Abstract
Cytokinin plays a critical role in plant growth and development by stimulating cell division and cell differentiation. Despite many years' research efforts, our current understanding of this hormone is still limited regarding both its biosynthesis and signaling. To genetically dissect the cytokinin pathway, we have used a functional screen to identify Arabidopsis gain-of-function mutations that enable shoot formation in the absence of exogenous cytokinins. By using a chemical-inducible activation tagging system, we have identified over 40 putative mutants, designated as pga (plant growth activators), which presumably were affected in key components of cytokinin biosynthesis and signaling pathway. Here, we report a detailed characterization of pga22, a representative mutant from this collection. A gain-of-function mutation in the PGA22 locus resulted in typical cytokinin responses. Molecular and genetic analyses indicated that PGA22 encodes an isopentenyl transferase (IPT) previously identified as AtIPT8. Plants of the pga22 mutant accumulated at remarkably higher levels of isopentenyladenosine-5′-monophosphate and isopentenyladenosine when analyzed by mass spectrometry, suggesting that AtIPT8/PGA22 is a functional IPT that may direct the biosynthesis of cytokinins in planta via an isopentenyladenosine-5′-monophosphate-dependent pathway.
Cytokinin plays an important role in many aspects of plant growth and development, such as regulating shoot and root growth, and controlling apical dominance and leaf senescence as well as flowering time. At the cellular level, it is generally believed that cytokinin executes its function by stimulating cell division and cell differentiation (Davies, 1995). Despite its critical role in plant growth and development, cytokinin is the least understood hormone among the so-called classical plant phytohormones with respect to its biosynthesis and signaling (Abel et al., 2000; Ross and O'Neill, 2001). Significant efforts have been made to elucidate the molecular and cellular mechanisms of cytokinin actions. For instance, extensive genetic screens, which have been carried out under conditions of high concentrations of exogenous cytokinins, have not yielded any mutations that are affected mainly in the cytokinin pathway. These results were presumably related to cross talk among different pathways, particularly between the ethylene and cytokinin pathways, or to the fact that cytokinin also partially evokes ethylene responses.
During the last several years, considerable progress has been made in efforts to elucidate the molecular mechanism of cytokinin signaling (for review, see D'Agostino and Kieber, 1999; Mok and Mok, 2001; Haberer and Kieber, 2002; Sheen, 2002). A major breakthrough was the use of an in vitro shoot formation bioassay to identify key components in cytokinin signal transduction (Kakimoto, 1996). A high cytokinin to auxin ratio has been shown to promote shoot formation from explants of certain species (Skoog and Miller, 1957; Sugiyama, 1999). In principle, an overactive key regulator may evoke cytokinin responses, thus leading to shoot regeneration in the absence of externally supplied cytokinins. This working hypothesis led to the identification of the Arabidopsis CKI1 (Cytokinin Independent 1) gene, which encodes a putative receptor-like His kinase (Kakimoto, 1996). Conversely, loss-of-function mutations in similar loci may render mutant plants or explants insensitive to exogenous cytokinins. By screening such cytokinin-insensitive mutants using the shoot formation assay, Kakimoto and colleagues identified the Arabidopsis cre1 (cytokinin receptor 1) mutant, which showed an attenuated response to the hormone (Inoue et al., 2001). Moreover, CRE1 is capable of binding to the phytohormone in yeast (Saccharomyces cerevisiae; Inoue et al., 2001) and bacterial (Suzuki et al., 2001; Yamada et al., 2001) cell-based bioassays. Thus, these results provided direct evidence that CRE1 is a bona fide cytokinin receptor.
Recent studies revealed that the cytokinin signal transduction pathway probably includes two branches: the hormone-dependent CRE1 branch and the hormone-independent CKI1 branch (Hwang and Sheen, 2001). In either case, the pathway or branch is presumably activated through a series of His-to-Asp phosphorelays, which are known to include several His-containing phosphotransfer factors and a large number of the so-called “response regulators” (Brandstatter and Kieber, 1998; Imamura et al., 1998). The Arabidopsis genome contains five His-containing phosphotransfer factors (AHP) and 22 response regulators (ARR; for review, see D'Agostino and Kieber, 1999; Schaller, 2000). Similar to Arabidopsis His kinases (AHKs), all AHPs and ARRs contain a highly conserved His residue in the kinase domain and an Asp residue in the receiver domain. It appears that an upstream signal is transmitted to and amplified by sequential transferring of phosphoryl groups between these conserved His and Asp residues located in AHKs, AHPs, and ARRs, leading to altered gene expression and eventually global physiological responses (Sakai et al., 2000, 2001; Hwang and Sheen, 2001; Suzuki et al., 2002; for review, see Haberer and Kieber, 2002; Sheen, 2002). However, little detail is known about this phosphorelay during cytokinin signaling, and other critical components of the pathway still remain to be identified.
Our current knowledge on cytokinin biosynthesis in plants is largely deduced from studies on a possibly analogous system in Agrobacterium tumefaciens. Cells of A. tumefaciens are able to infect certain plant species by inducing tumor formation in host plant tissues (Van Montagu and Schell, 1982; Hansen and Chilton, 1999). To do so, these A. tumefaciens cells synthesize and secrete cytokinins, which mediate the transformation of normal host plant tissues into tumors or calli. This process is facilitated by the A. tumefaciens tumor-inducing plasmid, which contains genes encoding the necessary enzymes and regulators for cytokinin biosynthesis (for review, see Van Montagu and Schell, 1982; Saito et al., 1992; Hansen and Chilton, 1999). Biochemical and genetic studies revealed that Gene 4 of the tumor-inducing plasmid encodes an isopentenyl transferase (IPT), which converts AMP and dimethylallyl-diphosphate (DMAPP) into isopentenyladenosine-5′-monophosphate (iPMP), the active form of cytokinins (Akiyoshi et al., 1984; Barry et al., 1984). Overexpression of the ipt gene in a variety of transgenic plants has been shown to cause an increased level of cytokinins and elicit typical cytokinin responses in the host plants (Hansen and Chilton, 1999). Therefore, it has been postulated that plant cells use machinery similar to that of A. tumefaciens cells for cytokinin biosynthesis. Although ipt-like enzymatic activity has been detected in crude extracts of various plant tissues (e.g. Horgan, 1975; Chen and Melitz, 1979), homologous genes from Arabidopsis were only identified recently by a bioinformatic approach (Kakimoto, 2001; Takei et al., 2001). These Arabidopsis homologs were designated as AtIPT1 through AtIPT9 (Kakimoto, 2001). With the exception of AtIPT2 and AtIPT9, recombinant proteins of the other seven AtIPT genes were able to catalyze the production of active cytokinins in Escherichia coli cells (Takei et al., 2001). Moreover, overexpression of AtIPT4 in transgenic Arabidopsis plants elevated cytokinin levels and elicited typical cytokinin responses in planta and under tissue culture conditions (Kakimoto, 2001). These phenotypes are similar to those observed with overexpression of the A. tumefaciens ipt gene.
