Abstract
A specialized database (DB) for Arabidopsis membrane proteins, ARAMEMNON, was designed that facilitates the interpretation of gene and protein sequence data by integrating features that are presently only available from individual sources. Using several publicly available prediction programs, putative integral membrane proteins were identified among the approximately 25,500 proteins in the Arabidopsis genome DBs. By averaging the predictions from seven programs, approximately 6,500 proteins were classified as transmembrane (TM) candidate proteins. Some 1,800 of these contain at least four TM spans and are possibly linked to transport functions. The ARAMEMNON DB enables direct comparison of the predictions of seven different TM span computation programs and the predictions of subcellular localization by eight signal peptide recognition programs. A special function displays the proteins related to the query and dynamically generates a protein family structure. As a first set of proteins from other organisms, all of the approximately 700 putative membrane proteins were extracted from the genome of the cyanobacterium Synechocystis sp. and incorporated in the ARAMEMNON DB. The ARAMEMNON DB is accessible at the URL http://aramemnon.botanik.uni-koeln.de.
Biological membranes constitute a chemical barrier to the environment and are thus the prerequisite for the establishment and maintenance of a controlled intracellular milieu, the cytoplasm. In eukaryotes, membranes are also responsible for the formation of chemically distinct intracellular compartments. The lipid bilayer membranes contain a great diversity of proteins that fulfill different functions and serve as an interface to the environment and between different compartments. Among these membrane proteins are receptors involved in signaling cascades and pathogen defense reactions, enzymes such as the apparatus for cell wall biosynthesis, and transporters responsible for the import and export of solutes and ions and the establishment of electrochemical gradients across membranes, thereby connecting the different metabolic pathways of the cellular compartments and organelles.
Many plant transport proteins were identified by complementation of yeast mutants that were deficient in certain transport or metabolic functions (Frommer and Ninnemann, 1995). Membrane proteins have a modular structure, consisting of hydrophobic domains and hydrophilic loops or termini that extend into the cytoplasm, the organelle, or point to the extracellular space. The hydrophobic transmembrane (TM) domains consist of amphipathic α-helices or β-barrels that pass across or dip into the hydrophobic membrane lipid bilayer. During recent years, the three-dimensional structures of more than 160 TM proteins or domains were determined at varying resolution, and it appears that modularity is a general feature of polytopic membrane proteins (http://www.rcsb.org/pdb/; http://www.ncbi.nlm.nih.gov:80/Structure/; Berman et al., 2002; Wang et al., 2002).
Arabidopsis is the first plant for which the genome has been deciphered completely (Arabidopsis Genome Initiative, 2000). Automatic gene predictions and annotations have been performed for the full genome and are continuously being improved at The Institute for Genomic Research (TIGR; http://www.tigr.org/tdb/e2k1/ath1/ath1.shtml), The Arabidopsis Information Resource (http://www.Arabidopsis.org/aboutarabidopsis.html), and the Munich Information Center for Protein Sequences (http://mips.gsf.de/proj/thal/db/about/about_frame.html). Gene predictions are sustained by expressed sequence tag analyses, full-length mRNA sequencing and individual research projects. Accordingly, the Arabidopsis genome offers the possibility to perform bioinformatic analyses and data mining that are not yet possible with other plant species. In the future, these analyses will also become possible for the recently completed rice (Oryza sativa) genome (Goff et al., 2002; Yu et al., 2002).
A couple of databases (DBs) specialized for membrane proteins are accessible on the internet. For Brewer's yeast (Saccharomyces cerevisiae) a transport protein DB has been established (http://alize.ulb.ac.be/YTPdb/; Andre, 1995; Van Belle and Andre, 2001). A comprehensive classification of transport systems and transport protein families from 20 bacterial, archaeal, and eukaryotic genomes has been established and is accessible at http://www-biology.ucsd.edu/∼msaier/transport/(Saier, 1999). Tables that summarize genomic comparisons of membrane transport systems are also published on the Web pages of the Paulsen laboratory (http://www.biology.ucsd.edu/∼ipaulsen/transport/). An Arabidopsis library of all TM candidate proteins containing more than one TM span has been published and was used to identify novel membrane protein families not known from other organisms (http://www.biosci.cbs.umn.edu/Arabidopsis; Ward, 2001). PlantsT is an Arabidopsis and yeast transporter DB with a focus on metal ion transporters from Arabidopsis (http://plantst.sdsc.edu; Mäser et al., 2001).
All of these membrane protein DBs use one or two algorithms for TM prediction. For a given protein, these particular method(s) used may generate an accurate prediction. However, in many cases, predictions by different programs vary with respect to the number of TM domains and their relative location in the polypeptide sequence, and it is not possible to know which prediction program will generate the most accurate prediction for a particular protein (Möller et al., 2001; Ikeda et al., 2002).
Here, we present a novel membrane protein DB, ARAMEMNON, which integrates features that are presently only available from separate sources, and thus should facilitate the interpretation of gene/protein sequence data. The major objectives of the ARAMEMNON DB are to provide (a) the possibility to directly compare the predictions of (currently) seven different TM span computation programs and (b) the predictions of subcellular localization by eight signaling peptide recognition programs, and (c) to identify protein families (“clusters”) that center around a user-selected protein. The ARAMEMNON DB is accessible on the Web at the URL http://aramemnon.botanik.uni-koeln.de.
