Abstract
Brassinosteroids (BRs) are steroidal plant hormones that are essential for growth and development. There is only limited information on where BRs are synthesized and used. We studied the organ specificity of BR biosynthesis in Arabidopsis, using two different approaches: We analyzed the expression of BR-related genes using real-time quantitative reverse transcriptase-polymerase chain reaction, and analyzed endogenous BRs using gas chromatography-mass spectrometry. Before starting this study, we cloned the second BR-6-oxidase (BR6ox2) gene from Arabidopsis and found that the encoded enzyme has the same substrate specificity as the enzyme encoded by the previously isolated 6-oxidase gene (BR6ox1) of Arabidopsis. Endogenous BRs and the expression of BR-related genes were detected in all organs tested. The highest level of endogenous BRs and the highest expression of the BR6ox1, BR6ox2, and DWF4 genes were observed in apical shoots, which contain actively developing tissues. These genes are important in BR biosynthesis because they encode the rate-limiting or farthest downstream enzyme in the BR biosynthesis pathway. The second highest level of endogenous BRs and expression of BR6ox1 and DWF4 were observed in siliques, which contains actively developing embryos and seeds. These findings indicate that BRs are synthesized in all organs tested, but are most actively synthesized in young, actively developing organs. In contrast, synthesis was limited in mature organs. Our observations are consistent with the idea that BRs function as the growth-promoting hormone in plants.
Since the discovery of brassinolide (BL; Grove et al., 1979), more than 40 natural analogs, collectively called brassinosteroids (BRs), have been isolated and characterized (Fujioka and Sakurai, 1997a, 1997b; Yokota, 1997; Fujioka, 1999). Exogenous application of BRs to plants at nanomolar to micromolar concentrations has a wide spectrum of physiological effects, including promotion of cell elongation and division, enhancement of tracheary element differentiation, retardation of abscission, enhancement of gravitropic-induced bending, promotion of ethylene biosynthesis, and enhancement of stress resistance, as reviewed by Clouse and Sasse (1998) and Sasse (1999). A number of BR-deficient mutants have been discovered in Arabidopsis, pea (Pisum sativum), and tomato (Lycopersicon esculentum; for review, see Clouse and Feldmann, 1999; Schumacher and Chory, 2000; Bishop and Koncz, 2002). These mutants exhibit dwarfism when grown in either light or dark conditions. Many of these mutants also have dark-green leaves, reduced fertility, a prolonged lifespan, and abnormal skotomorphogenesis. BR-insensitive mutants have been identified in Arabidopsis, pea, tomato, and rice (Oryza sativa; for review, see Müssig and Altmann, 2001; Bishop and Koncz, 2002).
In Arabidopsis, studies have reported BR biosynthesis and signal transduction mutants, including det2 (Li et al., 1996; Noguchi et al., 1999a), cpd (Szekeres et al., 1996), dwf4 (Choe et al., 1998), dwf1/dim (Klahre et al., 1998; Choe et al., 1999a), ste1/dwf7 (Choe et al., 1999b), sax1 (Ephritikhine et al., 1999), dwf5 (Choe et al., 2000), fackel (Jang et al., 2000; Schrick et al., 2000), bri1 (Clouse et al., 1996; Li and Chory, 1997), brs1 (Li et al., 2001), bin2/ucu1/dwf12 (Choe et al., 2002; Li and Nam, 2002; Perez-Perez et al., 2002), bzr1 (Wang et al., 2002), and bes1 (Yin et al., 2002). BAS1/CYP72B1, which encodes a repressor of a phyB mutant, catalyzes C-26 hydroxylation of BL (Neff et al., 1999). We recently showed that the BAS1 gene is induced by exogenous BRs (Goda et al., 2002). These observations strongly suggest that the BAS1 gene functions to maintain steady-state levels of endogenous BRs by inactivating active BRs. The ROT3/CYP90C gene was reported to be involved in polar cell elongation of leaf cells (Kim et al., 1998). We also demonstrated that the ROT3 gene and its homolog, CYP90D, are both repressed by exogenous BRs (Goda et al., 2002). These observations, together with the fact that the ROT3 and CYP90D genes are highly homologous to known BR-biosynthetic genes (CYP85As, CYP90A1, and CYP90B1), suggest that both ROT3 and CYP90D genes encode putative BR-biosynthetic P450 enzymes (Goda et al., 2002).
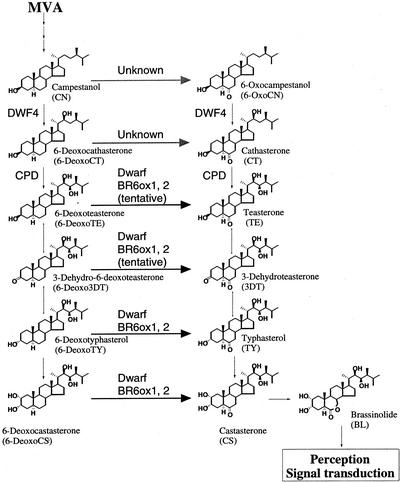
From the analysis of mutants and wild-type plants, we identified BL, castasterone (CS), typhasterol (TY), teasterone (TE), 6-oxocampestanol (6-OxoCN), 6-deoxocastasterone (6-DeoxoCS), 6-deoxotyphasterol (6-DeoxoTY), 3-dehydro-6-deoxoteasterone (6-Deoxo3DT), 6-deoxoteasterone (6-DeoxoTE), and 6-deoxocathasterone (6-DeoxoCT) as endogenous BRs in various Arabidopsis tissues, including shoots, siliques, and seeds (Fujioka et al., 1996, 1998, 2002; Noguchi et al., 1999b, 2000; S. Fujioka, unpublished data; Fig. 1). On the other hand, we studied the metabolism of deuterium-labeled BR intermediates in Arabidopsis and demonstrated the operation of the biosynthetic sequence: campestanol (CN) → 6-DeoxoCT → 6-DeoxoTE → 6-Deoxo3DT → 6-DeoxoTY → 6-DeoxoCS → 6α-hydroxyCS (6-OHCS) → CS → BL (Noguchi et al., 2000). We also showed the operation of the biosynthetic sequence: TE → 3-dehydroteasterone (3DT) → TY → CS → BL. We have also characterized the Arabidopsis BR6ox and tomato Dwarf gene, which were expressed in yeast (Saccharomyces cerevisiae), and the ability of the transformed yeast cells to metabolize 6-deoxo-BRs was tested. Both enzymes catalyze multiple steps in BR biosynthesis, 6-DeoxoTE to TE, 6-Deoxo3DT to 3-DT, 6-DeoxoTY to TY, and 6-DeoxoCS to CS (Shimada et al., 2001). These studies established the BR-biosynthetic pathway in Arabidopsis (Fig. 1).
Figure 1.
BR biosynthesis pathway in Arabidopsis. The proposed biosynthetic pathway for BL from mevalonate (MVA) is shown with identified enzymes.