By using genetic approaches, two mutations that affect cytokinin biosynthesis were recently identified from petunia (Petunia hybrida; Zubko et al., 2002) and Arabidopsis (Catterou et al., 2002). The petunia mutant sho (shooting), identified by a 35S enhancer tag, displayed the shooty phenotype characteristic of cytokinin responses. The sho mutant phenotype was found to be caused by increased levels of isopentenyladenosine (iPA) and derivatives in mutant plants (Zubko et al., 2002). Subsequent molecular analysis indicated that SHO encodes an ipt-like enzyme (Zubko et al., 2002). The Arabidopsis mutant hoc (high organogenic capacity) showed a phenotype similar to that of sho (Catterou et al., 2002). However, hoc appears to be a recessive mutation, whose wild-type (WT) allele, yet to be identified, may negatively regulate the cytokinin biosynthetic pathway.
To dissect the cytokinin signal transduction pathway, we have carried out a functional screening to identify key components of the hormone action. The screen, in principle, was similar to that developed by Kakimoto (1996), who identified the cki1 gain-of-function mutation. An improvement introduced by us was the use of a chemical-inducible promoter/enhancer (Zuo et al., 2000a; Zuo et al., 2002) rather than a constitutive enhancer; therefore, mutants that displayed severely abnormal plant growth and development or lethality could be recovered. In addition to the shooty phenotype, i.e. cytokinin-independent shoot formation, we also extend our screening criteria to include mutations that were capable of promoting embryonic callus formation. As reported previously, we collectively designated these two classes of mutants as pga (plant growth activators; Zuo et al., 2002). Among the pga mutants with a shooty phenotype, we expect that a mutation would affect either cytokinin biosynthesis or transduction of the hormonal signal. Here, we present genetic and molecular evidence showing that PGA22 may represent a functional IPT. Overexpression of PGA22 caused a massive increase in cytokinin levels in mutant plants, thus evoking typical cytokinin responses.
RESULTS
Identification of the plant growth activator 22(pga22) Mutant
To identify gain-of-function mutations related to cytokinin signaling and somatic embryogenesis, we carried out a functional screen by using a chemical-inducible activation-tagging vector (the LexA-VP16-estrogen receptor vector pER16; see Zuo et al., 2000a, 2002). In this screen, A. tumefaciens ABI cells carrying pER16 were used to transform Arabidopsis (the Wassilewskija [Ws] ecotype) root explants according to Koncz et al. (1989). Subsequently, the A. tumefaciens-infected root explants were cultured on the screening medium (SCM; containing auxin indole-3-acetic acid, kanamycin, and 17-β-estradiol without cytokinins). Under our screening conditions, WT explants were not able to generate shoots or embryogenic calli. However, a gain-of-function mutation may cause the activation of the cytokinin signal transduction pathway, thus producing green calli or shoots; alternatively, a gain-of-function mutation may promote a vegetative-to-embryogenic transition, thus leading to the formation of somatic embryos. In a large-scale screen, we have identified over 40 putative mutants, which displayed two distinctive phenotypes characterized as “shooty” or “embryogenic.” We collectively named these two classes of mutants as plant growth activators (see also Zuo et al., 2002). Results on the embryogenic mutants, represented by two mutant alleles of the PGA6/WUSCHEL (WUS) gene, have been published previously (Zuo et al., 2002). Here we report the characterization of pga22, a typical shooty mutant.
When cultured on the screen medium, WT root explants did not appear to undergo apparent cell divisions. Similar to most of the pga mutants, pga22 was identified by its capability to form green calli and subsequently develop into shoots on the SCM. Following standard tissue culture manipulations (for details, see Zuo et al., 2002), pga22 seeds were harvested from the mutant plants after being transferred to soil. To confirm the phenotype observed in the primary screening, putative pga22 seeds were germinated on Murashige and Skoog medium (Murashige and Skoog, 1962), and roots or leaves excised from the mutant seedlings were cultured on a 2,4-dichlorophenoxyacetic acid-containing medium to induce callus formation. The callus was subsequently transferred onto the SCM as described before. In the absence of the inducer (SCM without 17-β-estradiol), neither WT nor pga22 root explants showed any apparent cell division (Fig. 1A). When cultured on the SCM in the presence of the inducer, however, pga22 root explants were able to develop rapidly dividing green calli (Fig. 1A), which subsequently differentiated into shoots (Fig. 1B). These shoots were indistinguishable from those generated from WT root explants cultured on a standard shoot regeneration medium containing both auxin and cytokinin. Upon removal of the inducer, these shoots were able to grow and develop into morphologically normal plants that were fertile and set seeds (Fig. 1C). These observations indicated that an inducer-dependent gain-of-function mutation in the PGA22 locus was sufficient to activate the organogenesis pathway.
Figure 1.
Cytokinin-independent shoot formation of pga22 explants. A, Root explants derived from pga22 seedlings were cultured on the noninductive SCM (without cytokinins and 17-β-estradiol; left) or on the inductive SCM (containing 17-β-estradiol but without cytokinins; right) for 20 d. B, Shoots regenerated from pga22 root explants cultured on the inductive SCM were transferred onto a Murashige and Skoog medium (35 d). C, A 45-d-old plantlet derived from shoots shown in B was transferred into soil. Bar = 1 cm.
The pga22 Gain-of-Function Mutant Phenotype
To assay PGA22 functions during plant growth and development, pga22 seeds were germinated on Murashige and Skoog medium with or without the inducer. When germinated on the noninductive medium, no apparent growth and development abnormality was observed (Fig. 2A), suggesting that mutations in the pga22 genome did not appear to affect the normal function of the gene. On the inductive medium, however, pga22 mutant plants showed severe morphological abnormality. The mutant roots were extremely short, the cotyledons remained pale yellow after germination, and true leaf initiation was rarely observed (Fig. 2A). Shortly after germination, plant growth and development were completely arrested and mutant plants eventually died. Note that the above-described phenotype was routinely observed when pga22 mutant seeds were germinated in the presence of 17-β-estradiol in concentrations higher than 0.2 μm.
Figure 2.
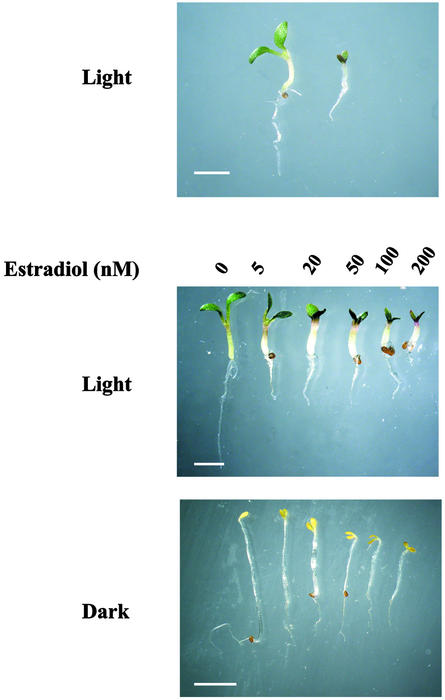
The pga22 gain-of-function mutant phenotype. Homozygous T2 pga22 seeds were germinated in the presence or absence of 17-β-estradiol in the light or dark as indicated. Photographs were taken 6 d after germination. A, Germinated in the absence (left) or presence of 10 μm 17-β-estradiol (right). B and C, Germinated in the absence (far left) or presence of different concentrations of 17-β-estradiol in the light (B) or dark (C), respectively, as indicated. Bar = 5 mm.
