Abstract
The cDNA for 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase of Arabidopsis encodes a polypeptide with an amino-terminal signal sequence for plastid import. A cDNA fragment encoding the processed form of the enzyme was expressed in Escherichia coli. The resulting protein was purified to electrophoretic homogeneity. The enzyme requires Mn2+ and reduced thioredoxin (TRX) for activity. Spinach (Spinacia oleracea) TRX f has an apparent dissociation constant for the enzyme of about 0.2 μm. The corresponding constant for TRX m is orders of magnitude higher. In the absence of TRX, dithiothreitol partially activates the enzyme. Upon alkylation of the enzyme with iodoacetamide, the dependence on a reducing agent is lost. These results indicate that the first enzyme in the shikimate pathway of Arabidopsis appears to be regulated by the ferredoxin/TRX redox control of the chloroplast.
The shikimate pathway is a series of enzyme-catalyzed reactions leading to chorismate, the precursor of Phe, Tyr, Trp, and numerous secondary metabolites derived from these aromatic amino acids. The first enzyme of this pathway, 3-deoxy-d-arabino-heptulosonate 7-phosphate (DAHP) synthase, catalyzes the condensation of phosphoenolpyruvate (PEP) and erythrose-4-phosphate (E4P), yielding DAHP and inorganic phosphate. The pathway is found only in microorganisms and plants (Herrmann and Weaver, 1999).
In bacteria, carbon flow into the pathway is regulated by transcriptional control and by feedback inhibition of DAHP synthase, both mediated by the aromatic amino acids; in vivo, feedback inhibition is the major form of control (Ogino et al., 1982).
In plants, DAHP synthase activity is regulated somewhat differently, which may not be surprising, because there is only about 20% sequence identity between the bacterial and the plant enzyme. Plants frequently must provide additional chorismate for the synthesis of aromatic secondary metabolites, for example, when mechanically wounded or attacked by insects. Under those conditions, mRNA encoding DAHP synthase increases within hours followed by a rise in enzyme activity (Dyer et al., 1989). Detailed northern-blot analyses showed that the syntheses of all the enzymes in the shikimate pathway are transcriptionally regulated (Bischoff et al., 1996, 2001). However, despite extensive experimentation, feedback inhibition of plant DAHP synthase by the aromatic amino acids has never been observed.
Plant DAHP synthase was first purified to homogeneity from carrot (Daucus carota) roots and potato (Solanum tuberosum) tubers (Suzich et al., 1985; Pinto et al., 1986). Polyclonal antibodies specific for this protein were used to obtain cDNA encoding DAHP synthase (Dyer et al., 1990). Sequence information from the potato cDNA facilitated the cloning of homologs from several other plants including, tobacco (Nicotiana tabacum), Arabidopsis, and tomato (Lycopersicon esculentum; Keith et al., 1991; Wang et al., 1991; Görlach et al., 1993). All cDNAs for plant DAHP synthases encode polypeptides with amino-terminal signal sequences that function in plastid import (Gavel and von Heijne, 1990). Signal sequences were also present in the other six enzymes of the shikimate pathway, providing evidence that the synthesis of chorismate in plant cells takes place in plastids (Herrmann and Weaver, 1999).
In chloroplasts, four enzymes of the Benson-Calvin cycle, spinach (Spinacia oleracea) phosphoribulokinase (Brandes et al., 1996), pea (Pisum sativum) glyceraldehyde-3-phosphate dehydrogenase (Li and Anderson, 1997) and Fru-1,6-bisphosphate phosphatase (Jacquot et al., 1995), and Arabidopsis sedoheptulose-1,7-bisphosphate phosphatase (Willingham et al., 1994) and the larger of two isoforms of Rubisco activase (Zhang and Portis, 1999), a H+-ATPase (Schwarz et al., 1997), and NADP-malate dehydrogenase (Miginiac-Maslow et al., 1997; Ocheretina et al., 2000) are regulated by light via the ferredoxin/thioredoxin (Fd/TRX) system (Buchanan et al., 1994; Ruelland and Miginiac-Maslow, 1999; Schürmann and Jacquot, 2000). In this paper, we show that Arabidopsis DAHP synthase requires reduced TRX for enzyme activity, thereby linking carbon flow into the shikimate pathway to electron flow from photosystem I. Thus, the biosynthesis of the three aromatic amino acids and of all aromatic secondary metabolites derived from Phe, Tyr, and Trp is apparently redox regulated by the Fd/TRX system.
