Abstract
It is generally recognized that a silencing-inducing locus can efficiently reduce the expression of genes that give rise to transcripts partially homologous to those produced by the silencing-inducing locus (primary targets). Interestingly, the expression of genes that produce transcripts without homology to the silencing-inducing locus (secondary targets) can also be decreased dramatically via transitive RNA silencing. This phenomenon requires primary target RNAs that contain sequences homologous to secondary target RNAs. Sequences upstream from the region homologous to the silencing inducer in the primary target transcripts give rise to approximately 22-nucleotide small RNAs, coinciding with the region homologous to the secondary target. The presence of these small RNAs corresponds with reduced expression of the secondary target whose transcripts are not homologous to the silencing inducer. The data suggest that in transgenic plants, targets of RNA silencing are involved in the expansion of the pool of functional small interfering RNAs. Furthermore, methylation of target genes in sequences without homology to the initial silencing inducer indicates not only that RNA silencing can expand across target RNAs but also that methylation can spread along target genes.
RNA silencing is a conserved mechanism that occurs in various eukaryotic organisms and leads to targeted degradation of RNA sequences homologous to the trigger (for review, see Matzke et al., 2001; Sharp, 2001; Zamore, 2001; Hutvágner and Zamore, 2002). The potency of double-stranded RNA (dsRNA) in activating RNA silencing was first demonstrated in Caenorhabditis elegans and was designated RNA interference (RNAi; Fire et al., 1998). Posttranscriptional gene silencing (PTGS; for reviews, see Kooter et al., 1999; Vaucheret et al., 2001; Voinnet, 2001), which is mechanistically related to RNAi, can also be efficiently elicited in plants upon introduction of gene constructs that give rise to dsRNA (Hamilton et al., 1998; Waterhouse et al., 1998; Chuang and Meyerowitz, 2000; Smith et al., 2000; Sijen et al., 2001a; Wesley et al., 2001; Stoutjesdijk et al., 2002) or upon viral infection of plants that initiates production of dsRNA replication intermediates (Ruiz et al., 1998; Baulcombe, 1999).
Sense and antisense small RNAs (approximately 20–25 nucleotides), homologous with posttranscriptionally silenced sequences, accumulate specifically in various PTGS systems in plants (Hamilton and Baulcombe, 1999). Studies of RNAi in fruitfly (Drosophila melanogaster) revealed that small RNAs result from symmetric processing of the dsRNA (Hammond et al., 2000). Hamilton and Baulcombe (1999) first proposed that these small RNAs correspond to specificity determinants in PTGS and RNAi. These small RNAs have been shown to guide a nuclease complex to cleave single-stranded RNA with complementary sequences in fruitfly embryo lysates (Elbashir et al., 2001a, 2001b). Therefore, the 21- to 23-nucleotide RNAs are referred to as small interfering RNAs (siRNAs) or guide RNAs. Target mRNAs are cut in the center of the region recognized by the complementary guide RNAs (Elbashir et al., 2001a), and mRNAs are cleaved only in the region corresponding to the dsRNA (Zamore et al., 2000).
The amplification of the siRNA signal during RNAi in C. elegans has been investigated and, in addition to trigger-coincident siRNAs, populations of small antisense RNAs have been detected that correspond to regions of the target RNA molecules located upstream of the initial trigger dsRNA, designated secondary siRNAs (Sijen et al., 2001b). The abundance of secondary siRNAs seems to decrease as a function of the distance from the region homologous to the primary trigger. Functionality has been demonstrated by means of a transitive RNAi assay (Sijen et al., 2001b), in which two targets for silencing are provided. Similarly, plant viral vectors carrying part of a transgene elicit the production of transgene-specific, secondary siRNAs upon infection. As a consequence, these plants are protected against infection by an unrelated virus that carries another part of the transgene (Vaistij et al., 2002).
Plant RNA silencing is frequently accompanied by DNA methylation in symmetrical and nonsymmetrical cytosines (Bender, 2001) in transcribed regions of the silenced genes (Ingelbrecht et al., 1994; English et al., 1996; Sijen et al., 1996; Kovařík et al., 2000; Van Houdt et al., 2000a), although its role is still unclear. Sequence-specific methylation signals consisting of RNA-DNA associations are believed to be involved in methylating silenced genes (Wassenegger, 2000). RNA-directed DNA methylation (RdDM) was first discovered in tobacco (Nicotiana tabacum) plants that contained multimeric genome-integrated copies of a viroid cDNA (Wassenegger et al., 1994). In these plants, specific de novo methylation that is restricted to the cDNA region was detected whenever viroids replicated autonomously (Pélissier et al., 1999). Further, viroid-infected plants accumulate small RNAs identical in size to those found in plants exhibiting PTGS of transgenes (Papaefthimiou et al., 2001). These results suggest that viroid-related RNAs induce methylation of the homologous cDNA copies. However, it remains controversial whether silencing-triggering dsRNA molecules, small guide RNAs, or intermediate RNA products are the signals for methylation of homologous DNA. PTGS induced by viral RNA that carries a short region homologous to the transgene leads to spreading of methylation throughout the transcribed region of the transgene (Jones et al., 1999; Thomas et al., 2001; Vaistij et al., 2002). Direct interaction between the input recombinant virus and the homologous transgene might lead to RdDM, and the progressive degradation of target mRNA (Zamore et al., 2000) could release more fragments, which additively direct methylation throughout the transcribed region of the transgene (Thomas et al., 2001). As an alternative, viral dsRNAs, synthesized by a putative plant RNA-dependent RNA polymerase (RdRp; Dalmay et al., 2000; Mourrain et al., 2000) or its derived siRNAs could mediate RdDM (Vaistij et al., 2002).
Here, we address the question of whether the in trans-silencing capacity of a silencing-inducing transgene locus can be transmitted to target RNA, subsequently able to silence secondary targets in trans. Therefore, we tested whether a posttranscriptionally silenced transgene locus can silence in trans a secondary target, which only produces nonhomologous transcripts; to this end, we created a stepwise homology between the silencing inducer and the secondary target by producing a transcript from a primary target with one region homologous to the silencing inducer and another region homologous to the secondary target mRNA. We investigated the production of small, approximately 22-nucleotide long RNAs, corresponding to target mRNAs and analyzed the methylation status of sequences silenced in trans in the region nonhomologous to the silencing inducer.
