Abstract
Plant resistance to glyphosate has been reported far less frequently than resistance to sulfonylurea and imidazolinone herbicides. However, these studies tend to be anecdotal, without side by side comparisons for a single species or natural isolate. In this study, we tested the frequencies of resistance of three herbicides in a controlled ethylmethanesulfonate (EMS) saturation mutagenesis experiment, allowing a direct comparison of the frequencies at which resistant mutant plants arise. The 100% growth inhibition dose rates of glyphosate, chlorsulfuron (a sulfonylurea herbicide), and imazethapyr (an imidazolinone herbicide) were determined for Arabidopsis. Populations of EMS-mutagenized M2 seedlings were sprayed with twice the 100% growth inhibition dose of glyphosate, chlorsulfuron, or imazethapyr, and herbicide-resistant mutants were identified. Although there were no glyphosate-resistant mutants among M2 progeny of 125,000 Columbia and 125,000 Landsberg erecta M1 lines, chlorsulfuron resistance and imazethapyr resistance each appeared at frequencies of 3.2 × 10−5. Given the observed frequency of herbicide resistance mutations, we calculate that there are at least 700 mutations in each EMS-mutagenized Arabidopsis line and that fewer than 50,000 M1 lines are needed to have a 95% chance of finding a mutation in any given G:C base pair in the genome. As part of this study, two previously unreported Arabidopsis mutations conferring resistance to imidazolinone herbicides, csr1-5 (Ala-122-Thr) and csr1-6 (Ala-205-Val), were discovered. Neither of these mutations caused enhanced resistance to chlorsulfuron in Arabidopsis.
Spontaneous herbicide resistance is generally thought to occur within weed populations as a consequence of the intense selective pressure exerted by a lack of diversity in weed management practices (Gressel and Segel, 1978). Factors such as extended residual soil activity, lack of rotation to other herbicidal modes of action, and specific managerial practices further discriminate between resistant and susceptible individuals within a population (Powles and Holtum, 1994). In addition, the rate and severity at which resistant weed infestations occur is influenced by genetic and ecophysiological determinants such as the mode of inheritance of a given resistance mechanism, the absence or presence of fitness penalties associated with resistance, and the reproductive habit of a given weed species (Gressel and Segel, 1978; Jasieniuk et al., 1996; Gardner et al., 1998). To date, more than 261 herbicide-resistant weed biotypes exist distributed among 52 different countries, involving at least 17 different herbicide modes of action (Heap, 2002). Because application rate and other factors vary greatly in the field, it is difficult to make a direct comparison of the frequencies at which weeds develop resistance to different herbicides. To circumvent this problem, we have used a controlled laboratory setting to compare the frequencies at which heavily mutagenized populations of Arabidopsis develop resistance to the herbicides glyphosate, chlorsulfuron, and imazethapyr.
Glyphosate is a broad-spectrum herbicide that has been used extensively for more than 25 years. The primary mode of action of glyphosate is the inhibition of 5-enolpyruvylshikimate 3-phosphate synthase (EPSPS; EC 2.5.1.19), an enzyme in the shikimate pathway leading to the formation of the aromatic amino acids Tyr, Phe, and Trp (Haslam, 1993; Franz et al., 1997). Although overexpression of the wild-type enzyme confers some resistance to glyphosate (Shah et al., 1986), this is not an acceptable level of tolerance for transgenic crops. Commercial resistance levels have been achieved using glyphosate-resistant EPSPS from Agrobacterium sp. strain CP4, which has low affinity for glyphosate and high catalytic efficiency (Barry et al., 1992; Padgette et al., 1996).
Four classes of herbicides, the sulfonylureas, imidazolinones, triazolopyrimidines, and pyrimidinyl oxybenzoates, inhibit the function of acetolactate synthase (ALS), the first enzyme in the biosynthesis of the branched-chain amino acids Ile, Leu, and Val (Coruzzi and Last, 2000). The ALS enzyme is a tetramer consisting of two catalytic (large) subunits and two regulatory (small) subunits (Lee and Duggleby, 2001). Plant resistance can occur because of reduced herbicide binding caused by mutations in the catalytic subunit (Saari et al., 1994). The DNA sequence changes conferring herbicide-resistant enzymes have been identified in many species, including cotton (Gossypium hirsitum), canola (Brassica napus), tobacco (Nicotiana tabacum), maize (Zea mays), Xanthium strumarium, Arabidopsis, and yeast (Saccharomyces cerevisiae; Chaleff and Ray, 1984; Haughn et al., 1988; Falco et al., 1989; Bernasconi et al., 1995; Hattori et al., 1995; Rajasekaran et al., 1996; Zhu et al., 1999). Resistance mutations tend to fall within five regions of the protein, ranging in size from four to 19 amino acids, that are highly conserved in plants and yeast (Boutsalis et al., 1999). Each of these domains contains at least one amino acid that, when altered, confers resistance to one or more ALS-inhibiting herbicides. Although activity of the regulatory (small) subunit of Arabidopsis ALS has been biochemically determined (Lee and Duggleby, 2001), there are no known plant mutations affecting this protein.
Arabidopsis, unlike many other plants, has only one gene encoding the catalytic subunit of ALS, CSR1. Selection for resistance to the sulfonylurea herbicide chlorsulfuron resulted in the identification of the resistance mutation csr1-1 causing the amino acid change Pro-197-Ser (Haughn et al., 1988). Selection for resistance to the imidazolinone herbicide imazapyr resulted in the identification of the resistance mutation csr1-2 causing the amino acid change Ser-653-Asn (Sathasivan et al., 1990). Intragenic recombination between csr1-1 and csr1-2 produced the novel allele csr1-4, which confers resistance to both sulfonylurea and imidazolinone herbicides (Mourad et al., 1994). Four additional herbicide-resistant mutations of Arabidopsis ALS (Met-124-Gln, Met-124-Ile, Arg-199-Ala, and Arg-199-Gln) have been identified using an in vitro approach (Ott et al., 1996).