RESULTS
Purification of Arabidopsis DAHP Synthase
The Arabidopsis gene DAHP synthase isoenzyme 1 (DHS1; Keith et al., 1991) encodes a DAHP synthase precursor that contains a putative amino-terminal sequence characteristic for plastid import (Gavel and von Heijne, 1990). The sequence is removed during the import of the polypeptide into isolated chloroplasts (Zhao, 1992). We cloned a partial cDNA sequence encoding the mature form of the enzyme into a pET vector and transformed Escherichia coli for heterologous expression of the plant protein. Induction of the resulting bacteria by isopropyl β-d-thiogalactoside yielded cell extracts, in which up to 5% of the soluble protein is DAHP synthase. In transformed E. coli strains grown in Luria broth, the bacterial DAHP synthase is repressed, and only the plant enzyme is detected. Moreover, bacterial and plant DAHP synthases are distinguishable, because the former enzyme is feedback inhibited by aromatic amino acids, and the two types of enzyme do not cross-react with heterologous antibodies raised against either the bacterial or the plant enzyme, nor do they copurify.
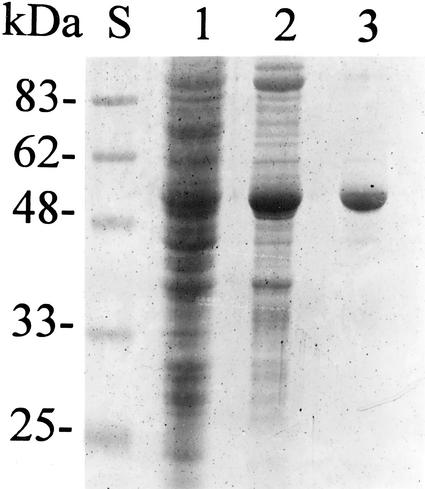
In Arabidopsis, DHS1 is preferentially expressed upon demand for increased carbon flow into the shikimate pathway (Keith et al., 1991; Zhao, 1992). Therefore, we have analyzed the encoded protein in some detail. From extracts of transformed E. coli cells, using standard techniques, we purified Arabidopsis DAHP synthase to apparent electrophoretic homogeneity. Figure 1 shows an SDS polyacrylamide gel of fractions from such a purification. The final product was judged to be better than 95% pure plant DAHP synthase and was not contaminated by the bacterial ortholog, because it was not inhibited by any of the three aromatic amino acids and had a strict requirement for a reducing environment. This preparation was used for all further experiments.
Figure 1.
Purification of Arabidopsis DAHP synthase heterologously expressed in E. coli. SDS-PAGE on a 10% (w/v) polyacrylamide gel. Lane S, Five microliters of prestained Mr standards (Bio-Rad, Hercules, CA); lane 1, 10 μL of E. coli BL21(DE3)/pET23d/DHS1 crude extract; lane 2, 10 μL of the phosphocellulose pool; and lane 3, 100 μL of the Sephacryl 300 pool.
Arabidopsis DAHP Synthase Is a Metallo Enzyme
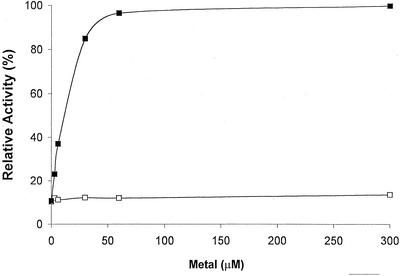
Like all other known DAHP synthases, the enzyme from Arabidopsis requires a metal ion for activity. The most effective cation is divalent Mn, which has an apparent dissociation constant of less than 10 μm (Fig. 2). Mg2+ can replace Mn2+, but millimolar concentrations are required. We also tested Co2+, Fe2+, and Ca2+. These ions failed to satisfy the metal requirement.
Figure 2.
Metal ion activation of Arabidopsis DAHP synthase. The relative enzyme activity is plotted as a function of the metal ion concentration in the assay. ▪, MnCl2; □, MgCl2. Before assay, the purified enzyme was passed over a gel-filtration column equilibrated with a buffer containing neither MnCl2 nor MgCl2. The reaction was then started by adding the indicated concentrations of metal ions.
Arabidopsis DAHP Synthase Requires Reduced TRX for Enzyme Activity
To obtain active DAHP synthase enzyme preparations from extracts of either Arabidopsis plants or E. coli expressing Arabidopsis DHS1, extraction buffers must contain reducing agents. The use of extraction buffers without reducing agents or the removal of the reducing agent after extraction yields enzymatically inactive proteins. We prepared pure Arabidopsis DAHP synthase in buffers containing reducing agent, removed the agent by buffer exchange, and added it back in increasing concentrations.