RESULTS
A Posttranscriptionally Silenced Inverted Repeat Transgene Locus Can Trigger Silencing of a Reporter Gene Producing Nonhomologous Transcripts
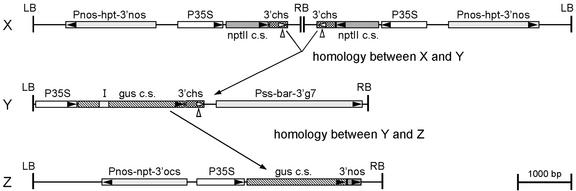
To determine whether the RNA-silencing activity of a silencing-inducing RNA can be transmitted to another RNA sequence by fusing this sequence to part of the silencing-inducing sequence in a single transcript, we studied transgenic tobacco plants with different combinations of three transgene loci (X, Y, and Z). Locus X (Fig. 1) harbors an inverted repeat about the right T-DNA border of T-DNA GVCHS287, carrying a neomycin phosphotransferase II (nptII) gene under control of the cauliflower mosaic virus 35S promoter (CaMV P35S) and the 3′-signaling sequences of the chalcone synthase gene (3′chs) of snapdragon (Van Houdt et al., 2000a, 2000b; Fig. 1). The two convergently transcribed nptII genes in locus X produce only very low amounts of the NPTII protein compared with those produced by a single-copy nptII transgene. The invertedly repeated nptII transgenes in locus X had been shown to be posttranscriptionally silenced and methylated in the 3′ one-half of the genes (Van Houdt et al., 2000a). Locus Y (Fig. 1) contains a single copy of the T-DNA GUSchsS and harbors a chimeric β-glucuronidase (gus) gene under control of P35S and 3′chs with an artificial intron in the 5′ region of the coding sequence. In tobacco plants hemizygous for locus Y, the gus expression levels are normal (Table I). Locus Z has two or more copies of T-DNA XD610 with a gus gene under control of P35S and the 3′-untranslated region (UTR) of the nopaline synthase gene (3′nos). In locus Z (Fig. 1), the gus expression is stable (Table I). Here, the in trans-silencing effects between these three transgene loci X, Y, and Z were studied in hybrid transgenic tobacco plants with any possible combination of these three loci under hemizygous condition, obtained by crossing the appropriate parental plants (“Materials and Methods”). In trans-silencing effects were revealed by a reduced GUS activity of particular loci in certain combinations. The results of the GUS activity measurements in protein extracts of different types of hybrid plants are summarized in Table I.
Figure 1.
Schematic outline of the T-DNA constructs (drawn to scale), present in silenced locus X, recombinant gene Y, and target gene Z (T-DNAs of pGVCHS287, pGUSchsS, and pXD610, respectively), and of the transcript homology between X, Y, and Z. The structure of locus X and Y is indicated; locus Z contains two or more copies of the XD610 T-DNA. Δ, 3′chs polyadenylation signal; 3′chs, 3′-UTR of the chalcone synthase gene of snapdragon (Anthirrinum majus); 3′g7, 3′-UTR of the Agrobacterium tumefaciens octopine T-DNA gene 7; 3′nos, 3′-UTR of the nopaline synthase gene; 3′ocs, 3′-UTR of the octopine synthase gene; bar, bialaphos acetyltransferase-coding sequence conferring phosphinothricin resistance; gus c.s., GUS-coding sequence; hpt, hygromycin phosphotransferase-coding sequence; I, artificial intron; LB, left T-DNA border; nptII c.s., neomycin phosphotransferase II-coding sequence; P35S, CaMV 35S promoter; Pnos, nopaline synthase promoter; Pss, promoter of the small subunit of ribulose-1,5-bisphosphate carboxylase; RB, right T-DNA border.
Table I.
GUS activity determination in protein extracts of leaf tissue harvested from tobacco plants containing different combinations of loci X, Y, and Z (Fig. 1)
| Genotype | GUS Activitya | No. of Plants Analyzedb |
|---|---|---|
| Units GUS mg−1 total soluble protein | ||
| X | Below detection | 1 |
| Y | 368 ± 165 | 9 |
| Z | 126 ± 30 | 10 |
| XY | 2 ± 1 | 4 |
| XZ | 139 ± 35 | 9 |
| YZ | 477 ± 101 | 10 |
| XYZc | 4 ± 3 | 22 |
Mean ± sd.
The axenically grown plants were analyzed 4 weeks after sowing on Murashige and Skoog medium supplemented with 1% (w/v) Suc in Falcon petri dishes (Becton Dickinson, Bedford, MA).
XYZ plants were grown in the presence of phosphinothricin (10 mg mL−1) and hygromycin (25 mg mL−1); under these conditions, both XYZ plants and YZ plants containing the pNE T-DNA (see “Materials and Methods”) were able to develop. A polymerase chain reaction with X-specific primers was performed to screen for the presence of X.
The silenced nptII genes in locus X have previously been demonstrated to silence in trans homologous nptII transgenes (Van Houdt et al., 2000a) and transiently expressed genes with partial transcript homology to locus X-derived nptII transcripts (Van Houdt et al., 2000b). We also observed that the stably expressed gus gene in locus Y, with partial transcript homology to the nptII transcripts of the silencing-inducing locus X (Fig. 1), was silenced efficiently in trans (compare XY with Y; Table I). The homology between the transcripts of X and Y was mainly situated in the 3′-UTR (206 nucleotides), but the 5′-UTR also had a small region of homology (29 nucleotides). This relatively short region of homology between locus X-derived nptII and locus Y-derived gus transcripts was sufficient to degrade very efficiently Y-derived transcripts. To assess the stability of in trans-silencing of Y in XY hybrids, a 4-week-old phosphinothricin-resistant progeny of a self-fertilized XY hybrid was analyzed. Loss of locus X in the progeny plants, as revealed by kanamycin sensitivity, correlated with reactivation of gus expression in locus Y in the expected 1:4 ratio, indicating that the in trans-silenced phenotype is not transmitted to the next generation when the silencing-inducing locus X is absent.