Reports of plant resistance to ALS-inhibiting herbicides in the field are far more common than reports of resistance to glyphosate. At least 72 weed species have developed resistance to ALS-inhibiting herbicides (Heap, 2002). In contrast, glyphosate resistance in the field is documented in only four species: rigid ryegrass (Lolium rigidum), Eleusine indica, Lolium multiflorum, and Conyza canadiensis (Powles et al., 1998; Pratley et al., 1999; Tran et al., 1999; Lee and Ngim, 2000; http://www.weedscience.org). While Arabidopsis mutant screens for herbicide-resistant ALS enzymes were successful in several laboratories (Haughn and Somerville, 1990; Sathasivan et al., 1990; Hattori et al., 1992; Mourad et al., 1993), large screens for glyphosate resistance in mutagenized Arabidopsis did not result in any resistant mutants (Haughn and Somerville, 1987; R.L. Last, unpublished data). Although these anecdotal data imply that glyphosate resistance occurs less readily than resistance to ALS-inhibiting herbicides, none of these studies involved side by side selection for resistance to both glyphosate and ALS-inhibiting herbicides under controlled conditions.
In this work, we describe a saturation mutagenesis with EMS and parallel screens in the M2 generation for resistance to twice the 100% growth inhibition (I100) concentration of glyphosate, chlorsulfuron, and imazethapyr. The best previous estimate of the number of Arabidopsis plants needed to obtain a saturation mutagenesis is from Haughn and Somerville (1987). On the basis of data available at that time, it was calculated that a population of 125,000 EMS-mutagenized M1 lines is needed to have a 95% chance of finding a mutation in any given base pair that can be mutated by EMS. Our mutant screen included M2 plants derived from 250,000 EMS-mutagenized M1 lines, 125,000 Arabidopsis ecotype Columbia (Col-0) and 125,000 Arabidopsis ecotype Landsberg erecta (Ler). We estimated the total number of EMS-induced mutations in the population, and we show that this was a saturation mutagenesis. Although mutations conferring resistance to chlorsulfuron and imazethapyr were found, there were no mutations conferring resistance to glyphosate.
RESULTS
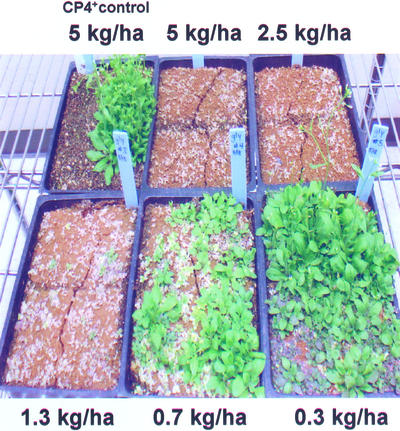
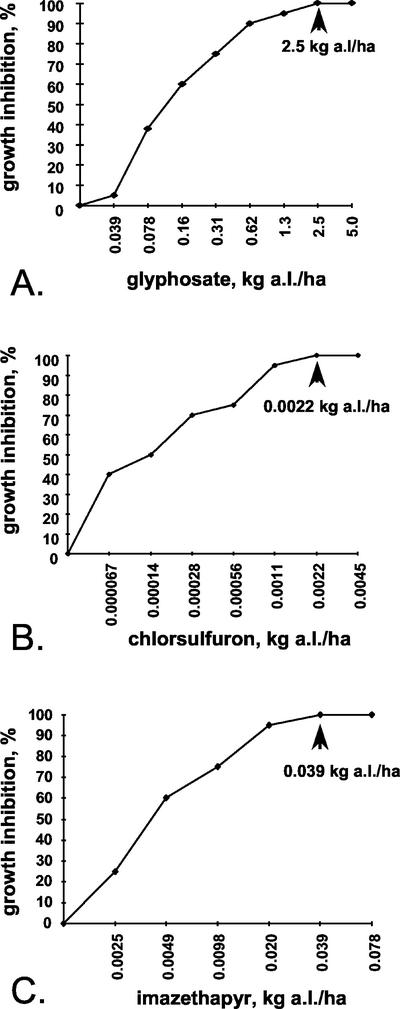
To determine the I100 concentration of glyphosate, chlorsulfuron, and imazethapyr for Arabidopsis, 7-d-old Col-0 seedlings were sprayed with a range of concentrations of each herbicide. In the case of glyphosate, Col-0 plants expressing EPSPS from Agrobacterium sp. strain CP4 were used as a positive control (Fig. 1). The percent lethality was assessed visually (Fig. 1), and the data were plotted on a graph to determine the I100 concentration (Fig. 2). From these experiments, the minimum herbicide concentrations required to kill 100% of the seedlings were determined to be 2.5 kg active ingredient (a.i.) ha−1 for glyphosate, 0.0022 kg a.i. ha−1 for chlorsulfuron, and 0.039 kg a.i. ha−1 for imazethapyr.
Figure 1.
Arabidopsis plants sprayed with 0.07, 0.28, 1.1, 2.2, and 4.5 lb a.i. acre−1 glyphosate to determine the I100 concentration. Resistant control plants are expressing the EPSPS from Agrobacterium sp. strain CP4.
Figure 2.
Sample growth inhibition curves for plants sprayed with glyphosate (A), chlorsulfuron (B), and imazethapyr (C). The I100 concentrations for each herbicide are marked with an arrow.
Herbicide-resistant Col-0 mutants were selected using twice the I100 concentration of glyphosate, chlorsulfuron, and imazethapyr. One million EMS-mutagenized M2 Col-0 seeds were planted in standard nursery flats and were sprayed with glyphosate (5.0 kg a.i. ha−1), chlorsulfuron (0.0045 kg a.i. ha−1), or imazethapyr (0.078 kg a.i. ha−1) 7 d later. Seeds were collected from plants that were herbicide resistant in the initial screen, and resistance was confirmed in the next generation using the same concentration of herbicide. No glyphosate-resistant mutants, one chlorsulfuron-resistant mutant, and five imazethapyr-resistant mutants were confirmed in the next generation. All herbicide-resistant mutants were crossed to wild-type Col-0, and the F1 generation plants were shown to be herbicide resistant, indicating that the mutations were dominant.