RESULTS
Arabidopsis Membrane Protein Predictions and Comparative Graphical Representation of TM Spans
The complete set of 25,492 predicted Arabidopsis protein sequences (January 2002) was screened for putative membrane proteins containing one or more TM domains. In the current version of the DB, the prediction of TM domains is based on seven different programs (Table I), and future versions will include additional predictions (see “Discussion”). Because of different approaches used, i.e. methods based on hidden Markov models, on the calculation of hydrophobicities, or on a DB of TM proteins, some programs recognize (or overlook) membrane spans that are predicted by others. For example, HmmTop 2.0, TmPred, TMap, and TopPred 2.0 classify more proteins as TM proteins compared with TmHMM 2.0, SosuiG 1.1, or Eiconda 0.9 (Table II). It should be noted that the overall number of predicted TM proteins differs significantly between the individual programs, and therefore the use of a single prediction algorithm does not allow high confidence level conclusions with respect to number and location of predicted TM spans.
Table I.
Transmembrane span prediction programs
| Program | Web Site | Reference |
|---|---|---|
| TmHMM 2.0 | http://www.cbs.dtu.dk/services/TMHMM/ | Sonnhammer et al. (1998) |
| HmmTop 2.0 | http://www.enzim.hu/hmmtop | Tusnady and Simon (1998, 2001) |
| SosuiG 1.1 | http://sosui.proteome.bio.tuat.ac.jp | Hirokawa et al. (1998) |
| Eiconda 0.9 | Not available | R. Schwacke (unpublished data) |
| TMPred | http://www.ch.embnet.org/software/TMPRED_form.html | Hofmann and Stoffel (1993) |
| TMap | http://www.mbb.ki.se/tmap/ | Persson and Argos (1994, 1996) |
| TopPred 2.0 | http://bioweb.pasteur.fr/seqanal/interfaces/toppred.html | von Heijne (1992); Claros and von Heijne (1994) |
Table II.
Predictions of TM spans in Arabidopsis proteins by different programs
| TM Spans | ARAMEMNON | TmHMM 2.0 | HmmTop 2.0 | SosuiG 1.1 | Eiconda 0.9 | TmPred | TMap | TopPred 2.0 |
|---|---|---|---|---|---|---|---|---|
| 1 | 2,011 | 2,957 | 6,182 | 3,302 | 1,747 | 7,511 | 6,368 | 7,344 |
| 2 | 1,759 | 845 | 1,881 | 1,441 | 1,977 | 4,051 | 2,768 | 3,523 |
| 3 | 729 | 310 | 916 | 441 | 966 | 1,985 | 1,083 | 1,452 |
| 4 | 434 | 336 | 456 | 363 | 560 | 1,075 | 574 | 782 |
| 5 | 223 | 195 | 287 | 221 | 285 | 481 | 283 | 365 |
| 6 | 249 | 211 | 246 | 213 | 235 | 337 | 229 | 282 |
| 7 | 184 | 166 | 198 | 159 | 190 | 262 | 193 | 191 |
| 8 | 123 | 130 | 100 | 153 | 161 | 151 | 154 | 161 |
| 9 | 165 | 140 | 120 | 169 | 160 | 183 | 198 | 194 |
| 10 | 220 | 224 | 212 | 177 | 198 | 216 | 175 | 176 |
| 11 | 158 | 129 | 118 | 163 | 140 | 196 | 132 | 149 |
| 12 | 126 | 134 | 225 | 93 | 141 | 125 | 93 | 95 |
| 13 | 39 | 22 | 77 | 49 | 35 | 52 | 32 | 39 |
| 14 | 28 | 23 | 28 | 24 | 29 | 40 | 16 | 20 |
| 15 | 10 | 10 | 27 | 9 | 11 | 17 | 10 | 4 |
| 16 | 17 | 18 | 35 | 15 | 22 | 28 | 23 | 12 |
| Total | 6,475 | 5,850 | 11,108 | 6,992 | 6,857 | 16,710 | 12,331 | 14,789 |
| False pos.a | 0 | 568 | 14 | 9 | 1185 | 513 | 561 | |
| False neg.b | 581 | 60 | 338 | 252 | 0 | 481 | 0 |
Number of TM proteins predicted only by this one program.
Number of TM proteins not predicted by this program but by all other programs.
The combination of the overlapping sets of TM proteins predicted by any one of the seven programs would result in the highly unlikely number of approximately 18,600 putative Arabidopsis membrane proteins. To select putative reading frames with a high probability to contain TM spans, the statistical median of all predicted TM regions was calculated for each protein. Only such reading frames were classified as membrane proteins by ARAMEMNON, for which at least three TM spans had been predicted by five or more of the seven programs, or one or two TM spans by at least six programs, resulting in an overall number of 6,047 proteins. In a subsequent step, among the disqualified proteins, those were selected and entered into the DB that share 30% or more sequence similarity with a member of the 6,047 preselected proteins and that are predicted by four programs to contain TM spans. This reiterative procedure increased the total number of putative membrane genes/proteins to 7,314.
Endoplasmatic reticulum targeting sequences of soluble proteins are often characterized by a hydrophobic core that may be recognized by prediction programs as a TM span (Nielsen et al., 1997a). To exclude such proteins from classification as integral membrane proteins, the 839 reading frames containing a single TM domain at the N terminus within a cleavable signal peptide, which is consistently predicted by TargetP, SignalP-HMM, SignalP-NN, and iPSORT (see below) were removed from the DB. In accordance, proteins supposedly having a non-cleavable signal sequence and thus remaining anchored to the membrane will be retained in the ARAMEMNON DB. However, as the recognition of (non-)cleavable signal sequences by the different programs is not always reliable, some actual membrane anchored proteins may have been excluded from the fraction of one-TM proteins, whereas some soluble proteins may have persisted. The excluded proteins are listed in the ARAMEMNON DB.