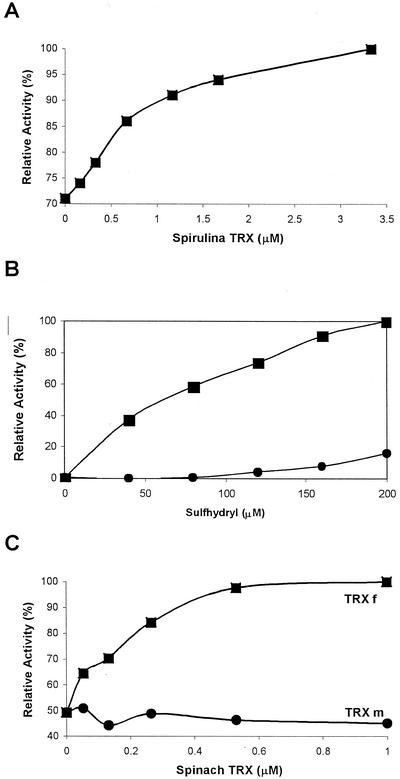
Figure 3 shows the activation of Arabidopsis DAHP synthase by reduced TRX. We used dl-dithiothreitol (DTT) to reduce TRX. DTT alone partially activates the enzyme as well (Fig. 3B). However, the dissociation constant for TRX to DAHP synthase is 50- to several 100-fold smaller than the corresponding constant of DTT to the enzyme, and maximal enzyme activity is only obtained with reduced TRX (Fig. 3). Plant TRX is a small protein containing two Cys residues that are readily oxidized by air and, in chloroplasts, reduced by Fd/TRX reductase. Spinach chloroplasts contain two isoforms, TRX f and m (Wolosiuk et al., 1979), originally identified as activators for Fru-1,6-bisphosphate phosphatase and malate dehydrogenase, respectively. Spinach TRX f and m were obtained in pure form from Dr. Peter Schürmann. Recombinant spinach TRX f (Aguilar et al., 1992) activates Arabidopsis DAHP synthase with an apparent dissociation constant of <0.2 μm (Fig. 3C). This value is close to the one reported for the interaction of TRX f with Fru-1,6-bisphosphatase (Aguilar et al., 1992). Recombinant spinach TRX m (Schürmann, 1995) at that concentration does not activate Arabidopsis DAHP synthase. Arabidopsis chloroplasts contain two TRX f isoforms (Sato et al., 1997). We have not yet shown which of these is the preferred reducing agent for DAHP synthase.
Figure 3.
Activation of Arabidopsis DAHP synthase by reducing agents. The relative enzyme activity is plotted as a function of the concentration of the reducing agent in the assay. Before all assays, the purified enzyme was passed over a gel-filtration column equilibrated with a buffer that contained no reducing agent. A, The indicated concentrations of Spirulina sp. TRX were preincubated with 10 mm DTT and enzyme for 15 min at 25°C. The reactions were started by addition of substrates as described in “Materials and Methods.” B, Activation of the enzyme by DTT (squares) or β-mercaptoethanol (circles). C, Activation of the enzyme by recombinant spinach TRX f (squares) or TRX m (circles).
Alkylated Arabidopsis DAHP Synthase Is Enzymatically Active without Reducing Agents
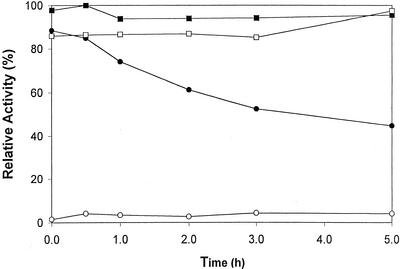
Arabidopsis DAHP synthase preparations stored in the absence of a reducing agent become permanently inactivated. When the enzyme is kept in the absence of a reducing agent for 5 h at 25°C and then assayed in the presence of DTT, only about 50% of the activity is recovered. The inactivation is faster at 37°C and much slower at 0°C. Thus, inactivation is time and temperature dependent. Figure 4 shows the data for the time-dependent inactivation at 25°C. Figure 4 also shows that the inactivation is prevented by treating the enzyme with iodoacetamide. Whereas the enzyme cannot be assayed in the presence of the alkylating agent, the enzyme is fully active after the removal of excess iodoacetamide. More importantly, the alkylated DAHP synthase is no longer dependent on a reducing agent for activity. Alkylation of native DAHP synthase requires DTT, a property previously observed for the redox-regulated NADP-malate dehydrogenase (Decottignies et al., 1988).
Figure 4.
Alkylation of Arabidopsis DAHP synthase eliminates the need for reducing agent. A sample of the purified enzyme was divided into two equal portions. One portion was alkylated with iodoacetamide and the other as a control treated with water. Then both were passed over a Sephadex G25 column equilibrated with buffer C. Alkylated (squares) and nonalkylated (circles) enzyme preparations were kept at room temperature for the indicated times without DTT and then assayed in the presence (black symbols) or absence (white symbols) of DTT.