In contrast to the intensive studies on the BR-metabolic pathway and BR-related genes, there is only limited information on where BRs are synthesized and function. BRs occur at very low concentrations in plants, and pollen and immature seeds are known to be the richest sources (Fujioka, 1999). The transcript abundance of downstream BR-metabolic genes, DWF4 (Noguchi et al., 1999a; Choe et al., 2001), ROT3 (Kim et al., 1998), BAS1 (Neff et al., 1999), and BR6ox (Shimada et al., 2001) are reported at extremely low levels. The signals are too weak to be detected on RNA gel blots, except for the CPD gene (Szekeres et al., 1996; Mathur et al., 1998). Therefore, studies have mainly used semiquantitative reverse transcriptase (RT)-PCR. There have been histochemical studies of the CPD (Mathur et al., 1998), ROT3 (Kim et al., 1999), BRI1 (Friedrichsen et al., 2000), and tomato dwarf/CYP85 (Pien et al., 2001) genes, but it is still unclear if the histochemical pattern of the reporter or in situ hybridization signal corresponds to the native gene expression, enzyme existence/activity, or endogenous BR accumulation.
Before starting to study organ-specific BR biosynthesis, we isolated and characterized the second CYP85A gene from Arabidopsis, which is homologous to the previously isolated BR6ox/CYP85A1 gene and the tomato Dwarf gene (Fig. 2). The BR6ox/Dwarf enzyme catalyzes an important biosynthetic step of BRs, 6-DeoxoCS to CS via 6-OHCS. This step is the farthest downstream step in BL biosynthesis for known mutations and enzymes. A defect in the Dwarf gene results in CS deficiency and causes dwarfism by suppressing stem elongation and leaf expansion (Bishop et al., 1996, 1999). These studies indicate that 6-deoxo BRs have very weak biological activity. It remains to be determined whether any or all of the 6-oxo BRs, e.g. TE, 3DT, TY, or CS, are active per se, or become active after being converted to BL. Intermediates of the late C-6 oxidation pathway are predominant over those of the early C-6 oxidation pathway in many species, including Arabidopsis, tomato, and pea (Choi et al., 1997; Yokota et al., 1997; Bishop et al., 1999; Noguchi et al., 1999b; Koka et al., 2000; Nomura et al., 2001). Especially high accumulation of 6-DeoxoCS has been recorded in many plants, suggesting that C-6 oxidation is a rate-limiting step. Such regulation seems to be canceled in BR-insensitive mutants. CS accumulates at aberrant levels in pea lka (Nomura et al., 1997, 1999) and tomato curl-3 (T. Nomura, T. Yokota, and G.J. Bishop, personal communication). Both CS and BL accumulate in the Arabidopsis bri1 mutant (Noguchi et al., 1999b). These observations indicate that C-6 oxidation is controlled by a feedback mechanism in steady-state conditions in wild-type plants. Interestingly, the presence of BL has not been demonstrated in tomato, indicating that CS may be a biologically active BR. The Dwarf enzyme may be a key enzyme governing the physiological role of BRs in tomato. Based on these observations, it is physiologically important to study how BR-6-oxidases regulate the biosynthesis of 6-oxo BRs, including BL.
Figure 2.
Sequence comparison of BR6ox2 and related genes. A, Sequence alignment of the BR6ox2, BR6ox1, and tomato Dwarf genes. Reverse contrast characters highlight identical amino acid residues and hatched characters indicate conserved residues. The GenBank/EMBL/DDBJ accession numbers of the BR6ox1 (CYP85A1) and the tomato Dwarf gene (CYP85A1) are AB035868 and U54770, respectively. The heme-binding signature sequence is underlined. B, Phylogenetic relationship between BR6ox2 and selected P450 genes. BR6ox2 belongs to a group consisting of Dwarf (tomato, CYP85A1), BR6ox1 (Arabidopsis, CYP85A1), CPD (Arabidopsis, CYP90A1 and X87367), ROT3 (Arabidopsis, CYP90C1 and AB008097), DWF4 (Arabidopsis, CYP90B1 and AF044216), and CYP90D (Arabidopsis, AB066286). BAS1/CYP72B1 (Arabidopsis, BAS1 and AC003105) is reported to be involved in BR inactivation. Dwarf3 (maize [Zea mays], CYP88A and U32579) and GA3 (Arabidopsis, CYP701A and AF047720) are reported to be involved in GA biosynthesis. The first P450 genes functionally identified from higher plants are CYP73A (Helianthus annuus cinnamate 4-hydroxylase, Z17369) and CYP75A (Petunia hybrida flavonoid-3′,5′-hydroxylase, D14588); both belong to the higher plant-specific group A of P450 genes. The accession numbers of the other members are D30718 (CYP8), M93133 (CYP7A), and X90458 (CYP86A).
In this study, we isolated and characterized the second BR-6-oxidase gene from Arabidopsis. Then, we studied organ-specific BR biosynthesis using two different approaches. First, we studied gene expression of BR-related genes, including genes encoding the downstream BR-biosynthetic enzymes (BR6ox1, BR6ox2, CPD [23-hydroxylase], and DWF4 [22-hydroxylase]), genes encoding the putative BR-biosynthetic enzymes (ROT3 and CYP90D), a gene encoding a degradation enzyme (BAS1 [26-hydroxylase]), and a gene encoding a critical component of the BR receptor (BRI1). We used a real-time quantitative (RTQ) RT-PCR to analyze transcripts at extremely low levels. Because BR-biosynthetic P450 genes have sequence similarities each other, care was taken to avoid cross detection/hybridization. The other approach was the biochemical quantitation of endogenous BRs by gas chromatography-mass spectrometry (GC-MS). These studies enable us to report the organ-specific distribution of BR biosynthesis for the first time, to our knowledge.
RESULTS
Cloning the Second BR-6-Oxidase from Arabidopsis
Recent progress in the Arabidopsis genome sequencing project (Arabidopsis Genome Initiative, 2000) has revealed that two Arabidopsis genes (CYP85A genes) are putatively orthologous to the tomato Dwarf gene. We previously cloned one of them, CYP85A1/At5g38970, and named it BR-6-oxidase (BR6ox; Shimada et al., 2001). Here, we isolate the second homolog, CYP85A2/At3g30180 from Arabidopsis, and examine functional overlap and differences between the two genes. An Arabidopsis cDNA library was screened using a PCR-based strategy and two cDNA clones were isolated. The entire nucleotide sequence of the two cDNAs revealed that they contained an identical open reading frame. Because one of these inserts had an in-frame stop codon in the 5′-upstream region of the open reading frame, we concluded that these cDNAs are the full length. The entire sequence of one of these cDNA clones, pGWB28, has been deposited to the GenBank/EMBL/DNA Data Bank of Japan (DDBJ) databases (accession no. AB087801; nucleotide sequence data not shown). We designated this gene BR6ox2 and renamed the previously reported BR6ox/CYP85A1 gene as BR6ox1. The deduced amino acid sequence of BR6ox2 has 82% identity with BR6ox1 and 68% identity with the tomato Dwarf gene (Fig. 1A). It was apparent that BR6ox2 belongs to the P450 gene superfamily because its amino acid sequence has characteristics of P450 genes, such as the heme-binding consensus sequence FxxGxxxCxG (lowercase x indicates variable amino acid residue; Nelson et al., 1996). A phylogenic relationship of P450 genes (Fig. 1B) indicates that BR6ox2 is nearer to the BR6ox1 than to the tomato Dwarf genes or any other Arabidopsis P450 genes. Comparison of the cDNA and genomic sequences revealed that the BR6ox2 gene consists of nine exons. The exon-intron structures are completely conserved in BR6ox1 and BR6ox2. These observations suggest that the BR6ox genes arose by recent gene duplication in this species.
