Abstract
Although cold and drought adaptation in cereals and other plants involve the induction of a large number of genes, inheritance studies in Triticeae (wheat [Triticum aestivum], barley [Hordeum vulgare], and rye [Secale cereale]) have revealed only a few major loci for frost or drought tolerance that are consistent across multiple genetic backgrounds and environments. One might imagine that these loci could encode highly conserved regulatory factors that have global effects on gene expression; therefore, genes encoding central regulators identified in other plants might be orthologs of these Triticeae stress tolerance genes. The CBF/DREB1 regulators, identified originally in Arabidopsis as key components of cold and drought regulation, merit this consideration. We constructed barley cDNA libraries, screened these libraries and a barley bacterial artificial chromosome library using rice (Oryza sativa) and barley Cbf probes, found orthologs of Arabidopsis CBF/DREB1 genes, and examined the expression and genetic map location of the barley Cbf3 gene, HvCbf3. HvCbf3 was induced by a chilling treatment. HvCbf3 is located on barley chromosome 5H between markers WG364b and saflp58 on the barley cv Dicktoo × barley cv Morex genetic linkage map. This position is some 40 to 50 cM proximal to the winter hardiness quantitative trait locus that includes the Vrn-1H gene, but may coincide with the wheat 5A Rcg1 locus, which governs the threshold temperature at which cor genes are induced. From this, it remains possible that HvCbf3 is the basis of a minor quantitative trait locus in some genetic backgrounds, though that possibility remains to be thoroughly explored.
When plants are exposed to environmental stress such as drought, cold, or high salt, they undergo physiological and biochemical adaptations (Bray, 1993; Ingram and Bartels, 1996; Thomashow, 1999). It has been established that plants have at least two major pathways, abscisic acid (ABA) dependent and ABA independent, for the induction of moisture deficit stress-inducible genes (Shinozaki and Yamaguchi-Shinozaki, 1997). ABA-independent gene activation often involves a cis-acting element called a dehydration response element (DRE; also known as a C repeat [CRT]) that responds to drought and low temperature (Baker et al., 1994; Yamaguchi-Shinozaki and Shinozaki, 1994) and has been found in many plants (Jiang et al., 1996; Dunn et al., 1998; Choi et al., 1999). Stockinger et al. (1997) identified a transcription factor that binds the DRE/CRT element. This protein, designated CBF1 (C-repeat binding factor 1), has a potential nuclear localization sequence (NLS), an AP2-DNA-binding domain, and an acidic activation domain. The Arabidopsis CBF (DREB1) genes are a small multigene family consisting of six paralogs that include three intensively studied genes (CBF1/DREB1B, CBF2/DREB1C, and CBF3/DREB1A) in an 8.7-kb region on chromosome 4 (Gilmour et al., 1998; Liu et al., 1998), and lesser studied genes on chromosome 5 (CBF4/DREBID; Nakamura et al., 1998; Thomashow et al., 2001) and chromosome 1 (DREB1E and DREB1F; Sakuma et al., 2002). The expression patterns of these genes have notable differences. For example, only the three CBF/DREB1 genes on chromosome 4 have been shown to be chilling induced (CBF/DREB1), and there are differences when comparing root versus shoot expression (Gilmour et al., 1998; Sakuma et al., 2002). The extent to which expression varies among alleles of individual CBF/DREB1 genes is not yet known. The CBF/DREB1 gene family seems not to be subject to autoregulation because no DRE/CRT element is present in the promoter region of CBF/DREB1 genes. Overexpression of CBF/DREB1 in Arabidopsis induces expression of several target genes and enhances freezing tolerance (Jaglo-Ottosen et al., 1998; Kasuga et al., 1999). Liu et al. (1998) isolated other trans-acting factors, DREB2A and DREB2B, which also bind the DRE/CRT element. The DREB2 proteins contain a Ser-/Thr-rich domain, and have no significant sequence similarity to CBF/DREB1 proteins, except for the presence of NLS and AP2 domains. The DREB2 genes are induced by dehydration and salt stress, but not cold stress (Liu et al., 1998; Nakashima et al., 2000). In summary, there are two different types of DRE-/CRT-binding factors, CBF/DREB1 and DREB2, keyed by at least somewhat separate signal transduction pathways.
We found several putative regulatory elements, including ABA response elements and DRE/CRTs, in the upstream regions of the barley (Hordeum vulgare) Dhn genes (Choi et al., 1999). The presence of these regulatory elements is consistent with Dhn gene expression patterns under dehydration, low temperature, and ABA treatment. These observations are consistent with prior observations made using the promoter region of a wheat (Triticum aestivum) Dhn gene (wcs120, an ortholog of barley Dhn5) that led others (Ouellet et al., 1998) to propose the existence of highly conserved cold response mechanisms in the tribe Triticeae and other plants. This hypothesis was confirmed by the appearance of numerous CBF/DREB1 cDNA sequences in public databases and recently reinforced by further cDNA characterization in several plant species by Jaglo et al. (2001). Here, we describe the methods that we employed to obtain barley genes encoding proteins that are highly similar to the Arabidopsis CBF/DREB1 family, and the sequence, expression characteristics, and genetic map location of the barley gene HvCbf3. Our work began with knowledge from publicly available rice (Oryza sativa) genome sequence data that there is a CBF1 ortholog in rice.
