Abstract
Rice (Oryza sativa) anther development is easily damaged by moderately low temperatures above 12°C. Subtractive screening of cDNA that accumulated in 12°C-treated anthers identified a cDNA clone, OsMEK1, encoding a protein with features characteristic of a mitogen-activated protein (MAP) kinase kinase. The putative OsMEK1 protein shows 92% identity to the maize (Zea mays) MEK homolog, ZmMEK1. OsMEK1 transcript levels were induced in rice anthers by 12°C treatment for 48 h. Similar OsMEK1 induction was observed in shoots and roots of seedlings that were treated at 12°C for up to 24 h. It is interesting that no induction of OsMEK1 transcripts was observed in 4°C-treated seedlings. In contrast, rice lip19, encoding a bZIP protein possibly involved in low temperature signal transduction, was not induced by 12°C treatment but was induced by 4°C treatment. Among the three MAP kinase homologs cloned, only OsMAP1 displayed similar 12°C-specific induction pattern as OsMEK1. A yeast two-hybrid system revealed that OsMEK1 interacts with OsMAP1, but not with OsMAP2 and OsMAP3, suggesting that OsMEK1 and OsMAP1 probably function in the same signaling pathway. An in-gel assay of protein kinase activity revealed that a protein kinase (approximately 43 kD), which preferentially uses myelin basic protein as a substrate, was activated by 12°C treatment but not by 4°C treatment. Taken together, these results lead us to conclude that at least two signaling pathways for low temperature stress exist in rice, and that a MAP kinase pathway with OsMEK1 and OsMAP1 components is possibly involved in the signaling for the higher range low-temperature stress.
Rice (Oryza sativa) is widely cultivated in a large number of different natural environments (Nishiyama, 1984). Compared with other cereal crops such as wheat (Triticum aestivum) and barley (Hordeum vulgare), rice is much more sensitive to low temperature as a result of its tropical origin. Male sterility is the most severe consequence among the many chilling-induced agronomic damages in rice production. The developmental stages from pollen formation to fertilization are the most vulnerable to low temperature throughout the life cycle of rice plants (Nishiyama, 1984). It has been reported that the young microspore stage in pollen development was the most sensitive to low temperature (Satake and Hayase, 1970). Exposure of rice plants at the tetrad stage to a moderately low temperature (12°C) for 4 d resulted in male sterility in 80% of spikelets (Satake and Hayase, 1970; Nishiyama, 1984). Microscopic observation of developing rice anthers suggested that one possible reason for the male sterility after low-temperature treatment was the failure of anther development. The observed abnormalities included the cessation of anther development, the arrest of pollen development, anthers remaining within the flowers after anthesis, and partial or no dehiscence (Satake, 1976). Cytological observation revealed a dilatation of tapetal layers in chilling-treated rice anthers (Nishiyama, 1976, 1984). The dilatation of tapetal layer was accompanied by a vigorous augmentation of cytoplasmic organelles such as mitochondria, proplastids, Golgi bodies, and endoplasmic reticula (Nishiyama, 1976, 1984). Chilling temperature treatment also affects the physiological status of anthers. Nonreducing sugar content was found to increase rapidly, whereas the acid phosphatase activity decreased in the moderately temperature-treated rice anthers (Nishiyama, 1984). Possible involvement of phytohormones such as gibberellin and auxin in the chilling-induced male sterility has been reported (Nishiyama, 1975; Yoshioka and Suge, 1996). However, it is still largely unknown how chilling temperature induces molecular events that result in male sterility in rice plants.
Signal transduction networks enable cells to perceive the variations in the extracellular environments and to mount an appropriate response. The mitogen-activated protein (MAP) kinase cascade pathway is among the most well-characterized signal transduction systems in animals, yeast, and plants (Jonak et al., 1994; Hirt, 1997; Mizoguchi et al., 1997; Robinson and Cobb, 1997; Schaeffer and Weber, 1999). The core of the MAP kinase cascade is constituted by MAP kinase (MAPK), MAPK kinase (MAPKK, also known as MEK) and MAPKK kinase (MAPKKK, also known as MEKK). Recently, MAPKKK kinases were identified in animal, yeast, and plant systems. The MAPK cascade was primarily found to be involved in regulating cell division, development, and differentiation, and in coordinating responses to stress stimuli in animals and yeast (Herskowitz, 1995; Robinson and Cobb, 1997; Schaeffer and Weber, 1999). In recent years, a variety of genes encoding MAPKs, MAPKKs, and MAPKKKs have been cloned from different plant species (Hirt, 1997; Mizoguchi et al., 1997; Ligterink, 2000; Calderini et al., 2001). An increasing body of evidence has shown that MAPKs play important roles in signal transduction in response to drought, reactive oxygen species, pathogen defense, wounding, and/or low temperature in plants (Seo et al., 1995; Jonak et al., 1996; Mizoguchi et al., 1996; Shinozaki and Yamaguchi-Shinozaki, 1996; He et al., 1999; Romeis, 2001; Ren et al., 2002). In alfalfa (Medicago sativa), an MAPK homolog, MMK4, has been linked with touch, drought, and salinity stresses (Bögre et al., 1996; Jonak et al., 1996). It has also been demonstrated that SIPK (an MAPK homolog) is activated by salicylic acid within 5 min in tobacco (Nicotiana tabacum) suspension cultures (Zhang and Klessig, 1997). In a similar manner, transcripts of WIPK, an MAPK homolog in tobacco, accumulate 1 min after mechanical wounding (Seo et al., 1995). In addition, WIPK is involved in jasmonate-based wounding signal transduction pathway (Seo et al., 1999).
In this study, we cloned two novel components of an MAPK pathway, OsMEK1 and OsMAP1, that are induced by 12°C treatment to elucidate the molecular responses of rice to a range of moderately low temperatures that eventually cause abnormal development of pollen. We characterized temperature dependence of OsMEK1 and OsMAP1 expression and an myelin basic protein (MBP) kinase activity. We conclude that there is a novel signal transduction pathway, distinct from the existing Lip19-involved pathway, for low-temperature responses in rice.
RESULTS
cDNA Subtraction, Cloning, and Sequence Analysis of Rice OsMEK1
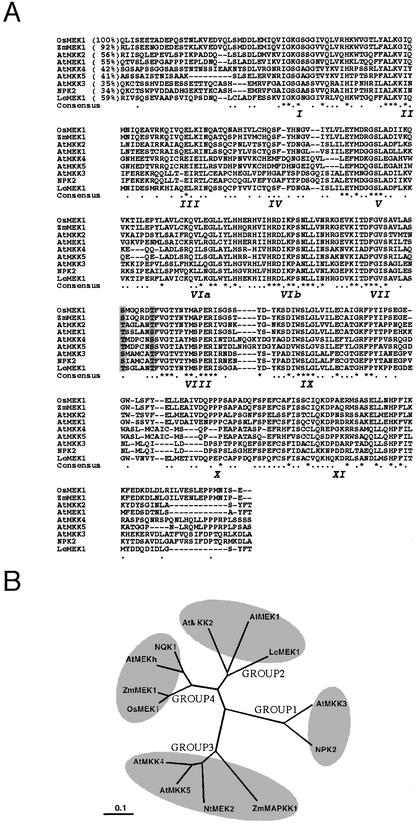
A PCR-based cDNA subtraction was used to isolate cDNA clones that are induced during 12°C treatment in rice anther. The cDNA synthesized from 12°C-treated rice anther poly(A)+ RNA and the cDNA from nontreated anther poly(A)+ RNA was used as the “tester” and “driver” of the subtraction, respectively. A cDNA fragment (550 bp) showing high homology to ZmMEK1 (Hardin and Wolniak, 1998) was isolated after subtraction. The 550-bp fragment was used as a probe to screen a cDNA library for isolation of a full-length cDNA, designated OsMEK1. A 1.4-kb cDNA was identified and found to contain a 1,064-bp open reading frame. The putative protein encoded by OsMEK1 cDNA has 355 amino acids with features characteristic of MAPKK. OsMEK1 is predicted to have an estimated molecular mass of 40 kD and a pI of 5.47. The putative OsMEK1 protein contains the 11 conserved catalytic subdomains that are typical of Ser/Thr protein kinases (Fig. 1A). A plant MEK-specific S/TXXXXXS/T motif was identified between subdomains VII and VIII in OsMEK1 (Fig. 1A). This motif differs from that of animal and yeast MEKs (SXXXS/T; Ichimura et al., 1998b). The putative OsMEK1 protein is closely related to the maize ZmMEK1 (overall 92% identity and 97% similarity at the amino acid sequence level; Hardin and Wolniak, 1998). The deduced amino acid sequence of OsMEK1 also exhibits extensive homology to other plant MEKs (Fig. 1A). Phylogenetic analysis of the amino acid sequences of the reported plant MEK homologs revealed that OsMEK1 and ZmMEK1 are grouped together with two dicot MEKs, AtMEKh (accession no. AB013392) and tobacco NQK1 (accession no. AB05514; Fig. 1B). They branched out of the subgroup 2 of plant MEKs (Mizoguchi et al., 1997) to form a novel subgroup, subgroup 4 (Fig. 1B).