Eventually, 6,475 (about 25%) of the nuclear encoded Arabidopsis proteins are classified as membrane proteins and listed in the ARAMEMNON DB (Table II). In addition, 78 membrane proteins of the organellar genomes were added. At present, approximately 40% of all Arabidopsis TM proteins are annotated in the TIGR DB as “unknown protein,” “hypothetical protein,” or “putative protein.”
The ARAMEMNON DB was primarily developed for the analysis of Arabidopsis membrane proteins. However, to augment comparative analyses of similar proteins, supplementary data sets of orthologs from other organisms will also be incorporated. As a first step, all 706 membrane proteins were extracted from the cyanobacterium Synechocystis sp. genome (3,165 genes; Kaneko et al., 1996) by applying the same procedure as with Arabidopsis protein sequences and included in the ARAMEMNON DB. For 500 Arabidopsis membrane proteins, ARAMEMNON finds 123 related Synechocystis sp. membrane proteins. ChloroP or PCLR remarkably predict only approximately 130 of these Arabidopsis proteins to be localized in the chloroplasts.
Table II shows the numbers and frequencies of Arabidopsis proteins with less than 17 TM spans in the ARAMEMNON DB in comparison with the numbers predicted by the individual programs. Major differences between the individual programs are obviously attributable to recognition of proteins containing one to three TM spans. Programs that identify more proteins containing one to three TM spans, i.e. HmmTop 2.0, TmPred, TMap, and TopPred 2.0, also classify the highest overall number of Arabidopsis proteins as TM proteins. TmPred, HmmTop, TopPred, and TMap also predict the highest fraction of “false positives” (proteins predicted only by this but no other program), whereas, except for TMap, the programs predicting a lower overall number of TM proteins tend to generate more “false negatives” (proteins not predicted by this program but by all other programs).
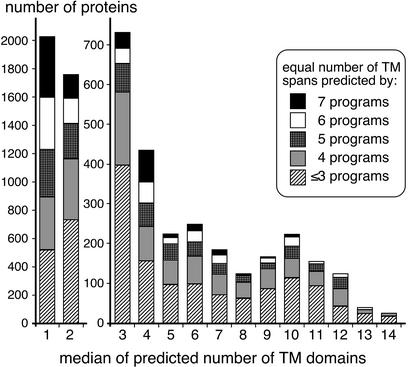
Because the TM predictions by individual algorithms sometimes deviate dramatically, the reliability of TM topology predictions can be significantly improved by combining the results from several prediction methods according to a “majority-vote” principle (Nilsson et al., 2000). Also for most Arabidopsis proteins, the programs generate deviating predictions. For each class of proteins with a median TM span number between one and 14, the proportion of proteins was determined; for that, the same number of TM domains is predicted by all seven programs used, or by six, five, four, or less than four programs, respectively (Fig. 1). For at least 40% of all proteins with a median of three or more TM spans, maximally three programs predict the same number of TM spans. Except for the 4-TM proteins, five or more programs predict the same TM span number only for less than one-third of the proteins. Moreover, it has to be pointed out that despite predicting the same number of TM spans, different programs frequently recognize putative TM domains in different locations (for example, see Fig. 2B).
Figure 1.
Uniformity of TM span predictions. For each class of proteins with a median TM span number between one and 14, the proportion of proteins indicated for that same number of TM domains is predicted by all seven programs used or by six, five, four, or less than four programs, respectively.
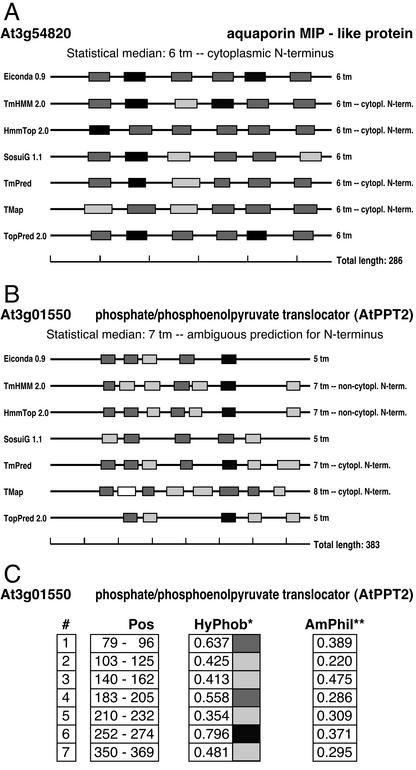
Figure 2.
Graphical representation of TM span predictions. ARAMEMNON displays plots of the TM predictions by seven programs. A, TM span predictions for an aquaporin MIP-like protein (At3g54820). All seven programs uniformly predict six TM spans at almost the same positions, and TmHMM 2.0, HmmTop 2.0, TmPred and TMap predict the same orientation within the membrane. The shading intensity of the membrane span candidate segments indicates the mean hydrophobicity range according to a normalized hydrophobicity scale (Eisenberg et al., 1984): white, 0 to 0.24; light gray, 0.25 to 0.49; dark gray, 0.50 to 0.74; and black, 0.75 to 0.99. B, TM span predictions for the PPT2 protein (At3g01550). The predictions differ in the number of TM spans, location of TM spans, and orientation of the protein within the membrane. C, For each prediction, the details are shown. #, Predicted TM spans starting from the N terminus; Pos, location of the TM span in the protein; HyPhob, mean hydrophobicity within the membrane span candidate segment (Eisenberg et al., 1984). The shading intensity correlates to that in A; AmPhil, relative maximal amphiphilicity within the membrane candidate segment.