RESULTS
Isolation of the Rice Ortholog of Arabidopsis CBF/DREB1
Using a tblastn search of the National Center for Biotechnology Information non-redundant database, we found a rice cv Nipponbare sequence (accession no. AB023482) that seemed to encode a polypeptide having significant amino acid sequence homology to Arabidopsis CBF/DREB1. From this sequence, Sasaki et al. (1999) predicted a reading frame consisting of four exons encoding a polypeptide of 423 amino acids. We analyzed this same sequence using DNASIS programs (Hitachi Software Engineering Ltd., San Bruno, CA) and found an open reading frame encoding a 27.7-kD polypeptide consisting of 252 amino acids with most significant amino acid sequence similarity to the Arabidopsis CBF/DREB1 gene CBF1. To amplify this DNA fragment from rice cv Somegawa genomic DNA, we designed oligonucleotides from the putative 5′- and 3′-untranslated regions flanking the open reading frame that we identified (Table I). The PCR product was purified and the sequence determined (Fig. 1A; GenBank accession no. AF243384). The deduced amino acid sequence of the PCR product from rice cv Somegawa was identical to that predicted from the rice cv Nipponbare sequence. We refer to this rice gene as OsCbf1.
Table I.
Gene-specific oligonucleotide sequences used in this work
| Gene | Primer Name | Sequence (5′ to 3′) |
|---|---|---|
| OsCbf1 | RCBF-1 | ACTGCTTGAGACGTCGCAC |
| RCBF-2 | GGTTCAGCTGCTGGACCG | |
| HvCbf3 | BCBF3-1 | GCACCATGCTCAGACTGTTC |
| BCBF3-2 | CAACATCTTCACTCTAAAAGAGGAA | |
| BCBF3-3 | CGAACGACGCTGCCATGCTC | |
| BCBF3-4 | GGACCCAGACGACGGAGATA | |
| BCBF3-7 | TGAAATGTTCAGGCTTGACTTGTT | |
| BCBF3-8 | TGTAGTACGAGCCCAGGTCCAT | |
| Dhn8 | Dc18-4-5 | GTGGAAGAGCCCGAGGTTAAG |
| Dc18-4-6 | CACCTCACCGTTCTCATCGA | |
| Dhn4 | Dc15-108-5 | CATGAGGGACGAGCACCAGACT |
| Dc15-108-10 | GATCTTCTCCTTGATGCCCTTCT | |
| 28S-rRNA | 28S-RNA-1 | GCGAAGCCAGAGGAAACT |
| 28S-RNA-2 | GACGAACGATTTGCACGTC |
Figure 1.
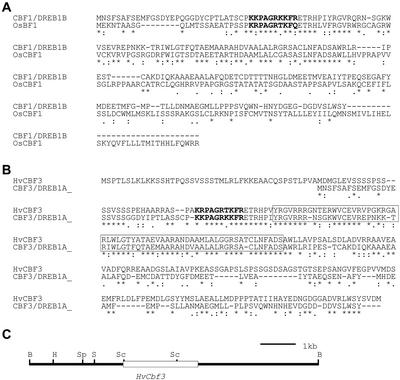
Comparison of CBF amino acid sequences from Arabidopsis, rice, and barley. Amino acids are designated in single-letter code. An asterisk indicates identical, a colon indicates closely related, and a period indicates somewhat related amino acid. Dashes indicate where a sequence has been expanded to optimize alignment. The NLS is in boldface and the AP2-DNA binding domain is boxed. Alignments were performed using Clustal W. A, Arabidopsis CBF1/DREB1B and rice OsCBF1. B, Arabidopsis CBF3/DREB1A versus barley HvCBF3. C, Restriction map of a fragment of bacterial artificial chromosome (BAC) clone 790P15 carrying HvCbf3. The white box indicates the location of the HvCbf3 gene. Restriction enzyme sites are: B, BamHI; H, HindIII; S, SalI; and Sc, SacI.
Isolation of Barley Cbf/DREB1 Gene
We produced cDNA libraries from drought- and cold-stressed barley cv Morex seedlings and from developing spikes of barley cv Morex. Using OsCbf1 as a probe of these libraries, seven positive clones were identified. Sequence data revealed that one clone (GenBank accession no. AF239616) among these seven encodes a polypeptide that is quite similar to Arabidopsis CBF/DREB1 protein CBF3 (Fig. 1A). We refer to this clone and gene as HvCbf3. DNA sequence analysis of this HvCbf3 cDNA suggested that the 5′ end is truncated, but it clearly carries sequences for the typical NLS, AP2-DNA-binding, and acidic domains present in Arabidopsis CBF/DREB1 proteins.