Figure 1.
Comparison of the deduced amino acid sequences of OsMEK1 and closely related plant MEKs. A, Alignment of amino acid sequence in the catalytic domain of OsMEK1 (accession no. AF216314) with that of other MEK homologs from plants: maize (Zea mays) ZmMEK1 (Hardin and Wolniak, 1998); Arabidopsis AtMAP2K (Jouannic et al., 1996), AtMKK2 (Ichimura et al., 1998b), AtMEK1 (Morris et al., 1997), AtMKK3 (Ichimura et al., 1998b), AtMKK4 (Ichimura et al., 1998b), and AtMKK5 (Ichimura et al., 1998a); tobacco NPK2 (Shibata et al., 1995), and tomato (Lycopersicon esculentum) LeMEK1 (Hackett et al., 1998). Sequences were aligned, and gaps (dashes) have been introduced to maximize the alignment. Numbers within parentheses indicate the percentage of identity to OsMEK1. Roman numerals in italics under the sequences indicate the 11 major conserved subdomains found in Ser/Thr protein kinases. In the consensus sequence, dots indicate conservative substitution of amino acid residues, and asterisks indicate the invariant residues in all MEKs sequences. The Ser and/or Thr residues in the conserved consensus motif S/TXXXXXS/T between subdomains VII and VIII of MEKs are indicated with gray background. B, A phylogenetic tree of plant MEKs was created with Clustal X and TreeView programs with 1,000 times boot strapping (Page, 1996; Thompson et al., 1997). The distance scale represents evolutionary distance expressed as the number of substitutions per amino acid.
Expression of OsMEK1 Is Responsive to 12°C Treatment But Not to 4°C Treatment
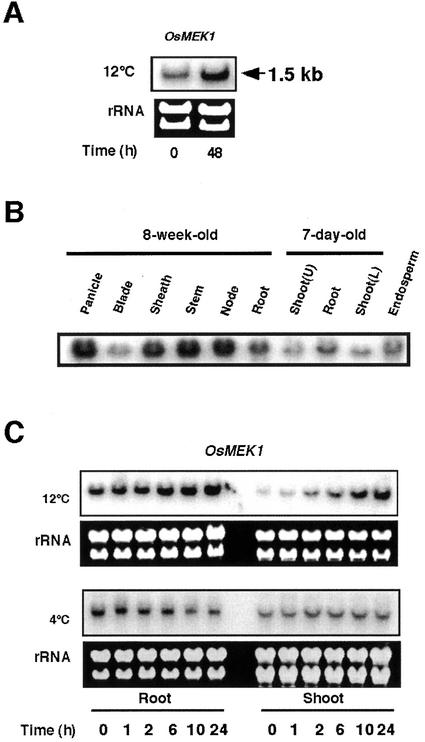
The full-length OsMEK1 cDNA hybridized to a single transcript of approximate 1.5-kb by northern-blot analysis of rice anther total RNA (Fig. 2A). The OsMEK1 mRNA was detected in anthers of nonstressed plants, however, 12°C treatment was found to dramatically increase the level of the OsMEK1 mRNA. In a similar manner, the OsMEK1 mRNA was also detected in nonstressed flowers, sheaths, stems, and nodes from 8-week-old rice plants. Lower levels were detected in 8-week-old leaf blades and mature seed endosperm (Fig. 2B). In 7-d-old seedlings, the level of the OsMEK1 mRNA was higher in roots than that in shoots (Fig. 2B).
Figure 2.
Tissue-specific and cold-regulated expression of OsMEK1. A, Anthers from chilling-stressed (12°C for 48 h) or nonstressed rice plants at a young microspore stage was collected and total RNA was isolated. The RNA blot was hybridized with the 32P-labeled entire OsMEK1 cDNA fragment. The ethidium bromide-stained rRNA reflects the uniform loading in each lane. B, Tissue-specific expression of OsMEK1 in rice plants under a nonstressed condition. Tissues from 8-week-old plants before heading, 7-d-old young seedlings, and mature seeds were collected and total RNA was used for hybridization. Shoot tissue of 7-d-old seedlings was separated into upper (U) and lower (L) parts. C, Time course of OsMEK1 expression in response to low-temperature treatments in roots and shoots of 7-d-old seedlings. Seedlings were subjected to 12°C or 4°C treatments for the indicated periods (hours).
To examine the expression of OsMEK1 in response to low temperature in detail, we used roots and shoots of 7-d-old rice seedlings. Northern-blot analyses revealed that the accumulation of the OsMEK1 mRNA was steadily induced in roots and shoots during a 24-h treatment at 12°C (Fig. 2C). The induction of OsMEK1 in roots and shoots was initiated within 2 h and steadily increased thereafter until 24 h subsequent to the 12°C treatment. The induction levels in shoots were higher than those in roots. It is interesting that the level of OsMEK1 mRNA was not induced in shoots and roots of 7-d-old seedlings that were subjected to a lower temperature (4°C; Fig. 2C). Instead, the levels of OsMEK1 mRNA decreased in roots during a 24-h period at 4°C. To clarify whether there was a rapid response to 4°C, OsMEK1 mRNA levels were analyzed within a short time course study (5, 10, 20, 40, and 60 min). Results indicated that OsMEK1 did not respond to 4°C treatment in a period of 1 h (data not shown). Thus, it could be concluded that OsMEK1 is induced by 12°C stress but not by 4°C stress. The data contrast previously identified cold-induced genes of rice, which have been shown to be responsive to 4°C to 6°C temperature treatments (Aguan et al., 1991; Binh and Oono, 1992; Saijo et al., 2000).
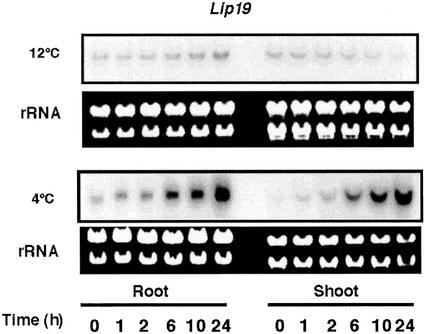
Expression of lip19 Is Responsive to 4°C Treatment But Not to 12°C Treatment in Rice Seedlings
The rice lip19 gene encoding a bZIP-type DNA-binding protein is inducible by low temperatures (5°C; Aguan et al., 1993). Thus, the Lip19 protein was suggested to be a transcription factor involved in a low-temperature signal transduction pathway. In this study, we compared the expression of lip19 at 12°C and 4°C. Northern blots showed that the accumulation of the lip19 transcript in roots and shoots of 7-d-old rice seedlings was substantially induced by 4°C treatment during a 24-h period (Fig. 3). However, the lip19 transcript levels were only slightly induced (shoots) or decreased (roots) in the 12°C-treated seedlings during the same time course (Fig. 3). These data suggest that the expression of lip19 is regulated by a signal transduction pathway that is activated within the 4°C temperature range. The results of the differential expression of OsMEK1 and lip19 in response to different ranges of low-temperature treatments implied that there are distinct signaling systems that perceive and transduce different temperature signals in rice.