DISCUSSION
During photosynthesis in chloroplast thylakoid membranes, electrons flow from water, via photosystems II and I, to NADP+, generating oxygen, ATP, and NADPH + H+. ATP and the reducing equivalents are used to drive carbon fixation. The first water-soluble intermediate in the flow of these electrons is Fd. Reduced Fd can donate electrons not only to NADP+ but also to TRX, a small protein containing a prominent disulfide bridge that is reduced to two thiols in the process. It is this Fd/TRX oxidation/reduction reaction that imposes light control on a number of critical pathways in the chloroplast. Reduced TRX reduces disulfide bridges in several enzymes, thereby changing the catalytic properties of these proteins. Foremost among the light-regulated polypeptides are several enzymes in the Benson-Calvin cycle and one in the malate-oxaloacetate shuttle, NADP-malate dehydrogenase. These enzymes are inactive in the oxidized form and are readily activated by reduced TRX, which is available only during active electron flow through the photosystems, i.e. exposure to light.
In vitro, all of these regulated enzymes can be activated by reduced TRX and most of them by DTT alone as well. For some of these enzymes, the specific Cys residues involved in this regulatory process have been identified. For example, the TRX f-regulated Fru-1,6-bisphosphate phosphatase has been analyzed by site-directed mutagenesis (Jacquot et al., 1995; Rodriguez-Suarez et al., 1997). Seven conserved Cys residues of the rapeseed (Brassica napus) enzyme were changed to Ser residues. In three of the seven mutant enzymes, sensitivity to DTT was strongly reduced, suggesting the involvement of two cystine bridges in the redox regulation of this enzyme. Although the crystal structure of the oxidized pea Fru-1,6-bisphosphate phosphatase shows the location of only one disulfide bridge, the structure does provide an explanation for the apparently conflicting data of the mutagenesis study (Chiadmi et al., 1999). The TRX m-regulated NADP malate dehydrogenase structure provides a basis for the mechanism of the redox activation for this enzyme (Carr et al., 1999; Johansson et al., 1999).
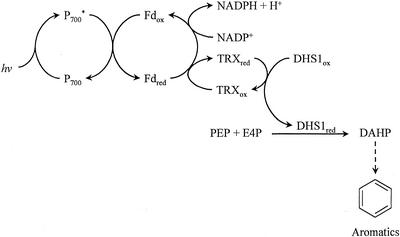
We show here that the first enzyme of the shikimate pathway should be included in this group of redox-regulated chloroplast enzymes (Fig. 5), specifically into the TRX f-activated anabolic enzymes. Purified Arabidopsis DAHP synthase is activated by reduced TRX. When the reducing agent is removed from reduced enzyme, all enzymatic activity is lost. Furthermore, in the absence of reducing agent, the enzyme apparently undergoes a conformational change in a time- and temperature-dependent manner. For example, we cannot reactivate an enzyme preparation left at 37°C for 1 h without reducing agent.
Figure 5.
Biosynthesis of aromatic compounds in chloroplasts. Electrons of photosystem I (P700*)-reduced Fd can either reduce NADP+ or TRX. Reduced TRX activates DAHP synthase, the first enzyme of the shikimate pathway.
When Arabidopsis DAHP synthase prepared in the presence of DTT is alkylated with iodoacetamide and, thereafter, the DTT and excess iodoacetamide are removed by buffer exchange, the resulting alkylated enzyme is fully active and no longer requires a reducing agent for enzyme activity. The alkylated enzyme is equally active in the absence or presence of the reducing agent, indicating that the Cys residues involved in the regulation by reduction are not directly required for catalysis.
Comparing the primary structures of redox-regulated enzymes with their unregulated orthologs from other cell compartments reveals additional Cys-containing peptide inserts in the regulated enzymes (Ruelland and Miginiac-Maslow, 1999). However, a consensus structure for such inserts cannot be deduced from the amino acid sequences. Redox-regulated plant DAHP synthases have about 170 more amino acid residues per chain than their prokaryotic homologs that do not require reducing agents for activity. Cys residues in these extra sequences may be involved in generating a redox-regulated enzyme.
Cys residues are also involved in the formation of the active site of DAHP synthase. The bacterial enzyme contains a CXXH motif involved in metal binding, a requirement for enzyme activity (Stephens and Bauerle, 1992). Even though the bacterial and plant enzymes are only about 20% identical in their primary structures, the CXXH motif is found in both, and the plant enzyme also requires a metal for enzyme activity. Based on the NADP-malate dehydrogenase model (Ruelland and Miginiac-Maslow, 1999), one can envision that the Arabidopsis DAHP synthase contains functionally different Cys residues, one that is subject to reduction/oxidation by the Fd/TRX regulatory system and one in the metal binding portion of the active site. Our alkylation experiments indicate that the regulatory Cys residue is solvent exposed. The regulatory Cys residue can be alkylated without destroying enzyme activity, and the resulting alkylated enzyme is no longer dependent on a reducing agent for activity. We are in the process of addressing the function of individual Cys residues by site-directed mutagenesis.