In contrast to the low GUS activity detected in XY hybrids, the GUS activity in XZ hybrids was normal and similar to that in Z plants (Table I). This observation allows us to conclude that the nptII genes of locus X could trigger neither transcriptional silencing of the gus genes in locus Z, although also driven by the CaMV P35S promoter, nor posttranscriptional silencing of the gus genes in locus Z, which was expected because both loci produce transcripts without significant homology (Fig. 1). These data, in addition to results of run-on transcription analyses of locus X-containing plants (Depicker et al., 1996; M. Fojtova and A. Kovařík, unpublished data), support that the in trans-silencing effects in XY hybrid tobacco plants are not triggered by P35S homology.
When Y and Z loci were combined in so-called YZ hybrids, the tobacco cells produced two types of gus transcripts with a 1,809-nucleotide homologous region in the gus-coding sequence. Both types of gus genes, however, remained normally expressed as reflected in the high GUS activity in YZ hybrids, and displayed a dosage effect, as expected for normally expressed genes (compare YZ with Y and Z; Table I). Thus, the RNA-silencing mechanism was not activated in YZ hybrid tobacco plants. Therefore, it is interesting to observe that upon creation of a stepwise homology between X and Z by the presence of locus Y, the gus expression in locus Z together with that of locus Y was reduced in XYZ hybrid tobacco plants (compare XYZ with YZ; Table I).
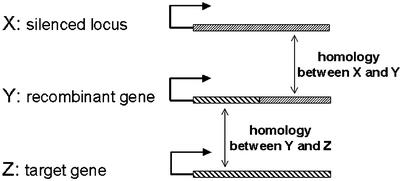
As shown schematically in Figure 2, silencing of the target could be triggered by a stepwise homology that was created between a silenced locus (X) and a nonhomologous target gene (Z) by introducing a chimeric recombinant gene (Y) with one region homologous to the silenced locus (X) and another homologous to the target (Z). We refer to this as a case of transitive silencing in plants in which the silencing inducer (X) and the primary and secondary targets (Y and Z) are all nuclearly expressed transgenes. The results imply that the silencing capacity of locus X is transferred to Y sequences upstream of the homology between X and Y.
Figure 2.
Schematic outline of homology between silenced locus X, recombinant gene Y, and target gene Z, resulting in silencing. The angular arrows and hatched boxes represent promoters and transcribed sequences, respectively.
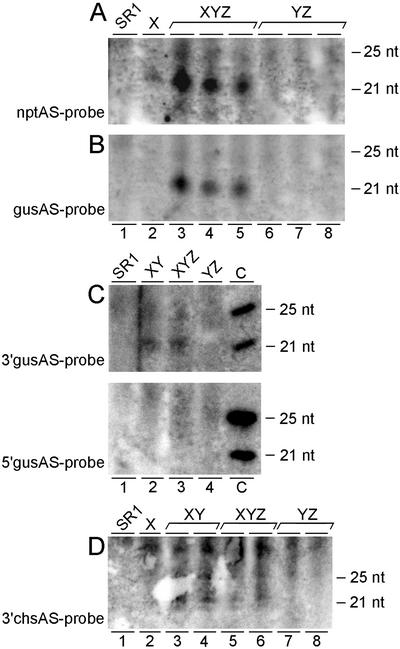
In Trans-Silenced Loci Give Rise to Approximately 22-Nucleotide Small RNAs
To understand the observation that locus Z is silenced only in XYZ hybrids through a transitive silencing effect and to confirm that silencing capacities of silencing-inducing loci can expand to target genes, we assessed the accumulation of sequence-specific small RNAs. First, we determined whether small RNAs specific for the silencing-inducing locus could be detected. RNA gel blots using hydrolyzed nptII transcripts as a probe (Fig. 3A) revealed locus X-specific nptII 22-nucleotide small RNAs in the low-Mr RNA fraction of plants hemizygous for locus X and of XYZ hybrid tobacco plants. Wild-type SR1 plants (Fig. 3A, lane 1), hybrid lines without the nptII transgene (Fig. 3A, YZ in lanes 6–8), and a transgenic line containing a normally expressed nptII transgene (HElo2; Van Houdt et al., 2000a; data not shown) did not accumulate small nptII RNAs.
Figure 3.
Detection of small RNAs. Low-Mr RNA fractions were isolated from leaf tissue of mature non-flowering tobacco plants, separated on polyacrylamide gels, blotted onto Hybond N+ membranes, and hybridized with hydrolyzed antisense riboprobes: nptII (full-length nptII-coding sequence; A), gus (full-length gus-coding sequence; B), 3′gus (most 3′ 800 bp of the gus-coding sequence; C, top panel), 5′gus (most 5′ 800 bp of the gus-coding sequence; C, bottom panel), and 3′chs (3′chs-UTR sequence; D). DNA oligomers were used as size controls (size indicated in nucleotides). Each numbered lane contains the low-Mr RNA fraction of another tobacco plant of the genotype indicated on top. For the wild-type tobacco SR1 and the normal gus-expressing hybrid plants YZ, no specific signal could be detected with either of the probes. A, Locus X-containing plants (X and XYZ) gave rise to small nptII-specific RNAs of approximately 22 nucleotides. B, The XYZ plants showing gus silencing accumulated approximately 22-nucleotide small gus-specific RNAs. C, In gus-silenced XY and XYZ plants, these small RNAs corresponding to the 5′gus probe do not accumulate to detectable levels (bottom panel), whereas those corresponding to the 3′gus probe give a detectable signal upon identical exposure time (top panel). C, The lanes indicated by c (controls) contain 3.3 pmol of 21-nucleotide long DNA and 2.5 pmol of 25-nucleotide long DNA with a GC content of 72% and 71.4%, respectively, and are shown for comparison of probe quality and quantity in top and bottom panel; the control DNA oligonucleotides in the upper and lower panels are 100% complementary to a stretch in the 5′gus probe and 3′gus probe, respectively. D, Approximately 22-nucleotide small 3′chs-specific RNAs were, although expected for all X-containing samples, only detected in XY and XYZ samples.