To reduce the chance of observing effects that are specific to Col-0, we looked for herbicide-resistant mutants among one million EMS-mutagenized M2 seedlings of Ler, a different isolate of Arabidopsis. Each flat was sprayed with glyphosate (5.0 kg a.i. ha−1), chlorsulfuron (0.0045 kg a.i. ha−1), or imazethapyr (0.078 kg a.i. ha−1) to select for herbicide-resistant mutants. These concentrations are 100% lethal to wild-type Ler seedlings (data not shown). In some cases, more than one herbicide-resistant plant was isolated from a given M2 pool, but they were assumed to be siblings derived from the same M1 mutant plant, and only one isolate was used for further work. All herbicide resistance phenotypes were confirmed by selection with the same concentration of herbicide in the next generation. As in the Col-0 selection, no glyphosate-resistant Ler plants were found, whereas seven independent chlorsulfuron-resistant mutants and three independent imazethapyr-resistant mutants were discovered. All resistant mutants were crossed to wild-type Ler, and the F1 generation plants were shown to be herbicide resistant, indicating that the mutations were dominant.
Mutations conferring resistance to more than one ALS-inhibiting herbicide have been found in plants (Bernasconi et al., 1995; Hattori et al., 1995). To determine whether any of the mutants isolated in this study showed cross-resistance, all chlorsulfuron-resistant mutants were tested for resistance to imazethapyr at twice the I100 concentration, and all imazethapyr-resistant mutants were tested for resistance to chlorsulfuron at twice the I100 concentration. In every case, mutant plants showed resistance only to the herbicide with which they had originally been isolated. In addition, all imazethapyr-resistant mutants were tested for resistance to imazapyr, another imidazolinone herbicide, by spraying each flat with imazapyr (0.052 kg a.i. ha−1). All imazethapyr-resistant mutants were also resistant to imazapyr, an indication that these mutations may provide general resistance to imidazolinone herbicides.
Resistance to sulfonylurea and imidazolinone herbicides in plants is caused by mutations in the catalytic subunit of ALS. This enzyme is encoded by the CSR1 gene in Arabidopsis, and we sequenced this gene from all of the mutant isolates. The DNA sequences of the mutant plants were compared with the respective Col-0 and Ler wild-type sequences. In each case, a 1-bp change was found (Table I). All eight of the chlorsulfuron-resistant mutants had the same nucleotide change, C589T resulting in the amino acid change Pro-197-Ser. This is the csr1-1 mutation, which has been previously described as causing chlorsulfuron resistance in Arabidopsis (Haughn et al., 1988). Two Ler and three Col-0 imidazolinone-resistant mutants had the base pair change G1957Ala (Ser-653-Asn), identical to the csr1-2 mutation found by Sathasivan et al. (1990). Two novel mutations causing imidazolinone resistance were found. One Col-0 mutant had the base pair change G364A (Ala-122-Thr), and we have named this allele csr1-5. One Col-0 mutant and one Ler mutant had the base pair change C614T (Ala-205-Val), and we have named this allele csr1-6.
Table I.
ALS mutations isolated in this study
| Allele | Mutation | Resistance | No. Found | Reference |
|---|---|---|---|---|
| Col-0 csr1-1 | Pro-197-Ser | Chlorsulfuron | 1 | Haughn et al. (1988) |
| Col-0 csr1-2 | Ser-653-Asn | Imazethapyr | 3 | Sathasivan et al. (1990) |
| Col-0 csr1-5 | Ala-122-Thr | Imazethapyr | 1 | This study |
| Col-0 csr1-6 | Ala-205-Val | Imazethapyr | 1 | This study |
| Ler csr1-1 | Pro-197-Ser | Chlorsulfuron | 7 | This study |
| Ler csr1-2 | Ser-653-Asn | Imazethapyr | 2 | This study |
| Ler csr1-6 | Ala-205-Val | Imazethapyr | 1 | This study |
DISCUSSION
We found a total of eight chlorsulfuron-resistant and eight imidazolinone-resistant Arabidopsis isolates in our mutant screens. Both the Col-0 and the Ler populations were derived from approximately 125,000 EMS-mutagenized M1 seeds. The one million M2 Col-0 seeds that we planted for each Col-0 screen represent an 8-fold sampling of seeds collected from 125,000 M1 plants (Lehle Seeds Web page; http://www.Arabidopsis.com). To generate an Ler mutant population, we mutagenized 250,000 seeds (5 g) and had a 50% survival rate in the M1 generation (data not shown). Thus, the Ler M2 seeds that we planted for each mutant screen were also derived from approximately 125,000 M1 plants. Herbicide resistance mutation frequencies, calculated by dividing the number of mutants isolated (Table I) by the number of lines in each M1 population (125,000), are: 4 × 10−5 (imazethapyr, Col-0), 2.4 × 10−5 (imazethapyr, Ler), 8 × 10−6 (chlorsulfuron, Col-0), and 5.6 × 10−5 (chlorsulfuron, Ler). The frequency of imazethapyr resistance was slightly higher in Col-0, and the frequency of chlorsulfuron resistance was 7-fold higher in Ler. These differences could be attributable to variation in the EMS mutagenesis, the spraying protocols, the genetic background of the Col-0 and Ler ecotypes, random variation in the experiment, or a combination of these factors. The total numbers of mutations in the CSR1 gene found in the two populations (six in Col-0 and 10 in Ler; Table I) are not significantly different. We have combined the two data sets in further analysis of mutation frequencies because mean data from two ecotypes are likely to give a better estimate of mutation frequency in Arabidopsis as a species.
As part of sequencing mutations in the Ler background, we also determined the wild-type CSR1 sequence of the Ler isolate of Arabidopsis (gi:22204127), which had not been previously published. There are two predicted amino acid changes in the sequence of the mature protein between Col-0 and Ler: Asn-555-Gln and Val-560-Ile. Not surprisingly, neither of these amino acid changes is in the highly conserved regions of the protein that are thought to be involved in herbicide binding.