Figure 3.
Northern-blot analyses of lip19 expression in response to low-temperature treatments. Seven-day-old seedlings were subjected to 12°C or 4°C treatments. Total RNA from roots and shoots was blotted and hybridized with the 32P-labeled cDNA fragment of lip19. The ethidium bromide-stained rRNA is shown as a loading control.
OsMAP1 Isolation and Expression in Response to Low-Temperature Treatment
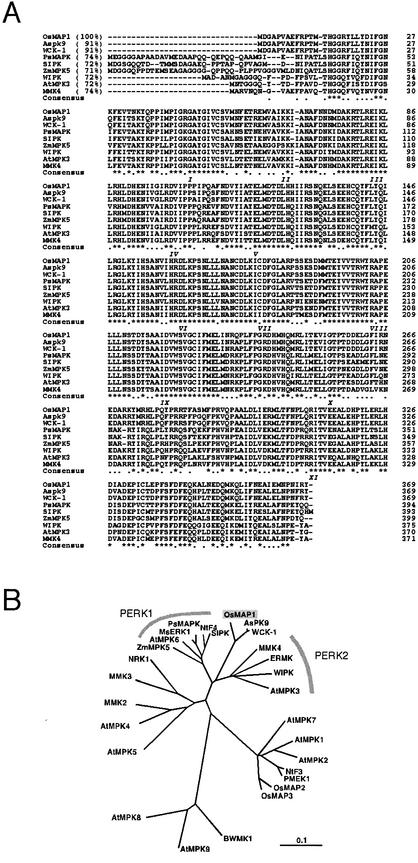
After cloning and characterizing the OsMEK1, we hypothesized that there might be an MAPK that functions in the same moderate temperature signal transduction pathway. Therefore, we attempted to isolate the MAPK gene by using expressed sequence tag (EST) sequences. Two cDNA fragments of 446 bp (M446) and 406 bp (M406) were amplified by PCR using two primer sets designed from rice EST clones (accession nos. C22363 and AU033195, respectively). Subcloned fragments were used as probes to screen a rice cDNA library. Three clones representing three different MAPKs were isolated. One of the clones screened with the probe M446, designated OsMAP1, was found to be induced by 12°C treatment. The other two clones (OsMAP2 and OsMAP3) screened with the probe M406 were not responsive to low temperatures (data not shown). The open reading frame of OsMAP1 encodes a putative protein (OsMAP1) that has 369 amino acid residues with an estimated molecular mass of 43.0 kD and a pI of 5.41 (Fig. 4A). OsMAP1 has the MAPK signature phosphorylation motif, TEY, between subdomains VII and VIII and thus belongs to the first subfamily of MAPKs (Seger and Krebs, 1995). OsMAP1 shares 91% identity with two other MAPK homologs, Aspk9 from oat (Avena sativa; Huttly and Phillips, 1995) and WCK-1 from wheat (Takezawa, 1999) at the amino acid sequence levels (Fig. 4A). A phylogenetic tree revealed that these monocot MAPKs constitute a novel subgroup of a plant MAPK superfamily (Fig. 4B). OsMAP1 also shows more than 70% identity to other stress-related plant MAPKs that belong to the PERK1 and PERK2 subgroups (Ligterink, 2000; Fig. 4B).
Figure 4.
Structural comparison of the deduced amino acid sequences of eight stress-responsive plant MAPKs and OsMAP1. A, Alignment of the deduced amino acid sequence of OsMAP1 (accession no. AF216315) with that of other MAPK homologs from plants: Aspk9 from oat (Huttly and Phillips, 1995), WCK-1 from wheat (Takezawa, 1999), PsMAPK from pea (Pisum sativum; Stafstrom et al., 1993), SIPK (Zhang and Klessig, 1997) and WIPK (Seo et al., 1995) from tobacco, ZmMPK from maize (Berberich et al., 1999), AtMPK3 from Arabidopsis (Mizoguchi et al., 1993), and MMK4 from alfalfa (Jonak et al., 1996). Sequences are aligned, and gaps (dashes) have been introduced to maximize the alignment. Numbers within parentheses indicate the percentage of identity to OsMAP1. Roman numerals in italics under the sequences indicate the 11 major conserved subdomains found in Ser/Thr protein kinases. In the consensus sequence, dots indicate conservative substitution of amino acid residues, and asterisks indicate the invariant residues in all nine MAPKs. The conserved TEY phosphorylation motif is underlined. B, A phylogenic tree of representative plant MAPKs. The tree was drawn with Clustal X (Page, 1996) and TreeView (Thompson et al., 1997) programs with 1,000 times boot strapping. The distance scale represents evolutionary distance expressed in the number of substitutions per amino acid.
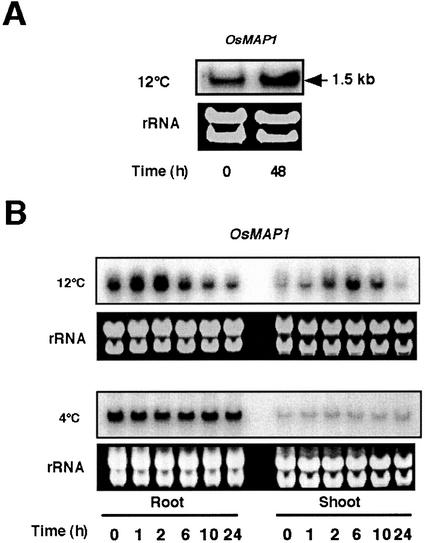
Expression of OsMAP1 Is Responsive to 12°C Treatment But Not to 4°C Treatment
Change in the levels of OsMAP1 mRNA was analyzed by northern-blot analysis. In anther tissue at the booting stage, a 12°C treatment for 48 h increased the OsMAP1 mRNA. However, OsMAP1 induction was less than that of OsMEK1 in response to 12°C (Fig. 5A). Seven-day-old seedlings were used to determine accumulation patterns of OsMAP1 in response to low temperatures (Fig. 5B). The data showed that the OsMAP1 mRNA rapidly increased in 2 h in root tissue, whereas a steady decrease in the transcript level was observed thereafter during a 24-h time period. A higher level of induction was observed in 12°C-treated shoots (6 h), although the induction was transient (Fig. 5B). In contrast, the OsMAP1 mRNA levels did not fluctuate in 4°C-treated roots and shoots during the same 24-h time period (Fig. 5B). The expression pattern of OsMAP1 contrasts those of cold-inducible MAPKs in PERK2 subgroup such as AtMPK3 (Mizoguchi et al., 1996) and MMK4 (Jonak et al., 1996), which are responsive to 4°C treatments. The observation that expression of OsMAP1 is induced by 12°C treatment and not by 4°C treatment is in good accordance with the OsMEK1 expression. These data suggest a possible involvement of OsMEK1 and OsMAP1 in moderately low-temperature signaling.
Figure 5.
OsMAP1 expression in response to low-temperature treatments. A, Anthers from chilling-stressed (12°C for 48 h) or nonstressed rice plants at a young microspore stage were collected and total RNA was isolated. The RNA blot was hybridized with the 32P-labeled entire cDNA fragment of OsMAP1. The ethidium bromide-stained rRNA is shown as a loading control. B, Seven-day-old seedlings were subjected to 12°C or 4°C treatments. Total RNA from roots and shoots was blotted and hybridized with the 32P-labeled entire cDNA fragment of OsMAP1. The ethidium bromide-stained rRNA is shown as a loading control.