Functional Analysis of BR6ox2
The sequence similarity between BR6ox1 and BR6ox2 does not necessarily indicate that the BR6ox enzymes catalyze the conversion of the same substrates in the BR-biosynthetic pathway. To establish the biochemical function of the BR6ox2 product, the gene was functionally expressed in yeast. The protein-coding region of the BR6ox2 gene was subcloned into a yeast expression vector, pYeDP60, and expressed in the yeast strain, WAT11, which carries Arabidopsis NADPH-P450-reductase (Urban et al., 1997). In this strain, both BR6ox2 and the P450 reductase gene were overexpressed in the presence of Gal (Pompon et al., 1996). An induced culture of the yeast transformant was incubated with 5 μg of deuterated [2H6]BRs. Products from the incubation were analyzed by GC-MS. The identity of the products was confirmed by a direct comparison of the relative abundance of characteristic ions of the metabolites and standard compounds (data not shown). When the yeast strain expressing BR6ox2 was incubated with [2H6]6-DeoxoCS, both [2H6]CS and [2H6]6-OHCS were identified as metabolites. Other 6-deoxo compounds were also fed to the yeast to determine whether they can also act as substrates for the BR6ox2 enzyme. It was found that [2H6]6-DeoxoTY, [2H6]6-Deoxo3DT, and [2H6]6-DeoxoTE were converted to [2H6]TY, [2H6]3DT, and [2H6]TE, respectively. However, [2H6]6-DeoxoCT and [2H6]CN were not converted to [2H6]cathasterone (CT) and [2H6]6-OxoCN, respectively. No conversion was detected in a yeast strain that was transformed only with the vector pYeDP60. From these results, we concluded that the BR6ox2 enzyme has the same substrate specificity as the BR6ox1 and tomato Dwarf enzymes, at least in our yeast expression system (summarized in Fig. 1).
Specificity of RTQ RT-PCR to Analyze BR-Related P450 Genes
To understand the organ specificity of BR biosynthesis, we analyzed downstream BR-biosynthetic genes, putative BR-biosynthetic genes, the BRI1, and the BAS1 genes. It has been technically difficult to quantify transcript abundance of BR-related genes because their transcript levels are extremely low. This study used the RTQ-RT-PCR to analyze transcript abundance. We tested the risk of cross amplification/detection before analyzing BR-related P450 genes, two CYP85 genes, four CYP90 genes, and one CYP72B1 gene, which share significant sequence similarity. We compared the SYBR Green and the Taq-Man methods. DNA solutions (101 ˜ 107 copies μL−1) of cDNA clones for BR-metabolic P450 genes were used as templates to draw standard curves. DNA solutions in this range of concentration were used as templates to estimate the rate of cross detection in each method. When the SYBR Green method was used, the CPD gene primers cross amplified ROT3 DNA (the efficiency of the amplification was less than 10−2 when compared with the amplification of the CPD DNA). This implies that if there are 100 times as many ROT3 transcripts as CPD transcripts, the abundance of CPD transcripts will be overestimated by a factor of two. Other primers generally cross detected nonspecific templates at low rate (<10−3) in SYBR Green method (data not shown). On the other hand, when the Taq-Man method was used for the analysis, Taq-Man probes cross detected nonspecific templates at lower rates (<10−4) compared with the SYBR Green method (Table I). We concluded that the rate of cross detection in the Taq-Man method does not affect the analysis of BR-related transcripts. The subsequent analyses were performed with the Taq-Man method.
Table I.
Cross detection of BR-related P450 gene expression using the TaqMan method
| Template | Primers and TaqMan Probes
|
||||||
|---|---|---|---|---|---|---|---|
| BR6ox1 | BR6ox2 | CPD | DWF4 | ROT3 | CYP90D | BAS1 | |
| BR6ox1 | – | <10−4 | NDa | ND | <10−5 | ND | ND |
| BR6ox2 | ND | – | ND | ND | ND | ND | ND |
| CPD | <10−5 | <10−5 | – | ND | ND | ND | ND |
| DWF4 | <10−4 | <10−4 | ND | – | ND | ND | ND |
| ROT3 | ND | <10−4 | ND | <10−4 | – | <10−4 | ND |
| CYP90D | ND | ND | ND | ND | <10−4 | – | ND |
| BAS1 | ND | <10−4 | ND | ND | ND | <10−4 | – |
ND, Not detected.
Feedback Regulation at the Transcript Level
It has been reported that the CPD (Mathur et al., 1998; Asami et al., 2001) and DWF4 (Noguchi et al., 1999a; Choe et al., 2001) genes are regulated by BRs in a feedback-regulatory manner. Here, we tested whether the BR6ox2 gene is regulated by BL. When 7-d-old det2 seedlings were treated with 10 nm BL, the transcript abundance of the BR6ox2 was decreased (Fig. 3). The repression was apparent 15 min after BL treatment and peaked at 6 h, at which time the transcript abundance was less than 2% of the initial level. The wild-type plants showed similar feedback response to BL but they were less sensitive than the det2 mutants (data not shown). We have reported recently that the BR6ox1 gene is also feedback regulated by BL, along with all CYP90 genes (Goda et al., 2002). The results of the RTQ-RT-PCR coincided with those of the GeneChip analysis. These observations provide good evidence that our system is reliable for analyzing rare transcripts of BR-biosynthetic genes.
Figure 3.
Kinetics of regulation of the BR6ox2 gene by BL. Light-grown det2 seedlings were treated with 10 nm BL for the indicated times. Transcript abundance was analyzed using the Taq-Man RTQ RT-PCR. Transcript abundance levels are given as relative values normalized to 18S ribosomal RNA levels. Data are shown as the means ± se from three different plant samples.
Organ-Specific Expression of the BR-Biosynthetic Genes
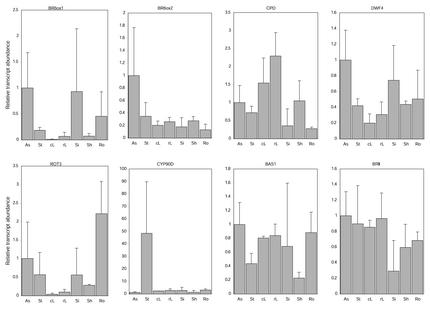
Organ-specific expression of the BR-related genes was analyzed by the RTQ-RT-PCR (Fig. 4). The genes analyzed were BR6ox1, BR6ox2, CPD (encoding a putative 23-hydroxylase), DWF4 (encoding a putative 22-hydroxylase), ROT3, CYP90D (encoding putative BR-biosynthetic enzymes), BAS1 (encoding a putative 26-hydroxylase, which inactivates BRs), and BRI1 (encoding the critical component of the BR receptor). Arabidopsis seedlings were grown in soil for 4 to 12 weeks. The aerial parts of the plants were divided into apical shoots including flower buds, inflorescent stems, rosette leaves, cauline leaves, and siliques. To compare roots and shoots, plants were germinated on agar medium for 7 d, and then cultured in liquid medium for 7 to 30 d, and divided into shoots and roots. Three independent plant samples were analyzed and the averages of transcript abundance are shown with variations (Fig. 4). The following common trends were found in gene expression, although the variations between different plant samples suggested that the expression of BR-biosynthetic genes can be influenced by factors such as age of the plant, the developmental stage of the organ, etc. In soil-grown plants, the expression of BR6ox1 was greatest in apical shoots and relatively high in siliques. The expression of BR6ox2 gene was greatest in apical shoots. CPD was expressed in all organs, except that the expression was low in siliques. DWF4 expression was highest in apical shoots, and high in siliques. ROT3 expression was relatively high in apical shoots. The expression of the CYP90D was predominant in inflorescent stems. The expression of BRI1 and BAS1 was similar; they were expressed ubiquitously in aerial organs, but in lesser amounts in siliques.