Our screen of progeny derived from 250,000 EMS-mutagenized M1 plants is likely to be a saturating mutagenesis. The frequencies of individual mutations, calculated by dividing the number of occurrences (Table I) by 250,000, are: Pro-197-Ser (3.2 × 10−5), Ser-653-Asn (2.0 × 10−5), Ala-122-Thr (4.0 × 10−6), and Ala-205-Val (8.0 × 10−6). The average frequency of the four mutations is 1.6 × 10−5 per M1 line. Given a genome size of 125 Mb and a 35% G:C content in Arabidopsis (Arabidopsis Genome Initiative, 2000), there are 4.4 × 107 bp that are susceptible to EMS mutagenesis, which causes primarily G:C to A:T transitions (Kreig, 1963). Assuming that all G:C base pairs are equally sensitive to EMS, we would expect approximately 700 mutations in each EMS-mutagenized M1 plant (1.6 × 10−5 × 4.4 × 107). The binomial distribution can be used to calculate the probability of finding at least one example of a mutation in any given G:C base pair in our M1 population. P = 1 − (1 − F)N, where P is the probability of finding the mutation, F is the mutation frequency per base pair, and N is the number of lines. Using a mutation frequency of 1.6 × 10−5 and N of 250,000, we calculate a 98% chance of finding at least one mutation in any given G:C base pair in the genome. Given the mutation frequency that we observed, a population of 45,000 M1 plants would be sufficient to have a 95% chance of finding a mutation in any given G:C base pair.
Factors that we cannot accurately assess influence the calculation that there are 700 mutations in each EMS-mutagenized line. A small and perhaps variable number of cells (estimates range from one to four) in an EMS-mutagenized seed contribute to the germline of an M1 plant (Feldman et al., 1994). Because of the uncertain number of effective germline cells in a mature seed, we have calculated mutation frequencies per M1 plant, rather than mutation frequency per M1 haploid genome or frequency per effective germline cell in a mutagenized seed. Some fraction of M1 lines carrying herbicide resistance mutations will not produce seed, for instance because of an independent mutation preventing pollen formation, and would not have been detected in our M2 herbicide resistance selection. Other herbicide resistance mutations may have been lost because we sampled an average of only eight M2 plants derived from each chimeric M1 line. We considered all mutants with the same herbicide resistance arising from the same seed pool to be siblings, even though there may have been more than one independent mutation in a given mutant pool. Overall, it is likely that we have underestimated the true mutation frequency and that there are actually more than 700 mutations in each M1 line.
Even though all of the herbicide resistance mutations that we found are in the CSR1 gene, it is unlikely that this is attributable to a hot spot for EMS mutagenesis. All chlorsulfuron and imidazolinone resistance mutations that have been isolated from other species have also been in the large subunit of ALS (Chaleff and Ray, 1984; Haughn et al., 1988; Falco et al., 1989; Bernasconi et al., 1995; Hattori et al., 1995; Rajasekaran et al., 1996; Zhu et al., 1999). Independent confirmation of the frequency of mutations in our EMS-mutagenized population comes from experiments in which we sequenced a total of 680 kb of DNA from 10 genes related to amino acid biosynthesis in 40 independent M5 EMS-mutagenized lines, with roughly equal numbers from Col-0 and Ler (G. Jander and M. Fraga, unpublished data). The sequenced Arabidopsis genes and their GenBank identifiers are: CSR1 (AT3G48560), anthranilate synthase α-subunit (AT1G19920), Asp kinase-home-Ser dehydrogenase (AT3G01120), cystathionine γ-synthase (AT3G01120), Thr synthase (AT4G29840), Asp kinase-Lys inhibited (AT5G14060), S-adenosyl-Met synthase (AT1G02500), Thr dehydratase/deaminase (AT3G10050), Met methyltransferase (AT5G49810), and dihydrodipicolinate synthase (AT3G60880). We found three differences, all G:C to A:T transitions with no discernable phenotype, between the mutant and wild-type DNA sequences: one each in CSR1, Asp kinase-home-Ser dehydrogenase, and Thr dehydratase/deaminase. This represents a mutation frequency of 1.3 × 10−5 for the 240,000 G:C bp that were sequenced. It is likely that heterozygous mutations, if they were present in our M5 population, would not have been detected by sequencing. Because we were studying individual plant lines rather than populations of herbicide-resistant mutants, it is possible to estimate the number of mutations per effective germline cell in an EMS-mutagenized seed. Assuming 4.4 × 107 G:C bp in the haploid Arabidopsis genome and a loss of 50% of the mutations because of segregation during propagation to the M5 generation, we estimate a frequency of 1,100 mutations in each germline cell of an EMS-mutagenized seed or 825 mutations (one-third homozygous) in an M2 plant. This is somewhat higher than our previous estimate of 700 mutations per genetically mosaic M1 plant (resulting in 130–525 mutations per M2 plant, depending upon the number of effective germline cells in the M1). However, given the approximations used in both of these calculations, the two results are consistent. Colbert et al. (2001) used the targeted induced local lesions in genomes (TILLING) approach to obtain an estimate of up to 1,000 EMS-induced mutations per M2 genome. Extrapolating from the frequency of gene knockout mutations in EMS-mutagenized populations and using less accurate information about the size and GC content of the Arabidopsis genome, Haughn and Somerville (1987) estimated 219 homozygous and heterozygous mutations per M2 plant.