The ARAMEMNON DB provides a function for comparative graphical representation of TM spans. It displays plots with TM spans predicted by the seven different prediction programs, that allow the immediate evaluation of the predictions. Figure 2A shows an example of a aquaporin-like MIP protein that is consistently predicted by all seven programs. In contrast, the overall numbers and locations of TM spans predicted by the alternative algorithms for the phosphate/phosphoenolpyruvate translocator PPT2 deviate considerably between each other (Fig. 2B). For each prediction, the TM details “location,” “mean hydrophobicity,” and “relative maximal amphiphilicity” can be displayed (Fig. 2C).
Prediction of Subcellular Localization
Subcellular localization predictions were performed by eight programs (Table III). However, only TargetP and iPSORT predict targeting to chloroplasts, to mitochondria, or to the secretory pathway. The other programs offer predictions for one or two specific compartments only. Overall, at least four predictions are available for each subcellular target (Table III). ARAMEMNON enables queries for proteins predicted to be located in plastids or mitochondria or to be secreted, if a absolute majority of programs predict one target compartment.
Table III.
Signal sequence prediction programs
| Program | Web Site | Prediction Targeta | Reference |
|---|---|---|---|
| TargetP 1.0 | http://www.cbs.dtu.dk/services/TargetP/ | CHP, MTC, SEC | Nielsen et al. (1997a); Emanuelsson et al. (2000) |
| ChloroP 1.1 | http://www.cbs.dtu.dk/services/ChloroP/ | CHP | Emanuelsson et al. (1999) |
| SignalP 2.0 HMM | http://www.cbs.dtu.dk/services/SignalP-2.0/ | SEC | Nielsen and Krogh (1998) |
| SignalP 2.0 NN | http://www.cbs.dtu.dk/services/SignalP-2.0/ | SEC | Nielsen et al. (1997b) |
| Predotar 0.5 | http://www.inra.fr/predotar/ | CHP, MTC | I. Small (unpublished data) |
| MitoProt II | http://www.mips.biochem.mpg.de/cgi-bin/proj/ medgen/mitofilter | MTC | Claros and Vincens (1996) |
| iPSORT | http://www.HypothesisCreator.net/iPSORT/ | CHP, MTC, SEC | Bannai et al. (2002) |
| PCLR | http://apicoplast.cis.upenn.edu/pclr/ | CHP | Schein et al. (2001) |
CHP, Plastids; MTC, mitochondria; SEC, secretory pathway.
The direct comparison of subcellular localization predictions implemented in the ARAMEMNON DB exemplifies that the reliability of the individual predictions is rather ambiguous (see also Emanuelsson and von Heijne, 2001). This is illustrated by proteins for which clear experimental data exist. For example, the plastidic localization of the xylulose-5-phosphate/phosphate translocator (At5g17630; M. Eicks, Universität zu Köln, personal communication) is correctly predicted by TargetP, ChloroP, Predotar, iPSORT, and PCLR, whereas the chloroplast inner envelope-localized triose phosphate translocator AtTPT (At5g46110; P. Niewiadomski, Universität zu Köln, personal communication) is predicted to be targeted to mitochondria (Table IV). The only hint toward a plastidic localization is the relatively high score generated by the PCLR program. Therefore, if experimental data for subcellular localization of a given protein were available (presently only few), this has been indicated in the membrane topology view and a reference to respective publication is provided.
Table IV.
Subcellular localization predictions
| Protein | Targeta | Predicted Targetb | TargetP | Predotar | iPSortc | ChloroPd | SignalP NN | SignalP HMM | MitoProt II | PCLR |
|---|---|---|---|---|---|---|---|---|---|---|
| AtXPT (At5g17630) | CHL | CHL | 0.901 | 0.983 | 1 | 0.7 | – | – | – | 0.758 |
| MIT | 0.206 | 0 | 0 | – | – | – | 0.909 | – | ||
| SEC | 0.003 | –e | 0 | – | 0.346 | 0.003 | – | – | ||
| AtTPT (At5g46110) | CHL | CHL | 0.316 | 0.005 | 0 | 0.445 | – | – | – | 0.548 |
| MIT | 0.607 | 0.984 | 1 | – | – | – | 0.904 | – | ||
| SEC | 0.013 | – | 0 | – | 0.253 | 0.002 | – | – |
CHL, Chloroplast localization was experimentally determined.
CHL, Chloroplasts; MIT, mitochondria; SEC, secretory pathway.
iPSort generates a 'yes' = 1/'no' = 0 prediction.
The prediction scores of ChloroP were normalized to a 0–1 scale.
–, No prediction is generated for this subcellular compartment.