That light may regulate DAHP synthase at the genetic (Henstrand et al., 1992) and enzyme level could suggest that light replaced the aromatic amino acids as regulators during evolution. If chloroplasts are of bacterial origin, one can assume that feedback-sensitive DAHP synthases were present in early plant cell organelles. Although today's chloroplast DAHP synthase has lost the sensitivity to aromatic amino acids, the enzyme still retains a binding site for those amino acids, because the enzyme is slightly activated by Trp (Suzich et al., 1985). However, light may be the main regulator of the plant enzyme. This finding raises a number of interesting questions. For example, is wound repair faster in the light? Or even more basic, do plants synthesize aromatic amino acids only during the day, or are the other DAHP synthase isoenzymes active in the dark? The shikimate pathway is one of the major routes of carbon flow in green plants. It now becomes a challenge to demonstrate experimentally that, in vivo, light regulation of DAHP synthase links carbon flow directly to the generation of fixed carbon by the carbon reactions of photosynthesis through sharing reduced Fd as a trigger for pathway activity.
MATERIALS AND METHODS
Chemicals and Biochemicals
N-(2-Hydroxymethyl)piperazine-N′-(3-propanesulfonic acid) (EPPS), DTT, MnCl2, MgCl2, isopropyl β-d-thiogalactoside, and Spirulina sp. TRX were from Sigma (St. Louis). PEP (Clark and Kirby, 1963), E4P (Sieben et al., 1966), and spinach (Spinacia oleracea) TRX (Wolosiuk et al., 1979) were prepared and assayed as described. Recombinant spinach TRX f and TRX m were generous gifts from Dr. Peter Schürmann (Aguilar et al., 1992; Schürmann, 1995). Sodium Chelex 100 resin, biotechnology grade, was from Bio-Rad. Vent polymerase was from New England Biolabs (Beverly, MA). Oligonucleotides were from the Purdue Laboratory for Macromolecular Structure. Escherichia coli BL21(DE3) and plasmid pET23d were from Novagen (Madison, WI). All other reagents were of the highest purity commercially available.
Expression of Arabidopsis DHS1 in E. coli
The open reading frame of the Arabidopsis DHS1 cDNA (Keith et al., 1991) encodes a DAHP synthase precursor with a putative signal sequence (Gavel and von Heijne, 1990) that is removed during plastid import. The actual cleavage site has been determined for potato (Solanum tuberosum) DAHP synthase (Zhao, 1992). From sequence alignments, we predicted that the Arabidopsis DAHP synthase cleavage site lays between Thr-48 and Ala-49. We used PCR to synthesize a DNA molecule encoding the mature form of Arabidopsis DAHP synthase beginning at Ala-49. The forward oligonucleotide was 5′-AACCTACCATGGGGGCTGTACACGCGGCTGAG- 3′, and the reverse oligonucleotide was 5′-AATCCGTCGACGACTCAAGACACACGCTGG-3′. The resulting fragment was cloned into the E. coli expression vector pET23d at the NcoI and SalI sites using standard techniques (Ausubel et al., 1987). The correctness of the construct was verified by sequencing both strands of the DNA. Expression of the resulting E. coli BL21(DE3)/pET23dDHS1 yields a DAHP synthase polypeptide with two additional residues (Met-Gly) at the amino terminus. Eight milliliters of an overnight culture of this strain were inoculated into 1 L of Luria broth (10 g L−1 bacto-tryptone, 5 g L−1 yeast extract, 10 g L−1 NaCl, and 1 mL of 1.0 n NaOH) containing 50 mg of ampicillin. The culture was grown at 37°C to an optical density of 0.6 at 600 nm. At that point, isopropyl β-d-thio-galactoside (final concentration 400 μm) was added, and incubation was continued for 4 h.
Purification of Arabidopsis DHS1
All operations were carried out at 4°C. The cells were harvested by centrifugation, washed, and resuspended in buffer A (50 mm potassium phosphate, pH 7.4, containing 1 mm PEP, 1.5 mm Trp, and 0.2 mm DTT). All buffers contain PEP, which stabilizes the enzyme, and Trp, which slightly activates plant DAHP synthases (Suzich et al., 1985). The cells were broken in a French pressure cell. The resulting cell extract was clarified by centrifugation for 20 min at 15,000g in an RC-5b centrifuge (Sorvall, Newton, CT). The supernatant was loaded onto a cellulose phosphate column equilibrated with buffer A. The protein was eluted with a linear gradient of 0.05 to 1 m potassium phosphate, pH 7.4, containing the buffer A supplements. Fractions containing enzyme activity were pooled and loaded onto a Sephacryl 300HR column equilibrated with buffer A. Elution was with buffer A. The final preparation with a specific activity of 15 units mg−1 protein is stable when stored at −20°C.