Given the saturating EMS mutagenesis and the repeated isolation of Pro-197-Ser, this might be the only mutation that confers chlorsulfuron resistance in Arabidopsis. However, another likely explanation for the lack of other mutations are the limited base pair changes (G:C to A:T transitions) that are generally produced by EMS mutagenesis. Several mutations that give rise to sulfonylurea herbicide resistance have been reported from other plants, and the high level of amino acid sequence conservation in ALS makes it possible to determine the equivalent Arabidopsis residue. For example, sulfonylurea resistance alleles have been described in canola, tobacco, and X. strumarium that are the equivalent of Trp-574-Leu in Arabidopsis (Lee et al., 1988; Bernasconi et al., 1995; Hattori et al., 1995). The equivalent of Arabidopsis Pro-197-His similarly yielded resistance in Lactuca serriola (Guttieri et al., 1992), and mutations analogous to Arabidopsis Trp-574-Cys and Trp-574-Ser gave resistance in cotton (Rajasekaran et al., 1996). Met-124-Arg was reported to confer chlorsulfuron resistance to Arabidopsis ALS in vitro (Ott et al., 1996). However, it should not be possible for EMS mutagenesis produce any of these amino acid changes in Arabidopsis because of a single-base change. Thus, the known plant alleles other than Pro-197-Ser should not have been found in the present study.
Two of our imazethapyr-resistance mutations (Ala-122-Thr and Ala-205-Val) have not been described previously in Arabidopsis. Neither of these Arabidopsis mutations showed resistance to the sulfonylurea herbicide chlorsulfuron at twice the I100 concentration. The equivalent mutations to Ala-122-Thr in X. strumarium (Bernasconi et al., 1995), maize (Siehl et al., 1996), and sugar beet (Beta vulgaris; Wright and Penner, 1998) also confer only imidazolinone resistance. In contrast, the equivalent changes in yeast ALS (Ala-117-Thr and Ala-200-Val) result in sulfonylurea resistance, but not imidazolinone resistance (Falco et al., 1989). The Ala-183-Val X. strumarium mutation (equivalent to Arabidopsis Ala-205-Val) confers resistance to all four classes of ALS-inhibiting herbicides (Woodworth et al., 1996). It appears that, despite the high level amino acid sequence conservation around the Ala-122 and Ala-205 residues, the sulfonylurea and imidazolinone herbicides have different inhibitory effects on the mutant ALS enzymes in yeast and various plant species.
In addition to the Ala-117-Thr and Ala-200-Val yeast mutations mentioned above, more than 80 other single-base changes in yeast ALS confer resistance to sulfonylurea herbicides (Yadav et al., 1986; Falco et al., 1989; Xie and Jimenez, 1996; Chang and Duggleby, 1998). As with yeast Ala-117-Thr and Ala-200-Val, the equivalent change in the Arabidopsis ALS for several of these mutations could result from single-base pair mutations induced by EMS. However, in a saturation screen for chlorsulfuron resistance in Arabidopsis, we only found the Pro-197-Ser mutation (the equivalent of yeast Pro-192-Ser). It is possible that a larger EMS mutant screen would find additional chlorsulfuron resistance mutations, but it seems more likely that other such mutations found in yeast do not give significant resistance to chlorsulfuron in Arabidopsis.
Despite the limitations of EMS mutagenesis, our results indicate that glyphosate resistance occurs less frequently than resistance to ALS-inhibiting herbicides in Arabidopsis. G:C to A:T transitions induced by EMS constitute one-sixth (17%) of all possible single-base changes. However, because Arabidopsis has a 35% G:C content (Arabidopsis Genome Initiative, 2000), only 12% of all possible mutations in the Arabidopsis genome are G:C to A:T transitions. We found one EMS mutation that gave chlorsulfuron resistance and three different mutations that gave imidazolinone resistance. If one assumes that all single-base changes are equally likely to produce herbicide resistance, then there should be approximately nine possible single-base mutations that give chlorsulfuron resistance and 26 possible single-base changes that give imidazolinone resistance in the Arabidopsis genome. On the other hand, if there were nine or 26 possible single-base mutations that give rise to glyphosate resistance in Arabidopsis, then there would be a 65% chance ([1 − (0.88)9] × 100) or 96% chance ([1 − (0.88)26] × 100), respectively, that at least one of them would be a G:C to A:T transition. If glyphosate resistance mutations were as common as imidazolinone and chlorsulfuron resistance mutations in Arabidopsis, then there is a high probability that we would have found them in our EMS saturation mutagenesis.
These results are the first direct comparison of the frequency of resistance to glyphosate and ALS-inhibiting herbicides in a plant. It is likely that no single-base change induced by EMS can produce glyphosate resistance to Arabidopsis. If two or more mutations are necessary to confer glyphosate resistance, then the frequency of resistant mutants should be much lower than that observed with chlorsulfuron and imazethapyr. A prohibitively large number of mutant plants would be needed to determine the actual frequency of such events. Since the pioneering work of Haughn and Somerville (1987), there has been no published attempt to calculate the number of plants needed for a saturating EMS mutagenesis in Arabidopsis. Here, we have used more complete information about the Arabidopsis genome and sequencing of EMS-induced missense mutations to calculate that there are at least 700 mutations in each M1 line. A relatively small number of lines (< 50,000) need to be screened to have a 95% chance of finding a mutation in any given G:C bp in the genome.
MATERIALS AND METHODS
Herbicides
Glyphosate was produced by the Monsanto Company (St. Louis). Imazethapyr, imazapyr, and chlorsulfuron were purchased from Chem Service (West Chester, PA).
Growth Conditions
Land race Col-0 plants were grown in growth chambers in standard nursery flats (approximately 20 × 40 cm) using Metromix 200 potting soil (Scotts, Marysville, OH) under cool-white fluorescent light at an intensity of 150 to 200 μmol m−2 s−1 PPFD and a 16-h/8-h day/night photoperiod. Land race Ler plants were grown under similar conditions but under continuous light.