To identify additional barley HvCbf cDNAs, we rescreened barley cDNA libraries using HvCbf3 as a probe and searched public sequence databases. Among the other positive clones that we found, one (GenBank accession no. AF298230) seems to be an ortholog of Arabidopsis CBF1/DREB1B. We also found a previously sequenced barley expressed sequence tag (GenBank accession no. BF631103) that has very high similarity to this clone. Although the existence of a Cbf multigene family in barley is interesting, here we confine our further considerations only to the HvCbf3 gene.
Isolation of the HvCbf3 Gene
Because the HvCBF3 cDNA sequence was less than full length, we wished to examine the sequence of a HvCbf3 genomic clone to gain a complete protein coding sequence and to examine the sequence of the 5′-untranslated region and flanking region. To identify HvCbf3 genomic clones, we screened a barley cv Morex BAC library (Yu et al., 2000). From screening 1.5-genome equivalents, we isolated six positive BAC clones (424E16, 424C17, 572K24, 745C23, 790P15, and 804E19), among which three were confirmed to carry the HvCbf3 gene by PCR using HvCbf3-specific oligonucleotides (Table I), and by blot hybridization of DNA restriction fragments of BAC DNA using the HvCbf3 cDNA as a probe at high stringency (data not shown).
To further isolate the HvCbf3 gene, we chose BAC clone 790P15 because it contained the smallest insert DNA fragment (approximately 98 kb). From BAC clone 790P15, we subcloned into pTZ18R a 7.5-kb BamHI fragment carrying the HvCbf3 gene. Figure 1C shows the restriction map of this fragment. SacI subfragments (1.2 and 4 kb) were then subcloned and sequenced (GenBank accession no. AF298321). These fragments encode a 31.0-kD polypeptide consisting of 293 amino acids (pI 7.22). The HvCBF3 polypeptide contains a high percentage of Ala (13.6%) and Ser (15.0%). The conserved NLS, AP2 DNA-binding domain, and acidic region are present in the HvCbf3 gene (Fig. 1B). Two additional Ser tracts, one in the N-terminal and one in the C-terminal acidic region, also are present. We examined the DNA sequence to about 500 bp upstream of the putative initiation codon of HvCbf3, but no cis-acting element (box I–IV) typical of the Arabidopsis DREB2 gene nor any DRE element was apparent. This suggests that the HvCbf3 gene may not be subject to autoregulation.
Expression of the HvCbf3 Gene
To quantitatively determine the expression pattern of the HvCbf3 gene, we used real-time reverse transcription PCR. Total RNA was isolated from barley seedling shoots sampled at different times throughout a chilling or ABA treatment. For comparison, mRNAs from the same RNA samples were amplified with primers specific for known chilling- and ABA-inducible barley genes, Dhn8 and Dhn4, respectively (Choi et al., 1999; Zhu et al., 2000). Dhn8 encodes a chilling-inducible, acidic SK3 dehydrin and is the ortholog of Arabidopsis COR47, which encodes an acidic SK3 cold-induced dehydrin. The wheat orthologs of barley Dhn8 and Arabidopsis COR47 are the WCOR410 genes (Danyluk et al., 1994), which also encode acidic SK3 cold-induced proteins. Dhn4 encodes a YSK2 dehydrin that is ABA inducible, prevalent during dehydration stress and embryo development, and is the ortholog of Arabidopsis RAB18, which is also an ABA-induced, dehydration-induced, and embryo YSK2 dehydrin (Close, 1997). The quantity of 28S rRNA was measured as a normalization control.
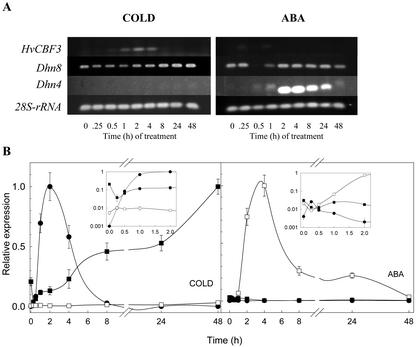
The results show that the HvCbf3 gene is transiently up-regulated by chilling treatment (Fig. 2). HvCbf3 transcripts began to rise after 15 min of cold treatment and reached a maximum level at 2 h, when the amount was at least 10 times the amount at 30 min. Later, HvCbf3 transcripts declined until they were below detection after 24 h. There was also a small, transient induction of HvCbf3 after ABA treatment (Fig. 2), but an effect of mechanical agitation from simply spraying the plants with water (Gilmour et al., 1998) was not ruled out as the cause of this small induction. A dehydration treatment also induced a rapid and transient increase of HvCbf3 transcripts (data not shown), but we also cannot rule out mechanical agitation as the cause of that effect. As expected, the control gene Dhn8 was strongly induced during the chilling treatment, and the Dhn4 gene was strongly induced after ABA application.
Figure 2.