In studies on pga6 and other pga mutants, we have observed that the penetration of a mutant phenotype was strictly dependent on the inducer dosage (Zuo et al., 2002). As highlighted above, this dosage dependency, however, was not apparent in pga22, within the range of inducer concentrations routinely used (0.2–10 μm). To further explore the effects of the pga22 gain-of-function mutation on plant growth and development, we carried out a titration experiment with inducer concentrations lower than routinely used. Under such test conditions, the pga22 mutant showed typical cytokinin responses, including shorter roots, enlarged hypocotyls, and dark-green cotyledons (Fig. 2B). When germinated in the dark, pga22 seedlings displayed a characteristic de-etiolated phenotype, including shorter hypocotyls and shorter roots, opened cotyledons, and the absence of apical hooks (Fig. 2C). The strength of phenotype was strictly dependent on inducer concentrations in both light and dark conditions. These observations suggested that a gain-of-function mutation in the PGA22 locus was able to evoke cytokinin responses, and that PGA22 is probably a key component in the cytokinin biosynthesis or signaling pathway. In addition, these data also suggested that the PGA22 gene was tagged by the OLexA-46 promoter of the pER16 vector.
PGA22 Encodes an IPT
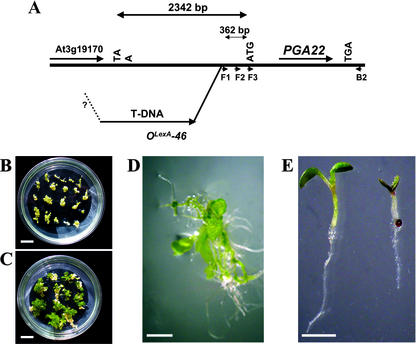
Genetic analysis showed that the 17-β-estradiol-dependent mutant phenotype and WT phenotype segregated in a ratio of 3:1, indicating that the mutation was dominant in a single genetic locus. Segregation analysis also suggested that the mutant genome contained a single T-DNA insertion. Because the pga22 gain-of-function phenotype was strictly dependent on the inducer, the PGA22 gene should be tagged by the OLexA-46 promoter of pER16 inserted in the mutant genome. Taking advantage of this, we cloned the OLexA-46 promoter-tagged mutant genomic sequences by thermal asymmetric interlaced-PCR (Liu et al., 1995). Sequence analysis indicated that the T-DNA inserted 362 bp upstream from the putative translation start codon of a putative open reading frame (ORF; Fig. 3A).
Figure 3.
The pga22 mutant genome and genetic confirmation of the PGA22 gene. A, Schematic diagram showing the insertion site of the T-DNA upstream from the AtIPT8/PGA22 gene (not in scale). Arrows indicate the directions of transcription. The insertion site of the right border (RB) was unclear (question mark). F1 through F3 and B2, Positions and orientations of primers used for PCR-amplification of the AtIPT8/PGA22 gene (see text for more details). B, WT root explants were transformed with pER10-AtIPT8 (the genomic DNA fragment spanned from the F2 to B2 region as shown in A) and cultured on the SCM without 17-β-estradiol for 15 d. C, WT root explants were transformed with pER10-AtIPT8 (the genomic DNA fragment spanned from the F2 to B2 region as shown in A) and cultured on the SCM containing 10 μm 17-β-estradiol for 25 d. D, A 40-d-old pER10-AtIPT8 shoot grown on Murashige and Skoog medium. E, T1 seeds of pER10-AtIPT8 transgenic plants were germinated in the absence (left) and presence of 10 μm 17-β-estradiol. Figure shows 7-d-old seedlings. Bar = 2 cm (B and C); bar = 1 cm (D and E).
To verify if this ORF represents the PGA22 gene, a genomic DNA fragment spanning the entire ORF and part of flanking 5′-untranslated region and 3′-untranslated region was cloned into pER10, a 17-β-estradiol-inducible expression vector (Zuo et al., 2000a, 2002). The resulting construct was used to transform root explants derived from WT plants. The transformed root explants were placed on the SCM or a control medium without cytokinins and 17-β-estradiol as described before. The explants formed neither green calli nor shoots on the control medium (Fig. 3B). In contrast, on the SCM, green callus formation could be observed after 10 to 15 d (Fig. 3C), and shoots were generated after 3 to 4 weeks (Fig. 3D). Similar results were obtained using a genomic fragment containing additional upstream sequences of the gene up to the T-DNA insertion site (see Fig. 3A), with a slightly lower regeneration efficiency (data not shown). All of the above-mentioned pga22 gain-of-function phenotypes were observed in the pER10-ORF1 transgenic plants in an inducer-dependent fashion (Fig. 3E). Taken together, these data demonstrated that the OLexA-46 promoter-tagged ORF represented the PGA22 gene.
The PGA22 gene, interrupted by an intron of 252 bp, encodes a polypeptide of 329 amino acids, with an estimated molecular mass of 37.2 kD and a pI of 8.03. A database search revealed that PGA22 was identical to the previously identified AtIPT8 (Arabidopsis IPT; GenBank accession nos. BAB02956 [Takei et al., 2001] and BAB59034 [Kakimoto, 2001]). AtIPT8 and several other IPT-like genes were identified by an in silica approach based on partial homology of these Arabidopsis genes with the A. tumefaciens ipt. Detailed annotation and phylogenetic studies on these AtIPT genes have been reported by Takei et al. (2001) and Kakimoto (2001). Hereafter, we will refer to the PGA22 gene/protein as AtIPT8 and the mutant/mutation as pga22.
AtIPT8 Is Mainly Expressed in Roots
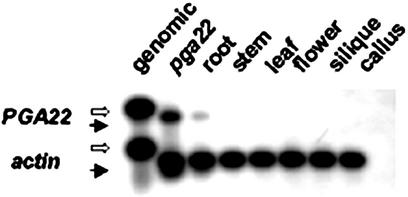
To better understand its function, we analyzed the AtIPT8 expression pattern in WT plants. Although the AtIPT8 expression was easily detected in pga22 treated with various concentrations of the 17-β-estradiol inducer, we were unable to detect its expression in WT plants by northern-blot analysis (see below). Therefore, we used reverse transcription-PCR (RT-PCR) followed by Southern-blot analysis to assess AtIPT8 expression in WT plants. Figure 4 shows that AtIPT8 could only be detected in roots under our assay conditions, suggesting that AtIPT8 is likely involved in cytokinin biosynthesis in roots.