DAHP Synthase Enzyme Assay and Protein Determination
DAHP synthase was assayed (Suzich et al., 1985) by measuring the A549 of the periodate degradation product of DAHP complexed with thiobarbiturate. The unit of activity is defined as the amount of protein catalyzing the appearance of 1 μmol DAHP min−1. Protein was determined by the method of Bradford (1976), with bovine serum albumin as a standard.
To analyze the metal ion requirements of the enzyme, all buffers and enzyme solutions were treated with Chelex 100 resin. The enzyme was passed through a Sephadex G25 column equilibrated with buffer B (25 mm EPPS, pH 8.4, containing 1 mm PEP, 1.5 mm Trp, and 0.2 mm DTT). The reaction mixture contained 50 mm EPPS, pH 8.6, 5 mm PEP, 2 mm E4P, 10 mm DTT, and suitably diluted enzyme in a total volume of 0.1 mL. The reaction was initiated by adding 50 μL of a solution containing various concentrations of metal ions. Incubation was at 37°C for 10 min. The reaction was stopped by the addition of 0.3 mL of 10% (w/v) trichloroacetic acid.
To analyze the enzyme for reducing agent requirement, the enzyme was passed through a Sephadex G25 column equilibrated with buffer C (25 mm EPPS, pH 8.4, containing 1 mm PEP and 1.5 mm Trp). The enzyme was then preincubated with various concentrations of TRX in 10 mm DTT or various concentrations of DTT alone (total volume of 50 μL), and the assay was started by the addition of 100 μL of reaction mixture containing 50 mm EPPS, pH 8.6, 5 mm PEP, 2 mm E4P, and 0.1 mm MnCl2.
Chemical Modification of DHS1
The enzyme was treated with iodoacetamide using a procedure developed for arginyl-tRNA synthetase (Liu et al., 1999). DAHP synthase was incubated at room temperature for 30 min in 0.45 m potassium phosphate, pH 7.4, containing 2 mm MnCl2, 1.5 mm Trp, 0.2 mm DTT, and 20 mm iodoacetamide. After incubation, excess iodoacetamide was removed by buffer exchange into buffer C using a Sephadex G25 column at room temperature.
ACKNOWLEDGMENTS
We thank Dr. Peter Schürmann for generous gifts of recombinant TRX f and m, and Drs. Peter Goldsbrough, Mark Hermodson, and Ronald Somerville for critical reading of the manuscript. This paper is dedicated to Dr. Nikolaus Amrhein for his unselfish support of our experimental efforts.
Footnotes
This is journal paper no. 16,461 of the Purdue University Agricultural Experiment Station.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.002626.
LITERATURE CITED
- Aguilar F, Brunner B, Gardet-Salvi L, Stutz E, Schürmann P. Biosynthesis of active spinach-chloroplast thioredoxin f in transformed E. coli. Plant Mol Biol. 1992;20:301–306. doi: 10.1007/BF00014497. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Siedman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York, NY: Green Publishing Associates and Wiley-Interscience; 1987. [Google Scholar]
- Bischoff M, Rösler J, Raesecke HR, Görlach J, Amrhein N, Schmid J. Cloning of a cDNA encoding 3-dehydroquinate synthase from a higher plant, and analysis of the organ-specific and elicitor-induced expression of the corresponding gene. Plant Mol Biol. 1996;31:69–76. doi: 10.1007/BF00020607. [DOI] [PubMed] [Google Scholar]
- Bischoff M, Schaller A, Bieri F, Kessler F, Amrhein N, Schmid J. Molecular characterization of tomato 3-dehydroquinate dehydratase-shikimate:NADP oxidoreductase. Plant Physiol. 2001;125:1891–1900. doi: 10.1104/pp.125.4.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brandes HK, Hartman FC, Lu TY, Larimer FW. Efficient expression of the gene for spinach phosphoribulokinase in Pichia pastoris and utilization of the recombinant enzyme to explore the role of regulatory cysteinyl residues by site-directed mutagenesis. J Biol Chem. 1996;271:6490–6496. doi: 10.1074/jbc.271.11.6490. [DOI] [PubMed] [Google Scholar]
- Buchanan BB, Schürmann P, Decottiginies P, Lozano RM. Thioredoxin: a multifunctional regulatory protein with a bright future in technology and medicine. Arch Biochem Biophys. 1994;314:257–260. doi: 10.1006/abbi.1994.1439. [DOI] [PubMed] [Google Scholar]