Expression of HvCbf3 in barley seedlings. Plant materials were 6-d-old seedlings growing with a 16-h photoperiod at 20°C or 2°C, and greenhouse-grown 6-d-old seedlings sprayed with 100 μm ABA. Total RNA was used for standard reverse transcription PCR (A, semiquantitative) and real-time reverse transcription PCR (B, quantitative). Gene-specific primers for HvCbf3, the cold-inducible gene Dhn8, the dehydration- and ABA-inducible gene Dhn4, and 28S rRNA were used (see “Materials and Methods”). A, PCR products were electrophoresed in 1.8% (w/v) agarose gels. B, The RNA amount is expressed relative to the highest value found for each gene and the values are normalized against the expression values of 28S rRNA used as an internal control (see “Materials and Methods”). Insets show the expression data on a logarithmic scale during the first 2 h. Bars are the sd of the mean of four amplifications. ●, HvCbf3; ▪, Dhn8; □, Dhn4.
Map Location of the HvCbf3 Gene
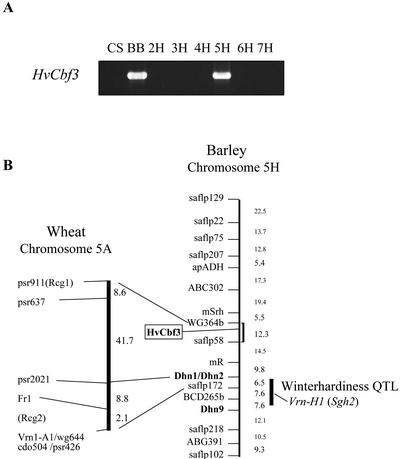
We designed HvCbf3 gene-specific primer sets from the DNA sequences, and used these to determine the chromosome location of HvCbf3 by PCR amplification. The PCR results using wheat-barley disomic addition lines indicated that HvCbf3 is on chromosome 5H (Fig. 3A). Because we identified allele-specific polymorphisms for the HvCbf3 gene, it was possible to determine the genetic map location of this gene using the barley cv Dicktoo × barley cv Morex doubled-haploid (DH) mapping population. Figure 3B shows the map location determined for the HvCbf3 gene. The HvCbf3 gene is located on 5H between the markers WG364b and saflp58.
Figure 3.
Map location of the HvCbf3 gene. A, PCR reactions were performed on genomic DNA from wheat cv Chinese Spring (CS), barley cv Betzes (BB), and six wheat-barley disomic addition lines (carrying 2H, 3H, 4H, 5H, 6H, or 7H) using HvCbf3 gene-specific primers (see “Materials and Methods”). Amplified DNAs were electrophoresed in a 1.2% (w/v) agarose gel. B, The genotypes of each HvCbf3 gene in the barley cv Dicktoo × barley cv Morex mapping population were compared with mapping data (available at http://wheat/pw.usda.gov/ggpages/DxM/dmsor.txt). The map location of HvCbf3 genes is boxed. The dark bar on chromosome 5H indicates a quantitative trait locus (QTL) for vernalization requirement and freezing tolerance (Pan et al., 1994). The same region of wheat chromosome 5A (Galiba et al., 1995; Vagujfalvi et al., 2000) is compared with barley chromosome 5H. Units are in cM.
DISCUSSION
The deduced amino acid sequence of the HvCBF3 protein has important similarities to the Arabidopsis CBF/DREB1 proteins. The NLS and AP2-DNA-binding domains and the Ala-rich acidic C-terminal region are present in all cases, and the “signature sequences” PKK/RPAGRxKFxETRHP and DSAWR (Jaglo et al., 2001) are present. There are also some notable differences between the barley and Arabidopsis proteins. Amino acid sequence identities are not extensive in the N- and C-terminal acidic regions, and there are additional Ser tracts in N- and C-terminal acidic regions of HvCBF3 relative to Arabidopsis.
The early and transient expression of HvCbf3 at chilling temperature is reminiscent of the expression of Arabidopsis CBF/DREB1 genes, and is generally consistent with the temporal induction of wheat and rye (Secale cereale) CBF genes at low temperature (Jaglo et al., 2001), with some caveats.
Chilling-induced barley HvCbf3 expression reached a maximum at about 2 h (Fig. 2), then declined to levels that were below detection by 24 h. In contrast, expression of the rye and wheat CBF genes (see Fig. 2 in Jaglo et al., 2001) seems in total to be less temporally constrained; rye CBF genes were expressed continuously from 30 min to 24 h, and wheat CBF genes were expressed in apparent surges in the 15-min to 24-h time frame. However, the northern-blot method used by Jaglo et al. (2001), which used a complete rye cDNA as a hybridization probe, seems unlikely to distinguish between individual CBF genes. In the highly inbred hexaploid wheat cv Norstar, one would expect 18 unique CBF genes if there are six paralogs for each of the three A, B, and D genome haplotypes. Similarly, because rye is an outcrossing species and therefore carries extensive heterozygosity, the diversity of CBF genes in the diploid rye cv Puma could include two allelic forms of each of six CBF paralogs for a total of as many as 12 distinct versions of CBF genes. Diversity in the expression patterns of these presumed 18 wheat cv Norstar or 12 rye cv Puma CBF genes would then be obscured within the signal averaging that is accomplished by the northern-blot method. It has been shown previously in Arabidopsis that different CBF paralogs have quite distinct differences in their spatial and temporal expression patterns (Gilmour et al., 1998; Sakuma et al., 2002). We note that Figure 2 of Jaglo et al. (2001) shows wheat CBF transcripts that appear to be of more than one size and that the different sizes of transcripts appear at different times. We interpret the results of Jaglo et al. (2001) to be evidence that there are differences in the expression patterns between CBF genes in Triticeae genomes.