Family Structure Analysis of TM Proteins
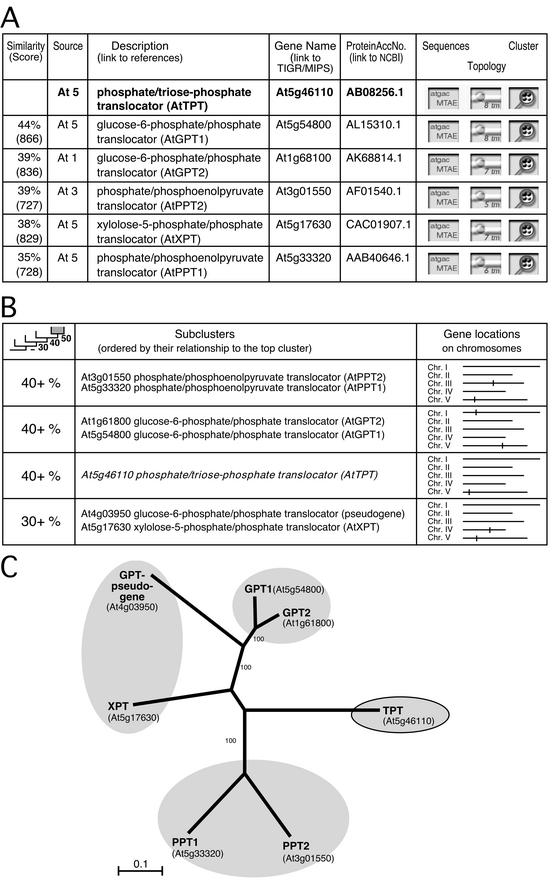
All proteins in the ARAMEMNON DB were subjected to pairwise local alignments. All pairs with a minimal similarity of 28% excluding gaps and a minimal Smith-Waterman score of 310 were registered. The ARAMEMNON DB provides this rather static view of membrane proteins related to a user-selected query sequence as a list ordered by the degree of similarity with a lower cut-off level of 28% (Fig. 3A).
Figure 3.
TPT-related PTs identified by ARAMEMNON DB. A, List-form display of TPT-related proteins. The columns show (from left to right): amino acid similarity excluding gaps and Smith-Waterman score; organism (At, Arabidopsis) and chromosome number; gene annotation (with links to relevant publications); gene name (with links to TIGR and Munich Information Center for Protein Sequences DBs); protein accession number in GenBank (with link to National Center for Biotechnology Information [NCBI]); button to call protein, cDNA, genomic DNA, 5′- and 3′-untranslated region sequence display; button to call the TM and signal sequence prediction display; button to call the family structure (cluster) display, as shown in B. B, Columns from left to right: average subcluster amino acid similarity levels; members of the subclusters; chromosomal location of genes. C, NJ tree based on a multiple alignment of PT sequences performed by ClustalX. The numbers beside branches indicate the frequency (%) with which the branch was found in 1,000 bootstrap replicas. The shaded branches correspond to the subclusters generated by ARAMEMNON DB.
To create a more dynamic representation of a protein family, especially with respect to subfamilies, a “cluster” generation function was implemented in the ARAMEMNON DB. For each protein with similarity to the query protein, all other related proteins are retrieved and the pairwise similarities between all sequences are determined. According to these similarities, the retrieved sequences are merged into a “cluster” that contains “subclusters” with similarity threshold levels of 28%, 40%, 50%, and an upper resolution limit of 70% (see “Materials and Methods” and Figs. 3–5). The proteins selected by this means have at least 28% sequence similarity to at least one other member of the cluster, but not necessarily to the original query sequence. Proteins of the cluster with less than 28% similarity to the query protein are marked in Figures 4 and 5 by “∼∼.”
Figure 5.
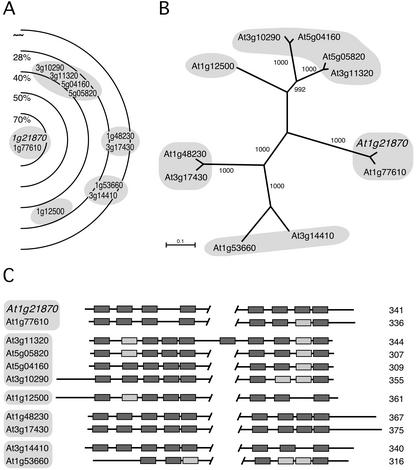
Family structure and TM domain topology of the putative K/VAG transporters. A, Graphical illustration the of At1g21870-related membrane protein family structure generated by the ARAMEMNON DB. B, NJ tree of the At1g21870-related proteins. C, The membrane topologies of the K/VAG transporters were predicted by using the TM protein prediction program Eiconda 0.9. Shading of the boxes that symbolize the TM spans is as in Figure 2. The graphical outputs are drawn to scale and were manually aligned. The length of the proteins in amino acids is indicated to the right.
Figure 4.
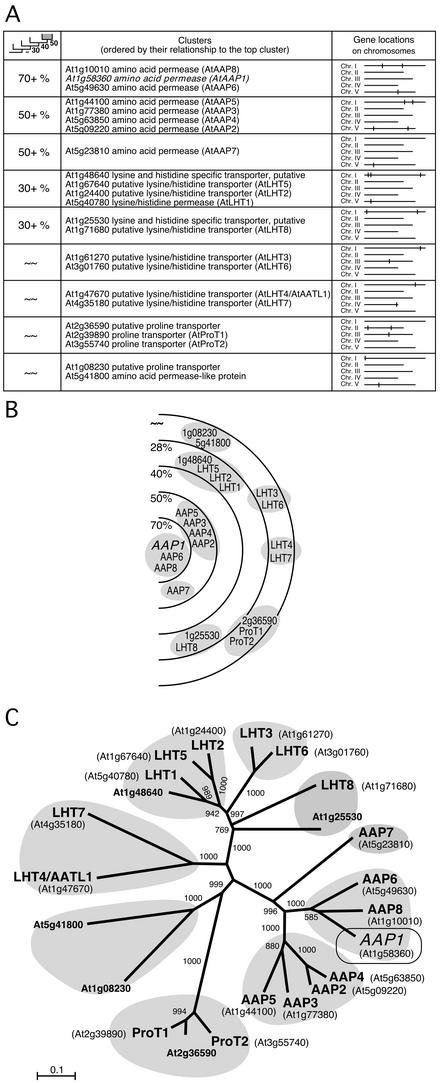
Amino acid permease (AAP) protein family structure generated by ARAMEMNON DB in comparison to the NJ tree. A, ARAMEMNON family structure (cluster) display of the AAP proteins. B, Schematic graphic to illustrate the relationships between the AAP protein subclusters listed in A. C, NJ tree of the AAP family, based on a multiple alignment of amino acid permease sequences performed by ClustalX.