We examined the ability of target loci to give rise to target-specific small RNAs. Therefore, we used hydrolyzed gus transcripts as a probe to detect small RNAs originating from the gus sequences that do not occur in the silencing-inducing locus X. Only XYZ hybrids (Fig. 3B, XYZ in lanes 3–5), which show transitive silencing, but not YZ hybrids, X-hemizygous plants, or wild-type SR1 (Fig. 3B, lanes 6–8, 2, and 1, respectively), accumulated small gus RNAs, although an identical set of gus genes was present in the XYZ and YZ hybrid lines. To determine which region of the gus transcripts is mainly giving rise to the detected small gus RNAs (Fig. 3B), we performed two RNA gel blots with identical RNA samples hybridized with different nonoverlapping partial gus transcripts as probes (Fig. 3C); one (5′GUS) spanned the most 5′ 800 bp of the gus-coding sequence, the other (3′GUS) the most 3′ 800 bp. A clear signal was obtained with the 3′GUS probe for XY and XYZ samples (Fig. 3C, top panel, lanes 2 and 3), but no signal was detected with the 5′GUS probe (Fig. 3C, bottom panel, lanes 2 and 3), although the controls (Fig. 3C, c in both panels) gave signals of comparable intensities with both probes, assuring similar probe quality and quantity. We conclude that small gus RNAs accumulate only upon silencing of gus genes in the presence of locus X and are mainly derived from the 3′ part of the gus transcripts. Further, a weak signal corresponding to 3′chs-specific small RNAs was detected in samples of XY and XYZ hybrids that showed gus silencing, whereas these molecules were not detected in samples of YZ hybrids with normal gus expression (Fig. 3D, lanes 3 and 4, 5 and 6, and 7 and 8, respectively). Therefore, we suggest that small RNAs corresponding to the 3′chs-UTR region may direct the formation of small RNAs of more upstream-located sequences.
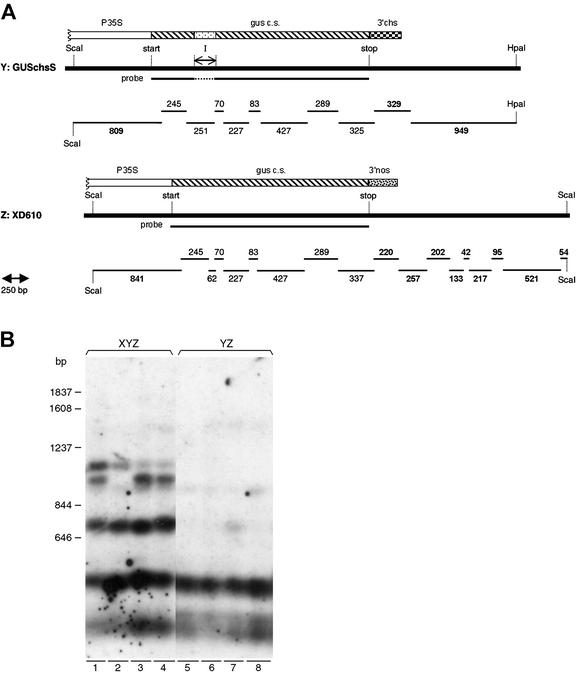
DNA Sequences That Are Nonhomologous to the Silencing-Inducing Locus X Become Methylated upon Inactivation
The silencing-inducing locus X contains two invertedly repeated, convergently transcribed nptII transgenes that are extensively methylated in the center of the repeat (Van Houdt et al., 2000a). In addition, a completely homologous nptII transgene becomes methylated upon in trans silencing by locus X (Van Houdt et al., 2000a). Does in trans methylation rely on sequence homology with the (initial) silencing-inducing locus or can target sequences silenced in trans be involved in producing the methylation signal? To address these questions, we examined the methylation status of several cytosines located in the gus-coding sequences in the genomic DNA of non-silenced YZ and silenced XYZ plants via DNA gel-blot analysis with the methylation-sensitive restriction enzyme HpaII. Figure 4A shows the gus-coding sequences in the T-DNAs of loci Y and Z, the location of non-methylation-sensitive restriction sites HpaI and ScaI, used to delimit the analyzed region of the T-DNA, and of the methylation-sensitive HpaII restriction sites, and the sizes of the digestion products of non-methylated gus transgenes. HpaII recognizes the sequence CCGG and will not cut this sequence in case at least one of the cytosines is methylated. Upon complete digestion with HpaII of the gus transgenes in locus Y and Z, only relatively small hybridizing DNA fragments will be detected, with the largest one being 427 bp (Fig. 4A). This result was obtained with different samples of YZ genomic DNA analyzed in a triple digest ScaI/HpaI/HpaII (Fig. 4B, lanes 5–8). However, the gus probe clearly revealed intense bands of higher Mr in the different XYZ genomic DNA samples than in the YZ samples, indicating strongly enhanced cytosine methylation upon silencing of the gus genes (Fig. 4B, compare lanes 1–4 with 5–8). The size of the hybridizing bands with higher molecular mass (0.7, 1.0, and 1.1 kb; Fig. 4B) suggests that at least three cytosines located in the 3′ one-half of the gus-transcribed sequences were methylated in XYZ tissues. As a confirmation, identical fragments of higher molecular mass were detected in the XYZ samples after the blot had been reprobed with a labeled gus fragment that spanned the most 3′-located 800 bp of the gus-coding sequence, whereas reprobing with the most 5′-located 800 bp did not reveal any fragments of higher molecular mass (data not shown). The data allow us to conclude that target genes become methylated upon in trans-silencing in a region without homology to the silencing-eliciting transgenes and that corresponds to the region giving rise to small gus RNAs.
Figure 4.