Our approach with HvCBF3 narrowed the gene expression analysis to one allele of a single CBF gene because we used fully homozygous diploid material (barley cv Morex) and gene-specific real-time reverse transcriptase-PCR for quantitative measurement of HvCBF3 transcripts. We can state with confidence that the barley cv Morex barley HvCBF3 gene is transiently cold induced, appearing only between the 15-min to 8-h time period during the treatment conditions that we employed. We suggest that it would be interesting to examine variation in the expression patterns of many more barley and other Triticeae CBF genes using gene-specific methods and to consider the consequences of allelic variation in regards to potential relationships of CBF genes to plant phenotypes.
The genetic picture so far, from cDNA sequencing (described in “Results”) and BAC clone contig analyses (data not shown), suggests that there are additional Cbf genes tightly linked to HvCbf3 in barley. We have shown in this work that the HvCbf3 locus lies between markers WG364b and saflp58 on chromosome 5H (Fig. 3B). The Triticeae group 5 chromosome contains a major QTL for winter hardiness that includes separate, but neighboring, loci for vernalization requirement (Vrn1 or Sh2) and freezing tolerance (Fr1; Fig. 3B; Pan et al., 1994; Galiba et al., 1995; Dubcovsky et al., 1998). However, this winter hardiness QTL is a considerable distance (approximately 40–50 cM) from the location of HvCbf3. This means that HvCbf3 certainly is not a component of the major winter hardiness QTL on barley chromosome 5H that was identified by Pan et al. (1994). However, HvCbf3 does map to an intriguing position. Vagujfalvi et al. (2000) identified two loci that are involved in the regulation of cor14b gene expression on wheat chromosome 5A, and it remains possible that one of these loci is HvCbf3, as follows. The cor14 genes are cold inducible and encode 14-kD polypeptides that are imported into the chloroplast. One cor14b regulatory locus (Rcg2) is located near markers psr2021 (also known as ABA2 or Dhn1) and Fr1, and the other (Rcg1) is tightly linked to psr911, which is 60 cM proximal to Fr1. Our HvCbf3 mapping data place it in the same general area as Rcg1, which leaves open the possibility that HvCbf3 may be the same locus as Rcg1. If that is the case, then it may eventually be shown that this or another tightly linked barley Cbf gene can, in certain genetic backgrounds, be a determinant of freezing tolerance. The Rcg1 locus has an effect on the threshold temperature at which cor14b and presumably other low-temperature-induced genes are induced, and, therefore, the threshold temperature of Triticeae Cbf gene expression would seem to be worthy of exploration. Careful analyses of germplasm variation, and consideration of epistatic and genotype x environment interactions, will help clarify the potential role of Cbf genes in freezing tolerance in barley and other Triticeae plants.
MATERIALS AND METHODS
Plant Material
Barley (Hordeum vulgare) cv Dicktoo (winter barley) and cv Morex (spring barley), and 100 F1-derived DH progeny from a cross between these two parents were obtained from Dr. Patrick Hayes (Oregon State University, Corvallis) and propagated at the University of California (Riverside). Seeds of wheat (Triticum aestivum) cv Chinese Spring, barley cv Betzes, and the six wheat-barley addition lines from these two parents (Islam et al., 1981) were provided by Dr. Adam Lukaszewski (University of California, Riverside). Plants were grown in a greenhouse and leaf tissues were cut off, rapidly frozen in liquid nitrogen, and stored at −80°C until use.
cDNA and Genomic Libraries
RNA was extracted from drought- and cold-treated barley cv Morex seedling shoots, and developing spikes at the white, green, and yellow anther stage. Total RNA was prepared using a hot phenol procedure described by Verwoerd et al. (1989). Poly(A+) RNA was purified using PolyATract mRNA Isolation System (Promega, Madison, WI). cDNAs with EcoRI on the 5′ and XhoI on the 3′ end were synthesized using a ZAP-cDNA synthesis kit (Stratagene, San Diego). cDNAs larger than 0.5 kb were selected by size fractionation via gel filtration and directionally cloned into the Uni-ZAP XR vector, and in vivo packaged using GigaPack III Gold packaging extract (Stratagene). About 5 × 104 plaques per primary library were lifted to nylon membrane and probed by standard methods (Sambrook et al., 1989).
The barley cv Morex BAC library has been described previously (Yu et al., 2000). Membranes spotted with BAC clones were obtained from Andris Kleinhofs (Washington State University, Pullman).