Figure 4.
Expression pattern of the AtIPT8/PGA22 gene. Five microliters of RT-PCR mixtures (see “Materials and Methods” for details) was separated by a 1.2% (w/v) agarose gel and then blotted onto a nylon membrane. The membrane was hybridized with an AtIPT8 (a genomic DNA fragment spanned the F3 and B2 region; see Fig. 3) probe and an actin 3 DNA probe. Fifty nanograms of Arabidopsis genomic DNA was used in the PCR to serve as a control (the “genomic” lane). RNA prepared from pga22 seedlings (T3 homozygous, 3 weeks old) treated with 5 μm 17-β-estradiol for 12 h was used for RT, and the PCR products of the primer pair F3/B2 were diluted 50 times before loading (the “pga22 ” lane). Other lanes: RNA prepared from various tissues/organs of WT plants was used for RT. Solid and white arrows indicate cDNA and genomic fragments, respectively.
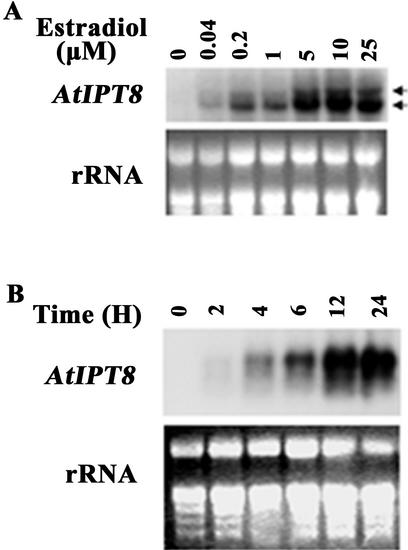
Consistent with the inducer dosage-dependent mutant phenotype (see Fig. 2), AtIPT8 expression was strictly dependent on the inducer concentration (Fig. 5A) as well as the time of induction (Fig. 5B). Similar to those observed in the LexA-VP16-estrogen receptor-green fluorescent protein transgenic lines (Zuo et al., 2000a), the ATIPT8 induction was saturated at 5 μm 17-β-estradiol and an incubation time of 12 to 24 h (see Fig. 5, A and B). We have reported previously that in the two alleles of pga6, the inserted OLexA-46 DNA segment could act as a functional promoter as well as an enhancer under our induction conditions (Zuo et al., 2002). Similarly, the two distinctive transcripts detected by the AtIPT8 probe (indicated by arrows in Fig. 5A) presumably represented those initiated from the OLexA-46 promoter (the longer transcript) and the native AtIPT8 promoter (the shorter transcript).
Figure 5.
Inducible expression of AtIPT8 in pga22 mutant plants. A, Dosage dependency of the AtIPT8 induction. Three-week-old pga22 seedlings were treated for 16 h with varying concentrations of 17-β-estradiol (indicated on the top). Ten micrograms of RNA prepared from the treated seedlings was used for northern-blot analysis by using AtIPT8 as a probe. Two transcripts with distinctive sizes were observed (indicated by arrows at the right side), presumably representing transcription initiation from two different sites (see text for detail). B, Time course of the AtIPT8 induction. Three-week-old pga22 seedlings were treated with 10 μm 17-β-estradiol for different time as indicated on the top of the photograph. See A for other technical details. Note that the two transcripts of slightly different sizes could be observed with a shorter exposure.
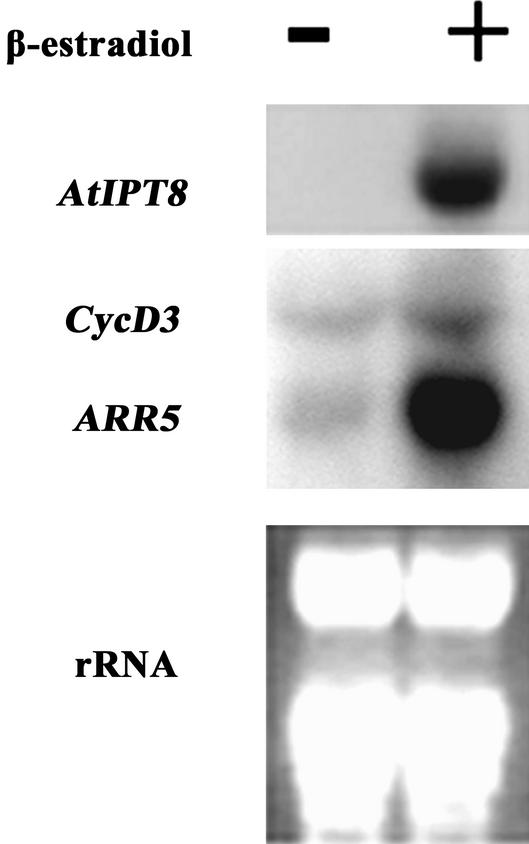
The pga22 Gain-of-Function Mutation Induces Expression of Type A ARR Genes
The cytokinin signal is believed to be transduced via a His-Asp phosphorelay pathway involving the cytokinin receptors CRE1 (Inoue et al., 2001) and, possibly, CKI1 (Kakimoto, 1996), and a series of conserved His kinases including AHPs and ARRs (for review, see Schaller, 2000; Haberer and Kieber, 2002; Sheen, 2002). One of the physiological consequences of the cytokinin action is to stimulate cell division (Davies, 1995), presumably mediated by CycD3, a D-type cyclin (Riou-Khamlichi et al., 1999). Although the precise functions of these proteins are not well understood, expression of type A ARRs has been shown to be induced by cytokinin applications (Brandstatter and Kieber, 1998; Imamura et al., 1998). To further characterize the pga22 mutant phenotype at the molecular level, we analyzed expression of several above-mentioned cytokinin marker genes. As shown in Figure 6, expression of ARR5, a type A ARR gene, was strongly induced in pga22 mutant plants upon induction of AtIPT8. Identical results were obtained with the expression of ARR6, another type A ARR gene. In contrast, expression of ARR1, a type B ARR gene, was not affected (data not shown). Moreover, expression of CycD3 was also increased in pga22 upon induction of AtIPT8. Along with morphological analyses of the mutant highlighted before, these data indicate that the pga22 gain-of-function mutation is sufficient to activate the cytokinin signal transduction pathway.
Figure 6.
Activation of cytokinin marker genes in pga22 conditional gain-of-function mutant. Three-week-old pga22 seedlings were cultured in the absence (−) or the presence (+) of 10 μm 17-β-estradiol for 12 h. Ten micrograms of RNA was used for northern-blot analysis by using AtIPT8, CycD3, and ARR5 as probes. See Figure 5 for other technical details.