Outline of the DNA gel blot to analyze the cytosine methylation status in the gus-coding sequences of XYZ and YZ plants. A, Representation of the analyzed restriction sites. The analyzed region of the gus-containing T-DNAs in locus Y and Z (GUSchs and LXD610, respectively) is represented by a thick black bar for each T-DNA. The relative location of the start and stop codons of the gus gene and the synthetic intron (I) are indicated above the bars. The recognition sites of the methylation-insensitive restriction enzymes HpaI and ScaI were used to border the segments for methylation analysis. The functional elements of the gus chimeric genes are indicated by boxes above the black bars (for abbreviations, see Fig. 1). The fragments drawn below the bars are the DNA fragments obtained upon full HpaII digestion of the ScaI/HpaI and ScaI/ScaI fragments of the T-DNAs in locus Y and Z, respectively (size in bp). The sequence used as probe (the full-length gus-coding sequence) is given as a bar just below the Y and Z T-DNA fragments. B, DNA gel blot of XYZ and YZ hybrids. Genomic DNA of four gus-silenced XYZ and four normal gus-expressing YZ tobacco hybrids (13 μg per lane) was cut in triple digests with two methylation-insensitive enzymes HpaI and ScaI and with the methylation-sensitive HpaII. The full-length gus-coding sequence was used as probe. The length of size markers is indicated in base pairs. Specifically in XYZ samples, hybridizing bands of higher Mr are detected.
DISCUSSION
According to the current model for PTGS or RNA silencing (Kooter et al., 1999; Matzke et al., 2001; Vance and Vaucheret, 2001), dsRNA molecules elicit the activation of the silencing response. Because locus X consists of two invertedly repeated T-DNAs and contains convergently transcribed nptII transgenes (Van Houdt et al., 2000a), we postulate that readthrough over the T-DNA right border sequences results in the production of double-stranded hairpin RNAs that trigger silencing of the nptII transgenes. We show that low-Mr RNA fractions of locus X-harboring tobacco plants and not similar fractions of transgenic plants with normal nptII expression contain approximately 22-nucleotide small nptII RNAs, which are probably the product of nptII dsRNA cleavage by a dicer-like RNase. The posttranscriptionally silenced transgene locus X can silence in trans the partially homologous gus transgene in locus Y (Van Houdt et al., 2000a; Table I). The mechanism of in trans silencing is most probably based on the presence of siRNAs that correspond to the region of homology between silencing inducer and target RNA, namely the 3′chs region in our analysis. Thus, the 3′chs-specific small RNAs seem to mark the locus Y-derived gus transcripts for degradation.
How can locus X direct degradation of locus Z-derived gus transcripts in XYZ plants? According to the current model of target RNA degradation (Elbashir et al., 2001a), the likely hypothesis is that siRNAs homologous to locus Z-derived transcripts would be involved. gus-specific small RNAs are readily detected in the low-Mr RNA fraction of XYZ plants, but not in similar fractions of non-silenced YZ plants. These molecules are candidates to function as siRNAs for sequence-specific degradation of gus transcripts, resulting in the low GUS activity detected in XYZ tobacco plants compared with that in Z plants. In a recent study, the spreading of RNA targeting upon virus-induced gene silencing (VIGS) in Nicotiana benthamiana and Arabidopsis has been described (Vaistij et al., 2002). The effect of VIGS spreads beyond the viral sequences inducing RNA silencing, because the single-stranded target transcripts are converted to dsRNA by the putative SDE1/SGS2 RdRp (Vaistij et al., 2002). This observation gives an insight into how the detected gus-specific small RNAs, corresponding to the most 3′ 800 bp of the gus-coding sequence, might be generated. RdRp-dependent synthesis of gus antisense RNA could be primed by locus X-derived 3′chs-specific small RNAs on a locus Y-derived transcript, in analogy with the extension of primer siRNAs into a dsRNA product in embryo extracts of fruitfly (Lipardi et al., 2001). As an alternative, because no priming is required in an RdRp-dependent polymerization reaction (Schiebel et al., 1993), a particular feature of the Y-derived gus transcript, such as the mere association with a siRNA-protein complex or partial degradation by an RNA-induced silencing complex (Hammond et al., 2000), could allow it to be recognized by an RdRp as template for synthesis. RNase III-like cleavage of the nascent dsRNA would subsequently give rise to the detected small gus RNAs, which are probably involved in the 3′-5′ spreading of silencing, as revealed by the reduced GUS activity in XYZ hybrids.
Several studies in plants indicate that silencing can also spread to transgene regions downstream of the primary target (5′-3′ spreading; Braunstein et al., 2002; Han and Grierson, 2002; Vaistij et al., 2002). This observation cannot be ascribed to siRNA-primed RdRp-directed synthesis of dsRNA on the basis of transgenic mRNA targets, nor can it be reconciled easily with unprimed RdRp synthesis, which appears to start preferentially at the 3′ terminus of the template (Schiebel et al., 1993).
Although several papers describe the production of secondary siRNAs and the spreading of RNA silencing induced upon viral infection (Braunstein et al., 2002; Vaistij et al., 2002) or by nuclearly expressed transgenes (Han and Grierson, 2002), target-specific siRNA production apparently does not occur by default. In tobacco plants transformed with a chimeric transgene comprising sequences encoding gus followed by satellite RNA (satRNA), there is no indication for spreading of siRNA production upon helper virus infection (Wang et al., 2001). Also, phytoene desaturase and ribulose-1,5-bisphosphate carboxylase endogenous transcripts do not serve as templates for secondary siRNA production upon VIGS in Arabidopsis and are therefore not involved in the spreading process of RNA silencing (Vaistij et al., 2002). In summary, the generality, requirements, and characteristics of secondary siRNA production in RNA silencing remain to be determined.