Sequence Analysis
DNA fragments were sequenced on both strands using the dideoxy chain termination method at Davis Sequencing, LLC (Davis, CA). The nucleotide and deduced amino acid sequences were analyzed with the DNASIS programs and compared with sequences in databases using BLAST. Amino acid sequence alignments were performed using Clustal W. From the barley Cbf gene sequences, we designed gene-specific oligonucleotides (Table I). Oligonucleotides were synthesized at Sigma-Genosys (The Woodlands, Texas).
Expression Analysis by Real-Time Reverse Transcription PCR
Barley seedlings were treated essentially as described previously (Choi et al., 1999; Choi and Close, 2000). For the cold treatment, 6-d-old seedlings growing in pots with soil in an illuminated growth chamber were used. The seedlings were first grown at 20°C, 70% relative humidity, and 16-h photoperiod, the cold treatment was initiated by changing the chamber temperature to 2°C, and green tissue from 20 seedlings was harvested at each sampling time. Seedlings were also grown in pots with soil in a greenhouse, sprayed 6 d after sowing with a solution of 100 μm ABA and 0.05% (v/v) Tween 20, then 20 or more seedling shoots were harvested at each sampling time for RNA extraction. In all cases leaf tissues were cut off, snap frozen in liquid nitrogen, and stored at −80°C until RNA extraction.
Total RNA was extracted from green tissue using the hot-phenol method (Verwoerd et al., 1989) and treated with RNAse-free DNase (Life Technologies, Gaithersburg, MD) at room temperature for 15 min in reaction buffer containing 20 mm Tris-HCl (pH 8.4), 2 mm MgCl2, and 50 mm KCl. DNaseI was inactivated by adding EDTA (2.5 mm final concentration) and heating to 65°C for 10 min.
A cDNA first strand was synthesized using Taq-Man Reverse Transcription Reagents (Applied Biosystems, Foster City, CA) following the manufacturer's directions. The reaction mixture contained: 20 ng μL−1 total RNA (DNase treated), 10 mm Tris-HCl (pH 8.3), 50 mm KCl, 5.5 mm MgCl2, 0.5 mm each dNTP, 2.5 μm random hexamers, 0.4 units μL−1 RNase inhibitor, and 1.25 units μL−1 MultiScribe Reverse Transcriptase. The mixture was pre-incubated at 25°C for 10 min and the reaction was completed at 48°C for 30 min. The enzyme was inactivated by incubation at 95°C for 5 min. The cDNA samples were stored at −20°C. Quantitative PCR was performed in an ABI Prism 7700 Sequence Detection System (Applied Biosystems) using the SYBR green I master mix (Applied Biosystems) containing optimized buffer, dATP, dGTP, dCTP, dUTP, and Amplitaq Gold DNA Polymerase. Each 25-μL reaction contained 2× SYBR green master mix, 50 ng of cDNA, and 100 nm forward and reverse primers (300 nm each primer for 28S rRNA amplifications).
The primers used for quantitative PCR were designed using Primers Design software (Applied Biosystems). Target sequences were barley cv Morex HvCbf3 (AF239616), Dhn8 (AF181458), Dhn4 (AF181454), and 28S rRNA. For 28S rRNA, the primers carried the same sequences used for a study of chicken Gallus gallus bursal disease virus (Moody et al., 2000) to measure chicken rRNA quantity; these primers can also prime the amplification of a fragment of a barley 28S rRNA sequence (BF616316), which is highly similar to chicken rRNA. The gene-specific oligonucleotides are shown in Table I.
Reaction conditions for thermal cycling were: 95°C for 10 min, 40 cycles of 95°C for 15 s, 60°C for 1 min, and 73°C for 15 s. Fluorescence data collection was done during the 73°C cycle step. Using this methodology, detection of PCR product is monitored by measuring the increase in fluorescence caused by the binding of SYBR green dye to dsDNA (Yin et al., 2001). For each gene, a standard curve was prepared using a serial dilution of an experimental cDNA sample. These samples were chosen upon the basis of having the largest amount of target cDNA according to gel electrophoresis analysis of nonquantitative PCR reactions. Data were analyzed with the Sequence Detector version 1.7 software (Applied Biosystems). The software calculated the threshold cycle from the plot of the increase in intensity of fluorescence of the reporter dye versus the cycle number. The quantity of cDNA was calculated from threshold cycle by interpolation from the standard curve. To account for differences in total RNA present in each sample, the cDNA amount calculated was normalized using the amount of 28S rRNA detected in the same sample.
Chromosome Assignment
Genomic DNA from wheat cv Chinese Spring, barley cv Betzes, and the six wheat-barley addition lines derived from these two parents was purified using DNAzol according to instructions provided by the manufacturer (Life Technologies). Genomic DNA amplifications were performed as described previously (Choi et al., 2000). In brief, PCR reactions were performed in a 50-μL volume containing 1.25 units of Taq DNA polymerase (Qiagen, Hilden, Germany), 1× PCR buffer (10 mm Tris-HCl, pH 8.3; 50 mm KCl2; and 1.5 mm MgCl2), 1× Q-solution, 200 μm of each dNTP, 0.3 μM of each oligonucleotide, and 100 ng of genomic DNA. PCR reactions were initiated at 95°C for 5 min, followed by 40 cycles at 95°C for 30 s, 61°C for 1 min, 72°C for 30 s, and terminated at 72°C for 10 min. Amplified DNAs were electrophoresed in a 1.2% (w/v) agarose gel.