AtIPT8 Directs iPMP Biosynthesis in Vivo
Previous studies suggested that many of the AtIPT genes (except AtIPT2 and AtIPT9) were able to catalyze cytokinin synthesis in E. coli cells (Takei et al., 2001) and in vitro (Kakimoto, 2001). Moreover, overexpression of AtIPT4 (Kakimoto, 2001) or AtIPT8 (i.e. the pga22 mutant, this study) was capable of inducing shoot formation independent of exogenous cytokinins. These results strongly suggested that AtIPTs play an important role in cytokinin biosynthesis. It has been demonstrated that ipt catalyzes the biosynthesis of isopentenyladenosine-5′-monophosphate (iPMP) from AMP and DMAPP and the corresponding nucleotide iPA (Taya et al., 1978), but there is as yet no experimental evidence to support the operation of a similar biosynthetic pathway in higher plants.
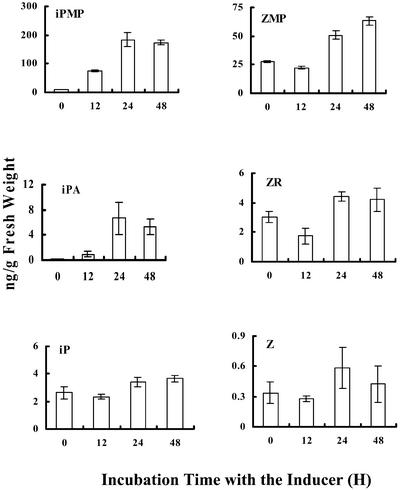
To test if AtIPT8 is a functional IPT in planta, we analyzed the cytokinin levels in the pga22 mutant as compared with those of WT plants. Upon inducer treatment, the iPMP and iPA levels increased more than 19- and 38-fold, respectively, in the mutant plants upon a 24-h induction. By contrast, we only observed a minor increase in levels of the zeatin type of ribosides and ribotides after induction (Fig. 7). Moreover, little alterations in the content of the free bases were observed during the course of the experiment.
Figure 7.
AtIPT8 catalyzes the production of iPMP and iPA. Cytokinin levels in pga22 mutant plants. Three-week-old pga22 seedlings were treated with 5 μm 17-β-estradiol for varying time periods. H, Hours, as indicated below the graph. Cytokinins extracted and purified from the frozen materials were measured by liquid chromatography/mass spectrometry analyses. The experiment was repeated three times, and the data shown here are mean values of the three experiments.
The above results suggest that AtIPT8 may act as a functional IPT, which directly catalyzes iPMP synthesis in planta. As a consequence, overexpression of the AtIPT8 gene in pga22 caused elevated iPMP and iPA levels, leading to cytokinin responses.
DISCUSSION
In a functional screen aimed at the dissection of cytokinin and auxin signaling pathways, we have identified two classes of novel mutants. Whereas the first class of mutants, represented by two alleles of pga6, appears to be involved in embryogenesis (Zuo et al., 2002), the second class of mutants is shown to be affected in key components in cytokinin signaling. Here, we have presented several lines of evidence showing that the Arabidopsis AtIPT8 locus is directly involved in cytokinin biosynthesis. First, a gain-of-function mutation in the PGA22 locus resulted in cytokinin-independent shoot formation, suggesting that the WT gene is a key component in biosynthesis or signaling. Second, overexpression of the AtIPT8 gene caused typical cytokinin responses, thus demonstrating its physiological function. Last, the AtIPT8 gene encodes a putative protein sharing amino acid sequence homology with the A. tumefaciens ipt enzyme, implying a possible role in cytokinin biosynthesis. Measurement of hormone levels in the mutant plants showed a remarkable increase of iPMP and iPA levels after induction, suggesting that AtIPT8 is likely a functional IPT.
Initially identified in Dictyostelium discoideum and several bacteria including A. tumefaciens, ipt has been demonstrated to be an enzyme that converts AMP and DMAPP into iPMP, a critical intermediate of cytokinin biosynthesis in these organisms (Taya et al., 1978). Although enzymatic activities similar to those of ipt have been detected in crude extracts prepared from various plant cells and tissues (Haberer and Kieber, 2002), it was only recently that ipt-like genes have been identified by an in silica approach upon the completion of the Arabidopsis genome (Arabidopsis Genome Initiative, 2000). These Arabidopsis homologs, designated as AtIPT1 through AtIPT9, appeared to have certain structural features of the bacterial ipt, based on phylogenetic analyses (Kakimoto, 2001; Takei et al., 2001). Except AtIPT2 and AtIPT9, recombinant proteins derived from the other seven AtIPT genes were capable of catalyzing cytokinin synthesis in vitro. In addition, overexpression of AtIPT4 was able to promote shoot regeneration in the absence of external cytokinins, strongly arguing that the gene product was involved in cytokinin biosynthesis. Interestingly, AtIPT4 appeared to catalyze the formation of isopentenyl-adenosine-5′-triphosphate (iPTP) and isopentenyl-adenosine-5′-diphosphate (iPDP) from ADP and ATP in an in vitro assay (Kakimoto, 2001). This finding is apparently inconsistent with the conventional view that ipt catalyzes the formation of iPMP by utilizing AMP and DMAPP as substrates. In addition, the lack of detectable alterations in iPMP levels in A. tumefaciens-infected plant tissues and in ipt overexpression transgenic plants (Åstot et al., 2000, and refs. therein) further argues that an iPMP-independent mechanism operates in plant cells, or that bacterial ipts function differently as compared with their plant counter-partners. A recent study on the petunia SHO gene revealed that the ipt-like gene was capable of promoting biosynthesis of iPA and derivatives (Zubko et al., 2002), which may potentially originate from not only iPMP but also from iPTP and/or iPDP (Kakimoto, 2001; Haberer and Kieber, 2002).
The fact that overexpression of AtIPT8 leads to the accumulation of high levels of iPMP and iPA in planta strongly suggests that plant IPTs are, at least in part, functionally analogous to bacterial ipt, which directs the biosynthesis of iPMP from AMP and DMAPP. Consistent with this notion, Km values of AMP and DMAPP (185 and 50 μm, respectively) for recombinant AtIPT1 are comparable with those of A. tumefaciens ipt; moreover, the plant enzyme catalyzed synthesis of iPMP (Takei et al., 2001). On the other hand, phylogenetic studies showed that AtIPT8, AtIPT4, and AtIPT1 are the most closely related members in the same subfamily (Kakimoto, 2001; Takei et al., 2001), making it difficult to understand why these three proteins have different enzymatic activities. The conflicting results obtained from these three studies are presumably because of different assay conditions used in the studies and the different genes investigated (AtIPT1, AtIPT4, and AtIPT8). Nevertheless, AtIPT8 is clearly involved in the synthesis of iPMP in planta, although it remains to be clarified whether or not the substrates of the ipt-like enzyme are AMP and DMAPP in vivo. In addition, it is also important to elucidate the potential involvement of iPDP and/or iPTP in this pathway.