Figure 4.
The expression of BR-related genes in various organs. Arabidopsis seedlings were grown in soil for 4 to 12 weeks. The aerial parts of the plants were divided into apical shoots (As), inflorescence stems (St), rosette leaves (rL), cauline leaves (cL), and siliques (Si). Alternatively, plants were germinated on agar medium for 7 d, then cultured in liquid medium for 7 to 30 d, and then divided into shoots (Sh) and roots (Ro). Transcript levels were analyzed by the Taq-Man RTQ RT-PCR. Transcript levels are given as relative values to As (the value of 1), after being normalized to the 18S ribosomal RNA levels. Data are shown as the means with variation bars (sd) from three different plant samples for As, St, rL, Si, Sh, and Ro, or from two different plant samples for cL.
When roots were compared with shoots in liquid-cultured seedlings, the expression of BR6ox1, ROT3, CYP90D, and BAS1 in roots was higher than in shoots. The expression of BR6ox2, DWF4, and BRI1 in roots was comparable with that in shoots. The expression of CPD was exceptional; the expression in roots was lower than that in shoots.
Organ-Specific Distribution of Endogenous BRs
Arabidopsis seedlings were grown in soil for 6 weeks to compare aerial organs. Alternatively, plants were germinated on agar medium for 7 d, and then cultured in liquid medium for 13 d to compare roots and shoots. They were then divided into organs as described for the gene expression analysis. The endogenous BRs were then extracted and analyzed by GC-MS (Table II). In soil-grown plants, apical shoots contained much higher levels of BRs than other organs, especially 24-methylenecholesterol, 6-DeoxoCT, 6-DeoxoCS, and CS. Siliques also contained more endogenous BRs, especially CN, 6-DeoxoCT, 6-DeoxoCS, and CS. Inflorescent stems, rosette leaves, and cauline leaves contained relatively less BRs. When roots are compared with shoots in liquid-cultured seedlings, roots contained more BRs, especially 6-DeoxoCT, 6-DeoxoTE, and 6-DeoxoTY. In contrast, shoots contained higher levels of further downstream components, 6-DeoxoCS and CS. BL was not detected in any of the organs tested. Repeated experiments using different plant samples gave similar results (data not shown).
Table II.
Endogenous BRs in different organs
| Soil-Grown Plants
|
Liquid-Cultured Plants
|
||||||
|---|---|---|---|---|---|---|---|
| Apical shoot (3.62 g) | Stem (44.7 g) | Cauline (56.0 g) | Rosette (11.0 g) | Silique (3.89 g) | Shoot (41.5 g) | Root (11.0 g) | |
| Sterol (μg g fresh wt−1) | |||||||
| 24-MCa | 38.7 | 2.11 | 3.27 | 2.91 | 7.77 | 0.64 | 0.85 |
| CRb | 72.7 | 27.1 | 28.9 | 18.8 | 47.9 | 14.6 | 40.4 |
| CN | 1.52 | 0.90 | 0.57 | 0.50 | 5.27 | 0.28 | 0.72 |
| 6-OxoCN | 0.030 | 0.018 | 0.013 | 0.018 | 0.036 | 0.012 | 0.035 |
| 6-DeoxoBRs (ng g fresh wt−1) | |||||||
| 6-DeoxoCT | 7.93 | 0.65 | 0.63 | 0.97 | 8.89 | 0.29 | 1.71 |
| 6-DeoxoTE | 0.72 | 0.14 | 0.20 | 0.19 | 0.36 | 0.08 | 0.22 |
| 6-DeoxoTY | 1.18 | 2.24 | 2.01 | 1.08 | 2.86 | 0.69 | 1.37 |
| 6-DeoxoCS | 6.01 | 2.64 | 4.33 | 2.85 | 5.64 | 0.73 | 0.23 |
| 6-OxoBRs (ng g fresh wt−1) | |||||||
| CT | NDc | ND | ND | ND | ND | ND | ND |
| TE | 0.03 | ND | ND | ND | ND | ND | ND |
| TY | 0.30 | 0.22 | 0.11 | 0.06 | 0.38 | 0.01 | 0.01 |
| CS | 2.02 | 0.40 | 0.31 | 0.13 | 0.94 | 0.12 | 0.05 |
| BL | ND | ND | ND | ND | ND | ND | ND |
a 24-MC, 24-Methylenecholesterol.
b CR, Campesterol.
c ND, Not detected.
DISCUSSION
In a previous report, we demonstrated that the BR6ox1 enzyme, expressed in yeast, catalyzed C-6 oxidation of 6-DeoxoCS, 6-DeoxoTY, 6-Deoxo3DT, and 6-DeoxoTE, whereas the enzyme did not catalyze C-6 oxidation of 6-DeoxoCT or CN (Shimada et al., 2001). We found naturally occurring 6-OxoCN in Arabidopsis (Choe et al., 2001). These two findings suggest that an unknown BR-6-oxidase catalyzing CN to 6-OxoCN (or 6-DeoxoCT to CT) exists in Arabidopsis (Shimada et al., 2001). In this manuscript, we isolated the second BR6ox gene, BR6ox2, from Arabidopsis and characterized the encoded enzyme in transgenic yeast, to test whether the isolated enzyme has the same or different substrate specificity as the BR6ox1 enzyme. The BR6ox2 enzyme catalyzed the same C-6 oxidation steps, 6-DeoxoCS to CS, 6-DeoxoTY to TY, 6-Deoxo3DT to 3DT, and 6-DeoxoTE to TE, but did not catalyze 6-DeoxoCT to CT and CN to 6-OxoCN. Therefore, the existence of an unknown BR-6-oxidase catalyzing CN to 6-OxoCN (or 6-DeoxoCT to CT) remains hypothetical, whereas the BR6ox1 and BR6ox2 genes are paralogous. The occurrence of the biosynthetic steps, 6-DeoxoCS to CS and 6-DeoxoTY to TY, has been reported in planta (Noguchi et al., 2000), but in planta conversion of 6-Deoxo3DT to 3DT and 6-DeoxoTE to TE is still tentative (Shimada et al., 2001). This is the first example of paralogs of BR-biosynthetic genes, and gene duplication may explain why no recessive mutant of the BR6ox genes is known in Arabidopsis, although a mutant has been identified in tomato (Bishop et al., 1999).
In the studies of aerial tissues of soil-grown seedlings, the two BR6ox genes showed different organ-specific expression patterns. The BR6ox1 expression predominates in apical shoots and siliques. In contrast, BR6ox expression is relatively ubiquitous, although it is most abundant in apical shoots (Fig. 4). The difference in organ specificity suggests that the two BR6ox2 genes play different physiological roles, especially in siliques. Further studies of the BR6ox1 gene may provide a clue to BR functions in siliques, embryo, and seed development. The analysis of endogenous BRs found that CS (the main product of the BR6ox enzymes) accumulated in apical shoots and siliques. Furthermore, the ratio of 6-DeoxoCS/CS in these organs was significantly lower than in other organs. These observations are consistent with the gene expression, and suggest that BR biosynthesis is active in apical shoots and siliques, especially the C-6 oxidation step. The RTQ-RT-PCR indicated that the transcript abundance of BR6ox1 and BR6xo2 roughly ranged between 5 × 10−6 to 10−8 and 5 × 10−5 to 3 × 10−7 copies, respectively, per copy of 18S ribosomal RNA.
