetic pathway into carotenoid-free rice endosperm. Science. 2000;287:303–305. doi: 10.1126/science.287.5451.303. [DOI] [PubMed] [Google Scholar]
- Yun D, Hashimoto JT. Metabolic engineering of medicinal plants: transgenic Atropa belladonna with an improved alkaloid composition. Proc Natl Acad Sci USA. 1992;89:11799–11803. doi: 10.1073/pnas.89.24.11799. [DOI] [PMC free article] [PubMed] [Google Scholar]