Mapping of the HvCbf3 Gene
To develop PCR product polymorphisms for the barley HvCbf3 gene, two forward and reverse primers were designed from the barley cv Morex HvCbf3 gene sequence using the program PRIMER-MASTER (Table I). All four possible combinations of PCR reactions were performed using genomic DNA from barley cv Dicktoo, cv Morex, and other parents of various barley DH mapping populations. A presence versus absence polymorphism between barley cv Dicktoo and cv Morex was observed for one of the four primer combinations. Using this presence versus absence polymorphism, the genotype of each DH line in the barley cv Dicktoo × barley cv Morex mapping population was determined, as described previously (Choi et al., 2000). The map location of the barley HvCbf3 gene was then determined by aligning our data with existing mapping data for the barley cv Dicktoo × barley cv Morex DH mapping population (available at http://wheat.pw.usda.gov/ggpages/DxM/dmsor.txt). The number of recombinants between the HvCbf3 locus and the nearest locus was divided by the number of individuals for which genotype data was available for both markers to give a percent recombination value.
ACKNOWLEDGMENTS
The authors thank Jorge Dubcovsky (University of California, Davis) for comments on genetic markers near the Rcg and Vrn1/Sh2 loci, and for first drawing our attention in March of 2000 to the similarity of the location of HvCbf3 and the Rcg1 locus. The authors also thank Raymond Fenton, Elena Turco, Saule Abugalieva, and Matthew Moscou (University of California, Riverside) for assistance with the barley seedling experiments.
Footnotes
This work was supported by the U.S. Department of Agriculture/Cooperative State Research, Education, and Extension Service (grant no. 95–37100–1595) and by the California Agricultural Experiment Station (Hatch grant no. 5306–H).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.003046.
LITERATURE CITED
- Baker SS, Wilhelm KS, Thomashow MF. The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol Biol. 1994;24:701–713. doi: 10.1007/BF00029852. [DOI] [PubMed] [Google Scholar]
- Bray EA. Molecular responses to water deficit. Plant Physiol. 1993;103:1035–1040. doi: 10.1104/pp.103.4.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D-W, Close TJ. A newly identified barley gene, Dhn12, encodes a YSK2 DHN, is located on chromosome 6H and has embryo-specific expression. Theor Appl Genet. 2000;100:1274–1278. [Google Scholar]
- Choi D-W, Koag MC, Close TJ. Map locations of barley Dhn genes determined by gene-specific PCR. Theor Appl Genet. 2000;101:350–354. [Google Scholar]
- Choi D-W, Zhu B, Close TJ. The barley (Hordeum vulgare L.) dehydrin multigene family: sequences, allele types, chromosome assignments, and expression characteristics of 11 Dhn genes of cv. Dicktoo Theor Appl Genet. 1999;98:1234–1247. [Google Scholar]
- Close TJ. Dehydrins: a commonality in the response of plants to dehydration and low temperature. Physiol Plant. 1997;100:291–296. [Google Scholar]
- Danyluk J, Houde M, Rassart E, Sarhan F. Differential expression of a gene encoding an acidic DHN in chilling sensitive and freezing tolerant Gramineae species. FEBS Lett. 1994;244:20–24. doi: 10.1016/0014-5793(94)00353-x. [DOI] [PubMed] [Google Scholar]
- Dubcovsky J, Lijavetzky D, Appendino L, Tranquilli G. Comparative RFLP mapping of Triticum monococcum genes controlling vernalization requirement. Theor Appl Genet. 1998;97:968–975. [Google Scholar]
- Dunn MA, White AJ, Vural S, Hughes MA. Identification of promoter element in a low-temperature-responsive gene (blt4.9) from barley (Hordeum vulgare L.) Plant Mol Biol. 1998;38:551–564. doi: 10.1023/a:1006098132352. [DOI] [PubMed] [Google Scholar]
- Galiba G, Quarrie SA, Sutka J, Morgounov A, Snape JW. RFLP mapping of the vernalization (Vrn1) and frost resistance (Fr1) genes in chromosome 5A of wheat. Theor Appl Genet. 1995;90:1174–1179. doi: 10.1007/BF00222940. [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Zarke DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 1998;16:433–442. doi: 10.1046/j.1365-313x.1998.00310.x. [DOI] [PubMed] [Google Scholar]
- Ingram J, Bartels D. The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:377–403. doi: 10.1146/annurev.arplant.47.1.377. [DOI] [PubMed] [Google Scholar]
- Islam AKMR, Shepherd KW, Sparrow DHB. Isolation and characterization of euplasmic wheat-barely chromosome addition lines. Heredity. 1981;46:161–174. [Google Scholar]