Recently, Catterou et al. (2002) identified an Arabidopsis mutant designated as hoc. The hoc recessive mutation rendered shoot formation from explants in the absence of exogenous cytokinins (Catterou et al., 2002), a phenotype similar to that of the dominant-positive pga22 mutant. Analysis of hormone levels revealed that an elevated cytokinin level in hoc plants caused the mutant phenotype. In contrast to pga22, in which the iPMP and iPA levels significantly increased but other cytokinins remained largely unchanged, hoc contained an elevated amount of most of the major cytokinins, particularly in roots. Because the iPMP concentration was not determined in the hoc mutant, it is unclear if HOC acts in the iPMP-dependent pathway (Catterou et al., 2002). Similar to that of sho (Zubko et al., 2002), the elevated amount of iPA and other derivatives may be derived from precursors other than iPMP. Nevertheless, the phenotypic differences between pga22 and hoc suggest that these two loci function differently in the cytokinin biosynthesis pathway. Considering the nature of the mutant (Catterou et al., 2002), it appears that HOC acts as a negative regulator other than a biosynthetic enzyme in the cytokinin biosynthesis pathway. Molecular characterization of the HOC gene will help to clarify its own function and aid our understanding of the cytokinin biosynthetic pathway.
In a previous study, we have shown that overexpression of A. tumefaciens ipt caused the accumulation of cytokinins via an iPMP-independent pathway, which leads to the formation of ZMP rather than iPMP (Åstot et al., 2000). In pga22 mutant plants, although the iPMP level showed a remarkable increase, the ZMP level increased only marginally, suggesting that AtIPT8 functions in an iPMP-dependent pathway. The different enzymatic activities of the A. tumefaciens ipt and AtIPT8 can be likely attributed to structural differences because the two proteins share only 12% homology. On the basis of the above-discussed results, we propose that Arabidopsis, perhaps other higher plants as well, may have several pathways for cytokinin biosynthesis, including iPMP-independent and -dependent pathways. On the other hand, it is reasonable to assume that the di- and tri-phosphates of both the iP and Z type of cytokinin are potential intermediates upstream of the ribotides (see also Kakimoto, 2001). Clearly, the role of iPDP and iPTP in cytokinin biosynthesis must be further elucidated to better understand the entire pathway.
AtIPT8 is expressed at a very low level, detectable only in roots by RT-PCR combined with Southern-blot analysis. As a consequence, no expressed sequence tag clone was found in all public databases. Although its precise expression pattern remains to be confirmed by in situ hybridization, AtIPT8 appears to be predominately expressed in roots where cytokinins are generally believed to be synthesized (Davies, 1995; Mok and Mok, 2001). As expected, overproduction of the hormone in pga22 resulted in the activation of the cytokinin signal transduction pathway, thereby leading to typical cytokinin responses of the mutant. Interestingly, we were unable to identify any T-DNA insertion mutants in the AtIPT8 gene by searching over 50,000 independent transgenic Arabidopsis lines from public databases and our own collections, implying that a knockout mutation in this gene may be embryo lethal, rendering it impossible to recover a loss-of-function pga22 mutation by conventional methods. A screen for the pga22 suppressor mutants is expected to identify weaker mutation alleles of the AtIPT8 gene and, possibly, important components in cytokinin biosynthesis, transport, and signal transduction, thus providing invaluable genetic tools to study AtIPT8 functions and the entire cytokinin biosynthetic and signaling pathway.
MATERIALS AND METHODS
Screening of pga Mutants
Screening of pga mutants has been described in detail in a previous report (Zuo et al., 2002). The pga22 mutant, in the Ws background, was out-crossed twice with Ws WT plants. Homozygous or heterozygous F2 or F3 progenies were used in all experiments.
Plant Materials, Growth Conditions, and Plant Transformation
The Ws and Columbia ecotypes of Arabidopsis were used. Plants were grown under a 16-h-light/8-h-dark cycle at 22°C on solid A medium (1× Murashige and Skoog salts, 3% [w/v] Suc, and 0.8% [w/v] agar) supplemented with the appropriate antibiotics and/or the inducer 17-β-estradiol. Transformation of root explants (derived from Ws or Columbia WT plants) was carried out according to Koncz et al. (1989). Treatment of plants with 17-β-estradiol was carried out as described previously (Zuo et al., 2000a).
Analysis of Cytokinin Concentrations
Three-week-old seedlings homozygous for the pga22 locus were sprayed with 5 μm 17-β-estradiol in 0.1% (v/v) Tween 20 and incubated for varying periods of time before the seedlings were immediately frozen in liquid nitrogen and stored at −80°C. The extraction and purification of cytokinins were essentially the same as described by Åstot et al. (1998). In brief, frozen tissues were grounded with mortar and pestle in liquid nitrogen. Fresh mass (500 mg) was extracted overnight in Bieleski solvent (Bieleski, 1964) in the presence of heavy labeled internal standards: 2H5-Z, 2H5-ZR, 2H5-Z9G, 2H6-iP, 15N, 2H5-ZMP, and 15N, 2H6-iPMP (Apex International, Haniton, UK) as internal tracers for quantification. After two rounds of ion-exchange chromatography steps (strong cation-exchange cartridge and DEAE-Sephadex combined with C18 cartridges), the samples were split into two fractions. The first fraction was directly loaded onto an immuno-affinity chromatography column (OlChemIm Ltd., Olomouc, Czech Republic) to purify cytokinin free bases, ribosides, glucosides, and the second ribotides. The second fraction was treated with alkaline phosphatase and subsequently immunopurified to obtain cytokinin ribotides. Derivatization (propionylation) was performed according to Åstot et al. (1998). All samples were evaporated in vacuo and stored at −20°C until further analysis.
Cytokinin levels were estimated by liquid chromatography/mass spectrometry analysis in selective reaction monitoring mode. Chromatographic separation was performed using a Symmetry Shield RP18 column (3.5 μm), 2.1× 150 mm. At a flow rate of 0.2 mL min−1, the following binary gradient was used: 0 to 3 min, isocratic elution of 10% (v/v) B; 3 to 20 min, a linear gradient to 90% (v/v) B; followed by a 2-min isocratic elution of 90% (v/v) B. Solvent A consisted of 1% (v/v) formic acid in water, and solvent B consisted of 1% (v/v) formic acid in acetonitrile. Effluents from the chromatographic column were introduced to a Micromass Quattro Ultima mass spectrometer (Jeol, Tokyo) via an electrospray ion source (capillary voltage + 3.2 kV, cone voltage + 60V, source temperature 110°C, desolvation temperature 250°C, cone gas flow 220 L h−1, desolvation gas flow 740 L h−1, collision energy 20 units, and dwell time 0.35 s).