Effects of Abscisic Acid (ABA), NaCl, and Drought Stresses on the Expression of OsMEK1 and OsMAP1
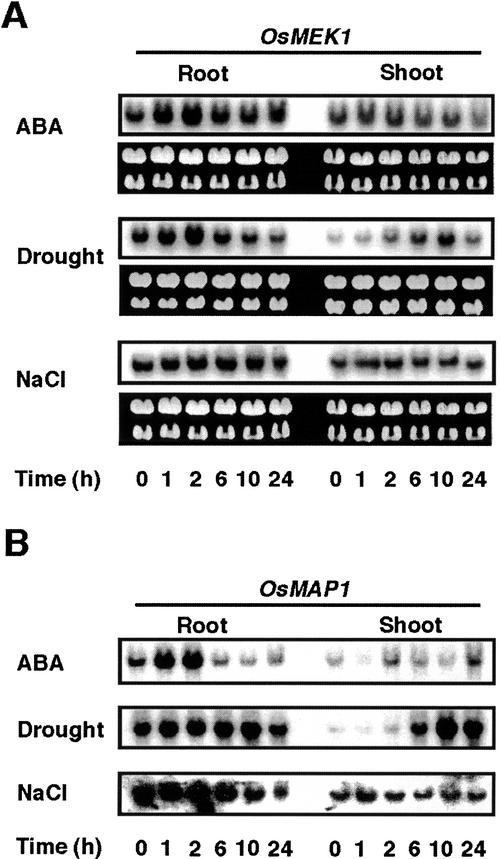
It has been widely reported that cold-inducible genes also respond to water deficit (Jonak et al., 1996; Shinozaki and Yamaguchi-Shinozaki, 1996). Evidence has also shown that an MAPK cascade is involved in several signal transduction pathways in plants (Hirt, 1997; Mizoguchi et al., 1997; Ligterink, 2000). Thus, we examined the effects of ABA, NaCl, and water deficit on the expression of OsMEK1 and OsMAP1. Figure 6A shows that the accumulation of OsMEK1 mRNA was observed in 7-d-old seedlings that are drought stressed. In roots, drought responsive induction of OsMEK1 was detected within 1 h of treatment and reached a peak at 2 h of treatment; thereafter, the levels of the transcript gradually declined (Fig. 6A). However, the response of OsMEK1 was found to be much slower in shoots (the induction peak appeared at 10 h of drought stress; Fig. 6A). The differential response is probably because roots desiccated prior to shoots in our experimental condition. OsMEK1 responded to exogenous ABA in a manner similar to drought stress in roots, whereas no clear induction was observed in ABA-treated shoots. OsMEK1 did not show clear responsiveness to the 0.2 m NaCl treatment during a period of 24 h (Fig. 6A). Accumulation of OsMAP1 mRNA was examined under the same conditions. The patterns of OsMAP1 induction were very similar to those of OsMEK1, except for the response of shoots to ABA. These data support the supposition that both genes are controlled by a similar regulatory mechanism.
Figure 6.
Accumulation of the OsMEK1 and OsMAP1 mRNAs in response to ABA, drought, and NaCl treatments. Seven-day-old seedlings were subjected to 50 μm ABA, drought, and 0.2 m NaCl treatments for the indicated periods (hours), and total RNA was isolated from roots and shoots. RNA blots were hybridized with the 32P-labeled entire cDNA fragment of OsMEK1 (A) or OsMAP1 (B). The ethidium bromide-stained rRNA is shown as a loading control.
OsMEK1 Interacts with OsMAP1 But Not with OsMAP2 and OsMAP3 in Yeast Cells
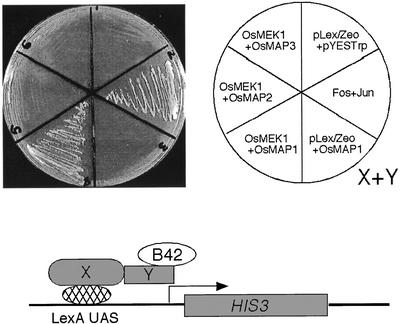
Similar responsiveness of OsMEK1 and OsMAP1 to environmental stimuli may suggest that the two proteins interact in vivo. To test this supposition, a yeast two-hybrid assay was used. Results in Figure 7 showed that OsMEK1 fused to the LexA DNA-binding domain interacted with OsMAP1 that was fused to the B42 activation domain. The interaction of OsMEK1 and OsMAP1 resulted in the expression of HIS3, which enabled the yeast cells to grow on the selective medium lacking Trp, Leu, and His. The yeast cells cotransformed with the LexA-fused OsMEK1 and the B42-fused OsMAP2 or OsMAP3 constructs did not grow on the selective medium, suggesting that there was no interaction among these proteins. A filter assay of β-galactosidase activity also confirmed the OsMAP1-specific interaction with OsMEK1 (data not shown). These results strongly suggest that OsMEK1 is a partner of OsMAP1 in rice.
Figure 7.
Yeast two-hybrid system demonstrating OsMEK1-OsMAP1 interaction. The entire OsMEK1 open reading frame was fused in-frame to the C terminus of the DNA-binding domain of LexA in pHybLex/Zeo (Invitrogen) and was used as a bait. The open reading frames of OaMAP1, OsMAP2, and OsMAP3 were individually fused in-frame to the C terminus of the transcriptional activation domain of B42 in pYES-Trp2 (Invitrogen) and were used as preys. The resulting bait- and prey-containing plasmids were cotransformed into yeast strain L40 according to the manufacturer's method. c-Fos and c-Jun are two proteins that are know to interact and are used as a positive control. The growth of streaked yeast cells transformed with different plasmid combination was prototrophically assayed on a selective medium lacking Trp, Leu, and His.
12°C Treatment Activates an Approximately 43-kD MAPK-Like Kinase in Rice Seedlings
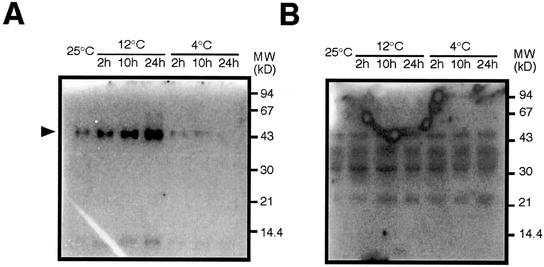
An in-gel kinase activity assay was used to search the kinase(s) involved in low temperature stress. Two artificial kinase substrates, MBP and casein, were independently embedded in the separating gel. Results showed that the 12°C treatment of rice seedlings activated an approximately 43-kD protein kinase that used MBP as a substrate. The kinase activity steadily increased during the 24 h of the cold stress. In contrast, the approximately 43-kD protein kinase activity steadily decreased in 4°C-treated seedlings during the same time period (Fig. 8A). These data support the idea that 4°C and 12°C treatment are perceived by different signaling pathways in rice. The 43-kD kinase activity was not detected when casein was used as a substrate, indicating that the 43-kD kinase preferentially uses MBP as a substrate (Fig. 8B). It is to be noted that although further analysis is needed, molecular mass and substrate preference of the 43-kD kinase are in good accordance with those of OsMAP1. The temperature-dependent patterns of the approximately 43-kD MBP kinase activation and OsMAP1 expression imply OsMAP1 is a possible candidate for the approximately 43-kD MBP kinase.
Figure 8.
12°C treatment of rice seedlings activates an approximately 43-kD protein kinase that preferentially uses MBP as a substrate. Seven-day-old rice seedlings were treated at 12°C or 4°C for the indicated times (hours). Untreated (25°C) seedlings were used as a control. Proteins extracted from treated or untreated shoots were separated by SDS-PAGE embedded with MBP (A) or casein (B) as a substrate. The protein kinase that is activated by 12°C treatment is indicated by an arrow in A. The size of molecular markers are shown in kilodaltons.