DWF4 had lowest transcript abundance among the analyzed BR-biosynthetic genes. The transcript abundance was roughly 2 × 10−6 to 10−7 copies per 18S ribosomal RNA from the RTQ-RT-PCR. This is consistent with the observation that the biggest gap between endogenous levels of BR precursors is at the step of 22-hydroxylation. For example, the CN content of inflorescent stems, rosette leaves, and cauline leaves was 0.90, 0.57, and 0.50 μg g−1 fresh weight, respectively, whereas that of 6-DeoxoCT (22-hydroxylated form of CN) was 0.65, 0.97, and 0.63 ng g−1 fresh weight (Table II). These findings suggest that the 22-hydroxylation step is one of the rate-limiting steps in BR biosynthesis and, therefore, is important for BR biosynthesis. The organ-specific expression pattern of the DWF4 gene was similar to that of BR6ox genes, but DWF4 expression was more ubiquitous than BR6ox1 expression and more abundant in siliques than BR6ox2 expression. CPD transcripts were extraordinarily abundant, ranging between 10−3 and 3 × 10−5 copies per the 18S ribosomal RNA. The organ-specific expression of CPD was also extraordinary; CPD was highly expressed in mature or maturing organs, inflorescence stems, cauline leaves, and rosette leaves (Fig. 4). These findings suggest that the CPD gene has an extraordinary function in the BR-biosynthetic pathway. For example, the CPD enzyme might have substrate specificity to sterols or upstream intermediates of BRs, which are more abundant than downstream BR intermediates. BAS1 transcripts were the most rare of the analyzed BR-related P450 genes. It was estimated to range between 10−6 and 5 × 10−8 copies per 18S ribosomal RNA. The organ-specific expression of BAS1 was generally higher where the expression of BR-biosynthetic genes was higher, suggesting that BR catabolism is active in organs where BR biosynthesis is active. BRI1 was expressed more ubiquitously than the BR-biosynthetic genes. This concurs with a previous GFP reporter study (Friedrichsen et al., 2000), and suggests that BR action in Arabidopsis is regulated at the level of biosynthesis rather than by the number of receptors. In contrast, the OsBRI1 gene expression predominates in shoot apex in rice (Yamamuro et al., 2000).
Only limited information is available on the sites of biosynthesis or action of BRs at the organ, tissue, and cell levels. Exogenously applied BRs are absorbed by plants, move acropetally, and promote cell elongation (Sasse, 1999), but long-distance transport of endogenous BRs is controversial. The variegated revertant phenotype of transposon-mutagenized tomato dwarf mutants (Bishop et al., 1996) suggests that BRs are synthesized in tissues adjacent to where they function. Therefore, it is very important to determine where BRs are synthesized to understand their sites of action. This study analyzed organ specificity in the BR biosynthesis, for the first time to our knowledge, using two different approaches. BR-biosynthetic gene expression was detected in all organs tested, as was the occurrences of endogenous BRs. Our findings indicate that BRs are synthesized in all organs. When aerial tissues of soil-grown plants were compared, apical shoots contained the highest levels of endogenous BRs and the greatest expression of BR6ox1, BR6ox2, and DWF4, which are important genes encoding the rate-limiting or farthest downstream enzyme in the BR-biosynthetic pathway. These observations strongly suggest that BRs are most actively synthesized, and, therefore, used in apical shoots, which contain actively developing tissues. Siliques contained the second highest levels of endogenous BRs and expression of BR6ox1 and DWF4. Siliques also contain developing embryos and seeds. Based on these observations, we propose that BRs are synthesized most actively in young developing organs in aerial tissues.
When roots were compared with shoots in liquid-cultured seedlings, the expression of BR6ox1, ROT3, CYP90D, and BAS1 was higher in roots than in shoots. The levels of midstream BR intermediates, 6-DeoxoCT, 6-DeoxoTE, and 6-DeoxoTY, were higher in roots than in shoots, whereas the levels of the downstream intermediates, 6-DeoxoCS and CS, were higher in shoots than in roots. Yokota et al. (2001) reported a very similar accumulation patter for C27 BRs in shoots and roots of tomato; 6-deoxo-28-norcathasterone and 6-deoxo-28-nortyphasterol levels were higher in roots than in shoots, whereas 6-deoxo-28-norcastasterone and 28-norcastasterone levels were higher in shoots than in roots. Therefore, Arabidopsis and tomato seems to share a common mechanism to maintain opposing levels of midstream and downstream BR intermediates, although the physiological or biochemical significance of this mechanism is not clear. One of our hypotheses is as follows. BR biosynthesis may be more active in roots than in shoots, as assumed from the biosynthesis gene expression pattern. On the other hand, from the BAS1 expression pattern, BR catabolism may also be more active in roots than in shoots. If the BAS1 enzyme acts more efficiently on downstream BRs (6-DeoxoCS and CS) as hypothesized in a previous report (Neff et al., 1999), the balance of biosynthetic enzymes and degradation enzymes could explain why more midstream intermediates accumulate in roots. The greater BAS1 enzyme activity in roots would maintain the lower levels of the furthest downstream, active BRs in roots because roots are more sensitive to active BRs than shoots.
Very recently, Bancos et al. (2002) reported endogenous BR levels in shoots and roots of Arabidopsis, pea, and tomato. In these plants, the levels of midstream BR intermediates were higher in roots than in shoots and the levels of downstream intermediates were higher in shoots than in roots. Their results, therefore, coincide well with our results. They also reported differential accumulation of Arabidopsis CYP85 and CYP90 genes in shoots and roots. Their results generally coincide with our results except for the BR6ox2/CYP85A2 gene, which was reported to be expressed preferentially in shoots. On the other hand, in our growth condition, the expression of BR6ox2 gene was not always higher in shoots than in roots (Fig. 4).
CONCLUSIONS
We isolated and characterized the second BR6ox gene from Arabidopsis. We then analyzed the organ specificity of BR biosynthesis using two different approaches: analyses of gene expression and analyses of endogenous BRs. BRs were synthesized in all organs tested, but were most actively synthesized in young actively developing organs. In contrast, synthesis was limited in mature organs. Our observation is consistent with the idea that BRs function as growth-promoting hormone in plants.
MATERIALS AND METHODS
Plant Materials
Arabidopsis ecotype Columbia was used as the wild type in this study. Wild-type seedlings were grown for 4 to 12 weeks to compare aerial organs. Alternatively, they were grown aseptically on one-half-strength Murashige and Skoog medium (Murashige and Skoog, 1962) supplemented with 1% (w/v) Suc with or without 0.8% (w/v) agar in a growth cabinet (at 22°C, continuous illumination of 100 μEm−2 s−1). Plants were grown under similar conditions and harvested at different times for gene expression analyses and BR quantification analyses. The growth conditions and BL treatment of det2-1 mutants were described previously (Goda et al., 2002).
GC-MS Analysis
GC-MS analysis was carried out on a mass spectrometer (JMS-AM SUN200, JEOL, Tokyo) connected to a gas chromatograph (6890A, Agilent Technologies, Wilmington, DE) with a capillary column DB-5 (0.25 mm × 15 m, 0.25-μm film thickness, J&W Scientific, Folsom, CA).