To test the quality of the clustering function, results obtained for three protein families were compared with family analysis performed by neighbor-joining (NJ) tree building based on ClustalX alignments. The chosen families were the plastidic phosphate translocators (PTs), the amino acid permeases of the ATF1 superfamily (Wipf et al., 2002), and a magnesium transporter family (Li et al., 2001).
The PT family consists of four different groups, (a) the triose phosphate/phosphate translocator (TPT), (b) the phosphoenolpyruvate/phosphate translocators (PPT), (c) the Glc 6-phosphate/phosphate translocators (GPT), and (d) the xylulose-5-phosphate/phosphate translocator (Eicks et al., 2002). Members of one group share 35% to 50% sequence identity with members of the other groups. The search in the ARAMEMNON DB for proteins similar to the TPT yields six hits (Fig. 3A). The cluster view initiated from the TPT suggests four similarity groups for these proteins, including an additional sequence in one group, a GPT pseudogene (Fig. 3B). Figure 3C shows a tree of the different Arabidopsis PTs that was calculated using ClustalX 1.8 (Thompson et al., 1997) for multiple alignment of the protein sequences (with gaps excluded) and the NJ method for tree building. The groups generated by the ARAMEMNON cluster representation closely resemble the tree calculated by commonly used approaches.
The second family analyzed was the larger family of Arabidopsis ATF1 amino acid permeases (Wipf et al., 2002). Again, the subfamilies found by the ARAMEMNON DB after clustering the amino acid transporters with similarity to the AAP1 amino acid permease (Fig. 4, A and B) closely resemble the branches of the NJ tree (Fig. 4C).
The third protein family analyzed were the AtMGT magnesium transporters (Li et al., 2001). Also in this case, the clusters generated by ARAMEMNON resemble the conventionally constructed NJ tree (data not shown). However, in addition to the published family members MGT1 to MGT10, ARAMEMNON detects another putative protein, At5g09710, with a high degree of similarity to MGT8.
The ARAMEMNON DB was used subsequently to analyze a recently described family of unknown proteins sharing weak homology with the phosphate transporters of the TPT group and that have been named K/VAG transporters (Knappe et al., 2003). By initializing a search with the putative At1g21870 protein, a distant relative to TPT, ARAMEMNON identified 11 putative membrane proteins in the Arabidopsis genome (Fig. 5A). The NJ tree confirmed these subgroups (Fig. 5B). The TM predictions for the K/VAG proteins consistently suggest a similar distribution of TM spans. Figure 5C shows the manually aligned predictions by the Eiconda 0.9 program. In contrast, the subcellular targeting predictions of the different programs were ambiguous for all K/VAG proteins (data not shown). Interestingly, the sequence alignment of the K/VAG transporters revealed that two members of the family, 3g10290 and 1g12500, may contain N-terminal hydrophilic extensions possibly representing transit peptides directing these proteins to plastids or/and to mitochondria. In 3g10290 and 1g12500, the most N-terminal TM span, located behind the putative signal peptide, aligns well with the TM spans at the N termini of eight other paralogs (Fig. 5C). Mitochondrial and plastidic signal sequences usually are hydrophilic and lack predicted TM spans, whereas secretory pathway signals may contain a TM domain. In accordance, the other proteins (except 1g53660) are supposedly located in the plasma membrane or the tonoplast or may be secreted.
DISCUSSION
Aims and Concept of the ARAMEMNON DB
A variety of bioinformatic tools are publicly available for the analysis and interpretation of gene and protein sequence data that were generated during the course of genome sequencing projects. For example, several TM span prediction programs have been published (Table I). However, in the past, TM protein identification frequently relied on a single program, although the predictions greatly differ for many proteins with respect to the number of predicted TM domains (Fig. 1) and their relative location in the polypeptide sequence (Fig. 2B). To compare the predictions generated by these programs for a given protein sequence, it is necessary to submit the sequence successively to each URL. Because the output formats of the programs differ, direct comparison is inconvenient. Similar inconveniences are encountered regarding the comparison of predictions of the subcellular localization of a specific protein or protein family.
A DB was created for Arabidopsis membrane proteins, named ARAMEMNON, that simplifies the identification, classification, and interpretation of membrane protein/gene sequences. The ARAMEMNON DB collects sequence data, predictions of TM regions and subcellular localization, and if available, also bibliographical data from different sources and displays them in an integrated format. For example, the ARAMEMNON DB combines TM predictions derived from seven different TM prediction programs and presents side-by-side the location of TM spans along the polypeptide sequence for each protein in a directly comparable, uniform graphical format. This feature is especially helpful in recognizing cases where predictions by individual programs deviate significantly. An evaluation of TM span prediction programs has indicated that frequently, but not always, the programs TmHMM and HmmTop, which are based on hidden Markov models, perform better than other methods (Möller et al., 2001, 2002). However, it has also been reported that a consensus prediction by using several programs achieves the best reliability (Nilsson et al., 2000).
The individual predictions for Arabidopsis proteins containing TM spans range from approximately 24% (TmHMM 2.0) to 65% (TmPred) of all proteins (Table II). After eliminating false positive reading frames (Table II) and open reading frames that contained a single TM domain at the extreme N terminus coinciding with a secretory pathway signal sequence, as is found in secreted soluble proteins, the ARAMEMNON DB classifies 6,475 proteins or 25% of the proteome as putative membrane proteins (Table II). This frequency is in the same range as had been estimated for several eubacterial, archaean, and eukaryotic organisms (Wallin and von Heijne, 1998; Mitaku et al., 1999; Stevens and Arkin, 2000).