DISCUSSION
By subtractive cDNA screening, we have cloned a cDNA, OsMEK1, encoding a putative MEK that interacts in vivo in yeast with an MAPK from rice. The putative OsMEK1 protein shares high homology (92% identity) with the maize ZmMEK1 and shows extensive homology with other MEK homologs identified in plants (Fig. 1). The deduced OsMAP1 protein shows 91% amino acid identity to Aspk9 (Huttly and Phillips, 1995) and WCK-1 (Takezawa, 1999), and more than 70% amino acid identity to other plant stress-responsive MAPK homologs (Fig. 5). OsMEK1 and OsMAP1 contain the 11 catalytic subdomains of Ser/Thr protein kinases. OsMEK1 also contains the conserved plant MEK-specific consensus motif (S/TXXXXXS/T; Fig. 1). Northern-blot analysis showed that OsMEK1 is ubiquitously expressed; however, the levels of mRNA accumulation were increased in anthers and seedlings during moderately low temperature (12°C) exposure (Fig. 2). The patterns of OsMEK1 and OsMAP1 expression in response to environmental stresses, including 12°C, 4°C, drought stress, salt stress, and ABA, were found to be similar, suggesting both genes are possibly operating in the same pathway under similar regulation (Figs. 2, 5, and 6).
Similar to MAPK cascade pathways in animals and yeast, plant MAPK cascades appear to play important roles in regulating cell division and coordinating responses to environmental stresses (Hirt, 1997; Asai et al., 2002). Studies on the kinase activity and transcript levels of components of MAPK pathways led to the connection between the involvement of plant MAPK cascades and the auxin-induced cell cycle reentry (Mizoguchi et al., 1994), leaf wounding (Stratmann and Ryan, 1997), pollen development (Wilson et al., 1997), and innate immunity (Asai et al., 2002). Evidence has shown that MAPK pathways are likely involved in signal transduction under drought and low temperature stresses (Jonak et al., 1996; Mizoguchi et al., 1996). In alfalfa, activation of the MMK4 (MAPK) protein was observed when cold stress was applied (4°C), which was accompanied by increased levels of the transcript (Jonak et al., 1996). Arabidopsis AtMPK3 expression is strongly induced by cold stress (Mizoguchi et al., 1996). Because the sequence of OsMAP1 is most similar to the subgroup 2 MAPKs of dicots, including MMK4 and ATMPK3, OsMAP1 may have a similar function. In some MAPKs in this group, environmental stimuli can activate MAPK at transcriptional and posttranslational levels (Seo et al., 1995; Jonak et al., 1996; Bögre et al., 1997). Coordinate induction of OsMEK1 and OsMAP1 by 12°C treatment suggests that OsMEK1 and OsMAP1 proteins may be involved in a moderate temperature-specific MAPK signaling pathway.
Stress-induced transcription factors have been studied extensively due to their possible involvement in stress signaling (Shinozaki and Yamaguchi-Shinozaki, 1996; Eckardt, 2001). Kusano and colleagues (Aguan et al., 1993) have isolated a cold-inducible gene (lip19) encoding a homolog of bZIP transcription factor from rice. Although direct evidence has not been shown, Lip19 is considered to be involved in the regulation of cold-induced gene expression because a maize ortholog (mLip15) was found to bind to the promoter region of a cold-inducible gene of maize (Kusano et al., 1995). Northern-blot analysis showed that lip19 expression is induced by 4°C but not by 12°C treatment (Fig. 3). These data suggest that the Lip19 protein is involved in a specific signal transduction pathway induced within the 4°C temperature range.
Molecular responses to low temperatures have been extensively studied in a variety of plants, and many cold-regulated genes have been identified (Hughes and Dunn, 1996; Thomashow et al., 1997). The general strategy for isolating cold-regulated genes has used a low temperature range from 2°C to 6°C. A number of cold-regulated genes have been isolated and found to be inducible within this range. The same temperature range has been used to screen cold-induced genes of rice (Aguan et al., 1991; Binh and Oono, 1992). As a result, it has been confirmed that rice has a signal transduction pathway for this low temperature range. On the other hand, male sterility and its related physiological and morphological changes occur at 12°C or even higher. Therefore, it is possible that rice contains an additional signaling pathway to perceive this moderate temperature range as well. A previous report suggested there are distinctive pathways for the two temperature ranges. An 18-kD polypeptide has been shown to accumulate when rice seedlings were treated at 5°C, whereas the protein was not induced at 15°C (Koga et al., 1991). In this study, we have shown that the transcripts of OsMEK1 and OsMAP1 significantly accumulate under 12°C treatment, but not under 4°C treatment, in roots and shoots of rice seedlings (Figs. 2C and 5B). In a converse manner, the expression of lip19 was strongly induced by 4°C but not by 12°C treatment (Fig. 4). Our expression analysis with OsMEK1, OsMAP1, and lip19 clearly demonstrated there is discrimination in signal transduction between the 12°C and 4°C temperature ranges in terms of gene expression. Furthermore, the activation of a approximately 43-kD protein kinase that preferentially uses MBP as a substrate was observed in 12°C-treated shoots but not in 4°C-treated shoots (Fig. 8). Although it needs to be determined if the approximately 43-kD protein kinase is identical to OsMAP1, it should be noted that discrimination of the two low temperature ranges can be observed at the transcriptional and posttranscriptional levels. It has been recently reported that a 56-kD calcium-dependent protein kinase (CDPK) is activated by the treatment of rice at 12°C (Martin and Busconi, 2001). Because activation of the 56-kD CDPK was observed after a longer (12 h) chilling period than that of the approximately 43-kD protein kinase, it is unlikely that the CDPK is an upstream component of the approximately 43-kD protein kinase. However, it is possible that several signaling pathways are involved in the signal transduction within the 12°C temperature range. It is worth noting that the activation of the approximately 43-kD protein kinase was not rapid and transient, but gradual during the treatment of 24 h, showing striking contrasts to the SIPK activation by salicylic acid (Zhang and Klessig, 1997) and WIPK by wounding in tobacco (Seo et al., 1995). SIPK and WIPK were rapidly and transiently activated within 5 min, and the activation could not be detected after 1 h of treatment, whereas the activation of the approximately 43-kD protein kinase lasted for 24 h.
It will be of great interest to elucidate the sensing mechanism for discriminating the two low temperature ranges. Reports show that a number of cold-inducible genes are also responsive to drought stress (Shinozaki and Yamaguchi-Shinozaki, 1996). Because OsMEK1 and OsMAP1 are induced by drought stress (Fig. 6), differences in the levels of dehydration may account for the differential expression of the genes by 12°C and 4°C treatments. However, our initial characterization suggested that 4°C-treated seedlings have slightly lower relative water contents than 12°C-treated seedlings within a 24-h time period, although both treatments reduce relative water contents (J.Q. Wen and R. Imai, unpublished data). ABA accumulation could also explain the differential expression because OsMEK1 and OsMAP1 were responsive to exogenous ABA application (Fig. 6). It was reported that ABA accumulated in rice seedlings under cold stress. The levels of ABA accumulation at 5°C were higher than that at 10°C (Lee et al., 1993). It is less feasible that higher levels of ABA accumulation occur at 12°C than at 4°C. Therefore, it is possible that the difference in the expression of OsMEK1 and OsMAP1 in response to 12°C and 4°C treatments is not due to different status of dehydration or ABA accumulation, but is due directly to the difference in the temperature. Thus, it is logical to consider that the rice plant has a mechanism to distinguish the two different ranges of low temperature, 12°C and 4°C, to elicit distinctive signals and thereafter activate specific responses.