- Jaglo KR, Kleff S, Amundsen KL, Zhang X, Haake V, Zhang JZ, Deits T, Thomashow MF. Components of the Arabidopsis C-repeat/dehydration-responsive element binding factor cold-responsive pathway are conserved in Brassica napus and other plant species. Plant Physiol. 2001;127:910–917. [PMC free article] [PubMed] [Google Scholar]
- Jaglo-Ottosen KR, Gilmour SJ, Zark DG, Schabenberger O, Thomashow MF. Arabidopsis CBF-1 overexpression induces Cor genes and enhances freezing tolerance. Science. 1998;280:104–106. doi: 10.1126/science.280.5360.104. [DOI] [PubMed] [Google Scholar]
- Jiang C, Iu B, Singh J. Requirement of a CCGAC cis-acting element for cold induction of the BN115 gene from winter Brassica napus. Plant Mol Biol. 1996;30:679–684. doi: 10.1007/BF00049344. [DOI] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nature Biotechnol. 1999;17:287–291. doi: 10.1038/7036. [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody A, Sellers S, Bumstead N. Measuring infectious bursal disease virus RNA in blood by multiplex real-time quantitative RT-PCR. J Virol Methods. 2000;85:55–64. doi: 10.1016/s0166-0934(99)00156-1. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Sato S, Asamizu E, Kaneko T, Kotani H, Miyajima N, Tabata S. Structural analysis of Arabidopsis thaliana chromosome 5: VII. Sequence features of the regions of 1,013,767 bp covered by sixteen physically assigned P1 and TAC clones. DNA Res. 1998;5:297–308. doi: 10.1093/dnares/5.5.297. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Shinwari ZK, Sakuma Y, Seki M, Miura S, Shinozaki K, Yamaguchi-Shinozaki K. Organization and expression of two Arabidopsis DREB2 genes encoding DRE-binding proteins involved in dehydration- and high-salinity-responsive gene expression. Plant Mol Biol. 2000;42:657–665. doi: 10.1023/a:1006321900483. [DOI] [PubMed] [Google Scholar]
- Ouellet F, Vazquez-Tello A, Sarhan F. The wheat wcs120 promoter is cold-inducible in both monocotyledonous and dicotyledonous species. FEBS Lett. 1998;423:324–328. doi: 10.1016/s0014-5793(98)00116-1. [DOI] [PubMed] [Google Scholar]
- Pan A, Hayes PM, Chen F, Chen THH, Blake T, Wright S, Karsai I, Bedo Z. Genetic analysis of the components of winter hardiness in barley (Hordeum vulgare L.) Theor Appl Genet. 1994;89:900–910. doi: 10.1007/BF00224516. [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun. 2002;290:998–1009. doi: 10.1006/bbrc.2001.6299. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sasaki T, Nagamura Y, Yamamoto K (1999) Oryza sativa genomic DNA, chromosome 6, clone P0680A03, complete sequence. GenBank accession no. AB023482
- Shinozaki K, Yamaguchi-Shinozaki K. Gene expression and signal transduction in water-stress response. Plant Physiol. 1997;115:327–334. doi: 10.1104/pp.115.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binding to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- Thomashow MF, Gilmour SJ, Stockinger EJ, Jaglo-Ottosen KR, Zarka DG. Role of Arabidopsis CBF transcriptional activators in cold acclimation. Physiol Plant. 2001;112:171–175. [Google Scholar]
- Vagujfalvi A, Crosatti C, Galiba G, Dubcovsky J, Cattivelli L. Two loci on wheat chromosome 5A regulate the differential cold-dependent expression of the cor14b gene in frost tolerant and sensitive genotypes. Mol Gen Genet. 2000;263:194–200. doi: 10.1007/s004380051160. [DOI] [PubMed] [Google Scholar]
- Verwoerd TC, Dekker BM, Hoekema A. A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 1989;17:2362. doi: 10.1093/nar/17.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin JL, Shackel NA, Zekry A, McGuiness PH, Richards C, Van der Putten K, McCaughan GW, Eris JM, Biship GA. Real-time reverse transcriptase-polymerase chain reaction (RT-PCR) for measurement of cytokine and growth factor mRNA expression with fluorogenic probes or SYBR Green I. Immunol Cell Biol. 2001;79:213–221. doi: 10.1046/j.1440-1711.2001.01002.x. [DOI] [PubMed] [Google Scholar]
- Yu Y, Tomkins JP, Waugh R, Frisch DA, Kudrna D, Kleinhofs A, Brueggeman RS, Muehlbauer GJ, Wise RP, Wing RA. A bacterial artificial chromosome library for barley (Hordeum vulgare L.) and the identification of clones containing putative resistance genes. Theor Appl Genet. 2000;101:1093–1099. [Google Scholar]
- Zhu B, Choi DW, Fenton RD, Close TJ. Expression of the barley dehydrin multigene family and the development of freezing tolerance. Mol Gen Genet. 2000;264:145–153. doi: 10.1007/s004380000299. [DOI] [PubMed] [Google Scholar]