Isolation of cDNA Clones
An Arabidopsis cDNA library (SUPERSCRIPT Arabidopsis cDNA Library, Invitrogen, Carlsbad, CA) was screened using a PCR-based strategy with gene-specific primers, BR6ox2–101F (5′ GGG GGA TCC ATG GGC ATA ATG ATG ATG ATT TTG3′) and BR6ox2–1509R (5′ GGG GGT ACC GAC AAA ACT AGT CAG TAA GGT GAA CAC TT3′). The isolated cDNA clone is a derivative of the pSPORT-P vector (Invitrogen).
DNA Sequence Analyses
DNA sequences were determined using an automated DNA sequencer (model 373A DNA Sequencing System, PE-Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. The nucleotide sequence was compiled and analyzed using GENETYX-Mac (Software Development Co., Ltd., Tokyo). The BLAST (Altschul et al., 1990) program was used to search for entries of homologous sequences in the DDBJ. The ClustalW program on the server at DDBJ was used to align the amino acid sequences and to draw phylogenic relationships using the neighbor-joining method (Saitou and Nei, 1987). The aligned sequences were shaded using the program Boxshade, available on the server at the European Molecular Biology Network.
Yeast (Saccharomyces cerevisiae) Expression Vector
The full-length cDNA of the BR6ox2 gene was subcloned from a cDNA clone pGWB28 to a vector, pDONR207, yielding pDONRB28 using the BP Clonase reaction of the Gateway system (Invitrogen). The cDNA was then subcloned into a yeast expression vector pYESDEST52 (Invitrogen) using the LR Clonase reaction (Invitrogen). The resulting plasmid was designated pYESB284. In this construct, the BR6ox2 gene is controlled under the yeast GAL1 promoter.
Yeast Functional Assay
pYESB284 vector was transformed to yeast strain WAT11 (Pompon et al., 1996) under selection using the URA3 gene. Yeast expression and functional assay of the BR6ox2 gene were performed as descried previously (Bishop et al., 1999; Noguchi et al., 1999b; Shimada et al., 2001). Products from the incubations were analyzed by GC-MS. The identity of the products was confirmed using their full-scan mass spectra.
RT-PCR Analysis
Total RNAs were extracted from Arabidopsis seedlings using the guanidine-hydrochloride method (Kawakami and Watanabe, 1988). The RNAs were then treated with DNase I. They were then converted to cDNAs with random primers using the Super Script First-Strand Synthesis System (Life Technologies/Gibco-BRL, Cleveland). Unless otherwise noted, quantitative RT-PCR was performed with the use of real-time-monitoring Taq-Man technology (Holland et al., 1991) with a model 7700 sequence detector and a Taq-Man Universal PCR Master Mix (PE-Applied Biosystems). 6-Carboxyfluorescein and 6-carboxytetramethylrhodamine were used for “Reporter” and “Quencher” to label Taq-Man primers, respectively. 6-Carboxy-X-rhodamine was used for “Passive Reference” as an internal standard to normalize the “Reporter” signal. Normalization is necessary to correct for well-to-well fluorescent fluctuations of “Reporter” because of changes in concentration or volume. The gene-specific primers were designed with care (e.g. not to be homologous to other Arabidopsis genes) using the Primer Express program (PE-Applied Biosystems) and the BLAST program. The primers and Taq-Man probes are listed in Table III. The 18S ribosomal RNA was analyzed as an internal control to monitor the efficiency of the RT reaction, and was used to normalize the transcript abundance in each sample. The 18S ribosomal RNA was chosen because it is the most abundant transcript in vivo. If the efficiency of the RT reaction of a sample was significantly lower than the efficiency of other samples, these samples were not used for further analysis. The same cDNA substrate was used for the quantifications of all genes, including the 18S ribosomal RNA. Because the RTQ-RT-PCR system is so sensitive that it can detect degradation of DNA solutions caused by freezing and thawing or absorption to tubes, the raw data of transcript abundance varies in each measurement. Therefore, we presented relative transcript abundance in results because relative values are reproducible regardless of measurements.
Table III.
Primers and TaqMan probes used for RT-PCR
| Gene Name | Forward Primer Sequence | Reverse Primer Sequence | TaqMan Probe Sequence |
|---|---|---|---|
| BR6ox1 | TGGCCAATCTTTGGCGAA | TCCCGTATCGGAGTCTTTGGT | ACCGAGTTTCTCAAACAAGGCCCCAAC |
| BR6ox2 | CAATAGTCTCAATGGACGCAGAGT | AACCGCAGCTATGTTGCATG | ACTTGTTGCCGGTTACCCGCAATCTATG |
| CPD | CCCAAACCACTTCAAAGATGCT | GGGCCTGTCGTTACCGAGTT | TCTGCCATCTCCAAGGGTTGAAAGTGC |
| DWF4 | GTGATCTCAGCCGTACATTTGGA | CACGTCGAAAAACTACCACTTCCT | CAGCAAAACAACGGAGCGTCATCG |
| ROT3 | ATTGGCGCGTTCCTCAGAT | CAAGACGCCAAAGTGAGAACAA | CTCACCTCAAAGACCGGATCACTCGAGA |
| CYP90D | CTCATTACCCTTGCCGTCAAA | CAGCTTCATGTTTTCTTCCGTTAG | CCTCTCTGATTCTCCTGCTGCCCTCAAT |
| BAS1 | TTGGCTTCATACCGTTTGGC | TTACAGCGAGTGTCAATTTGGC | CGGAGTTCGTACATGCATTGGTCAGAATC |
| BRI1 | GGTGAAACAGCACGCAAAACT | CACGCAACCGCAACTTTTAA | ACCCCGAGCTTATGAAGGAAGATCCAGC |
| 18S ribosomal RNA | CGGCTACCACATCCAAGGAA | GCTGGAATTACCGCGGCT | TGCTGGCACCAGACTTGCCCTC |
Quantification of BRs in Arabidopsis Seedlings
BR purification and quantification were carried out according to the method described by Noguchi et al. (1999a).
ACKNOWLEDGMENTS
We thank Drs. Philippe Urban and Denis Pompon for providing pYeDP60 and yeast strain WAT11. We thank Mr. Narumasa Miyauchi for technical assistance with the quantitative PCR and cDNA cloning. We also thank Ms. Masayo Sekimoto and Mr. Makoto Kobayashi for technical assistance with the endogenous BR analysis.
Footnotes
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.013029.