Moderate temperatures at 12°C or higher induce male sterility at the booting stage of rice. Exposure to 12°C for 4 d at the tetrad stage of anther development resulted in male sterility in 80% of spikelets (Satake and Hayase, 1970; Nishiyama, 1984). Physiological and morphological changes associated with the 12°C chilling treatment have been described (Nishiyama, 1976, 1984). The identification of two MAPK signaling components and their involvement in a moderate temperature signaling pathway will provide some new insights at the molecular level of male sterility in rice, although a profound study is needed to clarify the functions of OsMEK1 and OsMAP1.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Stress Treatment
Seeds of japonica rice (Oryza sativa cv Yukihikari) were surface-sterilized in 70% (v/v) ethanol for 5 min, followed by further sterilization in 1.5% (v/v) sodium hypochlorite for 25 min, and were finally washed in distilled water. The sterilized seeds were soaked in distilled water for 12 h for imbibition. Fully imbibed seeds were germinated for 1 d at 25°C in the dark. Germinated seeds were evenly placed onto a plastic mesh grid supported by a plastic container filled with water just to the base of the mesh grid. The container was kept in a growth chamber at 25°C under continuous illumination (256 μmol m−2 s−1). After growing for 7 d, rice seedlings were subject to environmental stress treatments described below.
Low-temperature treatment was conducted by transferring the mesh grid with 7-d-old seedlings onto a plastic container filled with water preequilibrated at 4°C or 12°C in a growth chamber for 24 h prior to the treatment. The seedlings were treated at 4°C or 12°C for 1, 2, 6, 10, and 24 h under continuous illumination.
In a similar manner, ABA or NaCl treatments were performed by transferring the mesh grid with seedlings onto the container filled with 50 μm ABA solution or 0.2 m NaCl solution. Drought treatment was performed by transferring the mesh grid with seedlings (seedling roots were blotted with a paper towel to remove the water prior to transferring) onto the container without water. The sampling time was the same as for low-temperature treatments. Seven-day-old seedlings were harvested as a control (0 h). Shoots and roots were collected separately, and samples were immediately frozen in liquid nitrogen. All samples were stored at −80°C until use.
To collect the flowers and anthers, rice plants were grown in pots with nutrient soil for 2 months in a phytotron room controlled at 25°C/19°C (day/night). Anthers and panicles at the tetrad stage of microspore development were collected and frozen immediately in liquid nitrogen. For chilling treatment, the pots with rice plants were transferred to a phytotron room that was precooled to 12°C. Anthers and panicles were collected at 48 h of 12°C treatment.
cDNA Subtraction and Subsequent Cloning of OsMEK1
Total RNA was isolated from nonstressed and cold-stressed (12°C for 2 d) rice anthers at the tetrad stage in microspore development using Trizol reagent (Invitrogen, Carlsbad, CA) as described by the manufacturer, and poly(A)+ RNA was purified from the total RNA using Dynabeads Oligo(dT)25 (Dynal, Oslo). Double-strand cDNA synthesis from poly(A)+ RNA and subsequent selective subtraction was carried out using the PCR-Select cDNA subtraction kit (CLONTECH Laboratories, Palo Alto, CA) according to the manufacturer's instructions. After a second amplification, the PCR products were directly cloned into pCR2.1 vector using TA Cloning kit (Invitrogen) and were transformed with One-Shot competent cells (Invitrogen). Approximately 50 separate colonies were used for amplification by PCR using the nested primers (CLONTECH Laboratories). PCR products were denatured and dot-blotted onto a Hybond-N+ membrane (Amersham Pharmacia Biotech, London). To search for low-temperature responsive genes, two copies of dot blots were hybridized with 32P-labeled control (nonstressed) and cold-stressed total cDNA. Plasmids from colonies with significant difference in dot hybridization were isolated and sequenced. One insert (550 bp) with high homology to ZmMEK1 was selected for further study and was used as a probe for cDNA library screening.
Screening of Rice cDNA Library
A cDNA library constructed from chilling-stressed rice roots (R. Imai, unpublished data) was used for screening of OsMEK1 and OsMAP1 cDNA clones. For OsMEK1 screening, the subtracted cDNA fragment (550 bp) was used as a probe. During the first screening, approximately 5 × 105 recombinant plaques were screened by the plaque hybridization method (Sambrook et al., 1989). Plaque lifts were hybridized with a 32P-labeled 550-bp fragment in Rapid Hyb buffer (Amersham Pharmacia Biotech) at 65°C and were washed once with 1× SSC and 0.1% (w/v) SDS for 15 min at 65°C, and twice with 0.1× SSC and 0.1% (w/v) SDS for 15 min at 65°C. Second and third screenings were conducted under the same conditions as the first screening. After the third screening, several independent plaques were then subjected to in vivo excision of the pBluescript phagemid from the Lambda ZAP phage using the helper phage ExAssist (Stratagene, La Jolla, CA) according to the manufacturer's instructions. Phagemids were isolated and purified with Quantum Prep kit (Bio-Rad, Hercules, CA).
Isolation of OsMAP1
After searching the EST database, a rice EST clone (accession no. C22363) was found to encode a peptide with high homology to oat MAPK homolog, Aspk9. Two primers, 5′-GAGTTCAGGCCGACGATGA-3′ and 5′-GCCGAGTGGATGTACTTGA-3′ were used to amplify the cDNA that was reverse transcribed from rice anther poly(A)+ RNA. The PCR product of approximately 450 bp was cloned into pCR2.1 vector using the TA Cloning kit (Invitrogen) and was then sequenced. A clone with the correct sequence was used as the probe for cDNA library screening. The labeling of probe and hybridization conditions were the same as described above. More than 15 positive clones were obtained by screening 5 × 105 plaques. Both strands of the clone with the longest insert were sequenced as described below.
Northern-Blot Analysis
Twenty micrograms of total RNA that was isolated from stressed or control samples using Trizol reagent (Invitrogen) was separated on 1.0% (v/v) formaldehyde denaturing agarose gels and then transferred onto Hybond-N+ membranes (Amersham Pharmacia Biotech) according to standard methods (Sambrook et al., 1989). RNA blots were hybridized with 32P-labeled OsMEK1 or OsMAP1 full-length cDNA at 65°C for 16 h and were washed once with 2× SSC and 0.1% (w/v) SDS for 15 min, and twice with 0.1× SSC and 0.1% (w/v) SDS for 20 min at 65°C. After washing, blots were exposed to MR x-ray film (BioMax; Kodak, New Haven, CT) with an intensifying screen at −80°C.
DNA Sequencing and Analysis
The cloned DNA insert was sequenced by the dideoxy method using a Thermo Sequenase version 2.0 kit (Amersham Pharmacia Biotech) with a DNA sequencer (model 373A; Applied Biosystems, San Jose, CA). A homology search in the DNA/protein databases was carried out using the BLAST program. Analyses of the DNA sequences were performed using DNASIS software (Hitachi Software Engineering, Yokohama, Japan) or GENETYX-MAC software (Software Development, Tokyo).
Yeast Two-Hybrid System
A Hybrid Hunter (Version D; Invitrogen) was used for the two-hybrid system analysis. Strain L40 of Saccharomyces cerevisiae was used in this study. For two-hybrid assays, competent yeast cells were cotransformed with two plasmids (bait and prey). The bait, pHybLex/Zeo, carried a LexA DNA-binding domain and a Leu prototrophic marker. The prey, pYES-Trp2, carried a B42 transcriptional activation domain and a Trp prototrophic marker. The entire OsMEK1 open reading frame was fused to the LexA DNA-binding domain in the bait plasmid. The open reading frames of OsMAP1, OsMAP2, and OsMAP3 were independently fused to the B42 activation domain in the prey plasmid. The ability to drive the HIS3 reporter gene was assessed by growing yeast transformants containing bait and prey plasmids on selective YC medium lacking Trp, Leu, and His. The activity of the lacZ reporter gene was measured by using the 5-bromo-4-chloro-3-indolyl-β-d-galactoside filter assay according to the manufacturer's method (Invitrogen).