PTGS in plants, resulting in the degradation of homologous RNAs, has frequently been associated with sequence-specific de novo methylation of transcribed sequences of silenced transgenes. Also, in trans-silenced transgenes homologous to a silencing inducer become extensively methylated (Van Houdt et al., 2000a; Béclin et al., 2002). The region of methylation of a silenced transgene, induced upon viral infection, has been confined to the region of homology between the viral genome and the transgene (Jones et al., 1998; Wang et al., 2001). However, in other studies, methylation of a silenced transgene induced upon viral infection spreads into transcribed sequences not corresponding to viral sequences (Jones et al., 1999; Thomas et al., 2001; Vaistij et al., 2002), which has been associated with maintenance of silencing in the absence of the viral inducer (Vaistij et al., 2002). Enhanced cytosine methylation of the gus-coding sequence in XYZ plants showing transitive silencing of the gus genes has been observed, whereas non-silenced gus genes in YZ plants remain hypomethylated. The detected methylation is confined to the 3′ one-half of the gus-transcribed sequences, coinciding with the region that mainly gives rise to the gus-specific small RNAs, probably because it might be the region to be copied first into dsRNA by an RdRp. Further, methylation is only partial, because fragments corresponding to hypomethylated molecules are detected. It remains unclear whether locus Y gus genes are predominantly methylated and thus Y and Z gus genes are discriminated as methylation targets or whether both Y and Z gus genes are partially methylated. Two possible types of interactions could be invoked for de novo methylation of the gus sequences. First, methylation could be RNA directed and induced by interactions of the Y and/or Z gus genes with gus dsRNAs or gus-specific small RNAs. In a second scenario, DNA-DNA interactions between the methylated 3′chs regions of the nptII genes of the silencing-inducing locus X and target Y could be a signal for methylation of the paired sequences followed by spreading of methylation into nonhomologous gus sequences.
The occurrence of transitive silencing will have to be taken into account in case RNA silencing or RNAi is the technique of choice in functional genomic studies to obtain a null mutant phenotype for any particular gene (Nishikura, 2001). It is possible that particular siRNAs produced by a silencing inducer correspond to an identical stretch of nucleotides in a family member of the studied gene, not targeted on purpose, that could initiate a process in which the partially homologous endogenous transcript is used as template to produce secondary siRNAs (Sanders et al., 2002). Therefore, transitive silencing of coordinately expressed genes with highly conserved domains can be anticipated. The design of a dsRNA trigger will be crucial to create a selected gene-specific mutant phenotype. On the other hand, we envision that transitive silencing can be applied in silencing technologies, circumventing laborious construction of inverted repeat transgenes.
MATERIALS AND METHODS
Transgene Tobacco Lines and Production of Hybrid Plants
The production of plants containing locus X and several characteristics of locus X have been described previously (locus 1 in Van Houdt et al., 2000a, 2000b). The locus Y-containing primary tobacco (Nicotiana tabacum) transformant GUSchsS29 was obtained via Agrobacterium tumefaciens cotransformation of tobacco cv Petit Havana SR1 leaf discs with the A. tumefaciens strains C58C1RifR(pGV2260, pNE) and C58C1RifR(pGV2260, pGUSchsS). The plasmids pNE, carrying a hygromycin resistance marker (De Buck et al., 1999), and pGUSchsS, with the phosphinothricin resistance marker (Van Houdt et al., 2000b), have been described previously. Transformant GUSchsS29 was obtained upon hygromycin selection and, in addition to the pNE T-DNA insert(s), contained an independently segregating locus, designated locus Y, harboring a single copy of the GUSchsS T-DNA. The tobacco leaf disc transformation in which the locus Z-containing primary transformant LXD610-2 was generated, has been described previously (De Loose et al., 1995).
Hemizygous X and Z plants were obtained as hybrid progeny by crossing tobacco plants homozygous for locus X (=HOlo1) and homozygous for locus Z (=LXD610-2/9) to wild-type SR1, respectively. Hemizygous Y plants originated from the cross between the hemizygous primary tobacco transformant GUSchsS29 and SR1 and by selecting for the presence of locus Y in the hybrid progeny with phosphinothricin (10 μg mL−1). Hemizygous plants XY and YZ were the hybrid progeny plants of the cross between HOlo1 and GUSchsS29 and between GUSchsS29 and LXD610-2/9, respectively, which were selected for the presence of Y. Hemizygous plants XZ were the hybrid progeny of the cross between HOlo1 and LXD610-2/9, and hemizygous plants XYZ were obtained by crossing hemizygous plants XY to LXD610-2/9, which is homozygous for locus Z, and selected for the presence of locus Y. Because the presence of locus X in this hybrid progeny could not be selected for, we screened for hemizygous plants XYZ through a locus X-specific PCR.
Enzymatic Assays
Protein extracts were prepared and GUS activity was measured as described by Van Houdt et al. (2000b).
DNA Gel-Blot Analysis
Genomic DNA from leaf tissue of mature tobacco plants was isolated with the DNeasy plant kit (Qiagen, Hilden, Germany). DNA gel-blot hybridization was mainly done as described previously (Van Houdt et al., 1997). Probes were labeled with the Gene Images random prime labeling kit (Amersham Biosciences, Little Chalfont, UK) and detected with the Gene Images CDP-Star module (Amersham Biosciences).
Small RNA Analysis
To detect small RNAs, the procedures described by Hamilton and Baulcombe (1999) and Mette et al. (2000) were adapted. Tobacco leaf tissue was frozen in liquid nitrogen, and total RNA was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA), according to the manufacturer's instructions. Most of the high-Mr RNAs were precipitated and the lower Mr RNAs were recovered from the supernatant as described. For the different samples analyzed, a similar amount of RNA of the lower Mr RNA fraction, as estimated on gel, was separated on gel (15% [v/w] polyacrylamide and 7 m urea) and transferred to Hybond N+ membranes (Amersham Biosciences) by electroblotting with a Kem en Tec semidry blotter II (BIOzym, Landgraaf, The Netherlands). As size and polarity controls, DNA oligomers were loaded on the same gels. 32P-labeled probes were synthesized in vitro from a linearized plasmid with an SP6/T7 transcription kit (Roche Diagnostics, Brussels) and [α-32P]CTP. The probe was hydrolyzed into fragments of approximately 50 nucleotides. Hybridization and washes were performed as described (Hamilton and Baulcombe, 1999; Mette et al., 2000) at 30°C. Labeled membranes were exposed to a phosphor imager screen (Amersham Biosciences).