- Carr PD, Verger D, Ashton AR, Ollis D. Chloroplast NADP-malate dehydrogenase: structural basis of light-dependent regulation of activity by thiol oxidation and reduction. Structure. 1999;7:461–475. doi: 10.1016/s0969-2126(99)80058-6. [DOI] [PubMed] [Google Scholar]
- Chiadmi M, Alvaza A, Miginiac-Maslow M, Jacquot J-P, Cherfils J. Redox signalling in the chloroplast: structure of oxidized pea fructose-1,6-bisphosphate phosphatase. EMBO J. 1999;18:6809–6815. doi: 10.1093/emboj/18.23.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark VM, Kirby AJ. An improved synthesis of phosphoenolpyruvate. Biochim Biophys Acta. 1963;78:732. [Google Scholar]
- Decottignies P, Schmitter JM, Miginiac-Maslow M, Le Marechal P, Jacquot JP, Gadal P. Primary structure of the light-dependent regulatory site of corn NADP-malate dehydrogenase. J Biol Chem. 1988;263:11780–11785. [PubMed] [Google Scholar]
- Dyer WE, Henstrand JM, Handa AK, Herrmann KM. Wounding induces the first enzyme of the shikimate pathway in Solanaceae. Proc Natl Acad Sci USA. 1989;86:7370–7373. doi: 10.1073/pnas.86.19.7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer WE, Weaver LM, Zhao J, Kuhn DN, Weller SC, Herrmann KM. A cDNA encoding 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase from Solanum tuberosum L. J Biol Chem. 1990;265:1608–1614. [PubMed] [Google Scholar]
- Gavel Y, von Heijne G. A conserved cleavage-site motif in chloroplast transit peptides. FEBS Lett. 1990;261:455–458. doi: 10.1016/0014-5793(90)80614-o. [DOI] [PubMed] [Google Scholar]
- Görlach J, Beck A, Henstrand JM, Handa AK, Herrmann KM, Schmid J, Amrhein N. Differential expression of tomato (Lycopersicon esculentum L.) genes encoding shikimate pathway isoenzymes: I. 3-Deoxy-d-arabino-heptulosonate 7-phosphate synthase. Plant Mol Biol. 1993;23:697–706. doi: 10.1007/BF00021525. [DOI] [PubMed] [Google Scholar]
- Henstrand JM, McCue KF, Brink K, Handa AK, Herrmann KM, Conn EE. Light and fungal elicitor induce 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase mRNA in suspension cultured cells of parsley (Petroselinum crispum L.) Plant Physiol. 1992;98:761–763. doi: 10.1104/pp.98.2.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann KM, Weaver LM. The shikimate pathway. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:473–503. doi: 10.1146/annurev.arplant.50.1.473. [DOI] [PubMed] [Google Scholar]
- Jacquot JP, Lopez-Jaramillo J, Chueca A, Cherfils J, Lemaire S, Chedozeau B, Miginiac-Maslow M, Decottignies P, Wolosiuk R, Lopez-Gorge J. High-level expression of recombinant pea chloroplast fructose-1,6-bisphosphatase and mutagenesis of its regulatory site. Eur J Biochem. 1995;229:675–681. doi: 10.1111/j.1432-1033.1995.tb20513.x. [DOI] [PubMed] [Google Scholar]
- Johansson K, Ramaswamy S, Saarinen M, Lemaire-Chamley M, Issakidis-Bourget E, Miginiac-Maslow M, Eklund H. Structural basis for light activation of a chloroplast enzyme: the structure of sorghum NADP-malate dehydrogenase in its oxidized form. Biochemistry. 1999;38:4319–4326. doi: 10.1021/bi982876c. [DOI] [PubMed] [Google Scholar]
- Keith B, Dong X, Ausubel FM, Fink GR. Differential induction of 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase genes in Arabidopsis thaliana by wounding and pathogenic attack. Proc Natl Acad Sci USA. 1991;88:8821–8825. doi: 10.1073/pnas.88.19.8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li AD, Anderson LE. Expression and characterization of pea chloroplastic glyceraldehyde-3-phosphate dehydrogenase composed of only the B-subunit. Plant Physiol. 1997;115:1201–1209. doi: 10.1104/pp.115.3.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Huang Y, Wu J, Wang E, Wang Y. Effect of cysteine residues on the activity of arginyl-tRNA synthethase from Escherichia coli. Biochemistry. 1999;38:11006–11011. doi: 10.1021/bi990392q. [DOI] [PubMed] [Google Scholar]
- Miginiac-Maslow M, Issakidis E, Lemaire M, Ruelland E, Jacquot JP, Decottignies P. Light-dependent activation of NADP-malate dehydrogenase: a complex process. Aust J Plant Physiol. 1997;24:529–542. [Google Scholar]