In-Gel Kinase Activity Assay
Crude protein extracts were prepared from control (0 h) and low-temperature-treated (12°C or 4°C) rice shoots. Approximately 0.5 g of plant samples was ground with 1 mL of extraction buffer (50 mm HEPES, pH 7.5, 5 mm EDTA, 5 mm EGTA, 10 mm dithiothreitol [DTT], 10 mm Na3VO4, 10 mm NaF, 50 mm β-glycero-phosphate, 1 mm phenylmethylsulfonyl fluoride, and one complete proteinase inhibitor mixture tablet [Amersham Pharmacia Biotech] 50 mL−1). After centrifugation at 13,000 rpm for 15 min, supernatants were further centrifuged at 55,000 rpm for 20 min. Aliquots of the supernatants were quickly frozen in liquid nitrogen and were stored at −80°C. The in-gel kinase assay was performed according to the procedures described previously (Zhang and Klessig, 1997) with slight modification. Ten micrograms of protein extracts was electrophoretically separated by 12% (w/v) SDS-polyacrylamide gels embedded with 0.5 mg mL−1 MBP or 1 mg mL−1 casein in the separating gel as a substrate for the kinase. After electrophoresis, SDS was removed from the gels by washing with 25 mm Tris-HCl, pH 7.5, 0.5 mm DTT, 0.1 mm Na3VO4, 5 mm NaF, 0.3 mg mL−1 bovine serum albumin, and 0.1% (v/v) Triton X-100 three times, each for 30 min at room temperature. The gel was renatured in the same buffer without bovine serum albumin and Triton X-100 at 4°C overnight with three changes of buffer. The gel was then equilibrated with the reaction buffer (25 mm Tris-HCl, pH 7.5, 2 mm EGTA, 10 mm MgCl2, 1 mm DTT, and 0.1 mm Na3VO4) at room temperature for 20 min. After changing to the fresh reaction buffer, the reaction was initiated by adding 200 nm ATP and 50 μCi [γ-32P]ATP (3,000 Ci mmol−1). The gel was incubated for 60 min at room temperature with shaking, and the reaction was terminated by transferring the gel into 5% (w/v) trichloroacetic acid and 1% (w/v) sodium pyrophosphate. The unincorporated [γ-32P]ATP was removed by washing with the same solution for 5 h with four changes. The gel was then dried and exposed to MR film (BioMax; Kodak).
ACKNOWLEDGMENTS
We thank Drs. John C. Walker and Dale Karlson for the comments on this manuscript. We also thank Dr. Tomonobu Kusano of Tohoku University for kindly providing a cDNA clone for lip19.
Footnotes
This work was supported by the Special Coordination Funds of the Science and Technology Agency of Japanese Government and the Cooperative System for Supporting Priority Research from Japan Science and Technology Cooperation (to R.I.). J.-Q.W. was supported by a Science and Technology Agency Fellowship from the Science and Technology Agency of Japan.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.006072.
LITERATURE CITED
- Aguan K, Sugawara K, Suzuki N, Kusano T. Isolation of genes for low-temperature-induced proteins in rice by a simple subtractive method. Plant Cell Physiol. 1991;32:1285–1289. [Google Scholar]
- Aguan K, Sugawara K, Suzuki N, Kusano T. Low-temperature-dependent expression of a rice gene encoding a protein with a leucine-zipper motif. Mol Gen Genet. 1993;240:1–8. doi: 10.1007/BF00276876. [DOI] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002;415:977–983. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- Berberich T, Sano H, Kusano T. Involvement of a MAP kinase, ZmMPK5, in senescence and recovery from low-temperature stress in maize. Mol Gen Genet. 1999;262:534–542. doi: 10.1007/s004380051115. [DOI] [PubMed] [Google Scholar]
- Binh LT, Oono K. Molecular cloning and characterization of genes related to chilling tolerance in rice. Plant Physiol. 1992;99:1146–1150. doi: 10.1104/pp.99.3.1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bögre L, Ligterink W, Heberle-Bors E, Hirt H. Mechanosensors in plants. Nature. 1996;383:489–490. doi: 10.1038/383489a0. [DOI] [PubMed] [Google Scholar]
- Bögre L, Ligterink W, Meskiene I, Barker PJ, Heberle-Bors E, Huskisson NS, Hirt H. Wounding induces the rapid and transient activation of a specific MAP kinase pathway. Plant Cell. 1997;9:75–83. doi: 10.1105/tpc.9.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderini O, Glab N, Bergounioux C, Heberle-Bors E, Wilson C. A novel tobacco mitogen-activated protein (MAP) kinase kinase, NtMEK1, activates the cell cycle-regulated p43Ntf6 MAP kinase. J Biol Chem. 2001;276:18139–18145. doi: 10.1074/jbc.M010621200. [DOI] [PubMed] [Google Scholar]
- Eckardt NA. Transcription factors dial 14-3-3 for nuclear shuttle. Plant Cell. 2001;13:2385–2389. doi: 10.1105/tpc.13.11.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett RM, Oh SA, Morris PC, Grierson D. A tomato MAP kinase kinase gene (Accession no. AJ000728) differentially regulated during fruit development, leaf senescence, and wounding (PGR98-151) Plant Physiol. 1998;117:1526. [Google Scholar]
- Hardin SC, Wolniak SM. Molecular cloning and characterization of maize ZmMEK1, a protein kinase with a catalytic domain homologous to mitogen- and stress-activated protein kinase kinases. Planta. 1998;206:577–584. doi: 10.1007/s004250050435. [DOI] [PubMed] [Google Scholar]
- He C, Fong SH, Yang D, Wang GL. BWMK1, a novel MAP kinase induced by fungal infection and mechanical wounding in rice. Mol Plant-Microbe Interact. 1999;12:1064–1073. doi: 10.1094/MPMI.1999.12.12.1064. [DOI] [PubMed] [Google Scholar]
- Herskowitz I. MAP kinase pathways in yeast: for mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- Hirt H. Multiple roles of MAP kinases in plant signal transduction. Trends Plant Sci. 1997;2:11–15. [Google Scholar]
- Hughes MA, Dunn MA. The molecular biology of plant acclimation to low temperature. J Exp Bot. 1996;47:291–305. [Google Scholar]
- Huttly AK, Phillips AL. Gibberellin-regulated expression in oat aleurone cells of two kinases that show homology to MAP kinase and a ribosomal protein kinase. Plant Mol Biol. 1995;27:1043–1052. doi: 10.1007/BF00037031. [DOI] [PubMed] [Google Scholar]
- Ichimura K, Mizoguchi T, Hayashida N, Seki M, Shinozaki K. Molecular cloning and characterization of three cDNAs encoding putative mitogen-activated protein kinase kinases (MAPKKs) in Arabidopsis thaliana. DNA Res. 1998a;5:341–348. doi: 10.1093/dnares/5.6.341. [DOI] [PubMed] [Google Scholar]
- Ichimura K, Mizoguchi T, Irie K, Morris P, Giraudat J, Matsumoto K, Shinozaki K. Isolation of ATMEKK1 (a MAP kinase kinase kinase)-interacting proteins and analysis of a MAP kinase cascade in Arabidopsis. Biochem Biophys Res Commun. 1998b;253:532–543. doi: 10.1006/bbrc.1998.9796. [DOI] [PubMed] [Google Scholar]
- Jonak C, Heberle-Bors E, Hirt H. MAP kinases: universal multi-purpose signaling tools. Plant Mol Biol. 1994;24:407–416. doi: 10.1007/BF00024109. [DOI] [PubMed] [Google Scholar]
- Jonak C, Kiegerl S, Ligterink W, Barker PJ, Huskisson NS, Hirt H. Stress signaling in plants: A mitogen-activated protein kinase pathway is activated by cold and drought. Proc Natl Acad Sci USA. 1996;93:11274–11279. doi: 10.1073/pnas.93.20.11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouannic S, Hamal A, Kreis M, Henry Y. Molecular cloning of an Arabidopsis thaliana MAP kinase kinase-related cDNA (accession No. Y07964) (PGR96-098) Plant Physiol. 1996;112:1397. [Google Scholar]