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
ACKNOWLEDGMENTS
We thank Sylvie De Buck, Gert Van der Auwera, and Frank Van Breusegem for critical reading of the manuscript and helpful comments, the partners of the European Union Biotech Project (no. QLRT–2000–00078) for stimulating discussions, Heidi Van Horebeke for generating transformants and performing crosses, Els Van Lerberge for technical assistance, Dries Brabant and Ruben Dario García Perez for analysis of particular hybrid lines, Pongsopee Attasart for her input in the small RNA analysis, and Martine De Cock and Rebecca Verbanck for help with the manuscript and the figures, respectively.
Footnotes
H.V.H. received a postdoctoral fellowship from the Instituut voor de aanmoediging van Innovatie door Wetenschap en Technologie in Vlaanderen.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.009407.
LITERATURE CITED
- Baulcombe DC. RNA makes RNA makes no protein. Curr Biol. 1999;9:R599–R601. doi: 10.1016/s0960-9822(99)80383-2. [DOI] [PubMed] [Google Scholar]
- Béclin C, Boutet S, Waterhouse P, Vaucheret H. A branched pathway for transgene-induced RNA silencing in plants. Curr Biol. 2002;12:684–688. doi: 10.1016/s0960-9822(02)00792-3. [DOI] [PubMed] [Google Scholar]
- Bender J. A vicious cycle: RNA silencing and DNA methylation in plants. Cell. 2001;106:129–132. doi: 10.1016/s0092-8674(01)00441-x. [DOI] [PubMed] [Google Scholar]
- Braunstein TH, Moury B, Johannessen M, Albrechtsen M. Specific degradation of 3′ regions of GUS mRNA in posttranscriptionally silenced tobacco lines may be related to 5′-3′ spreading of silencing. RNA. 2002;8:1034–1044. doi: 10.1017/s1355838202026080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang C-F, Meyerowitz EM. Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2000;97:4985–4990. doi: 10.1073/pnas.060034297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- De Buck S, Jacobs A, Van Montagu M, Depicker A. The DNA sequences of T-DNA junctions suggest that complex T-DNA loci are formed by a recombination process resembling T-DNA integration. Plant J. 1999;20:295–304. doi: 10.1046/j.1365-313x.1999.t01-1-00602.x. [DOI] [PubMed] [Google Scholar]
- De Loose M, Danthinne X, Van Bockstaele E, Van Montagu M, Depicker A. Different 5′ leader sequences modulate β-glucuronidase accumulation levels in transgenic Nicotiana tabacum plants. Euphytica. 1995;85:209–216. [Google Scholar]
- Depicker A, Ingelbrecht I, Van Houdt H, De Loose M, Van Montagu M. Post-transcriptional reporter transgene silencing in transgenic tobacco. In: Grierson D, Lycett GW, Tucker GA, editors. Mechanisms and Applications of Gene Silencing. Nottingham, UK: Nottingham University Press; 1996. pp. 71–84. [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001a;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001b;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- English JJ, Mueller E, Baulcombe DC. Suppression of virus accumulation in transgenic plants exhibiting silencing of nuclear genes. Plant Cell. 1996;8:179–188. doi: 10.1105/tpc.8.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Hamilton AJ, Brown S, Yuanhai H, Ishizuka M, Lowe A, Alpuche Solis A-G, Grierson D. A transgene with repeated DNA causes high frequency, post-transcriptional suppression of ACC-oxidase gene expression in tomato. Plant J. 1998;15:737–746. doi: 10.1046/j.1365-313X.1998.00251.x. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–295. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Han Y, Grierson D. The influence of inverted repeats on the production of small antisense RNAs involved in gene silencing. Mol Genet Genomics. 2002;267:629–635. doi: 10.1007/s00438-002-0696-z. [DOI] [PubMed] [Google Scholar]
- Hutvágner G, Zamore PD. RNAi: Nature abhors a double-strand. Curr Opin Genet Dev. 2002;12:225–232. doi: 10.1016/s0959-437x(02)00290-3. [DOI] [PubMed] [Google Scholar]
- Ingelbrecht I, Van Houdt H, Van Montagu M, Depicker A. Posttranscriptional silencing of reporter transgenes in tobacco correlates with DNA methylation. Proc Natl Acad Sci USA. 1994;91:10502–10506. doi: 10.1073/pnas.91.22.10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AL, Thomas CL, Maule AJ. De novo methylation and co-suppression induced by a cytoplasmically replicating plant RNA virus. EMBO J. 1998;17:6385–6393. doi: 10.1093/emboj/17.21.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L, Hamilton AJ, Voinnet O, Thomas CL, Maule AJ, Baulcombe DC. RNA-DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell. 1999;11:2291–2301. doi: 10.1105/tpc.11.12.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooter JM, Matzke MA, Meyer P. Listening to the silent genes: transgene silencing, gene regulation and pathogen control. Trends Plant Sci. 1999;4:340–347. doi: 10.1016/s1360-1385(99)01467-3. [DOI] [PubMed] [Google Scholar]
- Kovařík A, Van Houdt H, Holý A, Depicker A. Drug-induced hypomethylation of a posttranscriptionally silenced transgene locus of tobacco leads to partial release of silencing. FEBS Lett. 2000;467:47–51. doi: 10.1016/s0014-5793(00)01077-2. [DOI] [PubMed] [Google Scholar]
- Lipardi C, Wei Q, Paterson BM. RNAi as random degradative PCR: siRNA primers convert mRNA into dsRNAs that are degraded to generate new siRNAs. Cell. 2001;107:297–307. doi: 10.1016/s0092-8674(01)00537-2. [DOI] [PubMed] [Google Scholar]
- Matzke M, Matzke AJP, Kooter JM. RNA: guiding gene silencing. Science. 2001;293:1080–1083. doi: 10.1126/science.1063051. [DOI] [PubMed] [Google Scholar]
- Mette MF, Aufsatz W, van der Winden J, Matzke MA, Matzke AJM. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 2000;19:5194–5201. doi: 10.1093/emboj/19.19.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain P, Béclin C, Elmayan T, Feuerbach F, Godon C, Morel J-B, Jouette D, Lacombe A-M, Nikic S, Picault N et al. Arabidopsis SGS3 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- Nishikura K. A short primer on RNAi: RNA-directed RNA polymerase acts as a key catalyst. Cell. 2001;107:415–418. doi: 10.1016/s0092-8674(01)00581-5. [DOI] [PubMed] [Google Scholar]