LITERATURE CITED
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Asami T, Mizutani M, Fujioka S, Goda H, Min YK, Shimada Y, Nakano T, Takatsuto S, Matsuyama T, Nagata N et al. Selective interaction of triazole derivatives with DWF4, a cytochrome P450 monooxygenase of the brassinosteroid biosynthetic pathway, correlates with brassinosteroid deficiency in planta. J Biol Chem. 2001;276:25687–25691. doi: 10.1074/jbc.M103524200. [DOI] [PubMed] [Google Scholar]
- Bancos S, Nomura T, Sato T, Molnar G, Bishop GJ, Koncz C, Yokota T, Nagy F, Szekeres M. Regulation of transcript levels of the Arabidopsis cytochrome P450 genes involved in brassinosteroid biosynthesis. Plant Physiol. 2002;130:504–513. doi: 10.1104/pp.005439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop GJ, Harrison K, Jones J. The tomato Dwarf gene isolated by heterologous transposon tagging encodes the first member of a new cytochrome P450 family. Plant Cell. 1996;8:959–969. doi: 10.1105/tpc.8.6.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop GJ, Koncz C. Brassinosteroids and plant steroid hormone signaling. Plant Cell. 2002;14:S97–S110. doi: 10.1105/tpc.001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop GJ, Nomura T, Yokota T, Harrison K, Noguchi T, Fujioka S, Takatsuto S, Jones JD, Kamiya Y. The tomato DWARF enzyme catalyses C-6 oxidation in brassinosteroid biosynthesis. Proc Natl Acad Sci USA. 1999;96:1761–1766. doi: 10.1073/pnas.96.4.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Dilkes BP, Fujioka S, Takatsuto S, Sakurai A, Feldmann KA. The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22α-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell. 1998;10:231–243. doi: 10.1105/tpc.10.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Dilkes BP, Gregory BD, Ross AS, Yuan H, Noguchi T, Fujioka S, Takatsuto S, Tanaka A, Yoshida S et al. The Arabidopsis dwarf1 mutant is defective in the conversion of 24-methylenecholesterol to campesterol in brassinosteroid biosynthesis. Plant Physiol. 1999a;119:897–907. doi: 10.1104/pp.119.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Fujioka S, Noguchi T, Takatsuto S, Yoshida S, Feldmann KA. Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J. 2001;26:573–582. doi: 10.1046/j.1365-313x.2001.01055.x. [DOI] [PubMed] [Google Scholar]
- Choe S, Noguchi T, Fujioka S, Takatsuto S, Tissier CP, Gregory BD, Ross AS, Tanaka A, Yoshida S, Tax FE et al. The Arabidopsis dwf7/ste1 mutant is defective in the Δ7 sterol C-5 desaturation step leading to brassinosteroid biosynthesis. Plant Cell. 1999b;11:207–221. [PMC free article] [PubMed] [Google Scholar]
- Choe S, Schmitz RJ, Fujioka S, Takatsuto S, Lee M-O, Yoshida S, Feldmann KA, Tax FE. Arabidopsis brassinosteroid-insensitive dwarf12 mutants are semi-dominant and defective in a glycogen synthase kinase3 β-like kinase. Plant Physiol. 2002;130:1506–1515. doi: 10.1104/pp.010496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Tanaka A, Noguchi T, Fujioka S, Takatsuto S, Ross AS, Tax FE, Yoshida S, Feldmann KA. Lesions in the sterol Δ7 reductase gene of Arabidopsis cause dwarfism due to a block in brassinosteroid biosynthesis. Plant J. 2000;21:431–443. doi: 10.1046/j.1365-313x.2000.00693.x. [DOI] [PubMed] [Google Scholar]
- Choi Y, Fujioka S, Nomura T, Harada A, Yokota T, Takatsuto S, Sakurai A. An alternative brassinolide biosynthetic pathway via late C-6 oxidation. Phytochemistry. 1997;44:609–613. [Google Scholar]
- Clouse SD, Feldmann KA. Molecular genetics of brassinosteroid action. In: Sakurai A, Yokota T, Clouse S, editors. Brassinosteroids: Steroidal Plant Hormones. Tokyo: Springer-Verlag; 1999. pp. 163–190. [Google Scholar]
- Clouse SD, Langford M, McMorris TC. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111:671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD, Sasse JM. Brassinosteroids: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:427–451. doi: 10.1146/annurev.arplant.49.1.427. [DOI] [PubMed] [Google Scholar]
- Ephritikhine G, Pagant S, Fujioka S, Takatsuto S, Lapous D, Caboche M, Kendrick RE, Barbier-Brygoo H. The sax1 mutation defines a new locus involved in the brassinosteroid biosynthesis pathway in Arabidopsis thaliana. Plant J. 1999;18:315–320. doi: 10.1046/j.1365-313x.1999.00455.x. [DOI] [PubMed] [Google Scholar]
- Friedrichsen DM, Joazeiro CAP, Li JM, Hunter T, Chory J. Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 2000;123:1247–1255. doi: 10.1104/pp.123.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S. Natural occurrence of brassinosteroids in the plant kingdom. In: Sakurai A, Yokota T, Clouse SD, editors. Brassinosteroids: Steroidal Plant Hormones. Tokyo: Springer-Verlag; 1999. pp. 21–45. [Google Scholar]
- Fujioka S, Choi YH, Takatsuto S, Yokota T, Li J, Chory J, Sakurai A. Identification of castasterone, 6-deoxocastasterone, typhasterol and 6-deoxotyphasterol from the shoots of Arabidopsis thaliana. Plant Cell Physiol. 1996;37:1201–1203. doi: 10.1093/oxfordjournals.pcp.a029074. [DOI] [PubMed] [Google Scholar]
- Fujioka S, Noguchi T, Yokota T, Takatsuto S, Yoshida S. Brassinosteroids in Arabidopsis thaliana. Phytochemistry. 1998;48:595–599. doi: 10.1016/s0031-9422(98)00065-x. [DOI] [PubMed] [Google Scholar]
- Fujioka S, Sakurai A. Biosynthesis and metabolism of brassinosteroids. Physiol Plant. 1997a;100:710–715. [Google Scholar]
- Fujioka S, Sakurai A. Brassinosteroids. Nat Prod Rep. 1997b;14:1–10. doi: 10.1039/np9971400001. [DOI] [PubMed] [Google Scholar]
- Fujioka S, Takatsuto S, Yoshida S. An early C-22 oxidation branch in the brassinosteroid biosynthetic pathway. Plant Physiol. 2002;130:930–939. doi: 10.1104/pp.008722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Shimada Y, Asami T, Fujioka S, Yoshida S. Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 2002;130:1319–1334. doi: 10.1104/pp.011254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove MD, Spencer GF, Rohwedder WK, Mandava N, Worley JF, Warthen JD, Steffen GL, Flippen-Anderson JL, Cook JC. Brassinolide, a plant growth-promoting steroid isolated from Brassica napus pollen. Nature. 1979;281:216–217. [Google Scholar]
- Holland PM, Abramson RD, Watson R, Gelfand DH. Detection of specific polymerase chain-reaction product by utilizing the 5′ → 3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang JC, Fujioka S, Tasaka M, Seto H, Takatsuto S, Ishii A, Aida M, Yoshida S, Sheen J. A critical role of sterols in embryonic patterning and meristem programming revealed by the fackel mutants of Arabidopsis thaliana. Genes Dev. 2000;14:1485–1497. [PMC free article] [PubMed] [Google Scholar]
- Kawakami N, Watanabe A. Change in gene expression in radish cotyledons during dark-induced senescence. Plant Cell Physiol. 1988;29:33–42. [Google Scholar]