- Koga BY, Abe M, Kitagawa Y. Alteration in gene expression during cold treatment of rice plant. Plant Cell Physiol. 1991;32:901–906. [Google Scholar]
- Kusano T, Berberich T, Harada M, Suzuki N, Sugawara K. A maize DNA-binding factor with a bZIP motif is induced by low temperature. Mol Gen Genet. 1995;248:507–517. doi: 10.1007/BF02423445. [DOI] [PubMed] [Google Scholar]
- Lee T-M, Lur H-S, Chu C. Role of abscisic acid in chilling tolerance of rice (Oryza sativa L.) seedlings: endogenous abscisic acid levels. Plant Cell Environ. 1993;16:481–490. [Google Scholar]
- Ligterink W. MAP kinases in plant signal transduction: How many, and what for? In: Hirt H, editor. MAP Kinases in Plant Signal Transduction. Berlin: Springer-Verlag; 2000. pp. 11–27. [Google Scholar]
- Martin ML, Busconi L. A rice membrane-bound calcium-dependent protein kinase is activated in response to low temperature. Plant Physiol. 2001;125:1442–1449. doi: 10.1104/pp.125.3.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, Gotoh Y, Nishida E, Yamaguchi-Shinozaki K, Hayashida N, Iwasaki T, Kamada H, Shinozaki K. Characterization of two cDNAs that encode MAP kinase homologues in Arabidopsis thaliana and analysis of the possible role of auxin in activating such kinase activities in cultured cells. Plant J. 1994;5:111–122. doi: 10.1046/j.1365-313x.1994.5010111.x. [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Hayashida N, Yamaguchi-Shinozaki K, Kamada H, Shinozaki K. ATMPKs: a gene family of plant MAP kinases in Arabidopsis thaliana. FEBS Lett. 1993;336:440–444. doi: 10.1016/0014-5793(93)80852-l. [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Ichimura K, Shinozaki K. Environmental stress response in plants: the role of mitogen-activated protein kinases. Trends Biotechnol. 1997;15:15–19. doi: 10.1016/S0167-7799(96)10074-3. [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Irie K, Hirayama T, Hayashida N, Yamaguchi-Shinozaki K, Matsumoto K, Shinozaki K. A gene encoding a mitogen-activated protein kinase kinase kinase is induced simultaneously with genes for a mitogen-activated protein kinase and an S6 ribosomal protein kinase by touch, cold, and water stress in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1996;93:765–769. doi: 10.1073/pnas.93.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris PC, Guerrier D, Leung J, Giraudat J. Cloning and characterisation of MEK1, an Arabidopsis gene encoding a homologue of MAP kinase kinase. Plant Mol Biol. 1997;35:1057–1064. doi: 10.1023/a:1005963222768. [DOI] [PubMed] [Google Scholar]
- Nishiyama I. Male sterility caused by cooling treatment at the young microspore stage in rice plants: effects of some substances on sterility. Proc Crop Sci Soc Japan. 1975;44:397–402. [Google Scholar]
- Nishiyama I. Male sterility caused by cooling treatment at the young microspore stage in rice plants. Proc Crop Sci Soc Japan. 1976;45:254–262. [Google Scholar]
- Nishiyama I. Climate influence on pollen formation and fertilization. In: Tsunoda S, Takahashi N, editors. Biology of Rice. Amsterdam: Elsevier; 1984. pp. 153–171. [Google Scholar]
- Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Ren D, Yang H, Zhang S. Cell death mediated by MAPK is associated with hydrogen peroxide production in Arabidopsis. J Biol Chem. 2002;277:559–565. doi: 10.1074/jbc.M109495200. [DOI] [PubMed] [Google Scholar]
- Robinson MJ, Cobb MH. Mitogen-activated protein kinase pathways. Curr Opin Cell Biol. 1997;9:180–186. doi: 10.1016/s0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]
- Romeis T. Protein kinases in the plant defence response. Curr Opin Plant Biol. 2001;4:407–414. doi: 10.1016/s1369-5266(00)00193-x. [DOI] [PubMed] [Google Scholar]
- Saijo Y, Hata S, Kyozuka J, Shimamoto K, Izui K. Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J. 2000;23:319–327. doi: 10.1046/j.1365-313x.2000.00787.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Frisch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Satake T. Determination of the most sensitive stage to sterile-type cool injury in rice plants. Res Bull Hokkaido Natl Agric Exp Stat. 1976;113:1–43. [Google Scholar]
- Satake T, Hayase H. Male sterility caused by cooling treatment at the young microspore stage in rice plants: estimation of pollen developmental stage and most sensitive stage to coolness. Proc Crop Sci Soc Jpn. 1970;39:468–473. [Google Scholar]
- Schaeffer HJ, Weber MJ. Mitogen-activated protein kinases: specific messages from ubiquitous messengers. Mol Cell Biol. 1999;19:2435–2444. doi: 10.1128/mcb.19.4.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seger R, Krebs EG. The MAPK signaling cascade. FASEB J. 1995;9:726–735. [PubMed] [Google Scholar]
- Seo S, Okamoto M, Seto H, Ishizuka K, Sano H, Ohashi Y. Tobacco MAP kinase: a possible mediator in wound signal transduction pathways. Science. 1995;270:1988–1992. doi: 10.1126/science.270.5244.1988. [DOI] [PubMed] [Google Scholar]
- Seo S, Sano H, Ohashi Y. Jasmonate-based wound signal transduction requires activation of WIPK, a tobacco mitogen-activated protein kinase. Plant Cell. 1999;11:289–298. doi: 10.1105/tpc.11.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata W, Banno H, Ito Y, Hirano K, Irie K, Usami S, Machida C, Machida Y. A tobacco protein kinase, NPK2, has a domain homologous to a domain found in activators of mitogen-activated protein kinases (MAPKKs) Mol Gen Genet. 1995;246:401–410. doi: 10.1007/BF00290443. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Molecular responses to drought and cold stress. Curr Opin Biotechnol. 1996;7:161–167. doi: 10.1016/s0958-1669(96)80007-3. [DOI] [PubMed] [Google Scholar]
- Stafstrom JP, Altschuler M, Anderson DH. Molecular cloning and expression of a MAP kinase homologue from pea. Plant Mol Biol. 1993;22:83–90. doi: 10.1007/BF00038997. [DOI] [PubMed] [Google Scholar]
- Stratmann JW, Ryan CA. Myelin basic protein kinase activity in tomato leaves is induced systemically by wounding and increases in response to systemin and oligosaccharide elicitors. Proc Natl Acad Sci USA. 1997;94:11085–11089. doi: 10.1073/pnas.94.20.11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takezawa D. Elicitor- and A23187-induced expression of WCK-1, a gene encoding mitogen-activated protein kinase in wheat. Plant Mol Biol. 1999;40:921–933. doi: 10.1023/a:1006263607135. [DOI] [PubMed] [Google Scholar]
- Thomashow MF, Stockinger EJ, Gilmour SJ. Regulation of plant gene expression in response to low temperature. In: Li PH, Chen THH, editors. Plant Cold Hardiness. New York: Plenum Press; 1997. pp. 29–34. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C, Voronin V, Touraev A, Vicente O, Heberle-Bors E. A developmentally regulated MAP kinase activated by hydration in tobacco pollen. Plant Cell. 1997;9:2093–2100. doi: 10.1105/tpc.9.11.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka T, Suge H. Damage of seed fertility by cooling treatment and endogenous gibberellins in ears of rice plants (Oryza sativa L.) Breeding Sci. 1996;46:173–178. [Google Scholar]
- Zhang S, Klessig DF. Salicylic acid activates a 48-kD MAP kinase in tobacco. Plant Cell. 1997;9:809–824. doi: 10.1105/tpc.9.5.809. [DOI] [PMC free article] [PubMed] [Google Scholar]