- Ocheretina O, Haferkamp I, Tellioglu H, Scheibe R. Light-modulated NADP-malate dehydrogenases from mossfern and green algae: insights into evolution of enzyme's regulation. Gene. 2000;258:147–154. doi: 10.1016/s0378-1119(00)00409-1. [DOI] [PubMed] [Google Scholar]
- Ogino T, Garner C, Markley JL, Herrmann KM. Biosynthesis of aromatic compounds: 13C NMR spectroscopy of whole Escherichia coli cells. Proc Natl Acad Sci USA. 1982;70:5828–5832. doi: 10.1073/pnas.79.19.5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto JEBP, Suzich JA, Herrmann KM. 3-Deoxy-d-arabino-heptulosonate 7-phosphate synthase from potato tuber (Solanum tuberosum L.) Plant Physiol. 1986;82:1040–1044. doi: 10.1104/pp.82.4.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Suarez RJ, Mora-Garcia S, Wolosiuk RA. Characterization of cysteine residues involved in the reductive activation and the structural stability of rapeseed (Brassica napus) chloroplast fructose-1,6-bisphosphate phosphatase. Biochem Biophys Res Commun. 1997;232:388–393. doi: 10.1006/bbrc.1997.6242. [DOI] [PubMed] [Google Scholar]
- Ruelland E, Miginiac-Maslow M. Regulation of chloroplast enzyme activities by thioredoxins: activation or relief from inhibition? Trends Plant Sci. 1999;4:136–141. doi: 10.1016/s1360-1385(99)01391-6. [DOI] [PubMed] [Google Scholar]
- Sato S, Kotani H, Nakamura Y, Kaneko T, Asamizu E, Fukami M, Miyajima N, Tabata S. Structural analysis of Arabidopsis thaliana chromosome 5: I. Sequence features of the 1.6 Mb regions covered by twenty physically assigned P1 clones. DNA Res. 1997;4:215–230. doi: 10.1093/dnares/4.3.215. [DOI] [PubMed] [Google Scholar]
- Schürmann P. Ferredoxin:thioredoxin system. Methods Enzymol. 1995;252:274–283. doi: 10.1016/0076-6879(95)52030-9. [DOI] [PubMed] [Google Scholar]
- Schürmann P, Jacquot J-P. Plant thioredoxin systems revisited. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:371–400. doi: 10.1146/annurev.arplant.51.1.371. [DOI] [PubMed] [Google Scholar]
- Schwarz O, Schurmann P, Strotmann H. Kinetics and thioredoxin specificity of thiol modulation of the chloroplast H+-ATPase. J Biol Chem. 1997;272:16924–16927. doi: 10.1074/jbc.272.27.16924. [DOI] [PubMed] [Google Scholar]
- Sieben AS, Perlin AS, Simpson FJ. An improved preparative method for d-erythrose 4-phosphate. Can J Biochem. 1966;44:663–669. doi: 10.1139/o66-083. [DOI] [PubMed] [Google Scholar]
- Stephens CM, Bauerle R. Essential cysteines in 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase from Escherichia coli. J Biol Chem. 1992;267:5762–5767. [PubMed] [Google Scholar]
- Suzich JA, Dean JFD, Herrmann KM. 3-Deoxy-d-arabino-heptulosonate 7-phosphate synthase from carrot root (Daucus carota) is a hysteretic enzyme. Plant Physiol. 1985;79:765–770. doi: 10.1104/pp.79.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Herrmann KM, Weller SC, Goldsbrough PB. Cloning and nucleotide sequence of a complementary DNA encoding 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase from tobacco. Plant Physiol. 1991;97:847–848. doi: 10.1104/pp.97.2.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham NM, Lloyd JC, Raines CA. Molecular cloning of the Arabidopsis thaliana sedoheptulose-1,7-bisphosphatase gene and expression studies in wheat and Arabidopsis thaliana. Plant Mol Biol. 1994;26:3791–3795. doi: 10.1007/BF00040699. [DOI] [PubMed] [Google Scholar]
- Wolosiuk RA, Crawford NA, Yee BC, Buchanan BB. Isolation of three thioredoxins from spinach leaves. J Biol Chem. 1979;254:1627–1632. [PubMed] [Google Scholar]
- Zhang N, Portis AR., Jr Mechanism of light regulation of Rubisco: a specific role for the larger Rubisco activase isoform involving reductive activation by thioredoxin f. Proc Natl Acad Sci USA. 1999;96:9438–9443. doi: 10.1073/pnas.96.16.9438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J. Regulation of the first enzyme of the shikimate pathway in plants. PhD Thesis. West Lafayette, IN: Purdue University; 1992. [Google Scholar]