- Papaefthimiou I, Hamilton AJ, Denti MA, Baulcombe DC, Tsagris M, Tabler M. Replicating potato spindle tuber viroid RNA is accompanied by short RNA fragments that are characteristic of post-transcriptional gene silencing. Nucleic Acids Res. 2001;29:2395–2400. doi: 10.1093/nar/29.11.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pélissier T, Thalmeir S, Kempe D, Sänger H-L, Wassenegger M. Heavy de novo methylation at symmetrical and non-symmetrical sites is a hallmark of RNA-directed DNA methylation. Nucleic Acids Res. 1999;27:1625–1634. doi: 10.1093/nar/27.7.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz MT, Voinnet O, Baulcombe DC. Initiation and maintenance of virus-induced gene silencing. Plant Cell. 1998;10:937–946. doi: 10.1105/tpc.10.6.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders M, Maddelein W, Depicker A, Van Montagu M, Cornelissen M, Jacobs J. An active role for endogenous β-1,3-glucanase genes in transgene-mediated co–suppression in tobacco. EMBO J. 2002;21:5824–5832. doi: 10.1093/emboj/cdf586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiebel W, Haas B, Marinković S, Klanner A, Sänger HL. RNA-directed RNA polymerase from tomato leaves. II. Catalytic in vitro properties. J Biol Chem. 1993;268:11858–11867. [PubMed] [Google Scholar]
- Sharp PA. RNA interference—2001. Genes Dev. 2001;15:485–490. doi: 10.1101/gad.880001. [DOI] [PubMed] [Google Scholar]
- Sijen R, Vijn I, Rebocho A, van Blokland R, Roelofs D, Mol JNM, Kooter JM. Transcriptional and posttranscriptional gene silencing are mechanistically related. Curr Biol. 2001a;11:436–440. doi: 10.1016/s0960-9822(01)00116-6. [DOI] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RHA, Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001b;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- Sijen T, Wellink J, Hiriart J-B, van Kammen A. RNA-mediated virus resistance: role of repeated transgenes and delineation of targeted regions. Plant Cell. 1996;8:2277–2294. doi: 10.1105/tpc.8.12.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith NA, Singh SP, Wang M-B, Stoutjesdijk PA, Green AG, Waterhouse PM. Total silencing by intron-spliced hairpin RNAs. Nature. 2000;407:319–320. doi: 10.1038/35030305. [DOI] [PubMed] [Google Scholar]
- Stoutjesdijk PA, Singh SP, Liu Q, Hurlstone CJ, Waterhouse PA, Green AG. hpRNA-mediated targeting of the Arabidopsis FAD2 gene gives highly efficient and stable silencing. Plant Physiol. 2002;129:1723–1731. doi: 10.1104/pp.006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CL, Jones L, Baulcombe DC, Maule AJ. Size constraints for targeting post-transcriptional gene silencing and for RNA-directed methylation in Nicotiana benthamiana using a potato virus X vector. Plant J. 2001;25:417–425. doi: 10.1046/j.1365-313x.2001.00976.x. [DOI] [PubMed] [Google Scholar]
- Vaistij FE, Jones L, Baulcombe DC. Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell. 2002;14:857–867. doi: 10.1105/tpc.010480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houdt H, Ingelbrecht I, Van Montagu M, Depicker A. Post-transcriptional silencing of a neomycin phosphotransferase II transgene correlates with the accumulation of unproductive RNAs and with increased cytosine methylation of 3′ flanking regions. Plant J. 1997;12:379–392. [Google Scholar]
- Van Houdt H, Kovařík A, Van Montagu M, Depicker A. Cross-talk between posttranscriptionally silenced neomycin phosphotransferase II transgenes. FEBS Lett. 2000a;467:41–46. doi: 10.1016/s0014-5793(00)01076-0. [DOI] [PubMed] [Google Scholar]
- Van Houdt H, Van Montagu M, Depicker A. Both sense and antisense RNAs are targets for the sense transgene-induced posttranscriptional silencing mechanism. Mol Gen Genet. 2000b;263:995–1002. doi: 10.1007/pl00008700. [DOI] [PubMed] [Google Scholar]
- Vance V, Vaucheret H. RNA silencing in plants—defense and counterdefense. Science. 2001;292:2277–2280. doi: 10.1126/science.1061334. [DOI] [PubMed] [Google Scholar]
- Vaucheret H, Béclin C, Fagard M. Post-transcriptional gene silencing in plants. J Cell Sci. 2001;114:3083–3091. doi: 10.1242/jcs.114.17.3083. [DOI] [PubMed] [Google Scholar]
- Voinnet O. RNA silencing as a plant immune system against viruses. Trends Genet. 2001;17:449–459. doi: 10.1016/s0168-9525(01)02367-8. [DOI] [PubMed] [Google Scholar]
- Wang M-B, Wesley SV, Finnegan EJ, Smith NA, Waterhouse PM. Replicating satellite RNA induces sequence-specific DNA methylation and truncated transcripts in plants. RNA. 2001;7:16–28. doi: 10.1017/s1355838201001224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenegger M. RNA-directed DNA methylation. Plant Mol Biol. 2000;43:203–220. doi: 10.1023/a:1006479327881. [DOI] [PubMed] [Google Scholar]
- Wassenegger M, Heimes S, Riedel L, Sänger HL. RNA-directed de novo methylation of genomic sequences in plants. Cell. 1994;76:567–576. doi: 10.1016/0092-8674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- Waterhouse PM, Graham MW, Wang M-B. Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc Natl Acad Sci USA. 1998;95:13959–13964. doi: 10.1073/pnas.95.23.13959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang M, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA et al. Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 2001;27:581–590. doi: 10.1046/j.1365-313x.2001.01105.x. [DOI] [PubMed] [Google Scholar]
- Zamore PD. RNA interference: listening to the sound of silence. Nat Struct Biol. 2001;8:746–750. doi: 10.1038/nsb0901-746. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]