Molecular Manipulations
All molecular manipulations were performed according to standard methods (Sambrook et al., 1989). The OLexA-46 promoter-tagged genomic sequence in the pga22 genome was identified by thermal asymmetric interlaced-PCR as previously described (Liu et al., 1995; Zuo et al., 2002). The AtIPT8 genomic clones were obtained by PCR using the primer pairs pga22F1/pga22B2 (clone long [L]) and pga22F2/pga22B2 (clone short [S]). At the 5′ end, the clone L (1,638 bp) started exactly at the T-DNA insertion site, presumably in the AtIPT8 promoter region, whereas the clone S (1,482 bp) started 127 bp upstream from the putative ATG. The PCR products, amplified by PWO DNA polymerase (Roche Diagnostics Hong Kong, Hong Kong), were cloned into the SmaI site of a pBlueScript SK vector (Strategene, La Jolla, CA). Both clones, characterized by extensive restriction digests and DNA sequencing, were released by XhoI/SpeI digestion and inserted into the same sites of pER10 (Zuo et al., 2000a; 2002). The resulting constructs were transformed into Agrobacterium tumefaciens strain GV3101, which was used for subsequent root transformation experiments. Although both clones were functional for cytokinin-independent shoot formation, clone S appeared to be slightly more efficient.
Primers used in the PCR (non-plant sequences are in lower case and restriction sties used in cloning are underlined): pga22F1, 5′ ggactagtgtcgactcgagGACATTGTTAAGAGCATGAATGGTAAC; pga22F2, 5′ ggactagtgtcgactcgagCCGTATGAAATGTCCTTTGACATATCA; pga22F3, 5′ ATGCAAAATCTTACGTCCACATTC; pga22B2, 5′ tctagagtcgacgactagtAAATCGAGGTGCAAAAATCTTAAACATC; Act3F1, 5′ GTATGTGGCTATTCAGGCTGT; and Act3B1, 5′ CTGGCGGTGCTTCTTCTCTG. The pga22F3 primer starts at the putative translation start codon.
DNA Southern and RNA northern-blot analyses were carried out as previously described (Zuo et al., 2000a, 2001).
RT-PCR
RT-PCR was carried out as previously described with modifications (Zuo et al., 2000b). One microgram of total RNA was used for RT primed by oligo(dT). SuperScript II (Life Technologies/Gibco-BRL, Rockville, MD) was used for the RT reaction according to the manufacturer's instructions. After incubation with RNase H for 30 min at 37°C, 1 μL of the reaction mixture was used for subsequent PCR using appropriate primer pairs (pga22F3/pga22B2, see above; Act3F1/Act3B1). The reaction was cycled at 94°C for 30 s, 56°C for 1 min, and 72°C for 1.5 min 25 to 35 times. Five microliters of the PCR products was subjected to Southern-blot analysis using radioactive AtIPT8 and actin3 DNA fragments as probes. The AtIPT8 expression was undetectable if the PCR was cycled less than 30 times.
ACKNOWLEDGMENTS
We would like to thank the Arabidopsis Biological Resources Center (Columbus, OH) for providing seeds and DNA. We are grateful to Drs. Ted Klein, Bill Gordon-Kamm, Enno Krebbers, and Sheila Maddock for helpful discussions.
Footnotes
This work was supported by the E.I. DuPont de Nemours and Company (grant to Rockefeller University), the Swedish Natural Sciences Research Council (grant to G.S.'s laboratory), the National Natural Science Foundation of China (NSFC; grant no. NSFC 30270142 to J.Z.), and by the Ministry of Science and Technology of China (grant no. 2001AA225021 to J.Z.). J.Z. is a Bairen Jihua (Young Starter) fellow of the Chinese Academy of Sciences and a recipient of the NSFC Outstanding Young Investigator Award.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.011494.
LITERATURE CITED
- Abel S, Blazquez M, Dangl J, Deng X-W, Graham I, Harada J, Jones J, Nilsson O. Arabidopsisresearch 2000. Plant Cell. 2000;12:2302–2308. doi: 10.1105/tpc.12.12.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyoshi DE, Klee H, Amasino RM, Nester EW, Gordon MP. T-DNA of Agrobacterium tumefaciensencodes an enzyme of cytokinin biosynthesis. Proc Natl Acad Sci USA. 1984;81:5994–5998. doi: 10.1073/pnas.81.19.5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Åstot C, Dolezal K, Moritz T, Sandberg G. Precolumn derivatization and capillary liquid chromatographic/frit-fast atom bombardment mass spectrometric analysis of cytokinins in Arabidopsis thaliana. J Mass Spectrom. 1998;33:892–902. doi: 10.1002/(SICI)1096-9888(199809)33:9<892::AID-JMS701>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Åstot C, Dolezal K, Nordstrom A, Wang Q, Kunkel T, Moritz T, Chua N-H, Sandberg G. An alternative cytokinin biosynthesis pathway. Proc Natl Acad Sci USA. 2000;97:14778–14783. doi: 10.1073/pnas.260504097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry GF, Rogers SG, Fraley RT, Brand L. Identification of a cloned cytokinin biosynthetic gene. Proc Natl Acad Sci USA. 1984;81:4776–4780. doi: 10.1073/pnas.81.15.4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieleski RL. The problem of halting enzyme action when extracting plant tissues. Anal Biochem. 1964;9:431–442. doi: 10.1016/0003-2697(64)90204-0. [DOI] [PubMed] [Google Scholar]
- Brandstatter I, Kieber JJ. Two genes with similarity to bacterial response regulators are rapidly and specifically induced by cytokinin in Arabidopsis. Plant Cell. 1998;10:1009–1019. doi: 10.1105/tpc.10.6.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterou M, Dubois F, Smets R, Vaniet S, Kichey T, Onckelen HV, Sangwan-Norreel BS, Sangwan RS. hoc: an Arabidopsis mutant overproducing cytokinins and expressing high in vitroorganogenic capacity. Plant J. 2002;30:273–287. doi: 10.1046/j.1365-313x.2002.01286.x. [DOI] [PubMed] [Google Scholar]
- Chen CM, Melitz DK. Cytokinin biosynthesis in a cell-free system from cytokinin-autotrophic tobacco tissue cultures. FEBS Lett. 1979;107:15–20. doi: 10.1016/0014-5793(79)80452-4. [DOI] [PubMed] [Google Scholar]
- D'Agostino IB, Kieber JJ. Phosphorelay signal transduction: the emerging family of plant response regulators. Trends Biochem Sci. 1999;24:452–456. doi: 10.1016/s0968-0004(99)01465-6. [DOI] [PubMed] [Google Scholar]
- Davies PJ. Plant hormones: physiology, biochemistry, and molecular biology. Dordrecht, The Netherlands: Kluwer Academic Press; 1995. [Google Scholar]
- Haberer G, Kieber JJ. Cytokinins. New insights into a classic phytohormone. Plant Physiol. 2002;128:354–362. doi: 10.1104/pp.010773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen G, Chilton MD. Lessons in gene transfer to plants by a gifted microbe. Curr Top Microbiol Immunol. 1999;240:21–57. doi: 10.1007/978-3-642-60234-4_2. [DOI] [PubMed] [Google Scholar]