Mutant Seed
Two different land races were used in this study. EMS-mutagenized M2 seed of Arabidopsis Ler was produced as described by Redei and Koncz (1992). Five grams of Arabidopsis seed was imbibed overnight in 125 mL of water. Imbibed seeds were exposed to 0.2% (w/v) EMS (Sigma-Aldrich, St. Louis) in 1,250 mL of water on a shaker at room temperature for 16 h. Seeds were rinsed 10 times with 1 L of water, cold treated at 4°C for 3 d in 0.1% (w/v) agar, and sown uniformly on a total of 50 flats, with approximately 5,000 seeds per flat. There was 50% lethality among the mutagenized seeds. M1 plants were grown in a growth chamber under 24 h of continuous light, and progeny M2 seeds were harvested as 50 pools, one pool of 2,500 per flat. This was done to maximize the likelihood that herbicide-resistant mutants would represent independent selective events rather than siblings from a single mutagenic event. EMS-mutagenized M2 seed of Arabidopsis Col-0 was purchased from Lehle Seeds (Round Rock, TX) and were produced in a similar manner. In brief, Col-0 seeds were mutagenized by treatment with 0.13% to 0.25% (w/v) EMS for 12.5 h. M2 seeds were bulked from pools of 600 to 1,600 M1 plants (http://www.Arabidopsis.com).
I100 Determination
Col-0 seeds were planted at a density of approximately 10 seeds cm−2. Seven-day-old Col-0 seedlings were sprayed with active ingredient (glyphosate, imazethapyr, or chlorsulfuron) and surfactant (0.1% [w/v] Tween 20, Sigma-Aldrich) mixture using a pressurized track sprayer equipped with a 8002E TeeJet tip (Spraying Systems Co., Wheaton, IL), a boom height of 41 cm, delivering a volume of 360 L ha−1. A range of active ingredient concentrations was sprayed for each herbicide: glyphosate (0.039, 0.078, 0.16, 0.31, 0.62, 1.3, 2.5, and 5 kg a.i. ha−1), chlorsulfuron (0.000067, 0.00014, 0.00028, 0.00056, 0.0011, 0.0022, and 0.0045 kg a.i. ha−1), and imazethapyr (0.0025, 0.0049, 0.0098, 0.0120, 0.039, and 0.078 kg a.i. ha−1). Sixteen days after treatment, seedling viability was visually scored using digital images of flats with a previously determined kill rate for comparison, and percent lethality was estimated. The minimum herbicide concentration at which 100% of the seedlings were killed was determined to be the I100 concentration.
Selection of Resistant Seedlings
Growth conditions and spray treatments for EMS-mutagenized Col-0 seeds were identical to those used for the I100 determination, except all seedlings were sprayed with twice the I100 concentration of herbicide (glyphosate 5.0 kg a.i. ha−1), chlorsulfuron (0.0045 kg a.i. ha−1), or imazethapyr (0.078 kg a.i. ha−1). One million M2 seeds were planted at a density of 8,000 seeds per flat (125 flats). Plant viability was assessed after 16 d, and seeds were subsequently harvested from the surviving plants. Herbicide resistance was confirmed in the next generation with the same concentration of herbicide used in the selection.
Fifty pools of EMS-mutagenized Ler seeds were planted, with 20,000 seeds from each pool sown into a single flat in 3-fold replication (total of 60,000 seeds per pool) Plants were sprayed by hand using a mist sprayer. Each flat was sprayed evenly with 40 mL of water containing 1,000 mg a.i. L−1 glyphosate, 1 mg a.i. L−1 chlorsulfuron, or 15 mg a.i. L−1 imazethapyr, resulting in the final herbicide concentrations of glyphosate (5.0 kg a.i. ha−1), chlorsulfuron (0.0045 kg a.i. ha−1), and imazethapyr (0.078 kg a.i. ha−1). These herbicide concentrations are equivalent to twice the L100 concentrations determined for Col-0 plants. Seeds from surviving plants were collected, and herbicide resistance was confirmed in the next generation under the same spray conditions as was used in the original selection.
DNA Sequencing
DNA was extracted from leaf samples using a DNeasy 96 Plant Kit (Qiagen USA, Valencia, CA) according to the manufacturer's instructions. The DNA sequence of the coding region of the CSR1 gene and approximately 2,000 bp upstream and 500 bp downstream was amplified by PCR as 24 fragments of approximately 500 bp each, with an average overlap of approximately 150 bp between adjacent fragments. Each primer contained approximately 20 bp of homology to the CSR1 gene sequence and a 5′ tail of M13 universal primers: TGTAAAACGACGGCCAGT for forward primers and CAGGAAACAGCTATGACC for reverse primers. PCR was performed using Taq Gold polymerase (PerkinElmer Life Sciences, Boston), an MJ Tetrad PCR machine (MJ Research, Waltham, MA), and the following amplification protocol: 94°C for 10 min, 45 cycles (94°C for 15 s, 56°C for 15 s, and 72°C for 1 min 30 s), and 72°C for 10 min. M13 universal primers and an ABI3700 sequencing machine (Applied Biosystems, Foster City, CA) were used to determine the sequence of the amplified PCR fragments. The individual PCR sequences were assembled to form the complete gene using the Phred and Phrap software for UNIX (http://www.phrap.com). Differences between the mutant and wild-type sequences were viewed using the Consed program (http://www.phrap.org/consed/consed.html).
Genetic Analysis
To test whether chlorsulfuron- and imazethapyr-resistant mutations were dominant, F1 seeds were generated by pollinating emasculated flowers of wild-type plants with pollen from mutant plants. In every case, the wild-type plants used as the female parent in the cross were of the same land race (Col-0 or Ler) as the mutant plant. F1 seeds were planted and seedlings were sprayed with the same concentration of herbicide (1 mg a.i. L−1 chlorsulfuron or 15 mg a.i. L−1 imazethapyr) as was used in the original screen. Seeds from resistant F1 plants were harvested, and the phenotype was confirmed in the F2 generation.
ACKNOWLEDGMENTS
We thank Gerald M. Dill Jr. for helpful discussions and critical reading of the manuscript and Timothy P. Durrett for technical assistance.
Footnotes
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.010397.