The direct comparison of signal sequence predictions by eight programs implemented in the ARAMEMNON DB (Table III) shows that the prediction of subcelluar targeting is usually more ambiguous than TM span predictions. The predictions must occasionally fail, because some proteins do not have an exclusive destination but are dually targeted to plastids and mitochondria (Peeters and Small, 2001). Therefore, no attempt was made to extrapolate a “consensus” targeting information. Experimental data are presently the only reliable information about protein targeting to subcellular compartments.
The family structure analysis function implemented in the ARAMEMNON DB is superior to the simpler listing of similar paralogs that is also available in the DB. By assembling clusters of related proteins and determining the distance of the cluster to the query sequence, protein family members are detected that are more than 28% similar to at least one other member of the protein family, but not necessarily to the member with that the search was initiated. The clustering method implemented in ARAMEMNON DB is similar to the simple unweighted pair group method, which is less suitable for concise tree building as compared for example with NJ procedures on aligned protein sequences (Huelsenbeck, 1995). However, for several protein families, it was demonstrated that the results generated by the ARAMEMNON DB through clustering are comparable with NJ trees calculated from aligned protein sequences (see Figs. 3–5).
The ARAMEMNON DB will be further developed by incorporating new features that enhance functionality and support. Additional TM predictions will be incorporated in future ARAMEMNON versions (e.g. PSORT II/ALOM2 [Nakai and Kanehisha, 1992], PHDhtm [Rost et al., 1996]). The gene/protein models will be regularly updated, links to publications will be extended, and annotations will be improved.
MATERIALS AND METHODS
Sources of Sequence Information and Sequence Analysis Programs
The complete set of predicted Arabidopsis pseudochromosomes (genomic DNA, mRNA, and protein sequences) was downloaded from TIGR (http://www.tigr.org/tdb/e2k1/ath1/). From NCBI GenBank (http://www.ncbi.nlm.nih.gov), all Arabidopsis protein entries were extracted. Synechocystis sp. sequences were obtained from NCBI and the Kazusa DNA Research Institute (http://www.kazusa.or.jp/cyano/). All predicted Arabidopsis genes were subjected to TM span and subcellular targeting predictions using the programs shown in Tables I and III. All information was translated into a uniform, data-centric XML-vocabulary. XML data were compiled, reorganized, and finally mapped into a relational database.
Similarity Clustering
All Arabidopsis protein sequences were aligned pairwise to each other using the Smith-Waterman algorithm implemented in FASTA 3 (Pearson and Lipman, 1988; Pearson, 1996), yielding a table of pairwise distance values. For clustering, different similarity levels were chosen empirically: The maximal resolution is 70%, i.e. proteins with a higher degree of similarity are not subclustered. A lower threshold of 28% was chosen as the minimal similarity between two proteins to initiate a cluster, and two intermediate levels were chosen at 40% and 50%, respectively.
Different groups with equal similarity levels, that share at least one common protein, merge into a superordinate group (W. Martin, personal communication). Relationships between clusters are determined based on the distance between two clusters, which is defined as the average distance between pairs of sequences from each cluster (Sokal and Michener, 1958).
ACKNOWLEDGMENTS
We thank Prof. William Martin (Heinrich-Heine-University Düsseldorf) for inspiring discussions and Jochen Wiedmann (http://search.cpan.org/author/JWIED/) for providing EP, a flexible flavor of an embedded Perl scripting language.
Footnotes
This work was supported by Kleinwanzlebener Saatzacht AG (Einbeck, Germany), by Südzucker AG (Mannheim, Germany), and by the German Ministery for Education and Research-Genomanalyse in biologischen Systemen program.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.011577.