- Horgan R. A new cytokinin metabolite. Biochem Biophys Res Commun. 1975;65:358–363. doi: 10.1016/s0006-291x(75)80101-x. [DOI] [PubMed] [Google Scholar]
- Hwang I, Sheen J. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature. 2001;413:383–389. doi: 10.1038/35096500. [DOI] [PubMed] [Google Scholar]
- Imamura A, Hanaki N, Umeda H, Nakamura A, Suzuki T, Ueguchi C, Mizuno T. Response regulators implicated in His-to-Asp phosphotransfer signaling in Arabidopsis. Proc Natl Acad Sci USA. 1998;95:2691–2696. doi: 10.1073/pnas.95.5.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Higuchi M, Hashimoto Y, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Kakimoto T. Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature. 2001;409:1060–1063. doi: 10.1038/35059117. [DOI] [PubMed] [Google Scholar]
- Kakimoto T. CKI1, a histidine kinase homolog implicated in cytokinin signal transduction. Science. 1996;274:982–985. doi: 10.1126/science.274.5289.982. [DOI] [PubMed] [Google Scholar]
- Kakimoto T. Identification of plant cytokinin biosynthetic enzymes as dimethylallyl diphosphate: ATP/ADP isopentenyltransferases. Plant Cell Physiol. 2001;42:677–685. doi: 10.1093/pcp/pce112. [DOI] [PubMed] [Google Scholar]
- Koncz C, Martini N, Mayerhofer R, Koncz-Kalman Z, Korber H, Redei GP, Schell J. High-frequency T-DNA-mediated gene tagging in plants. Proc Natl Acad Sci USA. 1989;86:8467–8471. doi: 10.1073/pnas.86.21.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF. Efficient isolation and mapping of Arabidopsis thalianaT-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 1995;8:457–463. doi: 10.1046/j.1365-313x.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- Mok DW, Mok MC. Cytokinin metabolism and action. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:89–118. doi: 10.1146/annurev.arplant.52.1.89. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Riou-Khamlichi C, Huntley R, Jacqmard A, Murray JA. Cytokinin activation of Arabidopsiscell division through a D-type cyclin. Science. 1999;283:1541–1544. doi: 10.1126/science.283.5407.1541. [DOI] [PubMed] [Google Scholar]
- Ross J, O'Neill D. New interactions between classical plant hormones. Trends Plant Sci. 2001;6:2–4. doi: 10.1016/s1360-1385(00)01795-7. [DOI] [PubMed] [Google Scholar]
- Saito K, Yamazaki M, Murakoshi I. Transgenic medicinal plants: Agrobacterium-mediated foreign gene transfer and production of secondary metabolite. J Nat Prod. 1992;55:149–162. doi: 10.1021/np50080a001. [DOI] [PubMed] [Google Scholar]
- Sakai H, Aoyama T, Oka A. ArabidopsisARR1 and ARR2 response regulators operate as transcriptional activators. Plant J. 2000;24:703–711. doi: 10.1046/j.1365-313x.2000.00909.x. [DOI] [PubMed] [Google Scholar]
- Sakai H, Honma T, Aoyama T, Sato S, Kato T, Tabata S, Oka A. ARR1, a transcription factor for genes immediately responsive to cytokinins. Science. 2001;294:1519–1521. doi: 10.1126/science.1065201. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Manistis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schaller GE. Histidine kinases and the role of two-component systems in plants. Adv Bot Res. 2000;32:109–148. [Google Scholar]
- Sheen J. Phosphorelay and transcription control in cytokinin signal transduction. Science. 2002;296:1650–1652. doi: 10.1126/science.1071883. [DOI] [PubMed] [Google Scholar]
- Skoog F, Miller CO. Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp Soc Exp Biol. 1957;11:118–131. [PubMed] [Google Scholar]
- Sugiyama M. Organogenesis in vitro. Curr Opin Plant Biol. 1999;2:61–64. doi: 10.1016/s1369-5266(99)80012-0. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Ishikawa K, Yamashino T, Mizuno T. An Arabidopsis histidine-containing phosphotransfer (HPt) factor implicated in phosphorelay signal transduction: overexpression of AHP2in plants results in hypersensitiveness to cytokinin. Plant Cell Physiol. 2002;43:123–129. doi: 10.1093/pcp/pcf007. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Miwa K, Ishikawa K, Yamada H, Aiba H, Mizuno T. The Arabidopsissensor His-kinase, AHK4, can respond to cytokinins. Plant Cell Physiol. 2001;42:107–113. doi: 10.1093/pcp/pce037. [DOI] [PubMed] [Google Scholar]
- Takei K, Sakakibara H, Sugiyama T. Identification of genes encoding adenylate isopentenyltransferase, a cytokinin biosynthesis enzyme, in Arabidopsis thaliana. J Biol Chem. 2001;276:26405–26410. doi: 10.1074/jbc.M102130200. [DOI] [PubMed] [Google Scholar]
- Taya Y, Tanaka Y, Nishimura S. 5′-AMP is a direct precursor of cytokinin in Dictyostelium discoidum. Nature. 1978;271:545–547. doi: 10.1038/271545a0. [DOI] [PubMed] [Google Scholar]
- Van Montagu M, Schell J. The Ti plasmids of Agrobacterium. Curr Top Microbiol Immunol. 1982;96:237–254. doi: 10.1007/978-3-642-68315-2_13. [DOI] [PubMed] [Google Scholar]
- Yamada H, Suzuki T, Terada K, Takei K, Ishikawa K, Miwa K, Yamashino T, Mizuno T. The ArabidopsisAHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant Cell Physiol. 2001;42:1017–1023. doi: 10.1093/pcp/pce127. [DOI] [PubMed] [Google Scholar]
- Zubko E, Adams CJ, Machaekova I, Malbeck J, Scollan C, Meyer P. Activation tagging identifies a gene from Petunia hybridaresponsible for the production of active cytokinins in plants. Plant J. 2002;29:797–808. doi: 10.1046/j.1365-313x.2002.01256.x. [DOI] [PubMed] [Google Scholar]
- Zuo J, Niu Q-W, Chua N-H. An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 2000a;24:265–273. doi: 10.1046/j.1365-313x.2000.00868.x. [DOI] [PubMed] [Google Scholar]
- Zuo J, Niu Q-W, Frugis G, Chua N-H. The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. Plant J. 2002;30:349–359. doi: 10.1046/j.1365-313x.2002.01289.x. [DOI] [PubMed] [Google Scholar]
- Zuo J, Niu Q-W, Moller SG, Chua N-H. Chemical-regulated, site-specific DNA excision in transgenic plants. Nat Biotechnol. 2001;19:157–161. doi: 10.1038/84428. [DOI] [PubMed] [Google Scholar]
- Zuo J, Niu Q-W, Nishizawa N, Wu Y, Kost B, Chua N-H. KORRIGAN, an Arabidopsisendo-1,4-β-glucanase, localizes to the cell plate by polarized targeting and is essential for cytokinesis. Plant Cell. 2000b;12:1137–1152. doi: 10.1105/tpc.12.7.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]