LITERATURE CITED
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Barry G, Kishore G, Padgette S, Taylor M, Kolacz K, Weldon M, Re D, Eichholtz D, Fincher K, Hallas L. Inhibitors of amino acid biosynthesis: strategies for imparting glyphosate tolerance to crop plants. In: Singh BJ, Flores HE, Shannon JC, editors. Current Topics in Plant Physiology. 7: Biosynthesis and Molecular Regulation of Amino Acids in Plants. Rockville, MD: American Society of Plant Physiology; 1992. pp. 139–145. [Google Scholar]
- Bernasconi P, Woodworth AR, Rosen BA, Subraminian MW, Siehl DL. A naturally occurring point mutation confers broad range tolerance to herbicides that target acetolactate synthase. J Biol Chem. 1995;270:17381–17385. doi: 10.1074/jbc.270.29.17381. [DOI] [PubMed] [Google Scholar]
- Boutsalis P, Karotam J, Bowles SB. Molecular basis of resistance to acetolactate synthase-inhibiting herbicides in Sisymbrium orientale and Brassica tournefortii. Pestic Sci. 1999;55:507–516. [Google Scholar]
- Chaleff RS, Ray TB. Herbicide-resistant mutants from tobacco cell cultures. Science. 1984;223:1148–1151. doi: 10.1126/science.223.4641.1148. [DOI] [PubMed] [Google Scholar]
- Chang AK, Duggleby RG. Herbicide-resistant forms of Arabidopsis thaliana acetohydroxyacid synthase: characterization of the catalytic properties and sensitivity to inhibitors of four defined mutants. Biochem J. 1998;333:765–777. doi: 10.1042/bj3330765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbert T, Till BJ, Tompa R, Reynolds S, Steine MN, Yeung AT, McCallum CM, Comai L, Henikoff S. High-throughput screening for induced point mutations. Plant Physiol. 2001;126:480–484. doi: 10.1104/pp.126.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coruzzi G, Last RL. Amino acids. In: Buchanan BB, Gruissem W, Jones RL, editors. Biochemistry and Molecular Biology of Plants. Rockville, MD: American Society of Plant Physiologists; 2000. pp. 358–401. [Google Scholar]
- Falco SC, McDevitt RE, Chui C-F, Hartnett ME, Knowlton S, Mauvais CJ, Smith JK, Mazur BJ. Engineering herbicide-resistant acetolactate synthase. Dev Ind Microbiol. 1989;30:187–194. [Google Scholar]
- Feldman KA, Malmberg RL, Dean C. Mutagenesis in Arabidopsis. In: Meyerowitz EM, Somerville CR, editors. Arabidopsis. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. pp. 137–172. [Google Scholar]
- Franz JE, Mao MK, Sikorski JA. Glyphosate: A Unique Global Herbicide. Washington, DC: American Chemical Society; 1997. [Google Scholar]
- Gardner SN, Gressel J, Mangel M. A revolving dose strategy to delay the evolution of both quantitative vs major monogene resistances to pesticides and drugs. Int J Pest Manag. 1998;44:161–180. [Google Scholar]
- Gressel J, Segel LA. The paucity of plants evolving genetic resistance to herbicides: possible reasons and implications. J Theor Biol. 1978;75:349–371. doi: 10.1016/0022-5193(78)90340-5. [DOI] [PubMed] [Google Scholar]
- Guttieri MJ, Eberlein CV, Mallory-Smith CA, Thill DC, Hoffman DL. DNA sequence variation in domain A of the acetolactate synthase genes of herbicide-resistant and -susceptible weed biotypes. Weed Sci. 1992;40:670–676. [Google Scholar]
- Haslam E. Shikimic Acid: Metabolism and Metabolites. Chichester, UK: John Wiley & Sons; 1993. [Google Scholar]
- Hattori J, Brown D, Mourad G, Labbe H, Ouellet T, Sunohara G, Rutledge R, King J, Miki B. An acetohydroxy acid synthase mutant reveals a single site involved in multiple herbicide resistance. Mol Gen Genet. 1995;246:419–425. doi: 10.1007/BF00290445. [DOI] [PubMed] [Google Scholar]
- Hattori J, Rutledge R, Labbe H, Brown D, Sunohara G, Miki B. Multiple resistance to sulfonylureas and imidazolinones conferred by an acetohydroxyacid synthase gene with separate mutations for selective resistance. Mol Gen Genet. 1992;232:167–173. doi: 10.1007/BF00279993. [DOI] [PubMed] [Google Scholar]
- Haughn GW, Smith J, Mazur B, Somerville CR. Transformation with a mutant Arabidopsis acetolactate synthase gene renders tobacco resistant to sulfonylurea herbicides. Mol Gen Genet. 1988;211:266–271. [Google Scholar]
- Haughn GW, Somerville CR. Selection for herbicide resistance at the whole plant level. In: LeBaron HM, Mumma RO, Honeycutt RC, Duesing JH, editors. Applications of Biotechnology to Agricultural Chemistry. Washington DC: American Chemical Society; 1987. pp. 98–108. [Google Scholar]
- Haughn GW, Somerville CR. A mutation causing imidazolinone resistance maps to the Csr1 locus of Arabidopsis thaliana. Plant Physiol. 1990;92:1081–1085. doi: 10.1104/pp.92.4.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heap I (2002) The International Survey of Herbicide Resistant Weeds. Weed Science. www.weedscience.com
- Jasieniuk M, Brule-Babel AL, Morrison IN. The evolution and genetics of herbicide resistance in weeds. Weed Sci. 1996;44:176–193. [Google Scholar]
- Kreig D. Ethyl methanesulfonate-induced reversion of bacteriophage T4r II mutants. Genetics. 1963;48:561–580. doi: 10.1093/genetics/48.4.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YT, Duggleby RG. Identification of the regulatory subunit of Arabidopsis thaliana acetohydroxyacid synthase and reconstitution with its catalytic subunit. Biochemistry. 2001;40:6836–6844. doi: 10.1021/bi002775q. [DOI] [PubMed] [Google Scholar]