- Kim GT, Tsukaya H, Uchimiya H. The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Genes Dev. 1998;12:2381–2391. doi: 10.1101/gad.12.15.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim GT, Tsukaya H, Saito Y, Uchimiya H. Changes in the shapes of leaves and flowers upon overexpression of cytochrome P450 in Arabidopsis. Proc Natl Acad Sci USA. 1999;96:9433–9437. doi: 10.1073/pnas.96.16.9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klahre U, Noguchi T, Fujioka S, Takatsuto S, Yokota T, Nomura T, Yoshida S, Chua NH. The Arabidopsis DIMINUTO/DWARF1 gene encodes a protein involved in steroid synthesis. Plant Cell. 1998;10:1677–1690. doi: 10.1105/tpc.10.10.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koka CV, Cerny RE, Gardner RG, Noguchi T, Fujioka S, Takatsuto S, Yoshida S, Clouse SD. A putative role for the tomato genes DUMPY and CURL-3 in brassinosteroid biosynthesis and response. Plant Physiol. 2000;122:85–98. doi: 10.1104/pp.122.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- Li J, Lease KA, Tax FE, Walker JC. BRS1, a serine carboxypeptidase, regulates BRI1 signaling in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2001;98:5916–5921. doi: 10.1073/pnas.091065998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Nagpal P, Vitart V, McMorris TC, Chory J. A role for brassinosteroids in light-dependent development of Arabidopsis. Science. 1996;272:398–401. doi: 10.1126/science.272.5260.398. [DOI] [PubMed] [Google Scholar]
- Li JM, Nam KH. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science. 2002;295:1299–1301. doi: 10.1126/science.1065769. [DOI] [PubMed] [Google Scholar]
- Mathur J, Molnar G, Fujioka S, Takatsuto S, Sakurai A, Yokota T, Adam G, Voigt B, Nagy F, Maas C et al. Transcription of the Arabidopsis CPD gene, encoding a steroidogenic cytochrome P450, is negatively controlled by brassinosteroids. Plant J. 1998;14:593–602. doi: 10.1046/j.1365-313x.1998.00158.x. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiol Plant. 1962;15:473–498. [Google Scholar]
- Müssig C, Altmann T. Brassinosteroid signaling in plants. Trends Endocrinol Metab. 2001;12:398–402. doi: 10.1016/s1043-2760(01)00477-5. [DOI] [PubMed] [Google Scholar]
- Neff MM, Nguyen SM, Malancharuvil EJ, Fujioka S, Noguchi T, Seto H, Tsubuki M, Honda T, Takatsuto S, Yoshida S et al. BAS1: a gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc Natl Acad Sci USA. 1999;96:15316–15323. doi: 10.1073/pnas.96.26.15316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DR, Koymans L, Kamataki T, Stegeman JJ, Feyereisen R, Waxman DJ, Waterman MR, Gotoh O, Coon MJ, Estabrook RW et al. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics. 1996;6:1–42. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Tax FE, Feldmann KA. Biosynthetic pathways of brassinolide in Arabidopsis. Plant Physiol. 2000;124:201–209. doi: 10.1104/pp.124.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Yoshida S, Yuan H, Feldmann KA, Tax FE. Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 1999a;121:743–752. doi: 10.1104/pp.121.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Takatsuto S, Sakurai A, Yoshida S, Li J, Chory J. Arabidopsis det2 is defective in the conversion of (24R)-24-methylcholest-4-en-3-one to (24R)-24-methyl-5α-cholestan-3-one in brassinosteroid biosynthesis. Plant Physiol. 1999b;120:833–839. doi: 10.1104/pp.120.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Kitasaka Y, Takatsuto S, Reid JB, Fukami M, Yokota T. Brassinosteroid/sterol synthesis and plant growth as affected by lka and lkb mutations of pea. Plant Physiol. 1999;119:1517–1526. doi: 10.1104/pp.119.4.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Nakayama M, Reid JB, Takeuchi Y, Yokota T. Blockage of brassinosteroid biosynthesis and sensitivity causes dwarfism in garden pea. Plant Physiol. 1997;113:31–37. doi: 10.1104/pp.113.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Sato T, Bishop GJ, Kamiya Y, Takatsuto S, Yokota T. Accumulation of 6-deoxocathasterone and 6-deoxocastasterone in Arabidopsis, pea and tomato is suggestive of common rate-limiting steps in brassinosteroid biosynthesis. Phytochemistry. 2001;57:171–178. doi: 10.1016/s0031-9422(00)00440-4. [DOI] [PubMed] [Google Scholar]
- Perez-Perez JM, Ponce MR, Micol JL. The UCU1 Arabidopsis gene encodes a SHAGGY/GSK3-like kinase required for cell expansion along the proximodistal axis. Dev Biol. 2002;242:161–173. doi: 10.1006/dbio.2001.0543. [DOI] [PubMed] [Google Scholar]
- Pien S, Wyrzykowska J, Fleming AJ. Novel marker genes for early leaf development indicate spatial regulation of carbohydrate metabolism within the epical meristem. Plant J. 2001;25:663–674. doi: 10.1046/j.1365-313x.2001.01002.x. [DOI] [PubMed] [Google Scholar]
- Pompon D, Louerat B, Bronine A, Urban P. Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol. 1996;272:51–64. doi: 10.1016/s0076-6879(96)72008-6. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sasse J. Physiological actions of brassinosteroids. In: Sakurai A, Yokota T, Clouse SD, editors. Brassinosteroids: Steroidal Plant Hormones. Tokyo: Springer-Verlag; 1999. pp. 137–161. [Google Scholar]
- Schrick K, Mayer U, Horrichs A, Kuhnt C, Bellini C, Dangl J, Schmidt J, Jurgens G. FACKEL is a sterol C-14 reductase required for organized cell division and expansion in Arabidopsis embryogenesis. Genes Dev. 2000;14:1471–1484. [PMC free article] [PubMed] [Google Scholar]
- Schumacher K, Chory J. Brassinosteroid signal transduction: still casting the actors. Curr Opin Plant Biol. 2000;3:79–84. doi: 10.1016/s1369-5266(99)00038-2. [DOI] [PubMed] [Google Scholar]
- Shimada Y, Fujioka S, Miyauchi N, Kushiro M, Takatsuto S, Nomura T, Yokota T, Kamiya Y, Bishop GJ, Yoshida S. Brassinosteroid-6-oxidases from Arabidopsis and tomato catalyze multiple C-6 oxidations in brassinosteroid biosynthesis. Plant Physiol. 2001;126:770–779. doi: 10.1104/pp.126.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres M, Nemeth K, Koncz Kalman Z, Mathur J, Kauschmann A, Altmann T, Redei GP, Nagy F, Schell J, Koncz C. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85:171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- Urban P, Mignotte C, Kazmaier M, Delorme F, Pompon D. Cloning, yeast expression, and characterization of the coupling of two distantly related Arabidopsis thaliana NADPH-cytochrome P450 reductases with P450 CYP73A5. J Biol Chem. 1997;272:19176–19186. doi: 10.1074/jbc.272.31.19176. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Nakano T, Gendron J, He JX, Chen M, Vafeados D, Yang YL, Fujioka S, Yoshida S, Asami T et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell. 2002;2:505–513. doi: 10.1016/s1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- Yamamuro C, Ihara Y, Wu X, Noguchi T, Fujioka S, Takatsuto S, Ashikari M, Kitano H, Matsuoka M. Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell. 2000;12:1591–1605. doi: 10.1105/tpc.12.9.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin YH, Wang ZY, Mora-Garcia S, Li JM, Yoshida S, Asami T, Chory J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- Yokota T. The structure, biosynthesis and function of brassinosteroids. Trends Plant Sci. 1997;2:137–143. [Google Scholar]
- Yokota T, Nomura T, Nakayama M. Identification of brassinosteroids that appear to be derived from campesterol and cholesterol in tomato shoots. Plant Cell Physiol. 1997;38:1291–1294. [Google Scholar]
- Yokota T, Sato T, Takeuchi Y, Nomura T, Uno K, Watanabe T, Takatsuto S. Roots and shoots of tomato produce 6-deoxo-28-norcathasterone, 6-deoxo-28-nortyphasterol and 6-deoxo-28-norcastasterone, possible precursors of 28-norcastasterone. Phytochemistry. 2001;58:233–238. doi: 10.1016/s0031-9422(01)00237-0. [DOI] [PubMed] [Google Scholar]