LITERATURE CITED
- Andre B. An overview of membrane transport proteins in Saccharomyces cerevisiae. Yeast. 1995;11:1575–1611. doi: 10.1002/yea.320111605. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Bannai H, Tamada Y, Maruyama O, Nakai K, Miyano S. Extensive feature detection of N-terminal protein sorting signals. Bioinformatics. 2002;18:298–305. doi: 10.1093/bioinformatics/18.2.298. [DOI] [PubMed] [Google Scholar]
- Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, Feng Z, Gilliland GL, Iype L, Jain S et al. The protein data bank. Acta Crystallogr D Biol Crystallogr. 2002;58:899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- Claros MG, von Heijne G. TopPred II: an improved software for membrane protein structure predictions. Comput Appl Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- Eicks M, Maurino V, Knappe S, Flügge UI, Fischer K. The plastidic pentose phosphate translocator represents a link between the cytosolic and the plastidic pentose phosphate pathways in plants. Plant Physiol. 2002;128:512–522. doi: 10.1104/pp.010576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D, Schwarz E, Komaromy M, Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol. 1984;179:125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, von Heijne G. Prediction of organellar targeting signals. Biochim Biophys Acta. 2001;1541:114–119. doi: 10.1016/s0167-4889(01)00145-8. [DOI] [PubMed] [Google Scholar]
- Frommer WB, Ninnemann O. Heterologous expression of genes in bacterial, fungal, animal, and plant cells. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:419–444. [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- Hirokawa T, Boon-Chieng S, Mitaku S. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- Hofmann K, Stoffel E. TMbase: a database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler. 1993;374:166. [Google Scholar]
- Huelsenbeck JP. The robustness of two phylogenetic methods: Four-taxon simulations reveal a slight superiority of maximum likelihood over neighbor joining. Mol Biol Evol. 1995;12:843–849. doi: 10.1093/oxfordjournals.molbev.a040261. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Arai M, Lao DM, Shimizu T. Transmembrane topology prediction methods: a re-assessment and improvement by a consensus method using a dataset of experimentally characterized transmembrane topologies. In Silico Biol. 2002;2:19–33. [PubMed] [Google Scholar]
- Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S et al. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803: II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- Knappe S, Flügge U-I, Fischer K (2003) Analysis of the plastidic phosphate translocator (PT) gene family in Arabidopsis and identification of new PT-homologous transporters, classified by their putative substrate binding site. Plant Physiol (in press) [DOI] [PMC free article] [PubMed]
- Li L, Tutone AF, Drummond RS, Gardner RC, Luan S. A novel family of magnesium transport genes in Arabidopsis. Plant Cell. 2001;13:2761–2775. doi: 10.1105/tpc.010352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäser P, Thomine S, Schroeder JI, Ward JM, Hirschi K, Sze H, Talke IN, Amtmann A, Maathuis FJ, Sanders D et al. Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 2001;126:1646–1667. doi: 10.1104/pp.126.4.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitaku S, Ono M, Hirokawa T, Boon-Chieng S, Sonoyama M. Proportion of membrane proteins in proteomes of 15 single-cell organisms analyzed by the SOSUI prediction system. Biophys Chem. 1999;82:165–171. doi: 10.1016/s0301-4622(99)00116-7. [DOI] [PubMed] [Google Scholar]
- Möller S, Croning MDR, Apweiler R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 2001;17:646–653. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]
- Möller S, Croning MDR, Apweiler R. Evaluation of methods for the prediction of membrane spanning regions (erratum) Bioinformatics. 2002;18:218. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]
- Nakai K, Kanehisha M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997a;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int J Neural Syst. 1997b;8:581–599. doi: 10.1142/s0129065797000537. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Krogh A. Prediction of signal peptides and signal anchors by a hidden Markov model. Proc Int Conf Intell Syst Mol Biol. 1998;6:122–130. [PubMed] [Google Scholar]
- Nilsson J, Persson B, von Heijne G. Consensus predictions of membrane protein topology. FEBS Lett. 2000;486:267–269. doi: 10.1016/s0014-5793(00)02321-8. [DOI] [PubMed] [Google Scholar]
- Pearson WR. Effective protein sequence comparison. Methods Enzymol. 1996;266:227–258. doi: 10.1016/s0076-6879(96)66017-0. [DOI] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters N, Small I. Dual targeting to mitochondria and chloroplasts. Biochim Biophys Acta. 2001;1541:54–63. doi: 10.1016/s0167-4889(01)00146-x. [DOI] [PubMed] [Google Scholar]
- Persson B, Argos P. Prediction of transmembrane segments in proteins utilising multiple sequence alignments. J Mol Biol. 1994;237:182–192. doi: 10.1006/jmbi.1994.1220. [DOI] [PubMed] [Google Scholar]
- Persson B, Argos P. Topology prediction of membrane proteins. Protein Sci. 1996;5:363–371. doi: 10.1002/pro.5560050221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B, Fariselli P, Casadio R. Topology prediction for helical transmembrane proteins at 86% accuracy. Protein Sci. 1996;5:1704–1718. doi: 10.1002/pro.5560050824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH., Jr A functional-phylogenetic system for the classification of transport proteins. J Cell Biochem. 1999;Suppl:84–94. doi: 10.1002/(sici)1097-4644(1999)75:32+<84::aid-jcb11>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Schein AI, Kissinger JC, Ungar LH. Chloroplast transit peptide prediction: a peek inside the black box. Nucleic Acids Res. 2001;29:E82. doi: 10.1093/nar/29.16.e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal RR, Michener CD. A statistical method for evaluating systematic relationships. Univ Kans Sci Bull. 1958;28:1409–1438. [Google Scholar]
- Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- Stevens TJ, Arkin IT. Do more complex organisms have a greater proportion of membrane proteins in their genomes? Proteins. 2000;39:417–420. doi: 10.1002/(sici)1097-0134(20000601)39:4<417::aid-prot140>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusnady GE, Simon I. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J Mol Biol. 1998;283:489–506. doi: 10.1006/jmbi.1998.2107. [DOI] [PubMed] [Google Scholar]
- Tusnady GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- Van Belle D, Andre B. A genomic view of yeast membrane transporters. Curr Opin Cell Biol. 2001;13:389–398. doi: 10.1016/s0955-0674(00)00226-x. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Membrane protein structure prediction: hydrophobicity analysis and the “positive inside” rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- Wallin E, von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 1998;7:1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Anderson JB, Chen J, Geer LY, He S, Hurwitz DI, Liebert CA, Madej T, Marchler GH, Marchler-Bauer A et al. MMDB: Entrez's 3D-structure database. Nucleic Acids Res. 2002;30:249–252. doi: 10.1093/nar/30.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JM. Identification of novel families of membrane proteins from the model plant Arabidopsis thaliana. Bioinformatics. 2001;17:560–563. doi: 10.1093/bioinformatics/17.6.560. [DOI] [PubMed] [Google Scholar]
- Wipf D, Ludewig U, Tegeder M, Rentsch D, Koch W, Frommer WB. Conservation of amino acid transporters in fungi, plants and animals. Trends Biochem Sci. 2002;27:139–147. doi: 10.1016/s0968-0004(01)02054-0. [DOI] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]