- Lee KY, Townsend J, Teppetman J, Black M, Chui CF, Mazur B, Dunsmuir P, Bedbrook J. The molecular basis of sulfonylurea herbicide resistance in tobacco. EMBO J. 1988;7:1241–1248. doi: 10.1002/j.1460-2075.1988.tb02937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LJ, Ngim J. A first report of glyphosate-resistant goosegrass (Eleusine indica (L) Gaertn) in Malaysia. Pest Manag Sci. 2000;56:336–339. [Google Scholar]
- Mourad G, Haughn GW, King J. Intragenic recombination in the CSR1 locus of Arabidopsis. Mol Gen Genet. 1994;243:178–184. doi: 10.1007/BF00280315. [DOI] [PubMed] [Google Scholar]
- Mourad G, Pandey B, King J. Isolation and genetic analysis of a triazolopyrimidine-resistant mutant of Arabidopsis. J Hered. 1993;84:91–96. [Google Scholar]
- Ott K-H, Kwagh J-G, Stockton GW, Sidorov V, Kakefuda G. Rational molecular design and engineering of herbicide resistant crops by structure modeling and site-directed mutagenesis of acetohydroxyacid synthase. J Mol Biol. 1996;263:359–368. doi: 10.1006/jmbi.1996.0580. [DOI] [PubMed] [Google Scholar]
- Padgette SR, Re DB, Barry GF, Eichholtz DE, Delannay X, Fuchs RL, Kishore GM, Fraley RT. New weed control opportunities: development of soybeans with a Roundup Ready gene. In: Duke SO, editor. Herbicide-Resistant Crops: Agricultural, Economic, Environmental, Regulatory and Technological Aspects. Boca Raton, FL: CRC Press; 1996. pp. 53–84. [Google Scholar]
- Powles SB, Holtum JAM. Herbicide Resistance in Plants: Biology and Biochemistry. Boca Raton, FL: Lewis Publishers; 1994. [Google Scholar]
- Powles SB, Lorraine-Colwill DF, Dellow JJ, Preston C. Evolved resistance to glyphosate in rigid ryegrass (Lolium rigidum) in Australia. Weed Sci. 1998;46:604–607. [Google Scholar]
- Pratley JE, Urwin NAR, Stanton RA, Baines PR, Broster JC, Cullis K, Schafer DE, Bohn JA, Krueger RW. Resistance to glyphosate in Lolium rigidum: I. Bioevaluation Weed Sci. 1999;47:405–411. [Google Scholar]
- Rajasekaran K, Grula JW, Anderson DM. Selection and characterization of mutant cotton (Gossypium hirsutum L.) cell lines resistant to sulfonylurea and imidazolinone herbicides. Plant Sci. 1996;119:115–124. [Google Scholar]
- Redei GP, Koncz C. Classical Mutagenesis. In: Koncz C, Chua N-H, Schell J, editors. Methods in Arabidopsis Research. Singapore: World Scientific; 1992. pp. 16–82. [Google Scholar]
- Saari LL, Coterman JC, Thill DC. Resistance to acetolactate synthase inhibiting herbicides. In: Powles S, Holtum J, editors. Herbicide Resistance in Plants: Biology and Biochemistry. Boca Raton, FL: Lewis Publishers; 1994. pp. 83–139. [Google Scholar]
- Sathasivan K, Haughn GW, Murai N. Nucleotide sequence of a mutant acetolactate synthase gene from an imidazolinone-resistant Arabidopsis thaliana var. Columbia Nucleic Acids Res. 1990;18:2188. doi: 10.1093/nar/18.8.2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah D, Horsch R, Klee H, Kishore GM, Winter JA, Tumer NE, Hironaka CM, Sanders PR, Gasser CS, Aykent S et al. Engineering herbicide tolerance in transgenic plants. Science. 1986;233:478–481. doi: 10.1126/science.233.4762.478. [DOI] [PubMed] [Google Scholar]
- Siehl DL, Bengston AS, Brockman JP, Butler JH, Kraatz GW, Lamoreaus RJ, Subramanian MV. Patterns of cross-tolerance to herbicides inhibiting acetohydroxyacid synthase in commercial corn hybrids designed for tolerance to imidazolinones. Crop Sci. 1996;36:274–278. [Google Scholar]
- Tran M, Baerson S, Brinker R, Casagrande L, Faletti M, Feng Y, Nemeth M, Reynolds T, Rodriguez D, Schafer D et al. Proceedings 1(B) of the 17th Asian-Pacific Weed Society Conference. The Asian-Pacific Weed Science Society, Los Banos, Philippines. 1999. Characterization of glyphosate resistant Eleusine indica biotypes from Malaysia; pp. 527–536. [Google Scholar]
- Woodworth AR, Bernasconi P, Subraminian MV, Rosen BA. A second naturally occurring point mutation confers broad base tolerance to acetolactate synthase inhibitors. Plant Physiol. 1996;111:415. [Google Scholar]
- Wright TR, Penner D. Cell selection and inheritance of imidazolinone resistance in sugarbeet (Beta vulgaris) Theor Appl Genet. 1998;96:612–620. [Google Scholar]
- Yadav N, McDevitt RE, Benard S, Falco SC. Single amino acid substitutions in the enzyme acetolactate synthase confer resistance to the herbicide sulfometuron methyl. Proc Natl Acad Sci USA. 1986;83:4418–4422. doi: 10.1073/pnas.83.12.4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Jimenez A. Molecular cloning of a novel allele of SMR1 which determines sulfometuron methyl resistance in Saccharomyces cerevisiae. FEMS Microbiol Lett. 1996;137:165–168. doi: 10.1111/j.1574-6968.1996.tb08100.x. [DOI] [PubMed] [Google Scholar]
- Zhu T, Peterson DJ, Tagliani L, St. Clair G, Baszczynski CL, Bowen B. Targeted manipulation of maize genes in vivo using chimeric RNA/DNA oligonucleotides. Proc Natl Acad Sci USA. 1999;96:8768–8773. doi: 10.1073/pnas.96.15.8768. [DOI] [PMC free article] [PubMed] [Google Scholar]